Abstract
The green alga Chlamydomonas reinhardtii shows various light responses in behavior and physiology. One such photoresponse is the circadian clock, which can be reset by external light signals to entrain its oscillation to daily environmental cycles. In a previous report, we suggested that a light-induced degradation of the clock protein ROC15 is a trigger to reset the circadian clock in Chlamydomonas. However, light signaling pathways of this process remained unclear. Here, we screened for mutants that show abnormal ROC15 diurnal rhythms, including the light-induced protein degradation at dawn, using a luciferase fusion reporter. In one mutant, ROC15 degradation and phase resetting of the circadian clock by light were impaired. Interestingly, the impairments were observed in response to red and violet light, but not to blue light. We revealed that an uncharacterized gene encoding a protein similar to RAS-signaling-related leucine-rich repeat (LRR) proteins is responsible for the mutant phenotypes. Our results indicate that a previously uncharacterized red/violet light signaling pathway is involved in the phase resetting of circadian clock in Chlamydomonas.
Author summary
The unicellular green alga Chlamydomonas reinhardtii is used as a model system in many biological researches. Although blue light responses of this alga (e.g., phototaxis) are well known and well characterized, far less is understood about responses to other wavelengths. One such photoresponse is the circadian clock, which can be reset by various wavelengths of light, ranging from violet to red, to entrain its oscillation to daily environmental cycles. In this study, we identified a gene responsible for red and violet light responses of the circadian clock by a forward genetic screen. Our results shed light on a previously unrecognized red/violet light signaling pathway in green algae.
Introduction
Light is an important signal for most organisms, enabling them to interpret and respond to environmental conditions. In the green alga Chlamydomonas reinhardtii, various light responses are observed [1,2]. For example, blue and green light are sensed by channelrhodopsins, and regulate flagella activity for appropriate phototactic behaviors [3–5]. Blue light is also sensed by phototropins, and regulates sexual reproduction-related steps [6]. Moreover, red light regulates gene expression at the mRNA and protein levels [7–11]. Some responses to red light are mediated by aCRY, a homolog of animal cryptochromes, that can absorb not only blue light, but also a wide range of wavelengths, including red [11].
Circadian clocks are biological timekeepers that enable organisms to adapt to environmental daily cycles. The molecular mechanisms of the clocks have been studied in several model organisms, and "clock genes" have been revealed [12]. Clock genes are not conserved across the kingdom of organisms, but a common mechanism has been proposed [12]. In Chlamydomonas, several clock genes have been identified [13,14], and these genes revealed that the Chlamydomonas clock consists of land-plant-like and green-alga-specific components [15–18]. Because of its unique evolutionary position, Chlamydomonas can serve as an outstanding model organism for comparative and evolutionary studies of circadian clocks.
Although the circadian clocks can maintain their oscillation without any external cues, they need to be “reset” when a time lag between their oscillation and environmental daily cycles has occurred. A major cue that resets the circadian clock is light. In Chlamydomonas, the resetting in dark-adapted cells is induced by various wavelengths of light, ranging from violet to red [19,20]. A Chlamydomonas homolog of plant cryptochrome photoreceptors (CPH1) is known as a negative regulator for the resetting process by blue light [20]. In addition, aCRY is known to be involved in blue and red light responses of mRNA transcripts of several clock-related genes [11], although it remains to be determined to what extent these mRNA responses contribute to the phase resetting.
RHYTHM OF CHLOROPLAST 15 (ROC15), a clock gene in Chlamydomonas, encodes a transcription factor-like protein with a GOLDEN2/ARR/Psr1 (GARP) DNA-binding motif. Mutations in ROC15 affect period length and phase of the circadian clock [14]. Under the daily light/dark cycle, expression levels of ROC15 protein show a diurnal rhythm: ROC15 protein levels increase during the night, decline rapidly at dawn due to light-induced proteasomal degradation, and are kept low during the day [21]. Remarkably, red light is highly effective for degradation of ROC15, as well as the resetting of circadian clock [21]. In addition, since a ROC15 gene mutant shows severe defect in the resetting of circadian clock by light, the light induced degradation of ROC15 is thought to be a key event for the resetting of the Chlamydomonas circadian clock [21]. However, photoreceptors and signaling pathways for this process remain unidentified.
Herein, we performed a forward genetic analysis to identify genes involved in ROC15 diurnal regulation, including the light induced degradation of the protein at the dawn. Through analyses of an isolated mutant, we show that at least two light signaling pathways are involved in the resetting of the circadian clock in Chlamydomonas, and one is a red/violet light signaling pathway wherein an uncharacterized Chlamydomonas leucine-rich repeat (LRR) protein is involved.
Results
Screening for mutants of ROC15 diurnal rhythm
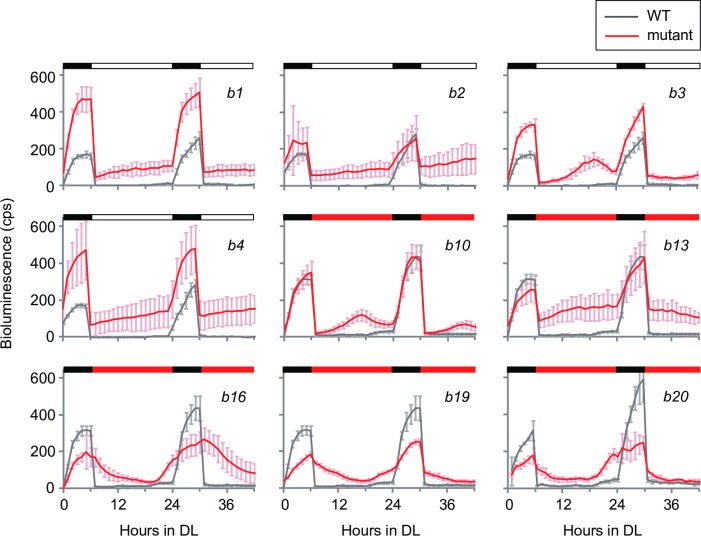
In a previous study, we developed a ROC15-LUC reporter strain that express a fusion protein of ROC15 and firefly luciferase to monitor fluctuations in ROC15 protein levels in vivo [21]. Taking advantage of the reporter strain and a high throughput bioluminescence monitoring system [22,23], we screened mutants showing abnormal diurnal rhythms through ROC15-LUC reporter bioluminescence. We employed a light regime consisting of two cycles of 6-h dark/18-h light. Under this regime, bioluminescence of the ROC15-LUC reporter increased during the 6 h dark period, declined immediately after light on, and was maintained at low levels throughout the 18 h light phase (S1 Fig, Wild-type [WT]). roc114-1, a clock mutant involved in the light induced degradation of ROC15 [21], showed an abnormal bioluminescence pattern (S1 Fig), indicating that this regime is appropriate for isolating mutants of ROC15 regulation. Insertional mutants were generated by random integration of a hygromycin-resistance gene (aph7”) [24] into the genome of the reporter strain. We screened 6,532 transformants under the regime, using white light as a light source for the day phase, and isolated 4 mutants, b1 –b4 (Fig 1). We further screened 7,672 transformants using red light as the day phase light, and isolated 5 mutants, b10, b13, b16, b19, and b20 (Fig 1).
Fig 1. Diurnal rhythms of bioluminescence of ROC15-LUC in isolated mutants.
Asynchronous TAP liquid cultures of mutants in white 96-well plates were subjected to two cycles of dark/light. White, red, and black bars represent white light, red light, and dark, respectively. Traces represent means ± standard deviation (SD) of bioluminescence levels from 4–6 independent cultures. A trace of the WT strain in the same assay is shown on each graph for comparison.
These nine mutants could be roughly divided into three types, 1–3. Type 1 (b1, b2, b4, and b13) showed insufficient decline of bioluminescence at light onset, and bioluminescence levels were maintained at relatively high levels during the light phase (Fig 1). This phenotype suggests that ROC15 synthesis or degradation is constantly high or low, respectively, in these mutants. Type 2 (b3 and b10) showed almost normal acute response at the light onset but failed to maintain a low bioluminescence level during the mid to late day phase (Fig 1), suggesting the existence of a phase-specific mechanism for maintaining low ROC15 levels. Type 3 (b16, b19, and b20) showed no or very weak acute responses at light onset (Fig 1), suggesting defects in the light-induced rapid degradation of ROC15. The acute response was almost completely absent in b16 in both the first and second cycles, but tended to be stronger in the second cycle compared to the first in b19 and b20 (Fig 1), indicating that b19 and b20 phenotypes, but not b16, are conditional.
Red and violet light responses of ROC15 are impaired in b16 mutant
To test whether the ROC15 acute light response phenotypes of b16, b19, and b20 are dependent upon light wavelength, these mutants were subjected to blue or red light pulses, and ROC15-LUC bioluminescence was measured. Although b19 and b20 showed similar defects for both light sources, b16 showed a different response (S2 Fig): Consistent with the observation in Fig 1, b16 showed no acute response to the red light pulse (S2 Fig). However, the bioluminescence level declined to the same extent as the WT in response to the blue light pulse (S2 Fig). To obtain more detailed characteristics of wavelength dependency, we compared an equal-quantum action spectrum of the acute response of ROC15-LUC in b16 mutant with that in WT. In WT strain, the light response of ROC15 was induced by blue (460 nm), green (510 nm), and especially red (660 nm) light pulses as we previously reported (Fig 2) [21]. In addition, we found that the light response of ROC15 was strongly induced by violet (410 nm) and ultraviolet (360 nm) light pulses, but not by far-red light pulses (710 nm and 760 nm) in the WT strain (Fig 2). In the b16 mutant, the response to red light pulse was almost completely abolished (Fig 2). In addition, the response to the violet light pulse was partially impaired (Fig 2). These results indicate that b16 has a defect in a pathway transmitting red and partial violet light information. On the other hand, the response to the blue light pulse (460 nm) was nearly normal or slightly stronger than that of the WT (Fig 2). This indicates that the blue light signal is transmitted by the other pathway. Taken together, these results indicate that there are multiple pathways (at least two) for transmitting light information to ROC15, and one is a red/violet light signaling pathway. Furthermore, the sensitization to blue light in b16 implies the existence of a crosstalk between these light signaling pathways. In the following experiment, we focused our attention on the b16 mutant.
Fig 2. Wavelength dependency of the acute light response of ROC15-LUC.
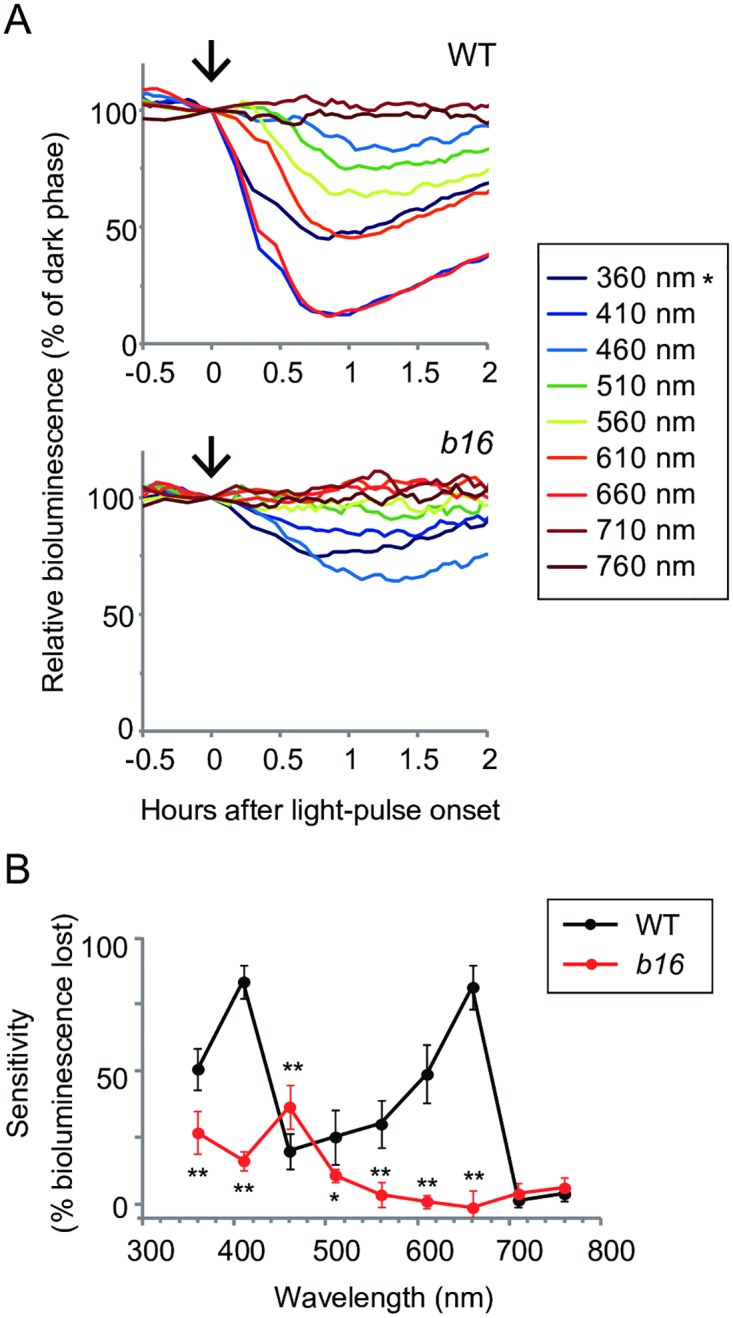
Asynchronous TAP cultures of WT and b16 cells in black 24-well plates were subjected to darkness for 3 h for accumulation of ROC15-LUC, and then light pulses were administered. (A) Representative traces of the response to various light qualities. 5 min light pulses were administered by a tunable monochromatic light source. Each monochromatic light was given at 0.5 μmol∙m-2∙s-1 at the surface of the culture plates (arrows). Note that the 360 nm light (*) was weaker than those of other wavelengths at the sample level because the lid of culture plates blocked 30% of 360 nm light. The bioluminescence level just before light pulse was set to 100. (B) Sensitivities of ROC15-LUC to light. The sensitivity was represented as the relative bioluminescence level lost after light pulse at the time point when the minimum bioluminescence was detected. Each point represents the mean ± SD of 4–6 independent cultures. * P < 0.05, ** P < 0.01 (Student’s t-test).
Photic phase-resetting of the circadian clock is impaired in b16
Because the rapid degradation of ROC15 by light has been suggested as a trigger for the phase-resetting mechanism in the circadian clock of Chlamydomonas [21], we tested photic phase-resetting in the b16 mutant. For precise analyses of circadian rhythm, the ROC15-LUC reporter gene in b16 was replaced by a genetic cross with the chloroplast tufA promoter-lucCP reporter gene of the CBR strain that shows a robust circadian bioluminescence rhythm [14,25]. Progenies harboring the b16 mutation showed a normal circadian rhythm in the chloroplast bioluminescence reporter (S3A and S3B Fig), indicating that the b16 mutation does not affect the circadian oscillation itself at least in constant darkness (DD). Next, we examined the effect of light pulses on the phase shift of the clock. Violet, blue, or red light pulse was given at the 34 h time point in DD (when the maximum phase advance occurs in this strain) [21]. Of all three light qualities tested, 1.6 to 1.9 h phase advances were observed in the WT (Fig 3A and 3B), whereas the amount of phase advance in b16 was significantly reduced in response to the red light pulse (Fig 3A and 3B). There were no significant differences between WT and b16 in the responses to blue and violet light pulse of these intensities tested (Fig 3A and 3B). Consistent with these results, the ROC15-LUC reporter strain showed a decline in bioluminescence in response to blue and violet but not red light in identical experimental conditions (S4 Fig).
Fig 3. Phase resetting of the circadian clock in the b16 mutant.
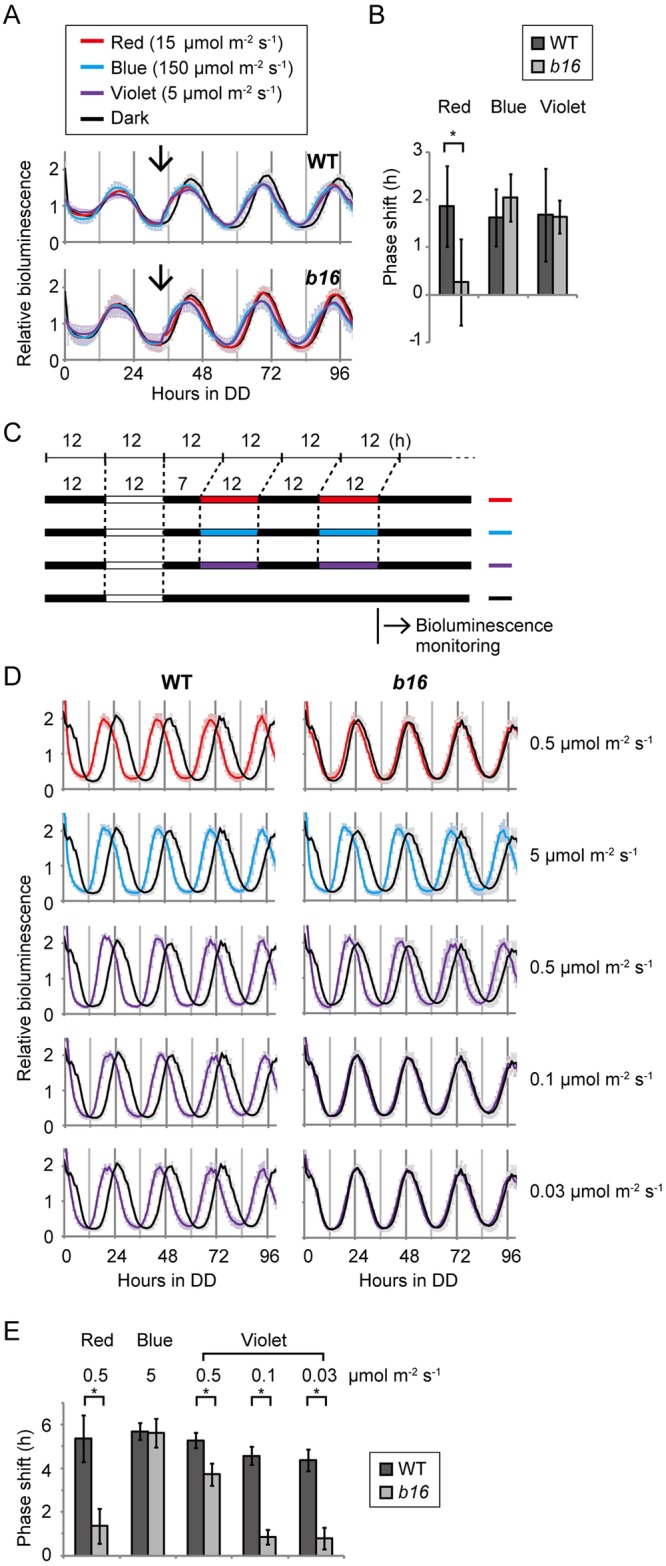
Bioluminescence rhythms of 5-day-old spot cultures of WT and b16 cells on HS agar media in white 96-well plates were measured. (A) Bioluminescence trace of a phase shifting experiment. Cells were exposed to a 12 h dark/12 h white light to entrain their circadian clock, and then bioluminescence rhythms were monitored in DD. 5 min light pulses were given at the 34 h time point in DD (arrows). Bioluminescence data are detrended by dividing by the 24 h moving average, and are shown as the mean ± SD of 10 independent cultures. (B) The amount of phase shift. Mean ± SD of the 10 independent cultures of the phase difference (h) between light-pulsed and dark control samples are shown. * P < 0.001 (Student’s t-test). (C) Schematic view of light conditions of the re-entrainment experiment. Cells were entrained by the first LD cycle of white light, and then re-entrained to a 5 h phase advanced LD cycles of red, blue, and violet light. Bioluminescence was monitored after release into DD. (D) Bioluminescence trace of re-entrainment experiment. Data are detrended by dividing by the 24 h moving average, and are shown as the mean ± SD of 10 independent cultures. (E) The amount of phase shift. The mean ± SD of 10 independent cultures of the phase difference (h) between light-entrained and dark control samples is shown. * P < 0.001 (Student’s t-test).
In addition, we tested re-entrainment of the circadian rhythm. For this purpose, light/dark (LD)-entrained cells were subjected to two cycles of 5 h phase-advanced light/dark cycles of violet, blue, or red light, and then released into DD for determination of their circadian phase (Fig 3C). In the WT strain, the bioluminescence rhythm was advanced for 4.1 to 5.4 h compared to dark controls in all light qualities tested (Fig 3D and 3E), indicating that the circadian clock re-entrained to new LD cycles. However, re-entrainment was clearly inhibited in the b16 mutant in red and weaker violet light conditions (Fig 3D and 3E). These results indicate that the phase-resetting of the circadian clock by red and violet light are impaired in the b16 mutant.
Light-induced phosphorylation of ROC15 is impaired in b16
Next, we tested the effect of b16 mutation on several molecular aspects of ROC15. ROC15 is a nuclear protein that accumulates at night and is phosphorylated gradually in the darkness and is phosphorylated further after light exposure [21]. To analyze ROC15 in a b16 background, we crossed b16 with a ROC15-HA strain that expresses a hemagglutinin (HA) epitope-tagged ROC15 [21]. ROC15-HA was detected in the nucleus even in the b16 background (Fig 4A), indicating that the b16 mutation does not affect subcellular localization of ROC15. Western blot analysis of ROC15-HA from b16 cells in the darkness detected a broad band, which is indicative of phosphorylated ROC15 [21] (Fig 4B, 0 min). However, no further mobility shift was observed in b16 after red and violet light pulses (Fig 4B, 5 and 10 min). These results suggest that light-induced phosphorylation of ROC15, but not dark phosphorylation, is impaired in b16. After the blue light pulse, however, the further mobility shift was observed in b16 as well as in the WT (Fig 4B). Taken together, these results indicate that b16 has a defect in a red and violet light signaling pathway at a step prior to or at ROC15 phosphorylation.
Fig 4. Subcellular localization and phosphorylation of ROC15.
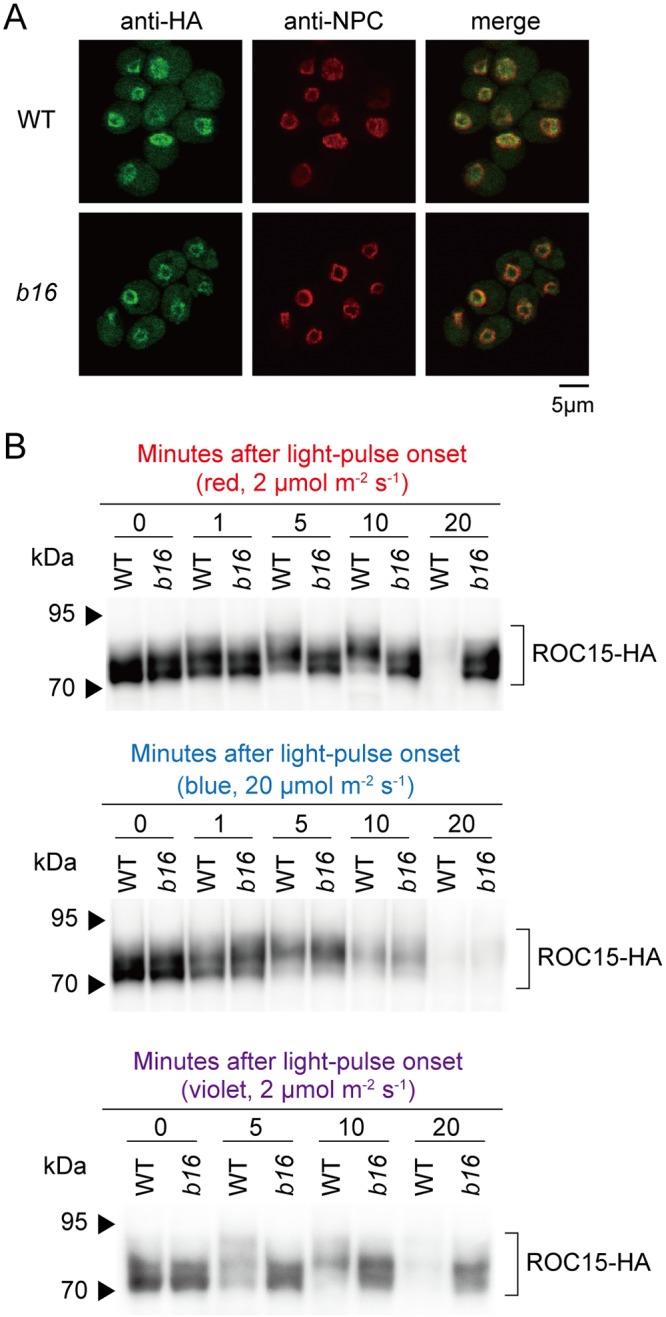
The b16 strain (mt+) was genetically crossed with the ROC15-HA strain (mt-). A progeny harboring b16 mutation and ROC15-HA transgene was selected by spot tests for antibiotic resistance and genomic PCRs. The hygromycin-resistance is genetically linked to b16 (see S5 Fig). LD-synchronized cultures of the progeny in HS liquid medium were used. (A) Cells were harvested at midnight and subjected to immunocytochemistry. The nuclear membrane was counterstained using an antibody against the nuclear pore complex (NPC). Photographs obtained under the same settings of microscope and detector are shown. (B) Cells were exposed to a 0.5 min pulse of red (660 nm, 2 μmol∙m-2∙s-1), blue (470 nm, 20 μmol∙m-2∙s-1), or violet light (405 nm, 2 μmol∙m-2∙s-1) at midnight, and total protein extracts were subjected to Western blot analysis.
In addition, the impaired ROC15-HA degradation in b16 was evident at the 20 min time point of red and violet light pulsed cells (Fig 4B). This excludes the possibility that the b16 phenotype in ROC15-LUC bioluminescence is an artifact caused by a defect in mechanisms related to bioluminescence.
mRNA regulations of ROC15 and other genes are essentially not affected by b16
The level of ROC15 mRNA is downregulated by light [11,21]. Thus, we examined mRNA downregulation by red light in the b16 mutant. As a result, decreased levels of mRNA transcripts were observed in b16 (Fig 5A). This indicates that the light signaling pathway for ROC15 mRNA downregulation is independent of b16.
Fig 5. Red light response of ROC15 and other mRNAs.
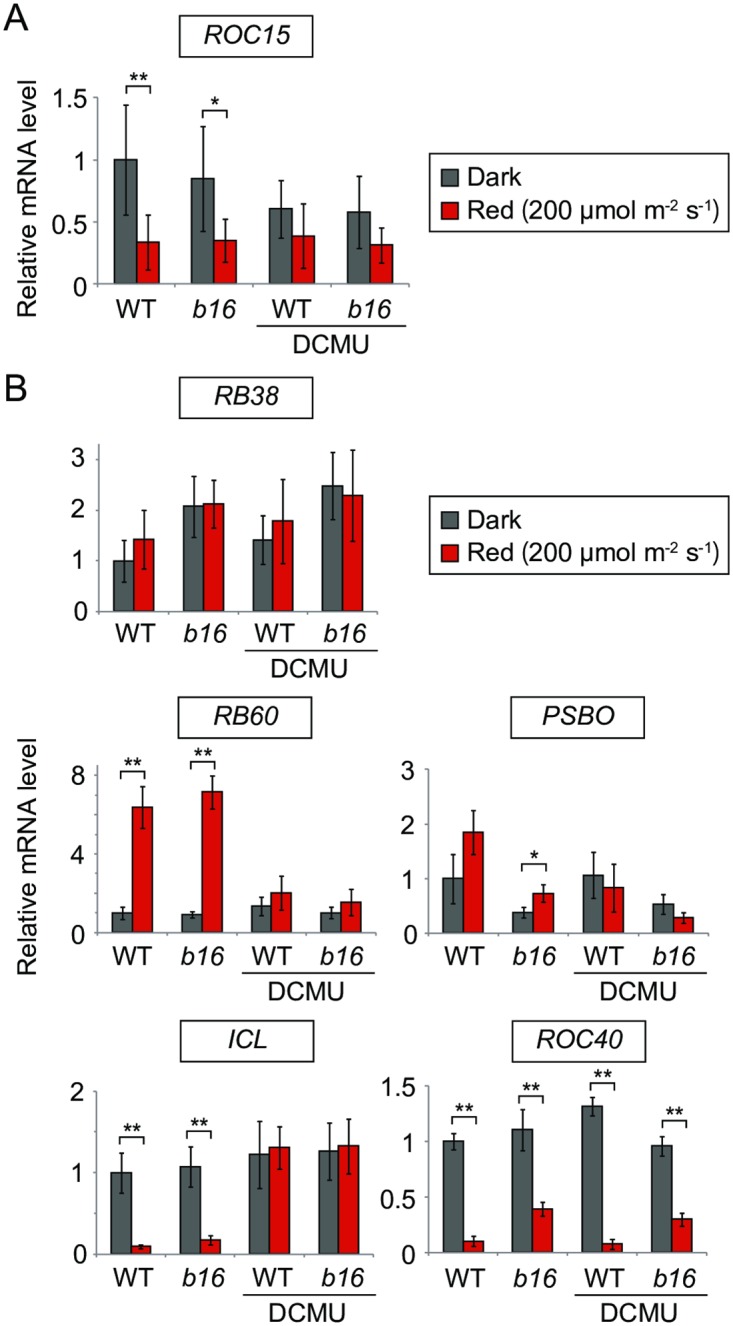
Early log phase TAP liquid cultures (approximately 5 x 105 cells/mL) of WT and b16 were adapted to darkness for one day, and then exposed to red light (200 μmol∙m-2∙s-1) for 2 h. DCMU was added to the cultures before dark adaptation at a final concentration of 10 μM. (A, B) RT-qPCR result of ROC15 (A) and other genes (B). Graphs represent the mean ± SD of 3–6 independent experiments. The transcript abundances relative to RCK1 were further normalized by dividing by the average for the dark control of the WT for easy comparison. * P < 0.05, ** P < 0.01 (Student’s t-test).
In addition, there are several reports describing red light responses on the mRNA level. Among them, we tested three red light up-regulated genes (RB38, RB60, and PSBO) and two down-regulated genes (ICL and ROC40) [7,10,11]. As in previous reports, obvious increases in RB60 and decreases in ICL and ROC40 expression were observed in our experimental conditions (Fig 5B). 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) blocked the response of RB60 and ICL (Fig 5B), indicating that these responses are dependent on photosynthetic electron transport. Essentially the same red light responses were observed in the b16 background (Fig 5B). These results indicate that the light signaling pathways for these genes are independent of b16. Although the decrease of ROC40 mRNA level was weaker in b16 (Fig 5B), b16 mutation should not be responsible for this phenotype, as this phenotype was not rescued in a complemented strain of b16 (see below).
Contrary to the previous report [10], little or no red-light-induction was observed in RB38 and PSBO mRNA levels (Fig 5B). This may be due to differences in the duration of dark adaptation (1 day [this study] vs. 1 week [10]) or the timing of sample collection after light-on (2 h [this study] vs. within 1 h [10]).
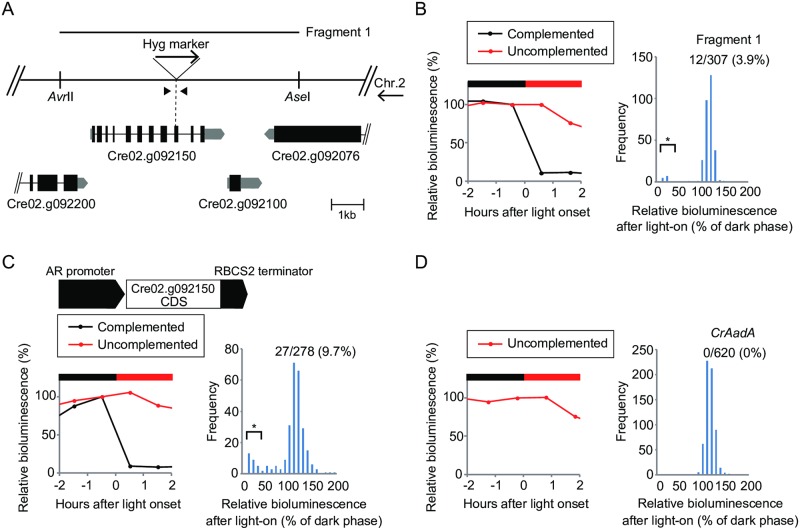
Identification of the disrupted gene in b16
To identify the disrupted gene in b16, we first examined genetic linkage between the b16 phenotype and hygromycin-resistance. Of progeny obtained in a genetic backcross to the parental strain, all hygromycin-resistant (HygR) and sensitive (HygS) progenies showed b16 and WT phenotypes in the ROC15 light response, respectively (S5A Fig). In addition, Southern blot analysis of the marker gene in the genomic DNA (gDNA) of b16 detected single bands (S5B Fig). These results indicate that b16 has a single insertion of the marker gene causing the b16 phenotypes. Thermal asymmetric interlaced PCR (TAIL-PCR) for the downstream of the marker gene amplified a DNA fragment (S5C Fig) corresponding to an uncharacterized gene on chromosome 2 (Cre02.g092150) (Fig 6A). PCR with a specific primer set bracketing the insertion site (Fig 6A, arrowheads) amplified a longer DNA fragment in b16 than in the WT (S5D Fig), and direct sequencing analysis of the fragment revealed that the marker gene was inserted into an exon of Cre02.g092150 (C. reinhardtii ver. 5.5, Phytozome, Joint Genome Institute) [26] with a small (8 bp) deletion (Fig 6A, Hyg marker). A gDNA fragment containing this gene (Fig 6A, Fragment 1) almost fully complemented the red light response phenotype of b16 (Fig 6B), and a WT transcript of Cre02.g092150 was restored in the complemented strains (S6 Fig). In addition, Cre02.g092150 cDNA driven by the HSP70A/RBCS2 fusion promoter (AR promoter) [27] complemented the phenotype again (Fig 6C). As a negative control, we transformed the b16 mutant with a selectable marker gene (CrAadA) alone and confirmed that no revertant appeared in this experimental condition (Fig 6D).
Fig 6. Identification of the disrupted gene in b16.
(A) Schematic representation of genomic region around the insertion site (shown in reverse orientation of the sequence for chromosome 2). The specific primers bracketing the insertion site are shown as arrowheads. (B–D), Complementation of the b16 phenotype. b16 mutant was transformed with a gDNA fragment (B, Fragment 1) and cDNA of the Cre02.g092150 gene (C). The left panels show ROC15-LUC bioluminescence of representative transformants monitored under the same conditions as Fig 1. Data before and after the light onset of the first DL cycle are shown. The bioluminescence level just before light onset was set to 100. The right panels are histograms representing the distribution of bioluminescence levels of all transformants after light on (relative to the dark phase). Asterisks indicate complemented transformants (the relative bioluminescence level after light on < 30). Numbers of complemented transformants and the rate of complementation are indicated in the graphs. A negative control experiment where b16 mutant cells were transformed by CrAadA marker only is shown (D).
The Cre02.g092150 is annotated as a part of large single gene fused with two upstream genes (Cre02.g092200 and Cre02.g092250) in GenBank/EMBL/DDBJ (XP_001701983) [26], presumably by an automatic annotation (S7A Fig). Cre02.g092200 encoding a protein motif that putatively acts as a binding site for pyrroloquinoline quinone (PQQ), a known redox cofactor [28]. Although, to our knowledge, there is no evidence that PQQ acts as a cofactor for light signaling molecules, the existence of this motif encouraged us to search for longer variants fused with the upstream genes. Through RT-PCR analysis, we could detect a transcript corresponding to Cre02.g092150, but longer variants were undetectable at least under our PCR conditions (S7B Fig). Furthermore, the complementation rates of the b16 phenotype were essentially identical between longer gDNA fragments covering the upstream genes and Cre02.g092150 gene alone (S7C Fig). These results indicate that Cre02.g092150 is sufficient to rescue the red light response of b16.
Cre02.g092150 is responsible for other phenotypes of b16
In addition to the red light response of ROC15-LUC, we tested whether Cre02.g092150 can restore the other b16 phenotypes or not. In the complemented strains (Fig 6B, S6 Fig), the weak response to violet light in ROC15-LUC bioluminescence (Fig 2) was also restored to the WT level at all light intensities tested (S8 Fig). To examine the re-entrainment phenotype (Fig 3C, 3D and 3E), the b16 mutant carrying chloroplast bioluminescence reporter (S3 Fig) was transformed with the Fragment 1 (Fig 6A). Some transformants showed a normal re-entrainment phenotype (S9A and S9B Fig) to the 5 h advanced red light cycle (Fig 3C). In addition, the restoration of re-entrainment was observed in not only red light but also violet light in these complemented strains (S9C Fig). To test the phenotype of ROC15 phosphorylation, we first complemented the b16 mutation by a FLAG-tagged version of Cre02.g092150 gene (Cre02.g092150-FLAG) (S10 Fig), and then the complemented strain was genetically crossed with a ROC15-HA strain [21] to obtain progenies that harboring the ROC15-HA gene and both of the b16 mutation and Cre02.g092150-FLAG gene. The phosphorylation of ROC15-HA by red and violet light pulses was restored in this strain (S11 Fig), indicating genetic complementation of the phosphorylation phenotype of b16 by Cre02.g092150-FLAG transgene. Taken together, these results indicate that Cre02.g092150 is responsible for almost all the b16 phenotypes in the ROC15 light response. On the other hand, the weak downregulation of ROC40 mRNA by red light (Fig 5B) was not rescued in the complemented strain (S12 Fig), suggesting that this phenotype is not caused by the mutation of Cre02.g092150 but rather by the genetic background of the strain.
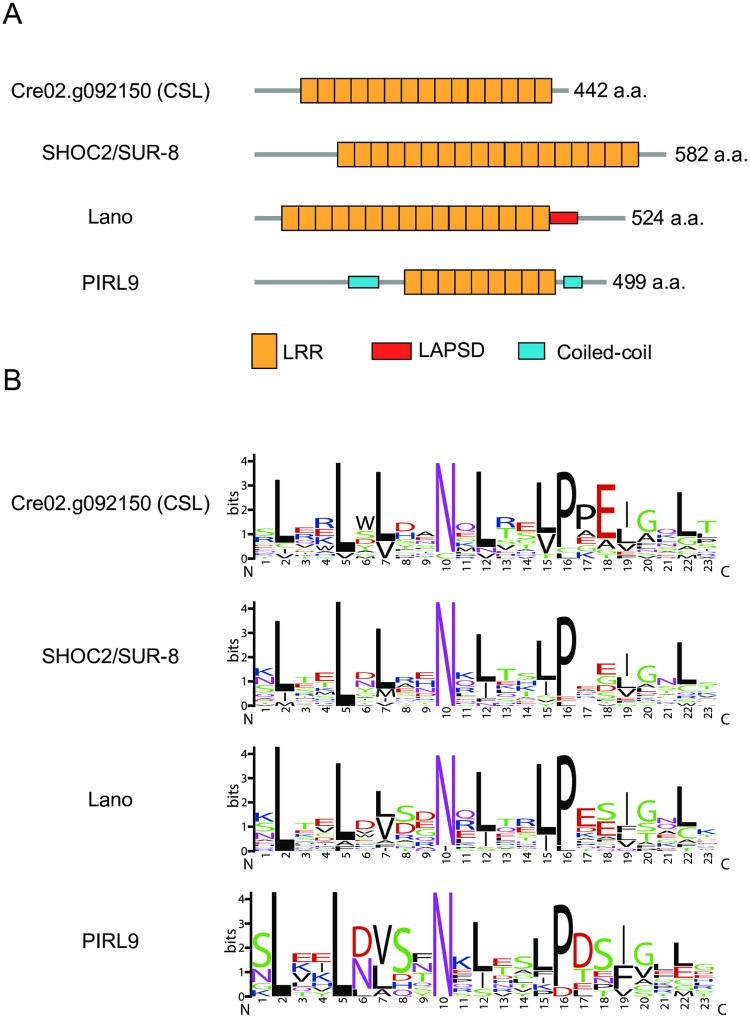
Cre02.g092150 encodes a LRR protein similar to RAS-signaling-related LRR proteins
Cre02.g092150 encodes a 442 amino acid protein comprised almost exclusively of LRR (Fig 7A). A homology search against protein sequences of major model organisms in GenBank/EMBL/DDBJ revealed that a hypothetical protein of Volvox carteri, a close relative of Chlamydomonas, was 76% identical to Cre02.g092150. In addition, many LRR proteins in a wide range of organisms, including animals and land plants, were detected, although their identities to Cre02.g092150 were low (< 40%). Among them, Cre02.g092150 showed relatively high similarity to animal SHOC2/SUR-8 proteins and LAP (LRR and PDZ [PSD-95/Disc Large/ZO-1]) family proteins [29–31], and their close relatives in land plants (PIRL [Plant Intracellular Ras-group LRR] family proteins) [32]. SHOC2/SUR-8 is known to act as a scaffold protein for interaction of RAS and RAF proteins [33]. In addition, LAP proteins are also known to associate with the SHOC2/SUR8-RAS complex [34,35]. SHOC2/SUR-8 and Cre02.g092150 are both composed almost extensively of LRRs [29,30] (Fig 7A). LAP family proteins have an N-terminal LRR domain followed by a LAP-specific domain (LAPSD) and a C-terminal PDZ domain, but Lano, a member of the family, lacks the PDZ domain [36] (Fig 7A). PIRL proteins have much shorter LRR domains and more extensive N-terminal regions [32] (Fig 7A). The LRR consensus amino acid sequences of these proteins, including Cre02.g092150, are highly similar (Fig 7B). The consensus sequences indicate that they belong to the plant-specific class of the LRR subfamily [37,38] but lack a glycine residue that is found in the extracellular LRR module of plant LRR-RLK (LRR Receptor-Like protein Kinase) family proteins [39] (Fig 7B, between positions 13 and 14). Collectively, these data suggest that the Chlamydomonas Cre02.g092150 protein is a close relative of RAS-signaling-related LRR proteins found in other organisms. Therefore, we hereafter refer to Cre02.g092150 gene and b16 mutant as CSL (Chlamydomonas SHOC2/SUR8-like LRR) gene and csl mutant, respectively.
Fig 7. Sequence analysis of Cre02.g092150.
(A) Schematic representation of domain architecture of Cre02.g092150 and other LRR proteins. PIRL9 is shown as a representative of the PIRL family. (B) Consensus LRR sequences visualized by the program WebLogo (http://weblogo.berkeley.edu/)[49].
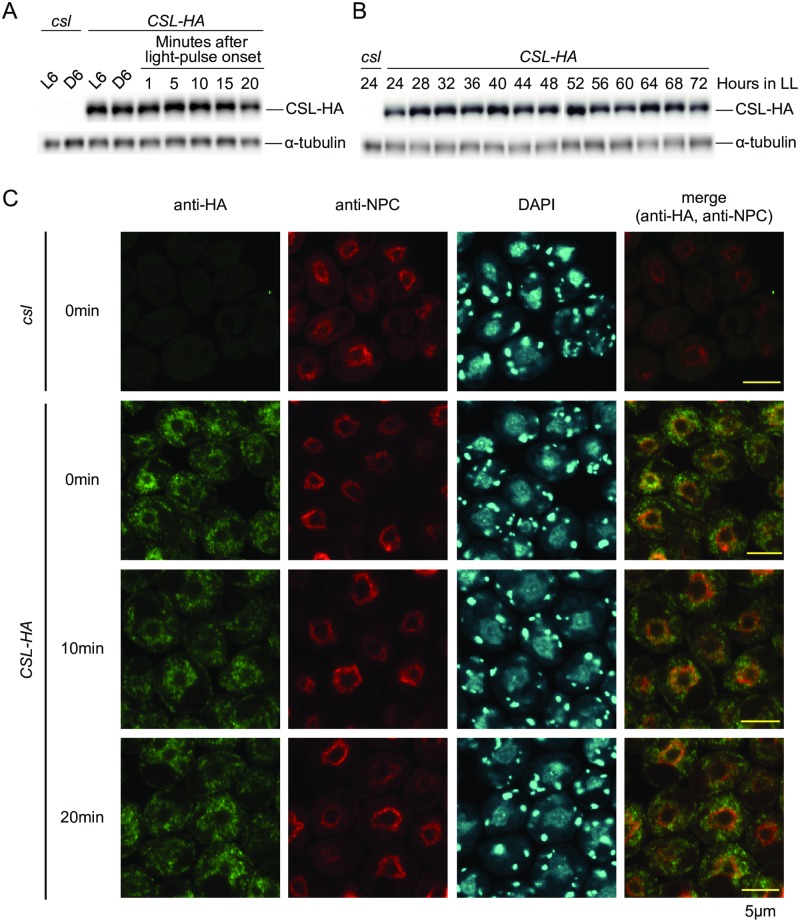
CSL is constitutively expressed and localized to the cytoplasm
To analyze expression of CSL gene product, we inserted a codon-adapted HA-epitope tag [21] at the end of the CSL coding sequence (CDS) of the gDNA fragment (Fragment 1; Fig 6A). This DNA fragment fully complemented the csl phenotype (S13 Fig), indicating that the tagged version of the CSL protein (CSL-HA) is functional. CSL-HA protein was expressed at almost the same level at midday and midnight in a 12-h light/dark cycle and after light pulse at midnight (Fig 8A). In addition, no obvious circadian rhythm was detected in the level of CSL-HA protein (Fig 8B). After immunocytochemical staining, CSL-HA signals were detected in perinuclear and cytoplasmic regions (Fig 8C), but no obvious changes in localization were detected after light pulse (Fig 8C). These results indicate that CSL protein is constitutively expressed and localized to the cytoplasm.
Fig 8. Expression analysis of CSL protein.
(A) Western blot analysis of CSL-HA in LD and after light pulse. Cell were harvested from LD-entrained HS liquid cultures at midday and midnight. A red light pulse (2 μmol∙m-2∙s-1, 0.5 min) was given at midnight, and cells were harvested at times indicated in the graph. 2 μg of total protein was loaded in each lane. (B) Western blot analysis of CSL-HA in a circadian condition. Mid-log-phase (approximately 2 x 106 cells/mL) HS liquid cultures at 24°C were transferred to 12 h darkness at 17°C to synchronize the circadian clock, and then released into a continuous light condition (2 μmol∙m-2∙s-1, white light, 17°C). 10 μg of total protein was loaded in each lane. Western blots for α-tubulin are shown as a reference protein (A,B). (C) Immunocytochemical staining of CSL-HA. Cells prepared as described in (A) were used. Cells were counterstained with NPC antibody and 4′,6-diamidino-2-phenylindole (DAPI). Photographs obtained under the same settings of microscope and detector are shown. csl mutant was used as negative controls (A,B,C).
Discussion
We previously demonstrated that the ROC15 gene is necessary for normal phase-resetting in Chlamydomonas through analysis of an insertional roc15 mutant expressing little or no ROC15 mRNA [14,21]. However, it remained unclear whether the light-induced degradation of ROC15 was important for phase resetting or the presence of ROC15 was essential for it. In this study, we showed that phosphorylation/nuclear localization of ROC15 and circadian rhythm in DD are normal in csl mutant (Fig 4, S3 Fig). This indicates that ROC15 functions normally in the oscillatory mechanism of the circadian clock. It seems that the light response of ROC15 protein was specifically affected in csl. This supports our previous suggestion that the light-induced degradation event in ROC15 protein is important for the phase-resetting mechanism of the Chlamydomonas circadian clock.
Our results clearly showed that blue and red/violet signals are transmitted separately in the light-induced degradation of ROC15 protein (Fig 2). On the other hand, b19 and b20, as well as roc114-1 [21], showed their phenotypes in both blue and red light (S2 Fig). This result suggests that these signals are integrated into a common mechanism. It is not clear whether they are integrated at the point of ROC15 or upstream (e.g., the kinase for ROC15). Further analysis of b19 and b20 may reveal the integration point of these light signaling pathways.
In addition, we showed that ROC15 mRNA downregulation by red light is independent of CSL (Fig 5). This finding suggests that there is another red light signaling pathway for mRNA regulation of ROC15. However, this is thought to be a supportive pathway in the phase resetting mechanism, because the red light response evident by the ROC15 protein level and phase resetting is largely abolished in csl, regardless of its normal mRNA response (Figs 2 and 3). ROC15 is a night phase expressed clock protein [21], and its mRNA level usually remains relatively high at the subjective dawn [14]. Thus, mRNA downregulation may be important to prevent re-accumulation of ROC15 protein and facilitate the night-to-day progression of the circadian clock.
CSL does not seem to be a light sensor because it does not possess known chromophore-binding domains; however, we do not exclude the possibility that its LRR may act as a novel chromophore binding site (Fig 7A). Although photoreceptor(s) in the CSL-related pathway remain unclear, it might be possible to speculate on the absorption spectrum from the equal-quantum action spectra in the WT and csl (Fig 2). S14 Fig shows a predicted contribution of the CSL pathway for ROC15 light response calculated by subtracting the sensitivity in the csl mutant from that in the WT. This perhaps reflects the in vivo action spectrum of CSL pathway. To our knowledge, there are no known photoreceptors in Chlamydomonas that show absorption spectra such as this. These spectral properties seem to be similar to some bilin-based photoreceptors, such as phytochromes and cyanobacteriochromes [40]. There are no obvious orthologs of them in the Chlamydomonas genome [26,41]; however, it is known that bilins (e.g. biliverdin [BV] and phycocyanobilin [PCB]) are synthesized in Chlamydomonas as retrograde signaling molecules from the chloroplast to the nucleus [42]. If an unknown bilin-based photoreceptor is involved in ROC15 light response, there is another possibility that CSL is related to bilin synthesis. However, this possibility is not probable given the fact that the csl phenotype was not rescued through the addition of bilins to the media (S15 Fig). In any case, CSL would be implicated in the red/violet signaling pathway. Further studies of CSL will shed light on a new light response mechanism of algae.
Materials and methods
Chlamydomonas strains and media
A reporter strain harboring ROC15-LUC gene in the nuclear genome was used as a WT strain [21]. In addition, CBR, roc114-1, and ROC15-HA strains [14,21] were also used. All strains are derivatives of CBR34 strain which shows a robust and stable bioluminescence rhythm [14]. Tris-acetate-phosphate (TAP) [43] and high-salt (HS) [44] media were used in this study.
Transformation
Nuclear transformation of Chlamydomonas by electroporation was performed as described previously [21,45]. For generation of insertional mutants, 15–30 ng of transforming DNA was used per electroporation of 2.5 x 107 cells.
Bioluminescence monitoring
Luciferin was added to culture media at the final concentration of 100 or 200 μM. Cell cultures in white 96-well and black 24-well plates were measured using the integrated automatic bioluminescence monitoring apparatus [22] and the highly sensitive bioluminescence monitoring apparatus (CL24-W and CL24A-LIC; Churitsu Electric Corp., Nagoya, Japan), respectively. For monitoring during light phases, cells were kept in darkness for 210 s just before bioluminescence measurement at each time point to decrease the delayed light emission of chlorophyll. Parameters of circadian rhythms were calculated with the Rhythm-Analyzing Program [23].
Light sources
We used fluorescence tubes (FLR 40S W/M; Panasonic) for white illumination and LEDs for violet (405 nm with a full-width at half maximum [FWHM] of 14 nm), blue (470 nm [FWHM of 27 nm]), and red (660 nm [FWHM of 24 nm]) illuminations (ISL-150×150-VV, ISL-150×150-BB, and ISL-150×150-RR, respectively; CCS, Kyoto, Japan). For screening, we used a larger size panel (30 × 60 cm) of red LED (660 nm [FWHM of 18 nm]) (LED Grow Light M; Home Grown, Yokohama, Japan). For the action spectra, we used a tunable monochromatic light source (PVL-3300; Asahi Spectra, Tokyo, Japan). Fluorescence rates were measured at the surface of plates or flasks using a light meter (Model LI-250; LI-COR, NE, USA) attached to a quantum probe (LI-190; LI-COR) or a multi-channel spectrometer (MC2100; Otsuka Electronics, Osaka, Japan). Spectrograms of light sources used in this study are shown in S16 Fig. We used an undetectable level of blue LED light as a safety light for harvesting the cells in darkness.
Western blot analysis
Western blot analysis was performed as described previously [21]. We used rat monoclonal anti-HA antibody (1:10,000–1:50,000; clone 3F10, Roche), mouse monoclonal anti-HA antibody (1:1,000; COVANCE, clone 16B12), and mouse monoclonal anti-FLAG antibody (1:1,000; Sigma, clone M2) as the primary antibody. Horseradish peroxidase (HRP) conjugated antibodies were used as the secondary antibody.
Immunocytochemistry
Immunocytochemistry was performed as described previously [21]. For detection of HA-tagged proteins, we used rat monoclonal anti-HA antibody (clone 3F10; 1:1,000; Roche) and Alexa488-conjugated goat anti-rat IgG (Thermo Fisher Scientific) as the primary and secondary antibodies, respectively. Fluorescence was observed with a laser-scanning confocal fluorescence microscope (FV10i-DOC, Olympus).
Genetic analyses for identification of disrupted gene
Genetic crosses, Southern blot analysis, and TAIL-PCR were performed as described previously [14].
RT-PCR and RT-qPCR
Total RNA was extracted using TRIzol reagent (ThermoFisher Scientific). RNA concentrations were determined using a spectrophotometer (UVmini-1240; SHIMADZU). For RT-PCR, reverse transcription was performed using SuperScript II (ThermoFisher Scientific) or ReverTra Ace qPCR RT Master Mix (TOYOBO), and following PCR amplification was performed using KOD FX neo (TOYOBO). For RT-qPCR, one microgram of total RNA was used in each reverse transcription reaction using ReverTra Ace qPCR RT Master Mix. Following qPCR was performed using KOD FX neo supplemented with SYBR® Green I Nucleic Acid Gel Stain (TaKaRa) and ROX dye (ThermoFisher Scientific) at final concentrations of 0.05x (200,000-fold dilution) and 0.5 μM, respectively. We used StepOnePlus (ThermoFisher Scientific) for qPCR at the following cycling conditions with standard ramp speed: 2 min at 94°C, 40 x (10 sec at 98°C, 1 min at 68°C). Relative transcript abundances were determined by the relative standard curve method of StepOnePlus using RCK1 as a reference gene for normalization [46]. Quantification standards for each target cDNA were obtained using band-purified PCR products as templates. In order to avoid amplifying contaminating genomic DNA, we designed all qPCR primers to anneal across a splicing junction of transcript, and to contain at least one intron between the primer pairs. Primers used for this study are provided in S1 Table. Target specificity was confirmed by melt curve method for all samples and by gel electrophoresis and sequencing for some representative samples.
Plasmid construction
Plasmids for complementation of csl by gDNA fragments were generated as follows: The paromomycin resistance cassette of pAC103PL [47] was replaced with the spectinomycin resistance cassette from pALM32 [48] by ligating the 3.3-kb NotI-BamHI fragment of pAC103PL and the 2.2-kb NotI-BamHI fragment of pALM32. The resulting plasmid termed pLaadA has a multicloning site from pAC103PL upstream of the spectinomycin resistance cassette. A bacterial artificial chromosome (BAC) clone (CRB1-063O12) carrying a csl locus was obtained from the genomic library described in the Kyoto Chlamydomonas Genome Database (KCGD; http://www.molecule.lif.kyoto-u.ac.jp/kcgd.html). Furthermore, 7.4-kb AvrII/AseI (Fragment 1), 10.8-kb AvrII/AseI (Fragment 2), and 17.4-kb BspEI/AseI (Fragment 3) fragments were cloned into the multicloning site of pLaadA. For HA- and FLAG-tagged construct, the stop codon of CSL of Fragment 1 was replaced with an FseI restriction site, and codon adapted HA [21] or FLAG (S10 Fig) tag sequences was inserted. To remove vector backbone sequences, we digested the plasmids with PacI, and the fragments carrying the gDNA linked to the spectinomycin resistance cassette were used for csl mutant transformation.
A plasmid for complementation by CSL CDS was generated as follows: The CSL CDS was amplified by DraI and EcoRI site attached primers just upstream and downstream of the CDS, respectively. The DraI-EcoRI fragment was cloned into the NaeI/EcoRI digested pAR [47] to generate pAR/CSL. The PacI fragment containing the CSL expression cassette was co-transformed with the spectinomycin resistant cassette [48].
Supporting information
A roc114-1 mutant strain with a ROC15-LUC reporter was used. Cultures of the strain were prepared as described in Fig 1 and their bioluminescence was monitored.
(TIF)
Asynchronous TAP liquid cultures of b16, b19, and b20 in black 24-well plates were subjected to darkness for 6 h for accumulation of ROC15-LUC, and then a 5 min light pulse (Red: 2 μmol∙m-2∙s-1; Blue: 20 μmol∙m-2∙s-1) was administered by blue and red LED panels (arrows).
(TIF)
To replace the ROC15-LUC reporter gene in b16 with the chloroplast reporter gene of CBR, the b16 strain (mating type minus [mt-]) was genetically crossed with the CBR strain (mt+). All 96 progenies showed bioluminescence due to uniparental inheritance of chloroplast DNA. We omitted paromomycin-resistance progenies because they possess ROC15-LUC introduced into the genome with the paromomycin-resistance APHVIII gene [21]. Genotype was confirmed definitively by genomic PCR. (A) Parameters of the bioluminescence rhythm of progeny. 5-day old spot cultures of progenies on HS agar in white 96-well plates were subjected to 12 h dark/12 h light to synchronize the circadian clock, and then the bioluminescence rhythm was monitored in DD. A scatter plot of period length and phase angle of circadian bioluminescence rhythms of hygromycin-resistant (HygR) and sensitive (HygS) progenies is shown. The resistance is genetically linked to b16 (see S5 Fig). (B) Representative trace of a progeny of WT and b16. Genotypes of some progenies were confirmed by genomic PCR, and representative WT and b16 progenies were subjected to a repeated rhythm assay. Data are the mean ± SD of relative bioluminescence (the average of the third peak was set to 100) from 10 independent cultures of the progenies.
(TIF)
Cells were treated as described in Fig 3A. Data before and after the light pulse (arrows) are shown. The bioluminescence level just before light pulse was set to 100. Each point represents the mean ± SD of 10 independent cultures.
(TIF)
(A) Co-segregation of the b16 and hygromycin-resistance phenotypes. The b16 mutant was backcrossed to a WT (ROC15-LUC) strain, and then the bioluminescence response to red light and hygromycin-resistance of progenies were tested. The left panel shows ROC15-LUC bioluminescence of progenies monitored under the same conditions as Fig 1. Data around the light onset of first DL cycle are shown. The bioluminescence level just before light pulse was set to 100. The right panel is a histogram representing the distribution of bioluminescence levels of progenies after light on (relative to the dark phase). (B) Southern blot analysis of b16 gDNA. Genomic DNA from b16 cells was digested by restriction enzymes without recognition (restriction) sites in the aph7” marker gene. The three lanes are distant lanes within the same gel. (C, D) TAIL-PCR products (C) and PCR product of specific primers overlapping the insertion site (D). These products were separated on an agarose gel and stained with ethidium bromide. The TAIL-PCR product used for sequencing analysis is indicated by an arrowhead.
(TIF)
(A) Schematic representation of primer locations. (B) RT-PCR result by using primers bracketing the insertion site (A, Red arrows). The solid and open triangle indicate RT-PCR products from a WT and mutant transcript, respectively. The mutant transcript was longer than that of WT probably due to insertion of Hyg maker. (C) Quantification of transcripts. The total amount of Cre02.g092150 transcripts was determined by RT-qPCR by using primers indicated in A as blue arrows. The transcript abundances relative to RCK1 were further normalized by dividing by the WT level for easy comparison.
(TIF)
(A) Schematic representation of the b16 locus (shown in reverse orientation of chromosome 2 sequence). Gene models and encoded protein motifs are shown: the FK506-binding-protein-type peptidyl-prolyl cis-trans isomerase (FKBP_C), tetratricopeptide repeat (TPR), pyrroloquinoline quinone (PQQ), and leucine-rich repeat (LRR) domains. Fourteen forward primers (1–14) and one reverse primer (15) for RT-PCR analysis are represented by red and blue arrows, respectively. The bars at the bottom indicate genomic DNA fragments for complementation analysis. (B) RT-PCR analysis of the b16 locus. RT-PCR products (25 cycles) were separated on an agarose gel and stained with ethidium bromide. (C) Complementation of the b16 phenotype by gDNA fragments. Data are collected and shown as Fig 6B, 6C and 6D.
(TIF)
Asynchronous TAP liquid cultures of WT, b16, and the complemented strains in black 24-well plates were subjected to darkness for 3.5 h for accumulation of ROC15-LUC, and then a 0.5 min violet light pulse of indicated intensities was administered by a violet LED panel (arrows). The graphs show representative bioluminescence traces. The bioluminescence level just before light pulse was set to 100.
(TIF)
The b16 mutant harboring the chloroplast bioluminescence reporter (S3 Fig) was transformed with the Fragment 1. Re-entrainment of transformants to the 5 h advanced light cycles was observed in experimental conditions essentially the same as in Fig 3C. (A) Bioluminescence traces. A representative trace of a complemented transformant is shown. WT and b16 traces are shown for comparison. Bioluminescence levels were detrended by dividing by the 24 h moving average. (B) Histograms representing the distribution of the amount of phase shift of all transformants (phase difference between transformants and a dark control of b16). Asterisk indicates complemented transformants (the phase shift > 4.5 h). Numbers of complemented transformants and the rate of complementation are indicated in the graphs. (C) The amount of phase shift to red, blue, and violet light cycles of the complemented transformants (Comp.1 and Comp2). Graphs show the mean ± SD of 10 independent cultures of the phase difference (h) between light-entrained and dark control samples. * P < 0.01, ** P < 0.001 (Student’s t-test).
(TIF)
(A) Nucleotide and amino acid sequences of a codon-adapted FLAG tag for C. reinhardtii nuclear genome. (B) A schematic representation of the FLAG-tagged Cre02.g092150 gene. Black and gray boxes represent the CDS and untranslated regions (UTRs), respectively. (C) Complementation of the b16 phenotype. Asynchronous cultures of transformants of b16 mutant with Cre02.g092150-FLAG were subjected to darkness for 3 h for accumulation of ROC15-LUC, and then a 0.5 min red light pulse (660 nm, 2 μmol∙m-2∙s-1) was administered (arrow). The left graph shows representative bioluminescence traces, and the right panel is a histogram representing the distribution of bioluminescence level of all transformants after light pulse (relative to the pre-light pulse level). Asterisk represents complemented transformants. Numbers of complemented transformants and the complementation rate are indicated in the graph. (D) Western blot analysis of Cre02.g092150-FLAG. Cells of asynchronous cultures of complemented strains were harvested and subjected to Western blot analysis with an anti-FLAG antibody. A protein sample of b16 mutant was loaded as a negative control.
(TIF)
The Cre02.g092150-FLAG strain was crossed with the ROC15-HA strain [21], and progenies harboring the ROC15-HA and both the b16 mutation and Cre02.g092150-FLAG transgenes (Cre02.g092150-FLAG/b16) were selected by spot tests for antibiotic resistance and genotyping by genomic PCR. Cell cultures were prepared as described in Fig 4B, and exposed to a 0.5 min pulse of red light (660 nm, 2 μmol∙m-2∙s-1) or violet light (405 nm, 2 μmol∙m-2∙s-1) at midnight. Total protein extracts were subjected to Western blot analysis using anti-HA antibody.
(TIF)
Samples of red light exposed cells were prepared as described in Fig 5. RT-qPCR results of ROC40 of the WT, b16, and the complemented strain (Comp1; Fig 6B, S6 Fig) are shown. The transcript abundances relative to RCK1 were further normalized by dividing by the dark control level of the WT for easy comparison.
(TIF)
(A) Schematic representation of the CSL-HA gene. Black and gray boxes represent the CDS and UTRs, respectively. (B) Complementation of the csl phenotype by CSL-HA. csl mutant was transformed with CSL-HA gene. Data are collected and shown as Fig 6B, 6C and 6D. (C) Western blot analysis of CSL-HA. Cells of asynchronous TAP liquid cultures of the complemented strains were harvested and subjected to Western blot analysis with an anti-HA antibody. A protein sample of csl mutant was loaded as a negative control. (D) Light dose dependency of the light response of ROC15-LUC in CSL-HA strain. Asynchronous cultures of the CSL-HA strains were subjected to darkness for 3 h for accumulation of ROC15-LUC, and then 2 min red light pulses of indicated intensities were administered. Data indicated are relative bioluminescence levels of CSL-HA-expressed strains against that of the csl mutant 30 min after light pulse onset.
(TIF)
The sensitivity of ROC15 light response remaining in csl was subtracted from that of WT. Negative values were regarded as 0.
(TIF)
Asynchronous TAP cultures of WT and csl in black 24-well plates were subjected to darkness for 3 h for accumulation of ROC15-LUC, and then a red light pulse (2 μmol∙m-2∙s-1) were administered for 0.5 min (arrows). Billins (Frontier Scientific, UT, USA) were added to the cultures just before dark adaptation at the final concentrations indicated in the graphs. Each trace is the mean of bioluminescence from two independent cultures. The bioluminescence level just before light pulse was set to 100.
(TIF)
(A) Spectrograms of monochromatic light emitted by the tunable monochromatic light source PVL-3300 (Asahi Spectra, Tokyo, Japan). (B) Emission spectra of the violet, blue, and red LED (ISL-150×150-VV, ISL-150×150-BB, and ISL-150×150-RR, respectively; CCS, Kyoto, Japan).
(TIF)
(XLSX)
Acknowledgments
We would like to thank Takafumi Tezuka (Nagoya University) for providing the spectrometer (MC2100) and helpful discussions, Yoko Kuroki (RIKEN), Atsushi Toyoda (National Institute of Genetics), Isao Fujiyama (National Institute of Genetics), and Yuji Kohara (National Institute of Genetics) for contributions to the BAC library construction and the end-sequence database development. We would also like to thank Editage for providing editorial assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Japan Society for the Promotion of Science (KAKENHI; 25440179 to TM and 25291075 to MI), Kato Memorial Bioscience Foundation (to TM) and Kurata Memorial Hitachi Science and Technology Foundation (to TM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hegemann P. Algal sensory photoreceptors. Annu Rev Plant Biol. 2008;59: 167–89. 10.1146/annurev.arplant.59.032607.092847 [DOI] [PubMed] [Google Scholar]
- 2.Kianianmomeni A, Hallmann A. Algal photoreceptors: in vivo functions and potential applications. Planta. 2014;239: 1–26. 10.1007/s00425-013-1962-5 [DOI] [PubMed] [Google Scholar]
- 3.Sineshchekov OA, Jung K-H, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2002;99: 8689–94. 10.1073/pnas.122243399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296: 2395–8. 10.1126/science.1072068 [DOI] [PubMed] [Google Scholar]
- 5.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100: 13940–5. 10.1073/pnas.1936192100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang K, Beck CF. Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2003;100: 6269–74. 10.1073/pnas.0931459100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petridou S, Foster K, Kindle K. Light induces accumulation of isocitrate lyase mRNA in a carotenoid-deficient mutant of Chlamydomonas reinhardtii. Plant Mol Biol. 1997;33: 381–92. [DOI] [PubMed] [Google Scholar]
- 8.Reisdorph NA, Small GD. The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol. 2004;134: 1546–54. 10.1104/pp.103.031930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Im C-S, Eberhard S, Huang K, Beck CF, Grossman AR. Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 2006;48: 1–16. 10.1111/j.1365-313X.2006.02852.x [DOI] [PubMed] [Google Scholar]
- 10.Alizadeh D, Cohen A. Red light and calmodulin regulate the expression of the psbA binding protein genes in Chlamydomonas reinhardtii. Plant & cell physiology. 2010. pp. 312–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beel B, Prager K, Spexard M, Sasso S, Weiss D, Müller N, et al. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell. 2012;24: 2992–3008. 10.1105/tpc.112.098947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11: 764–76. 10.1038/nrm2995 [DOI] [PubMed] [Google Scholar]
- 13.Iliev D, Voytsekh O, Schmidt E-M, Fiedler M, Nykytenko A, Mittag M. A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 2006;142: 797–806. 10.1104/pp.106.085944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M. A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 2008;22: 918–930. 10.1101/gad.1650408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulze T, Prager K, Dathe H, Kelm J, Kiessling P, Mittag M. How the green alga Chlamydomonas reinhardtii keeps time. Protoplasma. 2010;244: 3–14. 10.1007/s00709-010-0113-0 [DOI] [PubMed] [Google Scholar]
- 16.Matsuo T, Ishiura M. New insights into the circadian clock in Chlamydomonas. International Review of Cell and Molecular Biology. 2010. [DOI] [PubMed] [Google Scholar]
- 17.Matsuo T, Ishiura M. Chlamydomonas reinhardtii as a new model system for studying the molecular basis of the circadian clock. FEBS Letters. 2011. pp. 1495–1502. 10.1016/j.febslet.2011.02.025 [DOI] [PubMed] [Google Scholar]
- 18.Ryo M, Matsuo T, Yamashino T, Ichinose M, Sugita M, Aoki S. Diversity of plant circadian clocks: Insights from studies of Chlamydomonas reinhardtii and Physcomitrella patens. Plant Signal Behav. 2016;11: e1116661 10.1080/15592324.2015.1116661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo T, Johnson CH, Hastings JW. Action Spectrum for Resetting the Circadian Phototaxis Rhythm in the CW15 Strain of Chlamydomonas: I. Cells in Darkness. Plant Physiol. 1991;95: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes-Stovall J, Howton J, Young M, Davis G, Chandler T, Kessler B, et al. Chlamydomonas reinhardtii strain CC-124 is highly sensitive to blue light in addition to green and red light in resetting its circadian clock, with the blue-light photoreceptor plant cryptochrome likely acting as negative modulator. Plant Physiol Biochem. 2014;75: 14–23. 10.1016/j.plaphy.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niwa Y, Matsuo T, Onai K, Kato D, Tachikawa M, Ishiura M. Phase-resetting mechanism of the circadian clock in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2013;110: 13666–71. 10.1073/pnas.1220004110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto K, Onai K, Furusawa T, Ishiura M. A portable integrated automatic apparatus for the real-time monitoring of bioluminescence in plants. Plant Cell Environ. 2005;28: 1305–1315. [Google Scholar]
- 23.Okamoto K, Onai K, Ishiura M. RAP, an integrated program for monitoring bioluminescence and analyzing circadian rhythms in real time. Anal Biochem. 2005;340: 193–200. 10.1016/j.ab.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 24.Berthold P, Schmitt R, Mages W. An engineered Streptomyces hygroscopicus aph7”gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153: 401–12. 10.1078/14344610260450136 [DOI] [PubMed] [Google Scholar]
- 25.Matsuo T, Onai K, Okamoto K, Minagawa J, Ishiura M. Real-time monitoring of chloroplast gene expression by a luciferase reporter: evidence for nuclear regulation of chloroplast circadian period. Mol Cell Biol. 2006;26: 863–870. 10.1128/MCB.26.3.863-870.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science. 2007;318: 245–250. 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroda M, Beck CF, Vallon O. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant J. 2002;31: 445–55. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann C, Klinman JP. Pyrroloquinoline quinone: a new redox cofactor in eukaryotic enzymes. Biofactors. 1988;1: 41–9. [PubMed] [Google Scholar]
- 29.Selfors LM, Schutzman JL, Borland CZ, Stern MJ. soc-2 encodes a leucine-rich repeat protein implicated in fibroblast growth factor receptor signaling. Proc Natl Acad Sci U S A. 1998;95: 6903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sieburth DS, Sun Q, Han M. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell. 1998;94: 119–30. [DOI] [PubMed] [Google Scholar]
- 31.Saito H, Santoni MJ, Arsanto JP, Jaulin-Bastard F, Le Bivic A, Marchetto S, et al. Lano, a novel LAP protein directly connected to MAGUK proteins in epithelial cells. J Biol Chem. 2001;276: 32051–5. 10.1074/jbc.C100330200 [DOI] [PubMed] [Google Scholar]
- 32.Forsthoefel NR, Cutler K, Port MD, Yamamoto T, Vernon DM. PIRLs: a novel class of plant intracellular leucine-rich repeat proteins. Plant Cell Physiol. 2005;46: 913–22. 10.1093/pcp/pci097 [DOI] [PubMed] [Google Scholar]
- 33.Yoshiki S, Matsunaga-Udagawa R, Aoki K, Kamioka Y, Kiyokawa E, Matsuda M. Ras and calcium signaling pathways converge at Raf1 via the Shoc2 scaffold protein. Mol Biol Cell. 2010;21: 1088–96. 10.1091/mbc.E09-06-0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai P, Xiong WC, Mei L. Erbin inhibits RAF activation by disrupting the sur-8-Ras-Raf complex. J Biol Chem. 2006;281: 927–33. 10.1074/jbc.M507360200 [DOI] [PubMed] [Google Scholar]
- 35.Young LC, Hartig N, Muñoz-Alegre M, Oses-Prieto JA, Durdu S, Bender S, et al. An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth. Mol Cell. 2013;52: 679–92. 10.1016/j.molcel.2013.10.004 [DOI] [PubMed] [Google Scholar]
- 36.Santoni M-J, Pontarotti P, Birnbaum D, Borg J-P. The LAP family: a phylogenetic point of view. Trends Genet. 2002;18: 494–7. [DOI] [PubMed] [Google Scholar]
- 37.Kobe B, Kajava A V. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11: 725–32. [DOI] [PubMed] [Google Scholar]
- 38.Ng ACY, Eisenberg JM, Heath RJW, Huett A, Robinson CM, Nau GJ, et al. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci U S A. 2011;108 Suppl: 4631–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torii KU. Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int Rev Cytol. 2004;234: 1–46. 10.1016/S0074-7696(04)34001-5 [DOI] [PubMed] [Google Scholar]
- 40.Burgie ES, Vierstra RD. Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell. 2014;26: 4568–83. 10.1105/tpc.114.131623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittag M, Kiaulehn S, Johnson CH. The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 2005;137: 399–409. 10.1104/pp.104.052415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duanmu D, Casero D, Dent RM, Gallaher S, Yang W, Rockwell NC, et al. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc Natl Acad Sci U S A. 2013;110: 3621–6. 10.1073/pnas.1222375110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sueoka N. Mitotic replication of deoxiribonucleic acid in Chlamydomonas reinhardti. Proc Natl Acad Sci U S A. 1960;46: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1965;54: 1665–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimogawara K, Fujiwara S, Grossman A, Usuda H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics. 1998;148: 1821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem. 2007;282: 25475–25486. 10.1074/jbc.M701415200 [DOI] [PubMed] [Google Scholar]
- 47.Matsuo T, Iida T, Ishiura M. N-terminal acetyltransferase 3 gene is essential for robust circadian rhythm of bioluminescence reporter in Chlamydomonas reinhardtii. Biochem Biophys Res Commun. 2012;418: 342–346. 10.1016/j.bbrc.2012.01.023 [DOI] [PubMed] [Google Scholar]
- 48.Meslet-Cladière L, Vallon O. Novel shuttle markers for nuclear transformation of the green alga Chlamydomonas reinhardtii. Eukaryot Cell. 2011;10: 1670–8. 10.1128/EC.05043-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14: 1188–90. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A roc114-1 mutant strain with a ROC15-LUC reporter was used. Cultures of the strain were prepared as described in Fig 1 and their bioluminescence was monitored.
(TIF)
Asynchronous TAP liquid cultures of b16, b19, and b20 in black 24-well plates were subjected to darkness for 6 h for accumulation of ROC15-LUC, and then a 5 min light pulse (Red: 2 μmol∙m-2∙s-1; Blue: 20 μmol∙m-2∙s-1) was administered by blue and red LED panels (arrows).
(TIF)
To replace the ROC15-LUC reporter gene in b16 with the chloroplast reporter gene of CBR, the b16 strain (mating type minus [mt-]) was genetically crossed with the CBR strain (mt+). All 96 progenies showed bioluminescence due to uniparental inheritance of chloroplast DNA. We omitted paromomycin-resistance progenies because they possess ROC15-LUC introduced into the genome with the paromomycin-resistance APHVIII gene [21]. Genotype was confirmed definitively by genomic PCR. (A) Parameters of the bioluminescence rhythm of progeny. 5-day old spot cultures of progenies on HS agar in white 96-well plates were subjected to 12 h dark/12 h light to synchronize the circadian clock, and then the bioluminescence rhythm was monitored in DD. A scatter plot of period length and phase angle of circadian bioluminescence rhythms of hygromycin-resistant (HygR) and sensitive (HygS) progenies is shown. The resistance is genetically linked to b16 (see S5 Fig). (B) Representative trace of a progeny of WT and b16. Genotypes of some progenies were confirmed by genomic PCR, and representative WT and b16 progenies were subjected to a repeated rhythm assay. Data are the mean ± SD of relative bioluminescence (the average of the third peak was set to 100) from 10 independent cultures of the progenies.
(TIF)
Cells were treated as described in Fig 3A. Data before and after the light pulse (arrows) are shown. The bioluminescence level just before light pulse was set to 100. Each point represents the mean ± SD of 10 independent cultures.
(TIF)
(A) Co-segregation of the b16 and hygromycin-resistance phenotypes. The b16 mutant was backcrossed to a WT (ROC15-LUC) strain, and then the bioluminescence response to red light and hygromycin-resistance of progenies were tested. The left panel shows ROC15-LUC bioluminescence of progenies monitored under the same conditions as Fig 1. Data around the light onset of first DL cycle are shown. The bioluminescence level just before light pulse was set to 100. The right panel is a histogram representing the distribution of bioluminescence levels of progenies after light on (relative to the dark phase). (B) Southern blot analysis of b16 gDNA. Genomic DNA from b16 cells was digested by restriction enzymes without recognition (restriction) sites in the aph7” marker gene. The three lanes are distant lanes within the same gel. (C, D) TAIL-PCR products (C) and PCR product of specific primers overlapping the insertion site (D). These products were separated on an agarose gel and stained with ethidium bromide. The TAIL-PCR product used for sequencing analysis is indicated by an arrowhead.
(TIF)
(A) Schematic representation of primer locations. (B) RT-PCR result by using primers bracketing the insertion site (A, Red arrows). The solid and open triangle indicate RT-PCR products from a WT and mutant transcript, respectively. The mutant transcript was longer than that of WT probably due to insertion of Hyg maker. (C) Quantification of transcripts. The total amount of Cre02.g092150 transcripts was determined by RT-qPCR by using primers indicated in A as blue arrows. The transcript abundances relative to RCK1 were further normalized by dividing by the WT level for easy comparison.
(TIF)
(A) Schematic representation of the b16 locus (shown in reverse orientation of chromosome 2 sequence). Gene models and encoded protein motifs are shown: the FK506-binding-protein-type peptidyl-prolyl cis-trans isomerase (FKBP_C), tetratricopeptide repeat (TPR), pyrroloquinoline quinone (PQQ), and leucine-rich repeat (LRR) domains. Fourteen forward primers (1–14) and one reverse primer (15) for RT-PCR analysis are represented by red and blue arrows, respectively. The bars at the bottom indicate genomic DNA fragments for complementation analysis. (B) RT-PCR analysis of the b16 locus. RT-PCR products (25 cycles) were separated on an agarose gel and stained with ethidium bromide. (C) Complementation of the b16 phenotype by gDNA fragments. Data are collected and shown as Fig 6B, 6C and 6D.
(TIF)
Asynchronous TAP liquid cultures of WT, b16, and the complemented strains in black 24-well plates were subjected to darkness for 3.5 h for accumulation of ROC15-LUC, and then a 0.5 min violet light pulse of indicated intensities was administered by a violet LED panel (arrows). The graphs show representative bioluminescence traces. The bioluminescence level just before light pulse was set to 100.
(TIF)
The b16 mutant harboring the chloroplast bioluminescence reporter (S3 Fig) was transformed with the Fragment 1. Re-entrainment of transformants to the 5 h advanced light cycles was observed in experimental conditions essentially the same as in Fig 3C. (A) Bioluminescence traces. A representative trace of a complemented transformant is shown. WT and b16 traces are shown for comparison. Bioluminescence levels were detrended by dividing by the 24 h moving average. (B) Histograms representing the distribution of the amount of phase shift of all transformants (phase difference between transformants and a dark control of b16). Asterisk indicates complemented transformants (the phase shift > 4.5 h). Numbers of complemented transformants and the rate of complementation are indicated in the graphs. (C) The amount of phase shift to red, blue, and violet light cycles of the complemented transformants (Comp.1 and Comp2). Graphs show the mean ± SD of 10 independent cultures of the phase difference (h) between light-entrained and dark control samples. * P < 0.01, ** P < 0.001 (Student’s t-test).
(TIF)
(A) Nucleotide and amino acid sequences of a codon-adapted FLAG tag for C. reinhardtii nuclear genome. (B) A schematic representation of the FLAG-tagged Cre02.g092150 gene. Black and gray boxes represent the CDS and untranslated regions (UTRs), respectively. (C) Complementation of the b16 phenotype. Asynchronous cultures of transformants of b16 mutant with Cre02.g092150-FLAG were subjected to darkness for 3 h for accumulation of ROC15-LUC, and then a 0.5 min red light pulse (660 nm, 2 μmol∙m-2∙s-1) was administered (arrow). The left graph shows representative bioluminescence traces, and the right panel is a histogram representing the distribution of bioluminescence level of all transformants after light pulse (relative to the pre-light pulse level). Asterisk represents complemented transformants. Numbers of complemented transformants and the complementation rate are indicated in the graph. (D) Western blot analysis of Cre02.g092150-FLAG. Cells of asynchronous cultures of complemented strains were harvested and subjected to Western blot analysis with an anti-FLAG antibody. A protein sample of b16 mutant was loaded as a negative control.
(TIF)
The Cre02.g092150-FLAG strain was crossed with the ROC15-HA strain [21], and progenies harboring the ROC15-HA and both the b16 mutation and Cre02.g092150-FLAG transgenes (Cre02.g092150-FLAG/b16) were selected by spot tests for antibiotic resistance and genotyping by genomic PCR. Cell cultures were prepared as described in Fig 4B, and exposed to a 0.5 min pulse of red light (660 nm, 2 μmol∙m-2∙s-1) or violet light (405 nm, 2 μmol∙m-2∙s-1) at midnight. Total protein extracts were subjected to Western blot analysis using anti-HA antibody.
(TIF)
Samples of red light exposed cells were prepared as described in Fig 5. RT-qPCR results of ROC40 of the WT, b16, and the complemented strain (Comp1; Fig 6B, S6 Fig) are shown. The transcript abundances relative to RCK1 were further normalized by dividing by the dark control level of the WT for easy comparison.
(TIF)
(A) Schematic representation of the CSL-HA gene. Black and gray boxes represent the CDS and UTRs, respectively. (B) Complementation of the csl phenotype by CSL-HA. csl mutant was transformed with CSL-HA gene. Data are collected and shown as Fig 6B, 6C and 6D. (C) Western blot analysis of CSL-HA. Cells of asynchronous TAP liquid cultures of the complemented strains were harvested and subjected to Western blot analysis with an anti-HA antibody. A protein sample of csl mutant was loaded as a negative control. (D) Light dose dependency of the light response of ROC15-LUC in CSL-HA strain. Asynchronous cultures of the CSL-HA strains were subjected to darkness for 3 h for accumulation of ROC15-LUC, and then 2 min red light pulses of indicated intensities were administered. Data indicated are relative bioluminescence levels of CSL-HA-expressed strains against that of the csl mutant 30 min after light pulse onset.
(TIF)
The sensitivity of ROC15 light response remaining in csl was subtracted from that of WT. Negative values were regarded as 0.
(TIF)
Asynchronous TAP cultures of WT and csl in black 24-well plates were subjected to darkness for 3 h for accumulation of ROC15-LUC, and then a red light pulse (2 μmol∙m-2∙s-1) were administered for 0.5 min (arrows). Billins (Frontier Scientific, UT, USA) were added to the cultures just before dark adaptation at the final concentrations indicated in the graphs. Each trace is the mean of bioluminescence from two independent cultures. The bioluminescence level just before light pulse was set to 100.
(TIF)
(A) Spectrograms of monochromatic light emitted by the tunable monochromatic light source PVL-3300 (Asahi Spectra, Tokyo, Japan). (B) Emission spectra of the violet, blue, and red LED (ISL-150×150-VV, ISL-150×150-BB, and ISL-150×150-RR, respectively; CCS, Kyoto, Japan).
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.