Abstract
Despite advances in management, bladder cancer remains a major cause of cancer related complications. Characterisation of gene expression patterns in bladder cancer allows the identification of pathways involved in its pathogenesis, and may stimulate the development of novel therapies targeting these pathways. Between 2004 and 2005, cystoscopic bladder biopsies were obtained from 19 patients and 11 controls. These were subjected to whole transcript-based microarray analysis. Unsupervised hierarchical clustering was used to identify samples with similar expression profiles. Hypergeometric analysis was used to identify canonical pathways and curated networks having statistically significant enrichment of differentially expressed genes. Osteopontin (OPN) expression was validated by immunohistochemistry. Hierarchical clustering defined signatures, which differentiated between cancer and healthy tissue, muscle-invasive or non-muscle invasive cancer and healthy tissue, grade 1 and grade 3. Pathways associated with cell cycle and proliferation were markedly upregulated in muscle-invasive and grade 3 cancers. Genes associated with the classical complement pathway were downregulated in non-muscle invasive cancer. Osteopontin was markedly overexpressed in invasive cancer compared to healthy tissue. The present study contributes to a growing body of work on gene expression signatures in bladder cancer. The data support an important role for osteopontin in bladder cancer, and identify several pathways worthy of further investigation.
Keywords: bladder cancer, osteopontin, gene expression analysis, molecular classification, microarray, muscle invasive
Introduction
Carcinoma of the bladder is the most common urothelial malignancy, and is the 6th commonest cancer in the world by incidence (1). Despite significant improvements in management, bladder cancer remains one of the commonest causes of cancer related mortality, ranked 6th and 11th in the UK in 2008 in men and women, respectively (2). Additionally, curative surgery or radiotherapy commonly results in significant long-term morbidity.
Localised bladder cancer may be divided into either non-muscle invasive bladder cancer (NMIBC) or into muscle invasive bladder cancer (MIBC). While the former may be cured by transurethral resection, treatment of the latter requires radical local management using either cystectomy or radical radiotherapy (with or without concurrent chemotherapy). Approximately a third of NMIBC tumours will recur and progress to muscle invasive over time (3). Several clinicopathological factors including tumour grade, stage, size, presence of carcinoma in situ, previous recurrence (4), and p53 status are associated with increased likelihood of progression. However, these factors are not currently clearly sufficient to identify patients who need more radical therapy at the outset. Similarly, stratification of patients with muscle invasive cancer into those most likely to recur or progress following local therapy would enable a more personalised approach to treatment.
Previous studies have employed microarrays to investigate gene expression profiles in bladder cancer (5–8). The identification of differentially regulated genes and gene networks allows the dissection of pathways and processes that are dysregulated in bladder cancer. This, in turn, provides invaluable clues relating to pathogenesis and moreover, may provide novel targets for drug development.
Here, we describe a study in which we profiled gene expression in 20 bladder cancer samples and compared this to 11 healthy urothelial samples. In addition to defining gene expression signatures for different subgroups, the study identified several genes and gene networks which are significantly deregulated in bladder cancer. These include osteopontin and the classical complement pathway. In this study we used the Affymetrix Human Gene 1.0 ST array which is a whole transcript based array compared to many other studies which used arrays querying only the 3′ end of transcripts.
Materials and methods
Patients and tumour samples
Cystoscopic tissue biopsies were collected between May 2004 and November 2005. Prior to undergoing cystoscopy, participants gave written informed consent for additional biopsies to be taken for this study during their medical procedure. Patients eligible for inclusion were those aged 18–80 years with a diagnosis of histologically confirmed superficial (NMIBC) or muscle invasive (MIBC) transitional cell carcinoma (n=14 and n=6, respectively), and who had not received any prior therapy. Control (C) samples were obtained from uninvolved tissues, for which no abnormalities were observed upon histological examination (n=11). Ten of these were paired with bladder cancer biopsies (4 nonmuscle invasive and 5 muscle invasive), while the last was from a patient without a diagnosis of malignancy. Demographic characteristics of the patient cohort are presented in Table I. MIBC samples displayed a basal immuno-phenotype.
Table I.
Patient demographics.
| Demographics | Controls (n=11) | NMIBC (n=14) | MIBC (n=6) |
|---|---|---|---|
| Age in years | |||
| Median, range | 79.2 (64–85.8) | 70.8 (47.0–81.9) | 80.14 (65.3–85.8) |
| Gender | |||
| Male | 8 (72.2) | 11 (78.6) | 3 (60.0) |
| Female | 3 (27.2) | 3 (21.4) | 2 (40.0) |
| Grade | |||
| G1 | N/A | 6 (42.8) | 0 (0) |
| G2 | N/A | 1 (7.1) | 0 (0) |
| G3 | N/A | 7 (50.0) | 6 (100) |
| T stage | |||
| PT1 | N/A | 5 (35.7) | 0 (0) |
| PT1B | N/A | 1 (7.1) | 0 (0) |
| Pta | N/A | 8 (57.1) | 0 (0) |
| PT2 | N/A | 0 (0) | 6 (100) |
| Recurrence | |||
| No | N/A | 12 (92.3) | 0 (0) |
| Yes | N/A | 1 (7.7) | 5 (100) |
| Survival in months (95% CI) | N/A | 81.2 (73.3-not estimatable) | 12.26 (0–35.6) |
N/A, not applicable.
Tumour samples for IHC analysis were obtained from the Liverpool Bioinnovation Hub Biobank (Liverpool, UK). Pathologically assessed samples used healthy bladder tissue (n=3) and MIBC tumour tissue (n=3).
Ethical considerations
Approval for the present study was obtained from the South Birmingham Local Research Ethics Committee. All participants gave written informed consent for their respective biopsies to be utilized in this study. Approval for further anaylsis was granted by the Liverpool Tissue Bank Ethics Committee (application no. 12-09). All participants gave written informed consent for their respective biopsies to be used in biomedical analysis.
Tissue sample preparation
Tissue biopsy samples were snap-frozen and stored in liquid nitrogen prior to RNA extraction.
RNA isolation
Total RNA was extracted from each tissue using TRIzol (Invitrogen) prior to purification on silica (RNeasy Mini columns; Qiagen, Hilden, Germany) according to the manufacturer's recommendation. An Agilent Bioanalyser was used to confirm RNA integrity and only samples having a RIN of 6.0 or more were used.
cDNA probe synthesis
A total of 200 ng of total RNA was converted to cDNA including targeted depletion of rRNA transcripts using a WT Expression kit (Ambion, Cambridge, MA, USA) according to the manufacturer's procedures. Resultant cDNA was fragmented and biotin end labelled using an Affymetrix WT Terminal Labelling kit (Affymetrix, Santa Clara, CA, USA).
Microarray hybridisation analysis
Labelled cDNA was hybridised to Affymetrix Human Gene 1.0 ST whole transcript-based arrays followed by washing and staining on an Affymetrix FS450 fluidics station. The Human Gene 1.0 ST Array (Affymetrix) comprised 764,885 distinct 25-nt probes interrogating 28,869 annotated genes based on the March 2006 (UCSC hg18, NCBI Build 36) human genome sequence assembly with comprehensive coverage of RefSeq, Ensembl and putative complete CDS GenBank transcripts.
Signal collection and analysis
Arrays were scanned using an Affymetrix Scanner 3000 7G. Affymetrix Command Console was used for instrument control and data acquisition. All procedures were carried out according to the standard Affymetrix protocols. Raw microarray data were submitted to ArrayExpress (Accession: E-MTAB-1560).
Statistical analysis of microarray data
Data transformation
Robust multi-array averaging, adjusted for non-specific binding, (GCRMA) was used for normalisation and summarisation of the probe intensity signals (9,10). Data were subject to RMA background correction and quantile normalisation across all chips (to ensure comparable mean and standard deviation of the probe intensities), prior to log2 transformation and median polish probe set summarisation.
Array data quality estimation
The normalised unscaled standard error (NUSE) of each array was calculated to compare data variability between and within arrays (11). A NUSE value of 1.05 was set as the upper limit of acceptable quality.
Data analysis
Principal components analysis using a covariance dispersion matrix was performed to map high dimensional data of each sample to 3 dimensions for estimation of clustering according to classification and identification of outlying samples.
A mixed model analysis of variance (ANOVA) including methods of moments estimation was used to identify differential expression between non-invasive tumours, invasive tumours and healthy bladder tissue. Batch effects were taken into consideration within the model. ANOVA was conducted including 'patient' as a factor, taking the paired samples into consideration. Differences in gene expression between tumours of differing grade, and also differential expression between tumour samples were assessed using the t-test. To correct for multiple testing, genes were considered to be significantly differentially expressed if a false discovery rate (FDR) adjusted P<0.05 was obtained. The false discovery rate was calculated as previously described (12). Those genes whose fold change expression level was found to significantly differ by ±2× or greater were used for further analysis.
Unsupervised hierarchical agglomerative clustering (Euclidean distance with average linkage clustering) was used to identify samples with similar expression profiles.
MetaCore version 6.7 (GeneGo, Inc., St. Joseph, MI, USA), IPA (Ingenuity Systems, Redwood City, CA, USA) and DAVID Bioinformatics resource v 6.7 (13) were employed to identify canonical pathways and curated networks having statistically significant enrichment of differentially expressed genes. Pathways from MetaCore analysis filtered on FDR <0.05 are presented in Results, unless otherwise stated. Univariate Cox proportional hazards regression analysis was performed to evaluate the association between disease progression and differential gene expression.
Meta-analysis
Meta-analysis was conducted using publicly available gene expression data from Gene Expression Omnibus (14). Out of the possible 12 datasets identified (GSE88, GSE89, GSE7476, GSE30522, GSE24152, GSE3167, GSE5287, GSE12630, GSE27448, GSE19915, GSE13507 and GSE5479), we selected two datasets using the same microarray platform (Affymetrix Human Genome U133A array) with a large number of samples for further analysis. Raw CEL files from GSE3167 (15) with 60 samples and GSE5287 (16) with 30 samples were imported into Partek Genomic Suite (Partek, Inc., Chesterfield, MO, USA) and data were processed and differential genes identified following the same protocol described earlier for our own data. Clinico-pathological data for the datasets were kindly provided by the authors. The GSE3167 dataset contains superficial transitional cell carcinoma (NMIBC) with surrounding carcinoma in situ (CIS) (13 patients), without surrounding CIS lesions (15 patients), muscle invasive carcinoma (MIBC, 13 patients), CIS only (n=5) and healthy bladder (n=14). The GSE2587 dataset contains 30 samples from patients with muscle invasive bladder cancer.
Immunostaining
Western blotting
Western blotting was performed to validate the specificity of anti-OPN antibody (AF1433; R&D Systems, Inc., Minneapolis, MN, USA) for subsequent IHC analysis. Protein lysates were prepared in RIPA buffer from a MIBC cell line, 253-J and a NMIBC cell line, RT112. Bradford was used for protein quantification. Proteins were separated by SDS-PAGE prior to transferring onto a PDVF membrane. Blocking was performed for 1 h with 10% milk, prior to overnight incubation in 1:1,000 dilution of AF1433. Secondary antibody was applied for 1 h before visualising using ECL. β-actin (ab8227; Abcam) was used to demonstrate equal protein loading.
Immunohistochemistry
Conditions for IHC with AF1433 were optimised in clear cell renal carcinoma and kidney FFPE sections, using a no primary antibody control, a goat IgG isotype control (AB-108-C; R&D Systems) and a negative tissue control-breast skin. Antigen retreval was performed using the PT-link at pH 9.0, prior to peroxidase block (Dako), 1-h primary antibody (1:50 AF1433) incubation at RT, 1-h secondary antibody incubation at RT (1:100) prior to visualisation with DAB Chromagen (Dako). Staining intensity was scored by a consultant pathologist. Briefly, staining intensity was graded as 0 (negative), + (weak), ++ (moderate) and +++ (strong).
Results
Bladder cancer tissues exhibit specific gene signatures
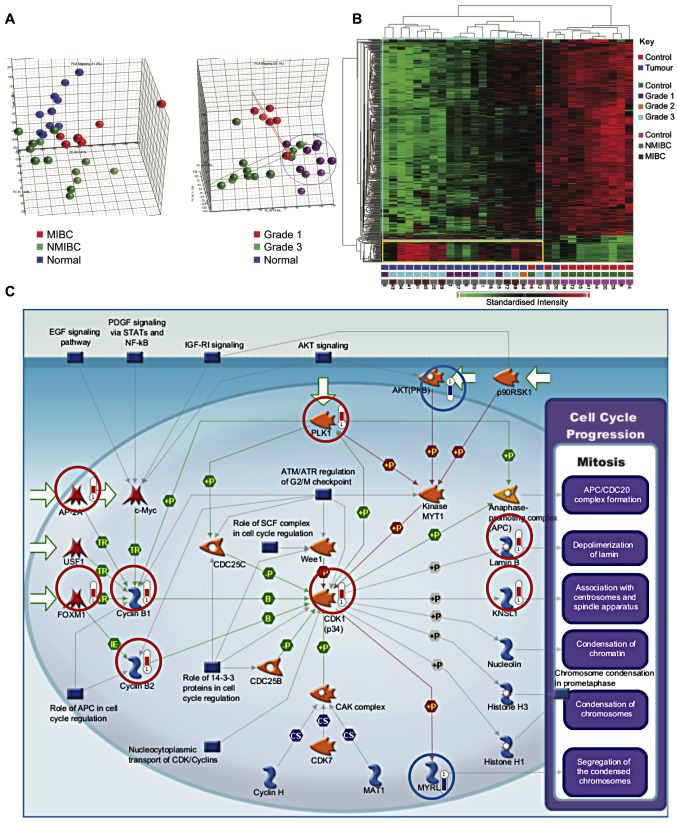
A total of 418 genes were differentially expressed between cancer and control samples, using a fold change of at least 2.5, and a false discovery rate (FDR) of 0.05. Of these, 368 genes showed reduced expression in tumours, while 50 genes showed increased expression in tumours. Principal components analysis demonstrated segregation of samples by tissue type and tumour grade (Fig. 1A). Similarly, hierarchical clustering segregated healthy bladder tissue and bladder cancer into two groups (Fig. 1B).
Figure 1.
Cell proliferation genes are upregulated in bladder cancer. (A) Principal components analysis. Segregation into groups by tissue type and tumour grade was assessed. (B) Hierarchical clustering segregates tumour samples from healthy tissue. Key shown on the right. The numbers at the bottom of each column correspond to patient serial numbers. Standardised intensity range -2 (green) to 2 (red), 0 (black) fold change. (C) Genes involved in the initiation of mitosis are upregulated in bladder cancer (P=2.59×10−9). Genes circled in red and blue were, respectively, upregulated or downregulated in cancer samples. The length of the bar is proportional to the degree of change in expression.
Multiple genes involved in proliferation were upregulated in bladder cancer samples as compared to healthy samples (P=2.59×10−9) (Fig. 1C). These included components of the cyclin-dependent kinase 1 (CDK1)/cydin B complex, activation of which is required to drive progression from G2- to M-phase. Initiation of cyclin B transcription is carried out by FOXM1. Moreover, the equilibrium between nuclear import and export of cyclin B1 is influenced by its phosphorylation status by PLK1, which was also upregulated. The substrates of CDKl/cyclin B include Lamin B, leading to depolymerization of the lamin nuclear-cytoskeleton, and Kinesin-related motor protein Eg5 (KNSL1), which associates with centrosomes and spindle apparatus.
Genes involved in myogenesis and epithelial-to-mesenchymal transition (EMT) are downregulated in cancer samples
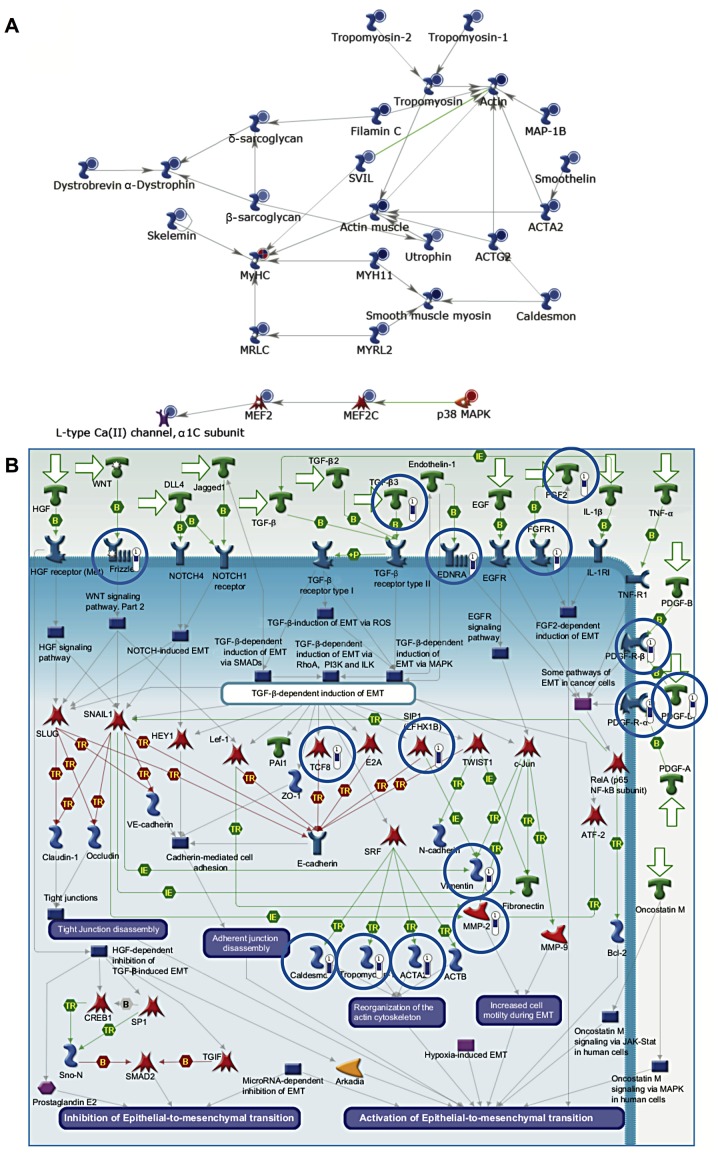
Genes downregulated in cancer (compared to healthy tissue) included several which are involved in myogenesis (P=1.22×10−10) (Fig. 2A). These included the myocyte enhancer factor, Mef2, which is expressed during myogenesis. Mef2 and MyoD interact leading to MyoD activation and hence the expression of myosin heavy and light chains. In addition, genes involved in EMT were also downregulated (P=1.41×10−9) (Fig. 2B). EMT is critical during myogenesis, particularly as the muscle precursor cells delaminate from the dermomyotome. Muscle precursors invade the developing tissue and differentiate into mature muscle cells.
Figure 2.
Genes involved in myogenesis and EMT are dysregulated in bladder cancer. (A) Genes involved in myogenesis were downregulated in cancer samples (P=1.22×10−10). The relative expression for each gene is depicted by the colour and intensity of the circle (blue, downregulation; red, upregulation). (B) Downregulation of genes involved in EMT in cancer samples (P=1.41×10−9). Genes circled in blue exhibited reduced expression within the tumour samples relative to healthy bladder tissue. The length of the blue bar is proportional to the extent of downregulation.
Comparison of NMIBC and healthy bladder tissue
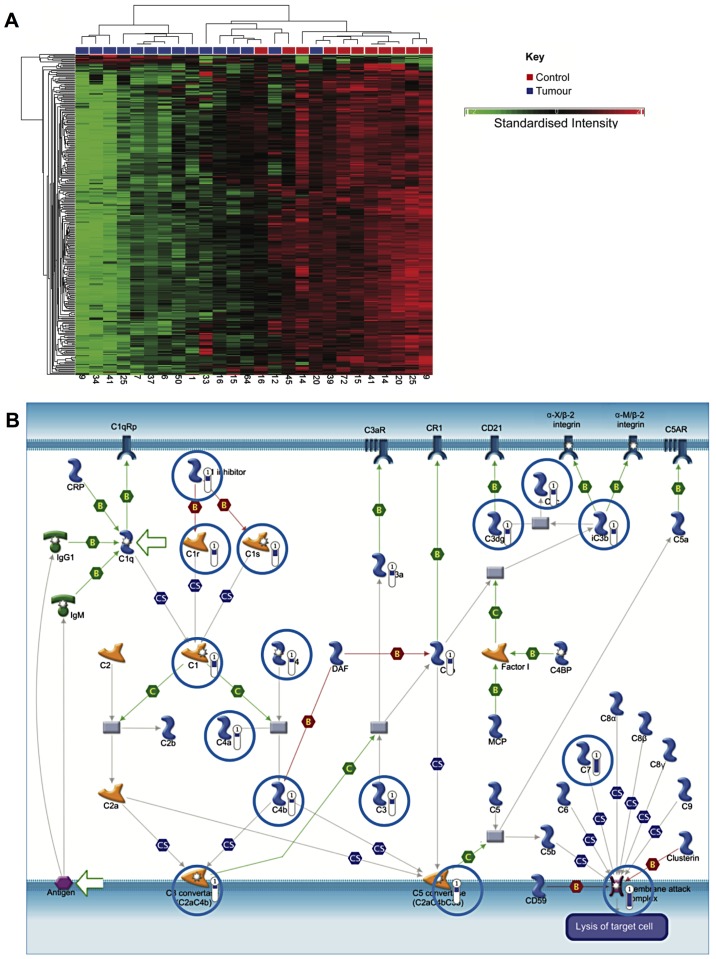
A specific comparison was also made between NMIBC and healthy tissue. This identified 410 genes, which were differentially expressed by ≥2.5-fold (P<0.05, false discovery rate). Of these, 393 were downregulated in cancer, while increased expression was observed in 17 (data not shown). Hierarchical clustering generally segregated into two groups, but with additional branching in one cluster as shown in Fig. 3A.
Figure 3.
Classical complement genes are downregulated in NMIBC. (A) Hierarchical clustering showing clustering of NMIBC and healthy tissue samples, though complete separation is not established. A key is given on the right of the figure. The numbers at the bottom of each column correspond to patient serial numbers. Standardised intensity range –2 (green) to 2 (red), 0 (black) fold change. (B) Genes associated with the classical complement pathway are downregulated in NMIBC (P=5.12×10−10).
Similar to the analysis comparing all bladder cancer samples to healthy samples, genes involved in smooth muscle development and EMT were downregulated (P=4.8×10−12). Notably, genes associated with the classical complement pathway were strongly downregulated in NMIBC samples, which may indicate a mechanism for immune evasion (P=5.12×10−10) (Fig. 3B).
MIBC vs. healthy tissue
Hierarchical clustering showed segregation of MIBC and healthy samples into two groups (Fig. 4A). There were 404 genes that were altered by at least 3-fold (P<0.05 false discovery rate). Out of these, 107 showed increased expression between invasive tumours and healthy samples, while 297 were decreased in expression. Pathways enriched for these differentially regulated genes are shown in Fig. 4B. Genes involved in chromosome condensation in prometaphase were upregulated in invasive bladder cancer (P=2.75×10−15). Similarly, several regulators of the anaphase promoting complex (APC) were also upregulated (P=3.47×10−12). These included the kinases, PLK1 and CDK1.
Figure 4.
Dysregulation of pathways in MIBC. (A) Hierarchical clustering of genes and samples showing segregation of muscle-invasive tumour samples and control samples into different clusters. A key is shown on the right of the figure. The numbers at the bottom of each column correspond to patient serial numbers. Standardised intensity range –1.9 (green) to 1.9 (red), 0 (black) fold change. (B) Pathway enrichment (MIBC vs. control samples).
Comparison of grade 1 and 3 NMIBC
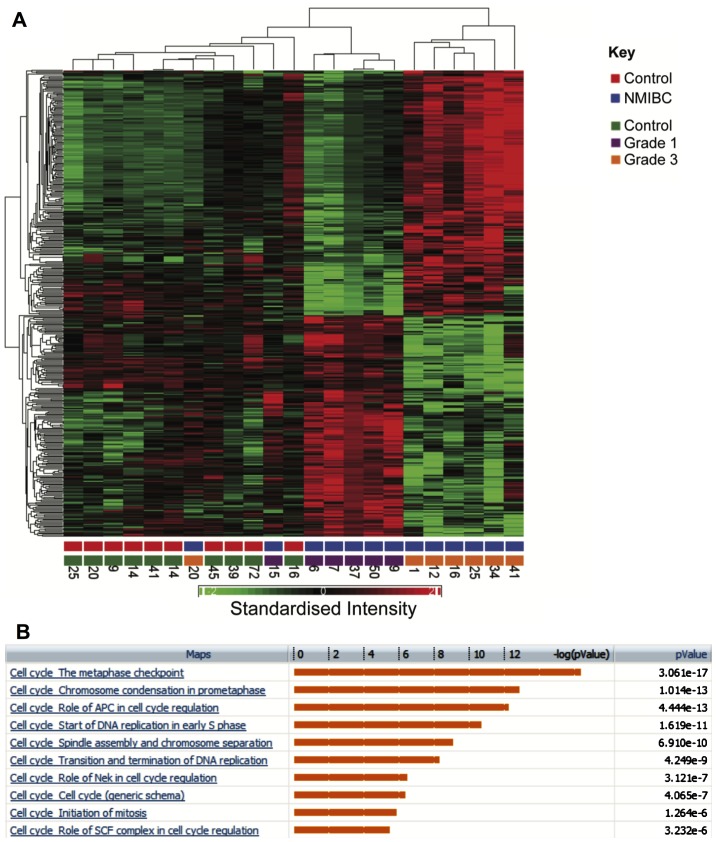
A further comparison was made between gene expression profiles in grade 1 and 3 bladder cancers, which identified 341 genes that were differentially regulated by at least 2-fold (FDR P<0.05). Hierarchical clustering clearly segregates between grade 1 and 3 (Fig. 5A). Enriched pathways are shown in Fig. 5B. Notably multiple pathways associated with cell cycle progression were upregulated in grade 3 tumours and multiple core cell cycle components were upregulated in grade 3 tumours. Genes involved in the metaphase checkpoint were particularly upregulated (P=3.06×10−17). A comparison was also made of genes that were upregulated both in grade 3 compared to grade 1 NMIBC and in MIBC compared to healthy samples.
Figure 5.
Comparing grade 3 and 1 NMIBC. (A) Hierarchical clustering segregates grade 1 and 3 NMIBC samples. The numbers at the bottom of each column correspond to patient serial numbers. Standardised intensity range –2 (green) to 2 (red), 0 (black) fold change. (B) Pathway enrichment (grade 3 vs. grade 1 NMIBC).
Comparing grade 3 NMIBC and MIBC
A total of 292 genes were differentially expressed 2 or more fold (P<0.05). Of these, 144 showed increased expression in MIBC while the remaining 148 exhibited decreased expression in MIBC. Hierarchical clustering resulted in clear segregation between the two groups. Table II lists genes that were differentially expressed by >5-fold in grade 3 NMIBC compared to MIBC. Among the genes most upregulated in MIBC were the proteinases. Overexpression and activation of proteinases, that destroy the connective tissue, are involved in various pathological processes including tumour progression. The most proteolytic of enzymes are the metalloproteinases. The matrix metalloproteinases (MMPs) comprise a family of enzymes that collectively can degrade all components of the extracellular matrix (ECM), these were upregulated in this study. For example, the matrix metalloproteinases MMP1 and MMP16 were upregulated 4.8- and 2.6-fold, respectively in MIBC compared to NMIBC.
Table II.
Genes exhibiting >5 fold differential expression between grade 3 and MIBC (FDR P<0.05).
| Gene | ID | RefSeq | P-value | Fold change (MIBC/NMIBC |
|---|---|---|---|---|
| Family with sequence similarity 83, member D | FAM83D | NM_030919 | 5.50E-06 | 7.87015 |
| Retinoic acid receptor responder (tazarotene induced) 1 | RARRES1 | NM_206963 | 0.001291 | 5.96921 |
| Centromere protein I | CENPI | NM 006733 | 1.35E-05 | 5.92042 |
| Fibronectin 1 | FN1 | NM_212482 | 0.000579 | 5.86325 |
| Centromere protein F, 350/400 ka (mitosin) | CENPF | NM_016343 | 0.000505 | 5.7622 |
| N mannosidase, α, class 1C, member 1 | MANICI | NM_020379 | 0.000401 | 5.52916 |
| Topoisomerase (DNA) II α 170 kDa | TOP2A | NM_001067 | 0.00091 | 5.32363 |
| TPX2, microtubule-associated, homolog (Xenopus laevis) | TPX2 | NM_012112 | 0.000892 | 5.27537 |
| Ras association (RalGDS/AF-6) domain family member 2 | RASSF2 | NM_014737 | 0.000644 | 5.04613 |
| Anillin, actin binding protein | ANLN | NM_018685 | 0.000489 | 5.01334 |
Further functional analysis using Database for Annotation, Visualization and Integrated Discovery (DAVID)
A further analysis of 143 genes, which were upregulated in MIBC compared to NMIBC was performed using the DAVID database. A summary of annotation clusters is given in Table III.
Table III.
DAVID functional annotation clusters.
| Annotation cluster | Enrichment score | Count | P-value |
|---|---|---|---|
| Extracellular glycoproteins | 6.47 | 37 | 2.7E-07 |
| Serine-type endopeptidase inhibitor activity | 3.29 | 7 | 7.90E-05 |
| Negative regulation of immune system processes | 2.21 | 6 | 4.60E-04 |
| Zymogen (proteolytic enzyme precursors) | 1.85 | 14 | 5.10E-04 |
| Cell migration | 1.33 | 9 | 1.40E-03 |
| EGF | 1.18 | 5 | 1.50E-02 |
| Response to wounding | 0.95 | 13 | 8.20E-04 |
Meta-analysis of gene expression data
In order to validate our results, we used a meta-analysis of a larger dataset consisting of 90 samples, generated by the combination of two publicly available bladder cancer gene expression datasets (15,16). When we followed the same criteria used with our dataset for classifying MIBC and NMIBC, there were 29 NMIBC samples and 42 MIBC samples in the meta-analysis dataset. There were also 14 healthy bladder tissue samples, while the rest of the samples (carcinoma in situ) were not used in analysis.
A total of 4513 genes were differentially expressed in NMIBC compared to healthy tissue, when a cut-off of FDR <0.05 and fold-change >2.5 were applied. When compared with differential genes from our data, there were 118 genes common in both analyses. Surprisingly except for one gene, PELO, all the other genes showed concordant expression. This included all the 8 genes upregulated in NMIBC and 109 genes downregulated in NMIBC compared to healthy bladder tissue (data not shown).
When MIBC samples were compared with healthy bladder samples, with a filter of FDR <0.05 and fold-change >3, 3905 genes were differentially expressed in the meta-analysis. A comparison with differential genes from our data under similar conditions and filtrations revealed 44 common genes. Similarly, the expressions of all except 2 genes (SERPINEB13 and CASC5) were concordant in both the analyses. All the 30 genes downregulated in MIBC compared to healthy tissue showed concordant expression, while 12 out of 14 genes highly expressed in MIBC in this study showed similar expression in the meta-analysis (data not shown).
Osteopontin protein is overexpressed in MIBC compared to healthy urothelium
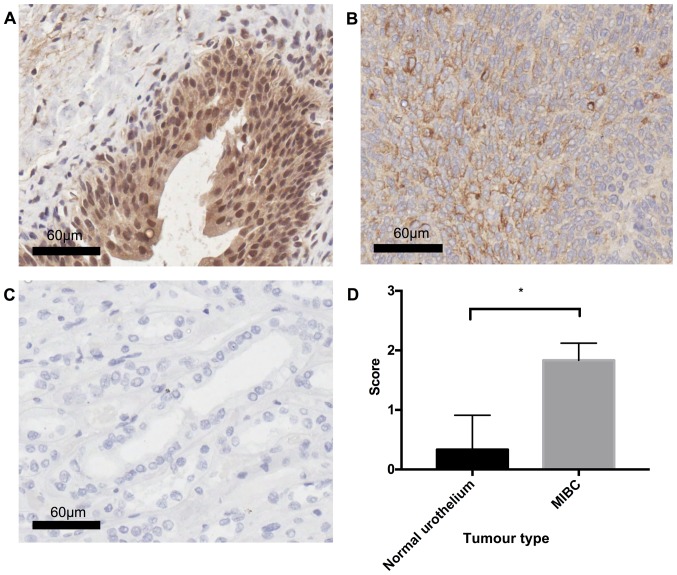
In order to determine the validity of the microarray data, OPN expression was examined in MIBC (n=3) and healthy urothelial tissue (n=3) using the IHC technique. Statistical analysis using Welch's unpaired t-test revealed a significant difference (P=0.0286) between MIBC and healthy tissue, with MIBC displaying diffuse cytoplasmic OPN staining of moderate (++) intensity, whilst healthy urothelium was typically OPN negative (−) (Fig. 6).
Figure 6.
Comparing OPN protein expression in MIBC and healthy urothelial tissue. (A) Healthy urothelium displaying negative OPN immunostaining, with the presence of non-specific binding in urothelial tissue; (B) MIBC tissue exhibiting moderate (++) diffuse OPN immunostaining; (C) Isotype control, OPN expressing kidney tissue with polyclonal goat IgG antibody; (D) graphical representation of staining results, results are average of staining from 3 healthy urothelial samples, and 3 MIBCs, statistical analysis was performed using Welch's unpaired t-test (P=0.0286). All images are representative of the cohort. Scoring was performed by a pathologist.
Discussion
The present study contributes to a body of work that uses microarray data to profile bladder cancer, and provides fresh insights into pathways involved in its pathogenesis. Distinctive patterns of gene expression differentiating between NMIBC and MIBC, and between high and low grade cancers were identified.
A striking upregulation in pathways involved in cell cycle control was observed in comparisons of MIBC and NMIBC, and between grade 3 and grade 1 NMIBC tumours. A significant overlap was observed between genes upregulated in grade 3 NMIBC tumours and those upregulated in MIBC compared to healthy tissue and these genes were predominantly mapped to cell cycle control pathways. This is likely to reflect the increased aggressiveness and likelihood of progression of grade 3 NMIBC. This panel of genes may therefore be useful in predicting progression of NMIBC.
Cell cycle genes which were upregulated included anillin (>8-fold upregulation), a substrate of APC, which is involved in cytokinesis (17). Anillin is upregulated in various cancers including pancreatic (18) and renal carcinoma (19). TPX2, which is involved in spindle assembly (20), was also upregulated.
This upregulation of cell cycle related genes in bladder cancer is not unexpected, and was similarly shown in a study by Zaravinos et al (21). Notably, several of the upregulated genes have been identified as potential therapeutic targets for bladder cancers. Inhibitors of TOP2A include anthracyclines and etoposide, two well-established chemotherapy agents. The anthracycline doxorubicin, for example, exhibits antibladder cancer effects and is a component of the commonly employed MVAC regimen. PLK1 inhibitors are currently in phase 1 and 2 trials, and preliminary studies suggest that PLK1 inhibitors may reduce proliferation of bladder cancer cell lines (22). However, results obtained from a recent phase 2 study evaluating the efficacy of the PLK1 inhbitor volasertib as a second-line treatment in patients with locally advanced or metastatic urothelial cancer, suggests that although volasertib was well-tolerated with an acceptable safety profile the antitumour activity of volasertib was not suffecient for further evaluation as a monotherapy (23). Several CDK inhibitors are also currently in phase 1 and 2 clinical trials.
The expression of osteopontin (OPN) 'secreted phosphoprotein 1', a matricellular protein, is strongly associated with malignancies (reviewed in ref. 24). Furthermore, OPN RNA-silencing in BC significantly attenuates tumour cell invasion (25). The finding that OPN expression was strikingly upregulated (>10-fold) in invasive bladder cancers compared with the healthy tissue is consistent with previous studies demonstrating upregulation of OPN in urothelial malignancy (22,26–28). Whilst OPN RNA expression data is well represented in BC tissue (28,29), protein expression data are limited (26,27). Therefore, OPN expression was examined between MIBC and healthy urothelium to determine whether commonality between RNA and protein expression exists in BC. OPN was found to be signifcantly (P=0.0286) overexpressed in MIBC compared to healthy urothelium. These data are consistent with obeservations in gastric (30), oesophageal (31), cervical (32) and prostate (33) cancers (26,34). Whilst a significant relationship between OPN and poor prognosis was noted in upper urinary tract urothelial cell carcinoma no correlation between stage and grades was found (27). Conversely, OPN expression is associated with improved survival in pancreatic adenocarcinoma (35).
Elevated levels of osteopontin have also been detected in the plasma of patients with advanced transitional cell carcinoma (TCC), suggesting that OPN could be a novel biomarker of disease stage (36,37). The utility of OPN expression as a prognostic marker (and/or predictor of treatment response), however, remains unproven (26,27,38). The differences in OPN expression (in blood and tissue) observed in studies could be explained, at least in part, by the expression of OPN isoforms. Specifically, OPN splice variants which exhibit divergent biological functions are differentially expressed in cancers. Intriguingly, CD44, the hyaluronic acid receptor, for which osteopontin is a key ligand (37), was found to be upregulated among invasive bladder cancers, further supporting a potential role for OPN in pathogenesis of TCC.
When comparing grade 3 NMIBC with MIBC, we observed an overexpression of the cell-matrix interaction network. In particular, there was upregulation in proteolytic enzymes such as the matrix metalloproteases (MMPs). This is consistent with the increased ability of MIBC to invade local tissues. It would be important to evaluate if overexpression of MMPs predicts NMIBC progression.
Genes involved in myogenesis and epithelial-to-mesenchymal transition (EMT) were downregulated in tumours, and this was particularly noted in NMIBC samples compared with the healthy tissue. This is intriguing, and could represent an inhibitory effect of bladder cancer on normal bladder muscle growth. EMT describes a process by which epithelial cells lose their characteristic apical-basal polarity, intercellular junctions and epithelial markers, while upregulating mesenchymal markers, acquiring front-rear polarity, and undergoing cytoskeleton reorganization into mesenchymal cells (39). EMT is associated with invasive cancers and metastases, portends a poor prognosis in patients with bladder cancer (40). In the present study, tumour samples were not micro-dissected, and the possible 'admixture' with normal muscle tissue within samples could have confounded the analysis of myogenesis and EMT genes.
One novel finding in this study is the downregulation of genes involved in the classical complement pathway within the NMIBC group. Previously, others have shown that complement inhibitors such as CD46 and complement factor H (CFH) are upregulated in bladder cancers (41). The aggregate data suggest that repression of complement activity is an important pathogenic mechanism in bladder cancer development. Intriguingly, CFH binds to the OPN protein and masks putative enzymatic-cleavage sites, and could thereby, alter OPN activity. Urinary levels of CFH were also investigated as markers of bladder malignancy (42,43), but were found to exhibit low sensitivity and specificity. This could be explained by changes in CFH levels during disease progression, and supports our findings that downregulation of complement genes occurred in NMIBC rather than MIBC samples.
ESRP1 and ESRP2 are upregulated in NMIBC (compared to healthy tissue), and are involved in the regulation of FGFR2 splicing (44). While decreased expression of FGFR2b (one of the FGFR2 splice variants) was observed in a subset of bladder cancers (45), upregulation of FGFR2c (another splice variant) was detected in a bladder cancer metastasis model (46). Consistent with reports on differential protein isoforms during cancer development, our findings support the hypothesis that differential expression of FGFR2 splice variants is an important pathogenic mechanism in bladder cancer. In agreement with data published from the Cancer Genome Atlas Reseach Network in 2013 (47) which comprehensively evaluated the molecular signature of 131 urothelial carcinomas and revealed that mutational spectrum was involved in cell cycle regulation, chromatin regulation, and kinase signalling pathways with potential therapeutic application, the present study highlights several dysregulated transcripts in BC that potentially could be targeted in BC treatment.
Whilst the long-term clinical outcome data would have allowed more robust conclusions to be drawn from the small cohort, meta-analysis of the microarray data together with the protein expression experiment highlight the importance of the presented data, in addition to the role of OPN in invasion (25). Furthermore, this study supplements a growing body of molecular data in bladder cancer allowing further understanding of this complex and heterogeneous disease.
In conclusion, this study complements the published literature, and gives credence to observed associations between patterns of gene signatures with the stage and grade of bladder cancers. Ultimately such signatures would be most clinically useful if they improve prognostication of disease, particularly among NMIBC, where a more aggressive therapeutic approach could be employed in cancers that were more likely to progress. The present study identified expression patterns, which may help this prediction (such as proteolytic enzymes), and these may be incorporated into future prospective studies. Significantly, we also identified several genes and pathways (such as OPN and the classical complement pathway) which were strikingly deregulated in bladder cancer, and which may be valuable drug targets.
Acknowledgments
We thank Sim Sihota for her skilled technical assistance in the extraction and microarray analysis of RNA.
Abbreviations
- NMIBC
non-muscle invasive bladder cancer
- MIBC
muscle invasive bladder cancer
- CDK1
cyclin-dependent kinase 1
- FOXM1
forkhead box protein M1
- PLK1
polo-like kinase 1
- KNSL1
kinesin family member 11
- Mef2
myocyte enhancer factor-2
- MyoD
myogenic differentiation 1
- EMT
epithelial to mesenchymal transition
- MMPs
matrix metalloproteinases
- MMP1
matrix metalloproteinase 1
- MMP16
matrix metalloproteinase 16
- PELO
protein pelota homolog
- SERPINB13
Serpin peptidase inhibitor, clade B (ovalbumin), member 13
- CASC5
cancer susceptiility candidate 5
- APC
anaphase promoting complex
- TPX2
targeting protein for Xklp2
- TOP2A
topoisomerase 2-alpha
- MVAC
methotrexate, vinblastine, adriamycin and cisplatin
- OPN
osteopontin
- TCC
transitional cell carcinoma
- CFH
compliment factor H
- ESRP1
epithelial splicing regulatory protein 1
- ESRP2
epithelial splicing regularoty protein 2
- FGFR2(b,c)
fibroblast growth factor receptor 2 (b isoform, c isoform)
- IHC
immunohistochemistry
References
- 1.IARC Cancer incidence and mortality Worldwide. IARC CancerBase No. 10. Journal. 2015 http://globocan.iarc.fr.
- 2.Cancer Research UK Bladder cancer statistics. 2015 http://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bladder-cancer.
- 3.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 4.Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, Newling DW, Kurth K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006:475–467. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 5.Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 6.Elsamman E, Fukumori T, Ewis AA, Ali N, Kajimoto K, Shinohara Y, Ishikawa M, Takahashi M, Nishitani MA, Baba Y, et al. Differences in gene expression between noninvasive and invasive transitional cell carcinoma of the human bladder using complementary deoxyribonucleic acid microarray: Preliminary results. Urol Oncol. 2006;24:109–115. doi: 10.1016/j.urolonc.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Riester M, Taylor JM, Feifer A, Koppie T, Rosenberg JE, Downey RJ, Bochner BH, Michor F. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18:1323–1333. doi: 10.1158/1078-0432.CCR-11-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 9.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A Model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. doi: 10.1198/016214504000000683. [DOI] [Google Scholar]
- 11.Bolstad BM, Collin F, Simpson KM, Irizarry RA, Speed TP. Experimental design and low-level analysis of microarray data. Int Rev Neurobiol. 2004;60:25–58. doi: 10.1016/S0074-7742(04)60002-X. [DOI] [PubMed] [Google Scholar]
- 12.Pawitan Y, Michiels S, Koscielny S, Gusnanto A, Ploner A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics. 2005;21:3017–3024. doi: 10.1093/bioinformatics/bti448. [DOI] [PubMed] [Google Scholar]
- 13.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyrskjøt L, Kruhøffer M, Thykjaer T, Marcussen N, Jensen JL, Møller K, Ørntoft TF. Gene expression in the urinary bladder: A common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 16.Als AB, Dyrskjøt L, von der Maase H, Koed K, Mansilla F, Toldbod HE, Jensen JL, Ulhøi BP, Sengeløv L, Jensen KM, et al. Emmprin and survivin predict response and survival following cisplatin-containing chemotherapy in patients with advanced bladder cancer. Clin Cancer Res. 2007;13:4407–4414. doi: 10.1158/1078-0432.CCR-07-0109. [DOI] [PubMed] [Google Scholar]
- 17.Zhao WM, Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J Biol Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- 18.Olakowski M, Tyszkiewicz T, Jarzab M, Krol R, Oczko-Wojciechowska M, Kowalska M, Kowal M, Gala GM, Kajor M, Lange D, et al. NBL1 and anillin (ANLN) genes over-expression in pancreatic carcinoma. Folia Histochem Cytobiol. 2009;47:249–255. doi: 10.2478/v10042-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 19.Gruss OJ, Vernos I. The mechanism of spindle assembly: Functions of Ran and its target TPX2. J Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronkainen H, Hirvikoski P, Kauppila S, Vaarala MH. Anillin expression is a marker of favourable prognosis in patients with renal cell carcinoma. Oncol Rep. 2011;25:129–133. [PubMed] [Google Scholar]
- 21.Zaravinos A, Lambrou GI, Volanis D, Delakas D, Spandidos DA. Spotlight on differentially expressed genes in urinary bladder cancer. PLoS One. 2011;6:e18255. doi: 10.1371/journal.pone.0018255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Zhang G, Kong C. High expression of polo-like kinase 1 is associated with the metastasis and recurrence in urothelial carcinoma of bladder. Urol Oncol. 2013;31:1222–1230. doi: 10.1016/j.urolonc.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Stadler WM, Vaughn DJ, Sonpavde G, Vogelzang NJ, Tagawa ST, Petrylak DP, Rosen P, Lin CC, Mahoney J, Modi S, et al. An open-label, single-arm, phase 2 trial of the Polo-like kinase inhibitor volasertib (BI 6727) in patients with locally advanced or metastatic urothelial cancer. Cancer. 2014;120:976–982. doi: 10.1002/cncr.28519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed M, Behera R, Chakraborty G, Jain S, Kumar V, Sharma P, Bulbule A, Kale S, Kumar S, Mishra R, et al. Osteopontin: A potentially important therapeutic target in cancer. Expert. Opin Ther Targets. 2011;15:1113–1126. doi: 10.1517/14728222.2011.594438. [DOI] [PubMed] [Google Scholar]
- 25.Xu ST, Guo C, Ding X, Fan WJ, Zhang FH, Xu WL, Ma YC. Role of osteopontin in the regulation of human bladder cancer proliferation and migration in T24 cells. Mol Med Rep. 2015;11:3701–3707. doi: 10.3892/mmr.2015.3202. [DOI] [PubMed] [Google Scholar]
- 26.Coppola D, Szabo M, Boulware D, Muraca P, Alsarraj M, Chambers AF, Yeatman TJ. Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res. 2004;10:184–190. doi: 10.1158/1078-0432.CCR-1405-2. [DOI] [PubMed] [Google Scholar]
- 27.Ke HL, Chang LL, Yang SF, Lin HH, Li CC, Wu DC, Wu WJ. Osteopontin overexpression predicts poor prognosis of upper urinary tract urothelial carcinoma. Urol Oncol. 2011;29:703–709. doi: 10.1016/j.urolonc.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Zaravinos A, Volanis D, Lambrou GI, Delakas D, Spandidos DA. Role of the angiogenic components, VEGFA, FGF2, OPN and RHOC, in urothelial cell carcinoma of the urinary bladder. Oncol Rep. 2012;28:1159–1166. doi: 10.3892/or.2012.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang ZQ, Zang WD, Chen R, Ye BW, Wang XW, Yi SH, Chen W, He F, Ye G. Gene expression profile and enrichment pathways in different stages of bladder cancer. Genet Mol Res. 2013;12:1479–1489. doi: 10.4238/2013.May.6.1. [DOI] [PubMed] [Google Scholar]
- 30.Imano M, Satou T, Itoh T, Sakai K, Ishimaru E, Yasuda A, Peng YF, Shinkai M, Akai F, Yasuda T, et al. Immunohistochemical expression of osteopontin in gastric cancer. JJ Gastrointest Surg. 2009;13:1577–1582. doi: 10.1007/s11605-009-0955-y. [DOI] [PubMed] [Google Scholar]
- 31.Kita Y, Natsugoe S, Okumura H, Matsumoto M, Uchikado Y, Setoyama T, Owaki T, Ishigami S, Aikou T. Expression of osteopontin in oesophageal squamous cell carcinoma. Br J Cancer. 2006;95:634–638. doi: 10.1038/sj.bjc.6603296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song JY, Lee JK, Lee NW, Yeom BW, Kim SH, Lee KW. Osteopontin expression correlates with invasiveness in cervical cancer. Aust N Z J Obstet Gynaecol. 2009;49:434–438. doi: 10.1111/j.1479-828X.2009.01027.x. [DOI] [PubMed] [Google Scholar]
- 33.Forootan SS, Foster CS, Aachi VR, Adamson J, Smith PH, Lin K, Ke Y. Prognostic significance of osteopontin expression in human prostate cancer. Int J Cancer. 2006;118:2255–2261. doi: 10.1002/ijc.21619. [DOI] [PubMed] [Google Scholar]
- 34.Weber GF, Lett GS, Haubein NC. Osteopontin is a marker for cancer aggressiveness and patient survival. Br J Cancer. 2010;103:861–869. doi: 10.1038/sj.bjc.6605834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins AL, Rock J, Malhotra L, Frankel WL, Bloomston M. Osteopontin expression is associated with improved survival in patients with pancreatic adenocarcinoma. Ann Surg Oncol. 2012;19:2673–2678. doi: 10.1245/s10434-012-2337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L, Wang Y, Qu N, Huang C, Chen L. Significance of plasma osteopontin levels in patients with bladder urothelial carcinomas. Mol Diagn Ther. 2012;16:311–316. doi: 10.1007/s40291-012-0005-4. [DOI] [PubMed] [Google Scholar]
- 37.Naor D, Sionov RV, Ish-Shalom D. CD44: Structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/S0065-230X(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 38.Ang C, Chambers AF, Tuck AB, Winquist E, Izawa JI. Plasma osteopontin levels are predictive of disease stage in patients with transitional cell carcinoma of the bladder. BJU Int. 2005;96:803–805. doi: 10.1111/j.1464-410X.2005.05716.x. [DOI] [PubMed] [Google Scholar]
- 39.Lim J, Thiery JP. Epithelial-mesenchymal transitions: Insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 40.Baumgart E, Cohen MS, Silva Neto B, Jacobs MA, Wotkowicz C, Rieger-Christ KM, Biolo A, Zeheb R, Loda M, Libertino JA, et al. Identification and prognostic significance of an epithelialmesenchymal transition expression profile in human bladder tumors. Clin Cancer Res. 2007;13:1685–1694. doi: 10.1158/1078-0432.CCR-06-2330. [DOI] [PubMed] [Google Scholar]
- 41.Varela JC, Atkinson C, Woolson R, Keane TE, Tomlinson S. Upregulated expression of complement inhibitory proteins on bladder cancer cells and anti-MUC1 antibody immune selection. Int J Cancer. 2008;123:1357–1363. doi: 10.1002/ijc.23676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babjuk M, Soukup V, Pesl M, Kostirova M, Drncova E, Smolova H, Szakacsova M, Getzenberg R, Pavlik I, Dvoracek J. Urinary cytology and quantitative BTA and UBC tests in surveillance of patients with pTapT1 bladder urothelial carcinoma. Urology. 2008;71:718–722. doi: 10.1016/j.urology.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 43.Cheng ZZ, Corey MJ, Pärepalo M, Majno S, Hellwage J, Zipfel PF, Kinders RJ, Raitanen M, Meri S, Jokiranta TS. Complement factor H as a marker for detection of bladder cancer. Clin Chem. 2005;51:856–863. doi: 10.1373/clinchem.2004.042192. [DOI] [PubMed] [Google Scholar]
- 44.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diez de Medina SG, Chopin D, El Marjou A, Delouvee A, LaRochelle WJ, Hoznek A, Abbou C, Aaronson SA, Thiery JP, Radvanyi F. Decreased expression of keratinocyte growth factor receptor in a subset of human transitional cell bladder carcinomas. Oncogene. 1997;14:323–330. doi: 10.1038/sj.onc.1200830. [DOI] [PubMed] [Google Scholar]
- 46.Chaffer CL, Brennan JP, Slavin JL, Blick T, Thompson EW, Williams ED. Mesenchymal-to-epithelial transition facilitates bladder cancer metastasis: Role of fibroblast growth factor receptor-2. Cancer Res. 2006;66:11271–11278. doi: 10.1158/0008-5472.CAN-06-2044. [DOI] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]