Abstract
Cancer cells are characterized by a pathological manifestation of uncontrolled proliferation, which results in tumor formation. Therefore, it is necessary to improve understanding of the underlying mechanism of cell cycle control. Here, we report that miR-150 is downregulated in nasopharyngeal carcinoma tissues and cells. Upregulation of miR-150 suppresses nasopharyngeal carcinoma (NPC) cell proliferation and induces G1/S arrest in vitro, and inhibits tumorigenesis in vivo. Conversely, silencing miR-150 yields the opposite effect. Our results further demonstrate that miR-150 retards nasopharyngeal carcinoma cell proliferation and G1/S transition via targeting multiple cell cycle-related genes, including CCND1, CCND2, CDK2 and CCNE2. Therefore, our results uncover a novel mechanistic understanding of miR-150-mediated tumor suppression in NPC, which will facilitate the development of effective cancer therapies against nasopharyngeal carcinoma.
Keywords: miR-150, proliferation, cell cycle, tumorigenicity, nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is a relatively rare malignancy worldwide, yet it is the most common cancer in some endemic areas, including north Africa, south-eastern Asia and a number of provinces in south-eastern China (1). Despite great progress in the systemic treatment, including primarily radiotherapy, chemotherapy or chemotherapy in combination with radiotherapy due to its intricate anatomical location, against stage I and II NPC over the past years, the 5-year survival rates of more than half of the patients with advanced NPC is still <50% (2). Extensive studies have indicated that uncontrolled proliferation ability of cancer cells is the most common momentum responsible for the progression of NPC, where multiple cell cycle-related genes have been implicated in the proliferation of tumor cells (3,4). Therefore, better understanding of the molecular mechanisms contributing to the uncontrolled proliferation of cancer cells will facilitate to improve the survival rate of NPC patients.
Normal cell growth and metabolism are under precise control of cell cycle in which protein kinase complexes composed of cyclins and cyclin-dependent kinase (CDK) play an important role and determine a cell's progression in a sequential fashion (5). From the molecular perspective, cyclins act as the regulatory subunits of an activated heterodimer and CDKs functions as the catalytic subunits, which orchestrates coordinated entry into the S phase of the cell cycle (5). Different cyclin-CDK combinations specifically determine the downstream targeted proteins that in turn promote the expression of cyclins and enzymes required for DNA replication (6). Previous studies showed that disregulation of the cell cycle components may cause the tumor cell to multiply uncontrollably, which finally leads to tumor formation (7). Importantly, therapies targeting CDK inhibitor present a favorable prospect in the treatment of a variety of cancers. O'Leary et al reported that phase III trials investigating palbociclib in patients with advanced-stage estrogen receptor-positive breast cancer have demonstrated a substantial improvement in progression-free survival, with a well-tolerated toxicity profile (8); furthermore, Kumar and colleagues demonstrated a single agent activity of dinaciclib, a novel potent small non-selective inhibitor of CDK1, CDK2, CDK5 and CDK9, in relapsed myeloma (9). These studies indicated that therapy targeting cell cycle related proteins exhibits broad-ranging efficacy in many cancer and could serve as a primary therapeutic avenue in the treatment of cancers.
MicroRNAs (miRNAs) are a diverse group of small non-coding RNAs composed of 19–25 nucleotides and mechanistically function by binding to the 3′-untranslated region of downstream mRNAs, leading to mRNA degradation or repression of translation (10,11). A growing body of evidence has demonstrated that miRNAs not only play crucial roles in many biological processes including proliferation, differentiation, cell cycle and apoptosis (11), but also regulate the progression and metastasis in various types of tumors (12–18). In recent studies, abnormal expression of miRNAs has been reported to be broadly implicated in the pathogenesis of NPC (19,20). Furthermore, several lines of evidence indicated that miRNAs have been reported to be critical regulators of cell proliferation in multiple human cancers, including NPC (21–23). For example, Zhao et al reported that miR-3188 regulates nasopharyngeal carcinoma proliferation through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN (24); moreover, a study from He and colleagues showed that miR-16 targeting fibroblast growth factor 2 inhibited NPC cell proliferation through PI3K/AKT and MAPK signaling pathways (25). Notably, numerous studies indicated that miRNAs regulated cancer cell proliferation via directly targeting single or several cell cycle-related genes, including cyclin D, cyclin E and cyclin-dependent kinase (CDK), which promoted the unlimited proliferation of cancer cells (26,27). Therefore, the above results imply that dysregulation of miRNAs promote the NPC cells proliferation, which contributes to the progression and recurrence of NPC.
In this study, we found that miR-150 expression is markedly decreased in NPC tissues and cells. Moreover, upregulation of miR-150 suppresses, while silencing miR-150 promotes nasopharyngeal carcinoma cell proliferation and cell cycle in vitro, as well as tumorigenesis in vivo. Our results further demonstrated that CCND1, CCND2, CDK2 and CCNE2 are the direct targets of miR-150; moreover, they mediate the regulation of miR-150 in NPC cells proliferation and cell cycle. Therefore, our results demonstrate that miR-150 inhibits nasopharyngeal carcinoma cells proliferation and cell cycle by directly targeting CCND1, CCND2, CDK2 and CCNE2 and indicate that miR-150 play a tumor-suppressive role by suppressing proliferation and cell cycle in nasopharyngeal carcinoma.
Materials and methods
Cell lines and cell culture
The human nasopharyngeal carcinoma cell lines CNE1, CNE2, C666-1, HNE1, HNE2, HONE1, SUNE1 and 5–8F were obtained from Department of Biochemistry and Molecular Biology, Guangdong Medical College (Zhanjiang, China) and were cultured in RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA) supplemented with penicillin G (100 U/ml), streptomycin (100 mg/ml) and 10% fetal bovine serum (FBS, Life Technologies). NP69 is an immortalized nasopharyngeal epithelium cell line which was cultured in defined keratinocyte serum-free medium supplemented with specific growth factors (Life Technologies). 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Life Technologies). All cells were incubated at 37°C in a humidified atmosphere with 5% CO2 and were routinely subcultured using 0.25% (w/v) trypsin-ethylenedi-aminetetraacetic acid solution.
Patients and tumor tissues
Paired tumor specimens and the matched adjacent normal tissues from 8 confirmed NPC patients were obtained from the Affiliated Hospital of Guangdong Medical College between January 2014 and December 2014. Tissue specimens were obtained by fiber optic nasopharyngoscopy directly on the tumor growth site and on the adjacent side with observed normal mucosal morphology for the corresponding non-tumor pair. Informed consent was obtained from the patients involved, and ethics approval for this study was obtained from the Institutional Research Ethics Committee. The median age was 48 years and the age range from 32 to 72 years. The histologic subtype of 8 NPC includes 7 undifferentiated non-keratinizing and 1 differentiated non-keratinizing. The EBV status of all 8 NPC patients were positive. The patient information is summarized in Table I.
Table I.
The basic information of 8 NPC patients for miR-150 expression analysis.
| Cases (n) | Percentage (%) | |
|---|---|---|
| Gender | ||
| Male | 5 | 62.5 |
| Female | 3 | 37.5 |
| Age | ||
| <50 | 5 | 62.5 |
| ≥50 | 3 | 37.5 |
| Histologic subtype | ||
| Undifferentiated non-keratinizing | 7 | 87.5 |
| Differentiated non-keratinizing | 1 | 12.5 |
| EBV status | ||
| Positive | 8 | 100.0 |
| Negative | 0 | 0.0 |
NPC, nasopharyngeal carcinoma; EBV, Epstein-Barr virus.
RNA extraction, reverse transcription, and real-time PCR
Total RNA from tissues or cells were extracted using TRIzol (Life Technologies) according to the manufacturer's instructions. Messenger RNA (mRNA) and miRNA were polyadenylated using a poly-A polymerase-based First-Strand Synthesis kit (Takara, Dalian, China) and reverse transcription (RT) of total mRNA was performed using a PrimeScript RT Reagent kit (Takara) according to the manufacturer's protocol. Complementary DNA (cDNA) was amplified and quantified on ABI 7500HT system (Applied Biosystems, Foster City, CA, USA) using SYBR Green I (Applied Biosystems). Table II lists the primers used in the reactions. Real-time PCR was performed according to a standard method, as described previously (28). Primers for U6 (no. MQP-0202) and miR-150 (no. miRQ0000451) were synthesized and purified by RiboBio (Guangzhou, China). U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as endogenous controls for miRNA or mRNA, respective. Relative fold expressions were calculated with the comparative threshold cycle (2−∆∆Ct) method.
Table II.
The primers used in the reactions for real-time RT-PCR.
| Gene | Sequence (5′-3′) |
|---|---|
| CCND1-up | GCCCTCGGTGTCCTACTTC |
| CCND1-dn | CTCCTCCTCGCACTTCTGTT |
| CCND2-up | GGTCGGGTTTTCAATCACAC |
| CCND2-dn | CCTCTTCACCTCCCTTCAACT |
| CCND3-up | TCCTCTCCCATTGTCCCTCT |
| CCND3-dn | CCACCAGCCTAAACCTTGC |
| CDK4-up | GTCTGGTTGTTTGTTGTGTGC |
| CDK4-dn | CGGTGTGTTTGTTGTTTGTCC |
| CDK6-up | GTCTGGTTGTTTGTTGTGTGC |
| CDK6-dn | CGGTGTGTTTGTTGTTTGTCC |
| CCNE1-up | CGGTATATGGCGACACAAGA |
| CCNE1-dn | ACATACGCAAACTGGTGCAA |
| CCNE2-up | AGGAAAACTACCCAGGATGTCA |
| CCNE2-dn | ATCAGGCAAAGGTGAAGGATTA |
| CDK2-up | CTGCTTCCTGTTGGCTCTTTCT |
| CDK2-dn | CTTTGTTTCTGCCTTCTCTCCT |
| GAPDH-up | GCACCGTCAAGGCTGAGAAC |
| GAPDH-dn | TGGTGAAGACGCCAGTGGA |
Plasmid, small interfering RNA and transfection
The human miR-150 expression plasmid was generated by cloning the genomic pre-miR-150 gene, with a 300-bp sequence on each flanking side, into retroviral transfer plasmid pMSCV-puro (Clontech Laboratories Inc., Tokyo, Japan) to generate plasmid pMSCV-miR-150. pMSCV-miR-150 was cotransfected with the pIK packaging plasmid into 293FT cells using the standard calcium phosphate transfection method, as previously described (29). Thirty-six hours after the cotransfection, supernatants were collected and incubated with cells to be infected for 24 h in the presence of polybrene (2.5 µg/ml). After infection, puro-mycin (1.5 µg/ml) was used to select stably transduced cells over a 10-day period. The luciferase reporter system of pE2F-luc (Clontech) and pRb-luc (Clontech) were used to examine the transcriptional activity of E2Fs and the transcriptional repression capability of Rb, respectively. The 3′-untranslated region (3′-UTR) regions of the human CCND1, CCND2, CDK2 and CCNE2 were PCR-amplified from genomic DNA and cloned into pmirGLO luciferase reporter vector (Promega, Madison, WI, USA), and the list of primers used in clone reactions is presented in Table III. The miArrest plasmids for anti-miR150 and negative control plasmids were constructed and cloned into pH1 plasmids by GeneChem (Shanghai, China). Small interfering RNA (siRNA) for CCND1, CCND2, CDK2 and CCNE2 knockdown were obtained from Santa Cruz (Dallas, TX, USA). Transfection of siRNAs and plasmids was performed using Lipofectamine 3000 (Life Technologies) according to the manufacturer's instructions.
Table III.
The primers used in the reactions for clone PCR.
| Gene | Sequence (5′-3′) |
|---|---|
| miR-150-up | TAGGCGCCGGAATTACTCCCCTGGAGCCTGTTCA |
| miR-150-dn | CTACCCGGTAGAATTGAGACGCCCCAACAATCAG |
| CCND1-3UTR-up | TAGTTGTTTAAACGAGCATTTTGATACCAGAAGGGAAA |
| CCND1-3UTR-dn | AGGTCGACTCTAGACTTGTCTTTTTGTCTTCTGCTGGA |
| CCND2-3UTR-up | TAGTTGTTTAAACGAGTTCTGTGACATCCTGCTTCTT |
| CCND2-3UTR-dn | AGGTCGACTCTAGACTACTTATCAGCACTTTCTAACATCC |
| CDK2-3UTR-up | TAGTTGTTTAAACGAGCCCTAATCTCACCCTCTCCT |
| CDK2-3UTR-dn | AGGTCGACTCTAGACCGTTAATAGCAAGAGCACTCAAGG |
| CCNE2-3UTR-up | TAGTTGTTTAAACGAAATTCACCAAGATTGGGTAGAAC |
| CCNE2-3UTR-dn | AGGTCGACTCTAGACAACAATGGGCTAAAAATAAACAGTA |
Cell counting kit-8 analysis and colony formation assay
For cell counting kit-8 analysis, cells (2×103) were seeded into 96-well plates and stained at the indicated time-point with 100 µl cell counting kit-8 (CCK-8; Dojindo, Japan) dye for 2 h at 37°C, followed by the absorbance measured at 450 nm, with 650 nm used as the reference wavelength. For colony formation assay, cells (0.2×103) were plated into 6-well plates and cultured for 10 days. Colonies were then fixed for 15 min with 10% formaldehyde and stained with 1.0% crystal violet for 30 sec.
Anchorage-independent growth ability assay
Cells (3×103) were suspended in 2 ml complete medium plus 0.3% agar (Sigma-Aldrich, St. Louis, MO, USA). The agar-cell mixture was plated as a top layer onto a bottom layer comprising 0.6% complete medium agar mixture. After 14-day culture, colony size was measured using an ocular micrometer and colonies >0.1 mm in diameter were counted.
Cell cycle analysis
Pretreatment and staining was performed using Cell Cycle Detection kit (KeyGen, China) according to the manufacturer's instructions. Cells (5×105) were harvested by trypsinization, washed in ice-cold phosphate-buffered saline (PBS) and fixed in 75% ice-cold ethanol in PBS. Before staining, cells were gently resuspended in cold PBS, and ribo-nuclease was added into cell suspension tube incubated at 37°C for 30 min, followed by incubation with propidium iodide (PI) for 20 min at room temperature. Cell samples (2×104) were then analyzed by FACSCanto II flow cytometer (Becton-Dickinson & Co., Franklin Lakes, NJ, USA) and the data were analyzed using FlowJo 7.6 software (TreeStar Inc., Ashland, OR, USA).
Tumor xenografts
Four-week-old BALB/c-nu female mice weighing 15–20 g were maintained in a standard pathogen-free environment where the animals were housed in sterile cages under laminar flow hoods in a 20–26°C temperature controlled room with a 12-h light/dark cycle and fed autoclaved chow and water. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Guangdong Medical College. BALB/c-nu mice at 4–6 weeks of age were randomly divided into four groups (n=5 per group) and indicated CNE-2 cells (2×106) were inoculated subcutaneously into the flanks of the nude mice. Tumor volume was determined using an external caliper and calculated using the equation (L x W2)/2. The mice were sacrificed by inhaling CO2 on day 36 after inoculation and the tumors were excised and subjected to pathologic examination.
Immunohistochemistry
The immunohistochemistry procedure and scoring of Ki67 expression levels were performed as previously described (12). The slides were incubated overnight at 4°C in a humidified chamber with the rabbit anti-human Ki67 antibody diluted 1:5,000 in PBS (no. ab16667. Abcam, Cambridge, MA, USA).
Dual luciferase report experiments
Cells (5×105) were plated in 60-mm cell culture dishes, at 60–80% confluence after 24 h of culture, and the reporter constructs were transfected into cells using Lipofectamine 3000. After 12-h incubation, the transfection medium was replaced; cells were harvested and washed with PBS, and lysed with passive lysis buffer (Promega). The cell lysates were analyzed immediately using Synergy™ 2 microplate system (BioTek, Winooski, VT, USA). Luciferase and Renilla luciferase were measured using a Dual-Luciferase Reporter assay system (Promega) according to the manufacturer's instructions. The luciferase activity of each lysate was normalized to Renilla luciferase activity. The relative transcriptional activity was converted into fold induction above the vehicle control value.
RNA immunoprecipitation
Cells (5×105) were plated in 60-mm cell culture dishes, at 60–80% confluence after 24 h of culture, and the pIRESneo-FLAG/HA-Ago2 plasma (10822; Addgene, Cambridge, MA, USA) was cotransfected into cells using Lipofectamine 3000. After 48-h transfection, cells were washed and lysed in radioimmunoprecipitation buffer (Sigma-Aldrich) containing 10% proteinase inhibitor cocktail (Sigma-Aldrich) and 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich). A fraction of the whole cell lysate was used for RNA isolation, and the remaining lysate was subjected to immunoprecipitation (IP) using an antibody against Ago2 (Abcam) or immunoglobulin G (IgG) (Abcam). RNA from whole cell lysates and RNA IP (RIP) fractions was extracted with TRIzol (Life Technologies) according to the manufacturer's instructions. The relative levels of mRNA were determined using real-time RT-PCR as described above. The relative mRNA enrichment in the RIP fractions was computed based on the ratio of relative mRNA levels in the RIP fractions and the relative mRNA levels in the whole cell lysates.
Western blotting
The proteins extracted from the cell lysates were loaded with 50 µg in each lane, which was further separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membranes were probed with antibodies against CCND1 (no. 3300; dilution, 1:1,000), CCND2 (no. 3741; dilution, 1:1,000), CDK2 (no. 2546; dilution, 1:1,000), and CCNE2 (no. 4132; dilution: 1:1,000) (Cell Signaling Technology, Beverly, MA, USA) overnight at 4°C, and then incubated with horseradish peroxidase-conjugated secondary antibodies (no. 7071; dilution, 1:3,000) (Cell Signaling Technology) for 1 h at room temperature. Immune complexes were detected by enhanced chemiluminescence (Cell Signaling Technology). α-tubulin (no. 12351; dilution, 1:1,000) (Cell Signaling Technology) was used to correct for differences in protein loading from the control and experimental groups.
Statistical analysis
All values are presented as means ± standard deviation (SD). Significant differences were determined using SPSS 19.0 software (SPSS, Chicago, IL, USA). A paired Student's t-test was used to analyze the paired control group (pMSCV-V or pH1-V) and treatment group (miR-150 or anti-miR-150) of in vitro experiments. An independent Student's t-test was used to analyze the paired control group (pMSCV-V or pH1-V) and treatment group (miR-150 or anti-miR-150) of in vivo experiments. Spearman's correlation tests were used to evaluate the pairwise expression correlation between miR-150 and targeted genes in NPC tissues. P<0.05 was considered statistically significant.
Results
miR-150 is downregulated in nasopharyngeal carcinoma tissues and cell lines
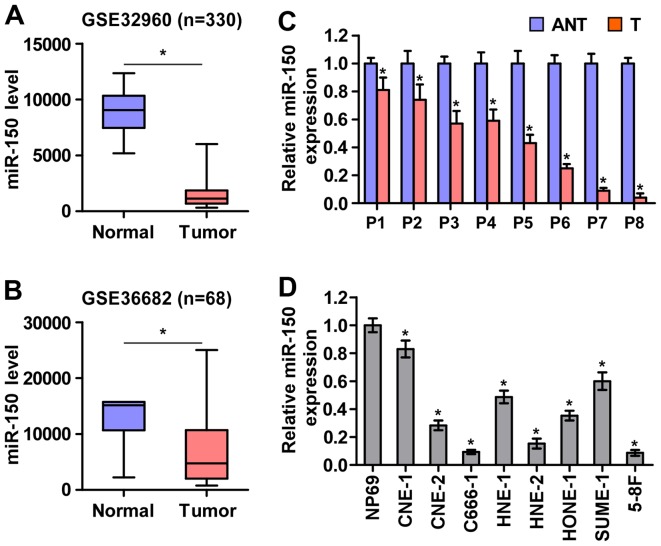
To screen the aberrant miRNA expression between NPC tissues and normal nasopharyngeal tissues, two microarray-based high-throughput datasets of NPC from GSE32960 (ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE32nnn/GSE32960/matrix/) and GSE36682 (ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE36nnn/GSE36682/matrix/) were analyzed and showed that miR-150 expression was downregulated in NPC tissues compared with normal nasopharyngeal tissues (Fig. 1A and B). To validate the miR-150 expression in NPC tissues, real-time PCR was performed on NPC clinical samples and cell lines. As shown in Fig. 1C and D, miR-150 expression was differentially downregulated in the primary NPC tissues from 8 individual patients and NPC cell lines compared with that in the matched adjacent normal tissues and immortalized nasopharyngeal epithelium cell line (NP69), respectively. Therefore, the published miRNA datasets and our results suggested that miR-150 is downregulated in NPC tissues and cells.
Figure 1.
miR-150 is downregulated in nasopharyngeal carcinoma tissues and cell lines. (A and B) miR-150 expression level was downregulated in the nasopharyngeal carcinoma datasets from GSE32960 and GSE36682. (C) Expression of miR-150 was downregulated in 8 paired nasopharyngeal carcinoma tissues compared with the matched adjacent normal nasopharyngeal carcinoma tissue samples. Each bar represents the mean values ± SD of three independent experiments. *P<0.05. (D) Real-time PCR analysis of miR-150 expression in NP69 and nasopharyngeal carcinoma cell lines. U6 was used as endogenous control in RT-PCR. Each bar represents the mean values ± SD of three independent experiments. *P<0.05.
Downregulation of miR-150 promotes NPC cell proliferation in vitro
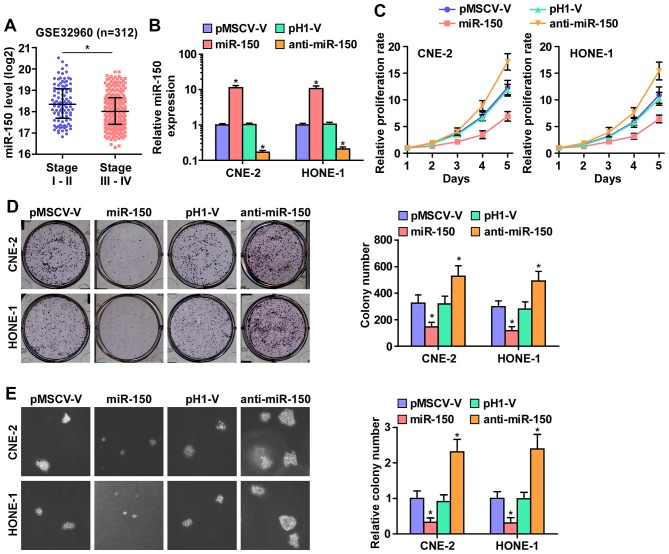
As downregulation of miR-150 was correlated with clinical stage in NPC clinical tissues (Fig. 2A), we examined whether miR-150 is involved in NPC cell proliferation. We first constructed miR-150-expressing stably HONE1 and CNE2 cells via exogenously overexpressing miR-150 and endogeneously silencing miR-150 via virus transduction (Fig. 2B). The reason why HONE1 and CNE2 cells were selected to study the biological roles of miR-150 in NPC is that miR-150 expression levels in CNE-2 and HONE1 cells were located in the intermediate level in the miR-150 expression spectrum of all NPC cell lines when we examined the miR-150 expression levels in 8 NPC cell lines. Furthermore, CNE-2 and HONE1 cell lines were the most common cell models in studying the proliferation and tumorigenesis of NPC in vitro and in vivo assays (30,31). CCK-8 assay was performed and the result indicated that miR-150 upregulation markedly decreased, while silencing miR-150 increased, the cell proliferation rate of CNE2 and HONE1 cells (Fig. 2C). Furthermore, colony formation assays and anchorage-independent growth assays revealed that miR-150 downregulation increased the colony-generating capability and anchorage-independent growth activity of CNE2 and HONE1 cells (Fig. 2D and E). Moreover, upregulated miR-150 drastically inhibited NPC cell growth, as indicated by the decreased colony numbers (Fig. 2D and E). These results indicated that miR-150 inhibits NPC cell proliferation ability in vitro.
Figure 2.
miR-150 inhibits the proliferation of nasopharyngeal carcinoma cells in vitro. (A) Downregulation of miR-150 positively correlated with clinical stages of nasopharyngeal carcinoma. (B) miR-150 expression was detected by real-time PCR analysis. miR-150 expression was examined in overexpressed miR-150 and anti-miR-150 NPC cells, and their respective control vectors. U6 was used as the loading control. Each bar represents the mean values ± SD of three independent experiments. *P<0.05. (C) CCK-8 assay revealed that overexpression of miR-150 decreased, while downregulation of endogenous miR-150 increased the proliferation rate in CNE-2 and HONE1 cells. Each bar represents the mean values ± SD of three independent experiments. (D) Colony formation assay revealed that overexpression of miR-150 decreased, while downregulation of endogenous miR-150 increased the colony-forming ability in CNE-2 and HONE1 cells. Each bar represents the mean values ± SD of three independent experiments. *P<0.05. (E) Anchorage-independent growth assays revealed that overexpression of miR-150 decreased, while downregulation of endogenous miR-150 increased the anchorage-independent growth ability in CNE-2 and HONE1 cells. Each bar represents the mean values ± SD of three independent experiments. *P<0.05.
miR-150 inhibits nasopharyngeal carcinoma cell tumorigenesis in vivo
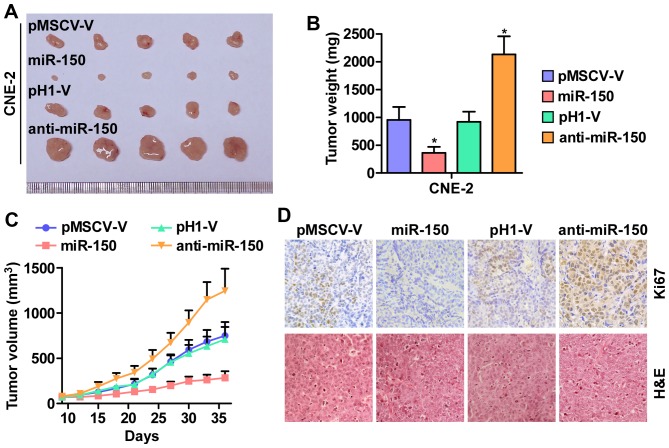
Furthermore, the effect of miR-150 on tumorigenesis of human nasopharyngeal carcinoma CNE-2 cells was tested in vivo. As shown in Fig. 3A–C, tumor volumes and weight were increased significantly in the miR-150-downregulation group compared with the control group. In contrast, the tumors formed by the miR-150-overexpressing cells were smaller and exhibited reduced tumor volume and weight compared to the control group. The analysis of IHC and H&E staining revealed that the miR-150-silencing tumor tissues displayed higher Ki67 proliferation indexes, whereas the miR-150-overexpressing tumor tissues exhibited reduced numbers of Ki67-positive cells (Fig. 3D). Taken together, these results indicated that miR-150 inhibits the tumorigenesis of NPC cells in vivo.
Figure 3.
miR-150 inhibits tumorigenesis of nasopharyngeal carcinoma cells in vivo. (A) Images of excised tumors from five BALB/c mice at 30 days after injection with the indicated cells. (B) Average weight of excised tumors from the indicated mice. Each bar represents the median values ± quartile values. *P<0.05. (C) Tumor volumes were measured every five days. Each bar represents the median values ± quartile values. (D) Representative images of sections sliced from the indicated tumors and stained with anti-Ki67 and H&E staining, respectively.
miR-150 downregulation promotes cell cycle progression of NPC cells
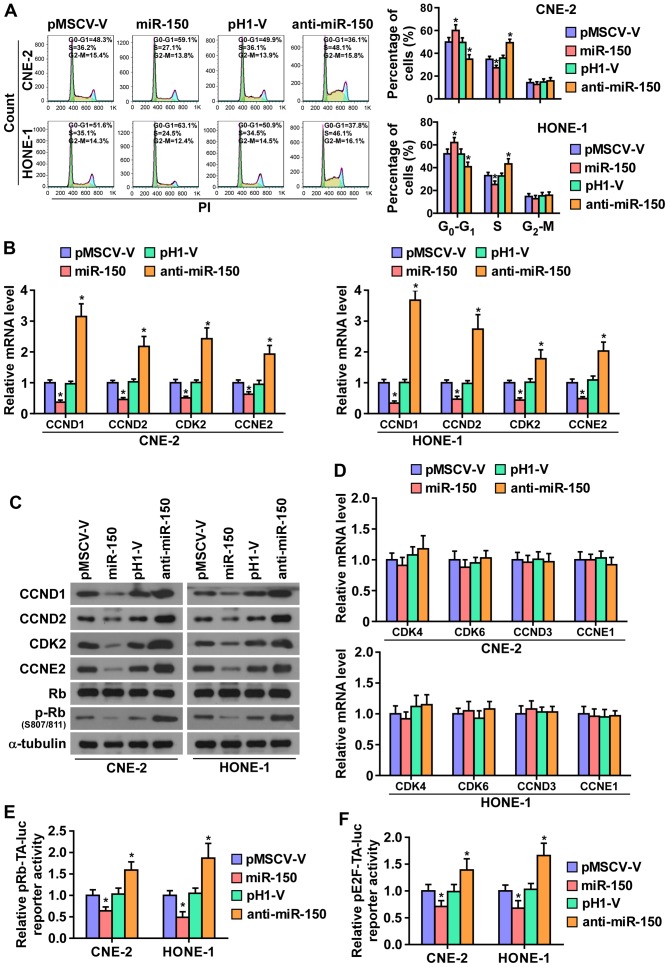
To further investigate the mechanism underlying the miR-150-mediated inhibition of NPC cell proliferation, the cell cycle progression was analyzed. As shown in Fig. 4A, flow cytometry showed that miR-150 upregulation markedly decreased the percentage of cells in the S phase and increased that of cells in the G1/G0 phase, while silencing miR-150 increased the percentage of cells in the S phase and decreased that of cells in the G1/G0 phase, suggesting that upregulation miR-150 might induce G1/S arrest in NPC cells. Furthermore, the expression levels of a number of critical cell cycle regulators in G1/S checkpoint were detected. As shown in Fig. 4B and C, real-time PCR assays and western blotting revealed that upregulating miR-150 decreased, while silencing miR-150 increased, the expression levels of cyclin D1 (CCND1), cyclin D2 (CCND2), cyclin-dependent kinase 2 (CDK2), and cyclin E2 (CCNE2) at both the protein and mRNA levels. Whereas, the mRNA levels of cyclin-dependent kinase 4 (CDK4), cyclin-dependent kinase 6 (CDK6), cyclin D3 (CCND3), and cyclin E1 (CCNE1) were not affected by miR-150 (Fig. 4D).
Figure 4.
miR-150 inhibits the cell cycle of nasopharyngeal carcinoma cells. (A) Flow cytometric analysis of the indicated nasopharyngeal carcinoma cells. Each bar represents the mean values ± SD of three independent experiments. *P<0.05. (B) Real-time PCR analysis of CCND1, CCND2, CKD2 and CCNE2 expression in the indicated cells. Transcript levels were normalized by GAPDH expression. Each bar represents the mean values ± SD of three independent experiments. *P<0.05. (C) Western blot analysis of CCND1, CCND2, CDK2, CCNE2 and p-Rb expression in the indicated cells. (D) Real-time PCR analysis of CDK4, CDK6, CCND3 and CCNE2 mRNA expression in overexpressed miR-150 and anti-miR-150 NPC cells, and their respective control vectors. U6 was used as the loading control. Each bar represents the mean values ± SD of three independent experiments. *P<0.05. (E and F) Relative Rb activity and E2F reporter activity in the indicated cells. Each bar represents the mean values ± SD of three independent experiments. *P<0.05.
It has been well documented that progression of cell cycle was promoted by E2F transcription factors. E2F can work with Rb, a negative regulator of the cell cycle, in regulating G1/S checkpoint. E2F activation or Rb inactivation can induce entry of cells into S phase (32,33). Thus, we hypothesized that miR-150 upregulation may suppress the activity E2F-Rb complexes. As shown in Fig. 4C, the level of phosphorylation of Rb (p-Rb) was markedly decreased in miR-150-overexpressing NPC cells, while its expression level is increased in the miR-150-silencing NPC cells. Moreover, the luciferase reporter analysis shows that upregulating miR-150 enhanced, while miR-150-silenced inhibited, the transcriptional repression capability of Rb (Fig. 4E). Moreover, the transcriptional activity of E2F was strongly repressed by miR-150 overexpression, whereas silencing miR-150 increased the activity (Fig. 4F). These results suggested that miR-150 inhibits the cell cycle progression in NPC cells.
miR-150 suppresses cell cycle progression by directly targeting CCND1, CCND2, CDK2 and CCNE2
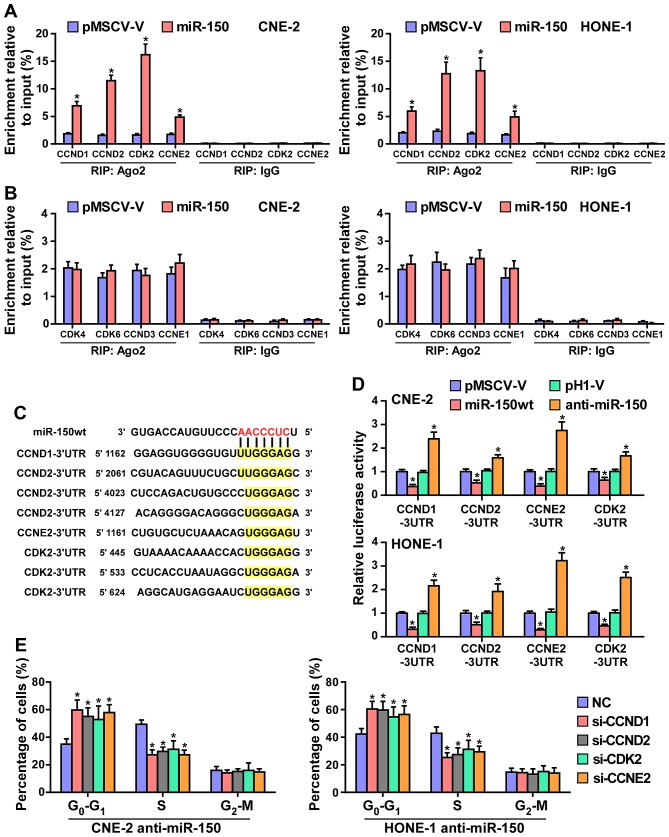
It has been confirmed that miRNAs regulate gene expression by targeting the 3′-UTR of mRNA, and the degradation of target mRNA was mediated by RNA-induced silencing complex (RISC). Therefore, we tested whether miR-150 mediated the RISC binding to the mRNA of critical cell cycle regulators in G1/S checkpoint using miRNP immunoprecipitation assay. As shown in Fig. 5A and B, miR-150 directly interacted with the mRNA of CCND1, CCND2, CDK2 and CCNE2, but did not affect the mRNA of CDK4, CDK6, CCND3 and CCNE1. Furthermore, publicly available algorithms (miRanda and TargetScan) were employed and showed that CCND1, CCND2, CDK2 and CCNE2 are potential targets of miR-150 (Fig. 5C). To examine whether miR-150-mediated CCND1, CCND2, CDK2 and CCNE2 downregulation occurs through a miR-150-binding in the 3′-UTR of CCND1, CCND2, CDK2 and CCNE2, the 3′-UTR of four genes were cloned into pmirGLO luciferase reporter vectors. As predicted, miR-150 overexpression reduced, whereas anti-miR-150 increased, the luciferase reporter activity of these four genes (Fig. 5D). Moreover, flow cytometry showed that downregulated CCND1, CCND2, CDK2 and CCNE2 significantly increased the percentage of NPC cells in G0 phase, while the percentage of cells in S phase was decreased in miR-150-silenced CNE2 and HONE1 cells (Fig. 5E). Taken together, these results indicated that miR-150 retards the cell cycle progression via directly targeting CCND1, CCND2, CDK2 and CCNE2, causing G1/S phase arrest.
Figure 5.
miR-150 inhibits the cell cycle progression by directly targeting CCND1, CCND2, CDK2 and CCNE2. (A) MiRNP IP (RIP) assay showing the association between miR-150 and CCND1, CCND2, CDK2 and CCNE2 transcripts in CNE-2 and HONE1 cells. Pulldown of IgG antibody served as the negative control. (B) RIP analysis show that CDK4, CDK6, CCND3 and CCNE1 were not affected by miR-150. RIP analysis, as assessed by immunoprecipitation of Ago2 in the indicated cells. IgG immunoprecipitation was used as a negative control. *P<0.05. (C) Predicted miR-150 targeting sequence in 3′-UTRs of CCND1, CCND2, CDK2 and CCNE2. (D) Luciferase assay of cells transfected with pmirGLO-3′-UTR reporter of CCND1, CCND2, CDK2 and CCNE2 in miR-150 overexpressing and silencing in CNE-2 and HONE1 cells, respectively. (E) Individual silencing of CCND1, CCND2, CDK2 and CCNE2 increased the percentage of NPC cells in G0 phase, while the percentage of cells in S phase was decreased in miR-150-silencing cells.
CCND1, CCND2, CDK2 and CCNE2 contribute to miR-150 downregulation-induced NPC cell growth
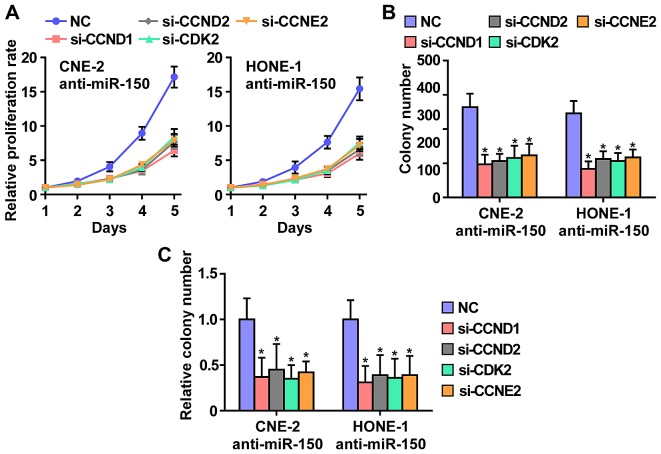
To further investigate whether CCND1, CCND2, CDK2 and CCNE2 contribute to the phenotypes induced by miR-150-downregulated in NPC cells, we used RNA interference to knock down the CCND1, CCND2, CDK2 and CCNE2. As shown in Fig. 6, individual silencing of CCND1, CCND2, CDK2 and CCNE2 significantly inhibited cell growth, cell colony number and cell colony numbers on soft agar of anti-miR-150 CNE2 and HONE1 cells. These results indicated that miR-150 inhibits NPC cells proliferation via silencing CCND1, CCND2, CDK2 and CCNE2.
Figure 6.
CCND1, CCND2, CDK2 and CCNE2 mediates the miR-150-downregulation-induced NPC cell growth. (A) Individual silencing of CCND1, CCND2, CDK2 and CCNE2 rescued the proliferation rate enhanced by anti-miR-150. (B) Individual silencing of CCND1, CCND2, CDK2 and CCNE2 rescued the colony-formation ability enhanced by anti-miR-150. (C) Individual silencing of CCND1, CCND2, CDK2 and CCNE2 rescued the anchorage-independent growth ability enhanced by anti-miR-150.
miR-150 levels clinically correlate with CCND1, CCND2, CDK2 and CCNE2 expression in NPC
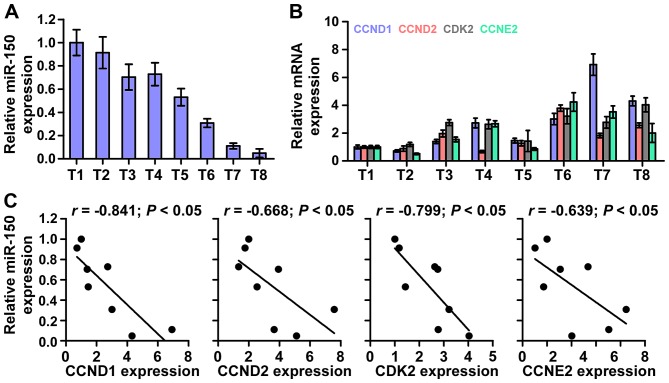
Finally, we investigated whether miR-150 correlated with CCND1, CCND2, CDK2 and CCNE2 expression in NPC clinical tissues. The rationale used to perform the correlation of miR-150 with CCND1, CCND2, CKD2 and CCNE2 expression in NPC tissues as follows: we first examined the miR-150 expression and the mRNA expression levels of CCND1, CCND2, CKD2 and CCNE2 via real-time PCR in 8 NPC tissues. Relative fold expressions of miR-150, CCND1, CCND2, CKD2 and CCNE2 in NPC tissue samples were calculated. Then, one tissue sample was randomly selected and numbered as T1, other 7 tissue samples were numbered in order as T2, T3, T4, T5, T6, T7 and T8. The expression levels of miR-150, CCND1, CCND2, CKD2 and CCNE2 in each NPC tissue were normalized by the corresponding miR-150, CCND1, CCND2, CKD2 and CCNE2 expression levels in T1 NPC tissue. The relative expression levels of miR-150, CCND1, CCND2, CKD2 and CCNE2 after normalization were used to perform the correlation analysis among miR-150 and CCND1, CCND2, CKD2 and CCNE2 expression. This analysis method of clinical correlation was widely used in multiple lines of studies (34,35). The analysis of clinical correlation revealed that a significant inverse correlation was found between miR-150 and CCND1 (r=−0.841; P<0.05), CCND2 (r=−0.668; P<0.05), CDK2 (r=−0.799; P<0.05), and CCNE2(r=−0.639; P<0.05) expression in NPC (Fig. 7) in eight fresh NPC tissues. Taken together, these results indicate that CCND1, CCND2, CDK2 and CCNE2 expression negatively correlate with miR-150 levels in NPC tissues.
Figure 7.
Clinical relevance of miR-150 with CCND1, CCND2, CDK2 and CCNE2 in nasopharyngeal carcinoma tissues. (A) miR-150 expression in eight freshly-frozen human nasopharyngeal carcinoma tissues. Transcript levels were normalized by U6 expression. One tissue sample was randomly selected and numbered as T1, other 7 tissue samples were numbered in order as T2, T3, T4, T5, T6, T7 and T8. The expression levels of miR-150 in each NPC tissue were normalized by the corresponding miR-150 expression in T1 NPC tissue. The relative expression levels of miR-150 after normalization were used to perform the correlation analysis among miR-150 and CCND1, CCND2, CKD2 and CCNE2 expression. (B) Analysis of CCND1, CCND2, CDK2 and CCNE2 expression in eight freshly-frozen human nasopharyngeal carcinoma tissues. Transcript levels were normalized by GAPDH expression. One tissue sample was randomly selected and numbered as T1, other 7 tissue samples were numbered in order as T2, T3, T4, T5, T6, T7 and T8. The expression levels of CCND1, CCND2, CKD2 and CCNE2 in each NPC tissue were normalized by the corresponding CCND1, CCND2, CKD2 and CCNE2 expression levels in T1 NPC tissue. The relative expression levels of CCND1, CCND2, CKD2 and CCNE2 after normalization were used to perform the correlation analysis among miR-150 and CCND1, CCND2, CKD2 and CCNE2 expression. (C) Correlation between miR-150 levels and CCND1, CCND2, CDK2 and CCNE2 expression in nasopharyngeal carcinoma tissues.
Discussion
This study found that miR-150 expression is markedly down-regulated in NPC tissues and cells, which is consistent with the analysis of the results of publicly available NPC datasets. Moreover, upregulation of miR-150 suppresses NPC cell proliferation and cell cycle in vitro and tumorigenesis in vivo. Conversely, silencing miR-150 displays the opposite effect on NPC cells. Our results further demonstrated that CCND1, CCND2, CDK2 and CCNE2 are the direct targets of miR-150 and importantly the stimulatory effects of downregulating miR-150 on the proliferation and cell cycle of NPC cells are reversed by individual silencing of these target proteins. Therefore, these results present our mechanistic understanding of miR-150-mediated tumor suppression in NPC.
Accumulating studies have indicated that several individual components of the cell cycle machinery are targeted by specific miRNAs and documented that the loss or gain of miRNA-mediated cell cycle control contributed to malignancy in a number of cancers. Several lines of evidence indicated that miRNAs regulate classic cell cycle control pathways by directly targeting the components of the cell cycle, including cyclins, CDKs, E2F transcription factors and Cdk inhibitors (CKIs) (36). For example, a large body of evidence indicated that upregulating let-7, a well-known tumor suppressor as well as a critical regulator of the cell cycle, induced a noted accumulation of G0 and G1 cell cycle stages. Microarray analysis and reporter assays revealed that multiple genes implicated in promoting G1/S transition, including CCND2, CDK6 and CDC25A, were the direct targets of let-7 (37). Moreover, Linsley and colleagues reported that miR-16 negatively regulated cellular growth and cell cycle progression. Through microarray profiling and functional screening, they found that the most downregulated transcripts were highly enriched for cell cycle- related genes, as indicated by gene ontology (GO) annotation, indicating that miR-16 simultaneously regulates these targets that may function in concert to control cell cycle progression (38). Therefore, these findings indicated that the identified cell cycle-related target genes of these miRNAs clarify well the molecular mechanisms by which miRNAs exert their effects on cell cycle control. Furthermore, miR-150-mediated tumor suppression via inhibiting cancer cell proliferation and cell cycle progression has been well established in several cancer (39–42). However, the expression level and biological role of miR-150 in NPC lack evidence. In this study, we found that miR-150 expression was decreased in NPC tissues and cells compared with normal NPC tissues and cell lines. Upregulation of miR-150 suppressed, whereas miR-150 downregulation promoted, the proliferation and cell cycle of NPC cells in vitro and tumorigenesis in vivo. To identify the mechanisms by which miR-150 regulates the proliferation and cell cycle pathways, we further examined the effects of miR-150 on the cell cycle-related genes, including CCND1, CCND2, CCND3, CCNE1, CCNE2, CDK2, CDK4 and CDK6. Of these, the mRNA and protein level of CCND1, CCND2, CDK2 and CCNE2 were decreased in the miR-150-overexpressing NPC cells and increased in the miR-150-silencing NPC cells. The expression of the other four of these genes were not influenced by miR-150. The analysis of the publicly available algorithms (miRanda and TargetScan) revealed that CCND1, CCND2, CDK2 and CCNE2 are the potential targets of miR-150. Importantly, the stimulatory roles of silencing miR-150 in the proliferation and cell cycle were attenuated by individual knockdown of the targeted genes. Therefore, our results uncover a novel mechanism responsible for the inhibitory effect of miR-150 on the proliferation and cell cycle in NPC cells.
The majority of the previous studies showed that miR-150 was downregulated in multiple human cancers, including hepatocellular carcinoma, lymphoma, colorectal cancer, pancreatic cancer which contributed to cancer cell proliferation, drug resistance and metastasis via varing mechanisms (39,42–46). Furthermore, the expression of miR-150 has also been reported to be upregulated in non-small cell lung cancer, gastric cancer and chronic lymphocytic leukemia (47–52). These findings indicated that miR-150 functions as both an oncomiR and tumor suppressive miRNA, depending on the tumor type. However, the expression level and specific role of miR-150 in NPC remains unknown. In this study, we found that miR-150 expression is downregulated in NPC tissues and cells compared with normal NPC tissues and cell lines. Upregulation of miR-150 suppressed, whereas miR-150 downregulation promoted, the proliferation and cell cycle of NPC cells in vitro and tumorigenesis in vivo. Furthermore, the inhibitory role of miR-150 in the proliferation and cell cycle progression in NPC were mediated by the cell cycle-related genes, including CCND1, CCND2, CDK2 and CCNE2. Intriguingly, Cao et al reported that miR-150 was significantly upregulated in lung cancer clinical specimens and high expression of miR-150 promoted the proliferation of lung cancer cells by targeting SRC kinase signalling inhibitor 1 (49). This evidence suggests that the complexity of the regulatory mechanisms by miR-150 might either promote or inhibit cellular proliferation, is environment- and tumor type-dependent.
In conclusion, this study revealed that tumor-suppressive miR-150 inhibits the proliferation and cell cycle progression via repressing cell cycle-related genes CCND1, CCND2, CDK2 and CCNE2. Thus, improved understanding of the specific mechanism of downregulation miR-150 in the pathogenesis of NPC will increase our knowledge of nasopharyngeal carcinoma development, which will help to develop new therapeutic measures against nasopharyngeal carcinoma.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81272434, 81500007), the Guangdong Provincial Medical Research Program (A2016395), the Dongguan Medical Research Program (no. 2016105101292), the Scientific and Technological Project of Zhanjiang (2015B01043), the Surface Cultivation Project of Guangdong Medical University (M2015010), and the Open Program of Guangdong Provincial Key Laboratory of Medical Molecular Diagnostics (FZZD201605).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Qu C, Liang Z, Huang J, Zhao R, Su C, Wang S, Wang X, Zhang R, Lee MH, Yang H. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle. 2012;11:785–796. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ai MD, Li LL, Zhao XR, Wu Y, Gong JP, Cao Y. Regulation of survivin and CDK4 by Epstein-Barr virus encoded latent membrane protein 1 in nasopharyngeal carcinoma cell lines. Cell Res. 2005;15:777–784. doi: 10.1038/sj.cr.7290347. [DOI] [PubMed] [Google Scholar]
- 4.Zhen Y, Fang W, Zhao M, Luo R, Liu Y, Fu Q, Chen Y, Cheng C, Zhang Y, Liu Z. miR-374a-CCND1-pPI3K/AKT-c-JUN feedback loop modulated by PDCD4 suppresses cell growth, metastasis, and sensitizes nasopharyngeal carcinoma to cisplatin. Oncogene. 2017;36:275–285. doi: 10.1038/onc.2016.201. [DOI] [PubMed] [Google Scholar]
- 5.Nigg EA. Cyclin-dependent protein kinases: Key regulators of the eukaryotic cell cycle. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 6.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champeris Tsaniras S, Kanellakis N, Symeonidou IE, Nikolopoulou P, Lygerou Z, Taraviras S. Licensing of DNA replication, cancer, pluripotency and differentiation: An interlinked world? Semin Cell Dev Biol. 2014;30:174–180. doi: 10.1016/j.semcdb.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 8.O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13:417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 9.Kumar SK, LaPlant B, Chng WJ, Zonder J, Callander N, Fonseca R, Fruth B, Roy V, Erlichman C, Stewart AK, Mayo Phase 2 Consortium Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood. 2015;125:443–448. doi: 10.1182/blood-2014-05-573741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khew-Goodall Y, Goodall GJ. Myc-modulated miR-9 makes more metastases. Nat Cell Biol. 2010;12:209–211. doi: 10.1038/ncb0310-209. [DOI] [PubMed] [Google Scholar]
- 12.Ren D, Wang M, Guo W, Huang S, Wang Z, Zhao X, Du H, Song L, Peng X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014;358:763–778. doi: 10.1007/s00441-014-2001-y. [DOI] [PubMed] [Google Scholar]
- 13.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren D, Wang M, Guo W, Zhao X, Tu X, Huang S, Zou X, Peng X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int J Oncol. 2013;42:1473–1481. doi: 10.3892/ijo.2013.1825. [DOI] [PubMed] [Google Scholar]
- 15.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Liu J, Zang D, Wu S, Liu A, Zhu J, Wu G, Li J, Jiang L. Upregulation of miR-572 transcriptionally suppresses SOCS1 and p21 and contributes to human ovarian cancer progression. Oncotarget. 2015;6:15180–15193. doi: 10.18632/oncotarget.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Ren D, Guo W, Wang Z, Huang S, Du H, Song L, Peng X. Loss of miR-100 enhances migration, invasion, epithelial-mesenchymal transition and stemness properties in prostate cancer cells through targeting Argonaute 2. Int J Oncol. 2014;45:362–372. doi: 10.3892/ijo.2014.2413. [DOI] [PubMed] [Google Scholar]
- 18.Huang S, Guo W, Tang Y, Ren D, Zou X, Peng X. miR-143 and miR-145 inhibit stem cell characteristics of PC-3 prostate cancer cells. Oncol Rep. 2012;28:1831–1837. doi: 10.3892/or.2012.2015. [DOI] [PubMed] [Google Scholar]
- 19.Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J, Wang S, Liu T, Cai H, Yao K, et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun. 2015;6:7353. doi: 10.1038/ncomms8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S, Long X, Jiang Q, Song Y, Cheng C, et al. Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma. Cell Death Dis. 2013;4:e872. doi: 10.1038/cddis.2013.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Y, Meng F, Venter J, Wu N, Wan Y, Standeford H, Francis H, Meininger C, Greene J, Jr, Trzeciakowski JP, et al. miR-34a-dependent overexpression of Per1 decreases cholangio-carcinoma growth. J Hepatol. 2016;64:1295–1304. doi: 10.1016/j.jhep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aakula A, Kohonen P, Leivonen SK, Mäkelä R, Hintsanen P, Mpindi JP, Martens-Uzunova E, Aittokallio T, Jenster G, Perälä M, et al. Systematic identification of microRNAs that impact on proliferation of prostate cancer cells and display changed expression in tumor tissue. Eur Urol. 2016;69:1120–1128. doi: 10.1016/j.eururo.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Z, Guo W, Wang Q, Chen Z, Huang S, Zhao F, Yao M, Zhao Y, He X. MicroRNA-124 reduces the pentose phosphate pathway and proliferation by targeting PRPS1 and RPIA mRNAs in human colorectal cancer cells. Gastroenterology. 2015;149:1587–1598.e11. doi: 10.1053/j.gastro.2015.07.050. [DOI] [PubMed] [Google Scholar]
- 24.Zhao M, Luo R, Liu Y, Gao L, Fu Z, Fu Q, Luo X, Chen Y, Deng X, Liang Z, et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-I3K/AKT-c-JUN. Nat Commun. 2016;7:11309. doi: 10.1038/ncomms11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Q, Ren X, Chen J, Li Y, Tang X, Wen X, Yang X, Zhang J, Wang Y, Ma J, et al. miR-16 targets fibroblast growth factor 2 to inhibit NPC cell proliferation and invasion via PI3K/AKT and MAPK signaling pathways. Oncotarget. 2016;7:3047–3058. doi: 10.18632/oncotarget.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jun GJ, Zhong GG, Ming ZS. miR-218 inhibits the proliferation of glioma U87 cells through the inactivation of the CDK6/cyclin D1/p21(Cip1/Waf1) pathway. Oncol Lett. 2015;9:2743–2749. doi: 10.3892/ol.2015.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 28.Guo W, Ren D, Chen X, Tu X, Huang S, Wang M, Song L, Zou X, Peng X. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J Cell Biochem. 2013;114:1606–1615. doi: 10.1002/jcb.24502. [DOI] [PubMed] [Google Scholar]
- 29.Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YM, Ting CM, Cheng YK, Fan TP, Wong RN, Lung ML, Mak NK. Mechanisms of 2-methoxyestradiol-induced apoptosis and G2/M cell-cycle arrest of nasopharyngeal carcinoma cells. Cancer Lett. 2008;268:295–307. doi: 10.1016/j.canlet.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Ghimire BR. Effects of zoledronic acid (ZOL) on proliferation of nasopharyngeal carcinoma (NPC) cell lines and synergistic effects of ZOL and cisplatin on NPC cell lines. J Clin Oncol. 2009;27:e13536. [Google Scholar]
- 32.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 33.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 34.Lin C, Liu A, Zhu J, Zhang X, Wu G, Ren P, Wu J, Li M, Li J, Song L. miR-508 sustains phosphoinositide signalling and promotes aggressive phenotype of oesophageal squamous cell carcinoma. Nat Commun. 2014;5:4620. doi: 10.1038/ncomms5620. [DOI] [PubMed] [Google Scholar]
- 35.Cui Y, Ma W, Lei F, Li Q, Su Y, Lin X, Lin C, Zhang X, Ye L, Wu S, et al. Prostate tumour overexpressed-1 promotes tumourigenicity in human breast cancer via activation of Wnt/β-catenin signalling. J Pathol. 2016;239:297–308. doi: 10.1002/path.4725. [DOI] [PubMed] [Google Scholar]
- 36.Chivukula RR, Mendell JT. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci. 2008;33:474–481. doi: 10.1016/j.tibs.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 38.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 40.Hornick NI, Doron B, Abdelhamed S, Huan J, Harrington CA, Shen R, Cambronne XA, Chakkaramakkil Verghese S, Kurre P. AML suppresses hematopoiesis by releasing exosomes that contain microRNAs targeting c-MYB. Sci Signal. 2016;9:ra88. doi: 10.1126/scisignal.aaf2797. [DOI] [PubMed] [Google Scholar]
- 41.Leoncini PP, Bertaina A, Papaioannou D, Flotho C, Masetti R, Bresolin S, Menna G, Santoro N, Zecca M, Basso G, et al. MicroRNA fingerprints in juvenile myelomonocytic leukemia (JMML) identified miR-150-5p as a tumor suppressor and potential target for treatment. Oncotarget. 2016;7:55395–55408. doi: 10.18632/oncotarget.10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun W, Zhang Z, Wang J, Shang R, Zhou L, Wang X, Duan J, Ruan B, Gao Y, Dai B, et al. MicroRNA-150 suppresses cell proliferation and metastasis in hepatocellular carcinoma by inhibiting the GAB1-ERK axis. Oncotarget. 2016;7:11595–11608. doi: 10.18632/oncotarget.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito M, Teshima K, Ikeda S, Kitadate A, Watanabe A, Nara M, Yamashita J, Ohshima K, Sawada K, Tagawa H. MicroRNA-150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6, in advanced cutaneous T-cell lymphoma. Blood. 2014;123:1499–1511. doi: 10.1182/blood-2013-09-527739. [DOI] [PubMed] [Google Scholar]
- 44.Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J, Liu W, Qin H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 45.Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, Singh AP. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32:1832–1839. doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wuerkenbieke D, Wang J, Li Y, Ma C. miRNA-150 down-regulation promotes pertuzumab resistance in ovarian cancer cells via AKT activation. Arch Gynecol Obstet. 2015;292:1109–1116. doi: 10.1007/s00404-015-3742-x. [DOI] [PubMed] [Google Scholar]
- 47.Yin QW, Sun XF, Yang GT, Li XB, Wu MS, Zhao J. Increased expression of microRNA-150 is associated with poor prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:842–846. [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K, Ren G, Su T, Pan Y, Feng B, et al. MiR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2. Biochem Biophys Res Commun. 2010;392:340–345. doi: 10.1016/j.bbrc.2009.12.182. [DOI] [PubMed] [Google Scholar]
- 49.Cao M, Hou D, Liang H, Gong F, Wang Y, Yan X, Jiang X, Wang C, Zhang J, Zen K, et al. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer. 2014;50:1013–1024. doi: 10.1016/j.ejca.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 50.Papakonstantinou N, Ntoufa S, Chartomatsidou E, Papadopoulos G, Hatzigeorgiou A, Anagnostopoulos A, Chlichlia K, Ghia P, Muzio M, Belessi C, et al. Differential microRNA profiles and their functional implications in different immunogenetic subsets of chronic lymphocytic leukemia. Mol Med. 2013;19:115–123. doi: 10.2119/molmed.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang M, Cui G, Ding M, Yang W, Liu Y, Dai D, Chen L. miR-935 promotes gastric cancer cell proliferation by targeting SOX7. Biomed Pharmacother. 2016;79:153–158. doi: 10.1016/j.biopha.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Zhang AX, Lu FQ, Yang YP, Ren XY, Li ZF, Zhang W. MicroRNA-217 overexpression induces drug resistance and invasion of breast cancer cells by targeting PTEN signaling. Cell Biol Int. 2015 doi: 10.1002/cbin.10506. Epub ahead of print. [DOI] [PubMed] [Google Scholar]