Abstract
MicroRNAs are a class of small non-coding, endogenous RNAs involved in cancer development and progression. MicroRNA-221 (mir-221) has been reported to have both an oncogenic and tumor-suppressive role in human tumors, but the role of miR-221 in ovarian cancer is poorly understood. In the present study, the expression levels of miR-221 and the apoptosis protease activating factor 1 (APAF1) protein in 63 samples of ovarian cancer tissues and the cell lines, IOSE25, A2780, OVCAR3, SKOV3 and 3AO were detected by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and western blot analysis, respectively. Cell proliferation was measured using Cell Counting kit-8 (CCK-8); cell migration and invasion were detected using a Transwell assay; cell apoptosis was evaluated by flow cytometry and Hoechst staining, and a luciferase assay was performed to verify a putative target site of miR-221 in the 3′-UTR of APAF1 mRNA. Expression of miR-221 was upregulated in ovarian cancer tissues. Patients with increased miR-221 expression levels had a reduced disease-free survival (P=0.0014) and overall survival (P=0.0058) compared with those with low miR-221 expression. Transfection of SKOV3 and A2780 cell lines with miR-221 inhibitor induced APAF1 protein expression, suppressed cell proliferation and migration and promoted tumor cell apoptosis. In conclusion, the APAF1 gene was confirmed as a direct target of miR-221 and overexpression of APAF1 suppressed ovarian cancer cell proliferation and induced cell apoptosis in vitro. These findings indicate that miR-221-APAF1 should be studied further as a potential new diagnostic or prognostic biomarker for ovarian cancer.
Keywords: ovarian cancer, miRNA-221, prognosis, apoptosis protease activating factor 1, cell proliferation, apoptosis
Introduction
Ovarian cancer is a malignancy that is associated with a high mortality. Worldwide, this form of cancer represents ~3% of all cancers in women and is the fifth leading cause of death in the female population (1). Despite developments in cancer therapy in the past two decades, the mortality from ovarian cancer has remained constant (1). Early diagnosis and improved prognostic markers are required to improve patient survival rates for ovarian cancer (2). The identification of new molecular markers may also lead to more effective and targeted therapies for ovarian cancer.
MicroRNAs (miRNAs) are a class of small, non-coding, endogenous RNAs that are ~22 nucleotides in length and play important roles in post-transcriptional regulation (3). miRNAs can inhibit target gene expression by binding to the 3′-untranslated region (3′-UTR) of target mRNA, which results in either mRNA degradation or prevention of translation to protein expression (4,5). miRNAs have several roles in normal cellular processes involving cell proliferation, apoptosis, cell differentiation and stress response (6–8). In recent years, miRNAs have received attention in cancer research, as 98 miRNAs are located at genomic regions involved in malignancy, indicating that miRNAs could be used as diagnostic and prognostic biomarkers (9). However, miRNAs have been reported to have both tumor suppressor and oncogenic activities (10).
In particular, miR-221 has been reported to have both tumor suppressor and oncogenic roles in different types of human cancer, including breast cancer (11,12) and hepatocellular carcinoma (13). Recently, miR-221 has been shown to have as tumor suppressor role in non-small cell lung cancer (14). miRNA-221 is encoded by a gene located on chromosome Xp11.3 and acts as an oncogene in tumors of epithelial origin and as an oncosuppressor in hemopoietic malignancies (12–14). The increasing awareness of the role of miRNAs in human malignancy has raised the possibility of their future role as diagnostic and prognostic biomarkers (15,16). Recent studies have also indicated that miRNAs may be potential therapeutic targets in human malignancy (17,18).
In 2007, a study in human prostate cancer cell lines demonstrated that miR-221 mediated its effects on the control of cell proliferation by targeting p27Kip1 (20). It is now known that miR-221 exerts its oncogenic effects through the inhibition of the cyclin-dependent kinase (CDK) inhibitors p27Kip1 and p57, upregulating ZEB2, an epithelial-to-mesenchymal transition (EMT)-inducing gene through TRPS1 (19,20). Overexpression of miR-221 in malignant glioblastoma cells has been shown to promote the cell cycle through G1 into S phase and to promote cell apoptosis (21). In addition, miR-221 has been shown to be downregulated in Kaposi's sarcoma-associated herpes virus-associated cancers, including Kaposi sarcoma and primary effusion lymphoma (PELs) (22). A recent study has shown that the overexpression of miR-221 enhances chemosensitivity to gemcitabine in cholangiocarcinoma cells (23). These data demonstrate that miR-221 has a bimodal function in the tumorigenesis of human cancers.
Apoptotic protease activating factor 1 (APAF1) is a crucial component of the apoptosome that is present in humans and mice (24,25) and is assembled in response to cellular stress, including DNA damage, hypoxia and oncogene activation (26–28). APAF1 is closely related to several oncogenes and tumor suppressor genes, including B-cell lymphoma-2 (Bcl-2) and p53 (29,30). A series of studies have shown that APAF1 protein is an important apoptosis factor that is abnormally expressed in cancer tissues (27,31). APAF1 is downregulated in human colorectal cancer, and its expression is associated with adverse patient prognosis (32,33).
Since the role of miR-221 in the development and progression of ovarian cancer remains unknown, the present study was done using both established tumor cell lines and tumor tissue samples from patients with histopathologically confirmed primary ovarian carcinoma.
Materials and methods
Patients studied and tissue collection
Sixty-three patients with histopathologically diagnosed ovarian cancer had tumor tissue sampled as part of their routine diagnosis with adjacent normal tissue samples. All patients underwent surgery at the First Affiliated Hospital of Sun Yat-sen University between 2008 and 2010. The diagnosis of primary ovarian cancer was based on histopathological evaluation according to the International Federation of Obstetrics and Gynecology (FIGO) criteria. Clinical pathology information was available for all patient samples, as shown in Table I. No other treatments were conducted in these patients before surgery and tissue sampling. Informed patient consents were obtained from every study participant. The present study was approved by the Research Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University. All tissues were collected and immediately frozen in liquid nitrogen and stored at −80°C for further study.
Table I.
Clinicopathological characteristics and GAS5 expression in 63 patient samples of ovarian cancer.
| Clinical parameters | No. of cases (%) |
|---|---|
| Age (years) | |
| <55 | 29 (46.03) |
| >55 | 34 (53.96) |
| Size (cm) | |
| >5 | 33 (52.38) |
| <5 | 30 (47.62) |
| Histologic differentiation | |
| Well | 5 (7.94) |
| Moderate | 23 (36.51) |
| Poor | 28 (44.44) |
| Undifferentiated | 7 (11.11) |
| Invasion depth | |
| T1 | 15 (23.81) |
| T2 | 16 (25.40) |
| T3 | 18 (28.57) |
| T4 | 14 (22.22) |
| FIGO stages | |
| I | 8 (12.70) |
| II | 22 (34.92) |
| III | 27 (42.86) |
| IV | 6 (9.52) |
| Lymphatic metastasis | |
| Yes | 30 (47.62) |
| No | 33 (52.38) |
| Distant metastasis | |
| Yes | 8 (12.70) |
| No | 55 (87.30) |
| Expression of miR-221 | |
| Low expression | 31 (50.79) |
| High expression | 32 (49.21) |
Cell culture
Human ovarian surface epithelial cells (IOSE25) were immortalized and cultured as previously described (34). Four human ovarian cancer cell lines, A2780, OVCAR3, SKOV3 and 3AO were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and the American Type Culture Collection (ATCC; Manassas, VA, USA), respectively.
The ovarian cancer cell lines were maintained according to the vendor's instructions. Briefly, A2780 and 3AO cell lines were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA). SKOV3 cells were cultured in McCoy's 5A Modified Medium (ATCC) with 10% FBS (Gibco). OVCAR3 cells were cultured in RPMI-1640 medium (ATCC) with 20% FBS (Gibco). All the media contained 1% penicillin-streptomycin (100 U/ml penicillin and 100 µg/ml streptomycin). The ovarian cancer cells were cultured and maintained in a humidified incubator at 37°C and supplemented with 5% CO2.
Plasmid construction
The apoptotic protease activating factor 1 (APAF1) 3′-untranslated region (3′-UTR) containing putative binding sites for microRNA-221 (miR-221) was cloned downstream of the psi-CHECK2 vector (Promega, Madison, WI, USA) and named as APAF1-3′UTR-WT. APAF1 mutant 3′-UTR recombinant plasmid was generated by QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA), which generated a mutation of 7 bp from AUGUAGC to UACGCGU in the predicted miR-221 target binding site, named as APAF1-3′UTR-MUT. The full-length cDNA encoding human APAF1 was amplified and the recombinant plasmid, pcDNA3.1-APAF1 was constructed. All plasmids were confirmed by DNA sequencing.
Cell transfection
The miR-221 mimics, miR-221 inhibitor and scrambled sequence pre-miR negative control (NC) were purchased from a commercial manufacturer (Guangzhou RiboBio, Co., Ltd., Guangzhou, China). Ovarian cancer cells (100 µl) were seeded in 24-well plates (1×106 cells/ml) and incubated for 24 h, then cells were transfected with miRNA mimics/inhibitor (50 mM) or vectors (2 µg) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in serum-free medium in accordance with the manufacturer's instructions.
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
TRIzol reagent (Invitrogen) was used to extract total RNA from tissue samples or cultured cells. For quantitative real-time polymerase chain reaction (qRT-PCR), 2 µg of total RNA was used for the reverse transcription reaction and cDNA synthesis by using a Reverse Transcription kit (Takara, Dalian, China). Quantitative real-time PCR analysis was performed with SYBR-Green Real-Time Master Mix (Toyobo, Co., Ltd, Osaka, Japan). Results were normalized to a constitutive expression gene, GAPHD. The relative expression of miRNA-221 was detected using a SYBR PrimeScript miRNA RT PCR kit (Takara) in accordance with the manufacturer's instructions and U6 was used as an internal control. The gene-specific primers were synthesized by Sangon Biotech, Co., Ltd. (Shanghai, China) (Table III). qRT-PCR and data collection were performed on an Applied Biosystems 7500 Sequence Detection system (Applied Biosystems, Foster City, CA, USA). The relative expression of miR-221 and APAF1 were calculated and normalized using the 2−ΔΔCt method.
Table III.
Primer sequences used for miRNA and mRNA expression analysis.
| Name | Primer sequence (5′-3′) |
|---|---|
| miR-221-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGGGTATT |
| U6-RT | CGCTTCACGAATTTGCGTGTCAT |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| miR-221-F | ACACTCCAGCTGGGTGTCAGTTTGTCAA |
| Universal-R | CTCAACTGGTGTCGTGGA |
| APAF1-F | TTGCTGCCCTTCTCCATGAT |
| APAF1-R | TCCCAACTGAAACCCAATGC |
| GAPDH-F | TGTTCGTCATGGGTGTGAA |
| GAPDH-R | ATGGCATGGACTGTGGTCAT |
F, forward primer; R, reverse primer; RT, reverse transcription primer.
Protein extraction and western blot analysis
Total proteins were extracted from tissue samples or cultured cells with SDS lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) on ice for 20 min and the protein concentrations were determined using BCA protein assay kit (Pierce, Rockford, IL, USA). Equal amounts of total proteins were separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) at 120 V for 2 h, transferred to 0.22 µm polyvinylidene difluoride membranes (PVDF) (Millipore, Billerica, MA, USA) and incubated with APAF1 antibodies (1:1,000, ab32372; Abcam). Proteins were detected by enhanced chemiluminescence (ECL) as described by the manufacturer (Beyotime Institute of Biotechnology) and the intensity of the bands was quantified by densitometry (Quantity One software; Bio-Rad Laboratories, Hercules, CA, USA). Results were normalized to a constitutive expression gene, GAPHD (1:2,000, ab9485; Abcam).
Luciferase assays
For the reporter assay, 1×105 cells were cultured in 24-well plates one day before transfection. A total of 100 ng APAF1-3′UTR-WT or -MUT vectors were co-transfected with 50 nM miR-221 mimics or negative control into cells using Lipofectamine 2000 reagent. After 48 h of transfection, luciferase activities were measured using Dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Cell proliferation assays
Cell Counting kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) was used to examine the cell proliferation, according to the manufacturer's instruction. Briefly, 2×104 cells were seeded into a 96-well plate. Cell viability was evaluated with CCK-8 at daily intervals from the next 24, 48 and 72 h after seeding. Following the CCK-8 assay at 37°C for 1 h, ovarian cancer cells were used to measure the absorbency at 450 nm using a microplate reader Thermo Plate (Rayto Life and Analytical Science, Co., Ltd., Hamburg, Germany).
Cell migration and invasion assays
A Transwell migration assay and a Matrigel invasion assay were performed separately using 24-well Transwell inserts with 8-µm pore size (Corning Costar Corp., Corning, NY, USA). For the Transwell migration assay, 2×104 ovarian cancer cells were plated in 100 µl corresponding culture medium without FBS into the upper chamber of a Transwell insert with a non-coated membrane. For the Matrigel invasion assay, 4×104 ovarian cancer cells were suspended in 100 µl serum-free corresponding culture medium were loaded in the upper Matrigel-coated chamber. In both assays, 500 µl of culture medium containing 20% FBS was added to the lower chamber. Cells were then allowed to migrate or invade for 24 h at 37°C. The cells that migrated or invaded into the bottom chamber were fixed with 100% methanol for 30 min and stained using 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) for 20 min, and the permeating cells were counted under a phase-contrast microscope (Olympus Corp., Tokyo, Japan).
Cell apoptosis
Cell apoptosis was studied using an Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) apoptosis detection kit (BestBio, Shanghai China) with flow cytometry and in accordance with the manufacturer's instruction. Briefly, 1×106 cells were harvested and resuspended in cold PBS, and then stained using the Annexin V FITC/PI apoptosis detection kit according to the manufacturer's instructions. Samples were analyzed using Becton-Dickinson flow cytometer. Annexin V(+)/PI(−) and Annexin V(+)/PI(+) represented the ovarian cancer cells in early and late apoptosis or necrosis, respectively.
Hoechst 33342 nuclear staining method was performed to detect cell apoptosis. Briefly, cells were cultured with Hoechst 33342 blue fluorescent nuclear dye (Sigma-Aldrich) in 6-well cell culture plates for 30 min. The nuclear morphology was examined using fluorescence microscopy with a filter for Hoechst 33342 at 365 nm.
Statistical analysis
All results were expressed as the mean ± standard deviation (SD). Statistical analysis was performed using the SPSS 18.0 (SPSS, Inc., Chicago, IL, USA). Statistical significance between the groups was determined using the Student's t-test or the Chi-square test. Survival analysis was performed using the Kaplan-Meier method. The log-rank (Mantel-Cox) test was used to compare the differences between the patient groups.
Results
miR-221 upregulation in human ovarian cancer tissues
Fig. 1A shows that miR-221 was upregulated in ovarian cancer tissues compared with matched adjacent normal tissues in 63 cases, using quantitative reverse transcription polymerase chain reaction (qRT-PCR). In tumor specimens, miR-221 expression levels were greater than in adjacent normal tissues (median ratio of tumor to normal of 3.68).
Figure 1.
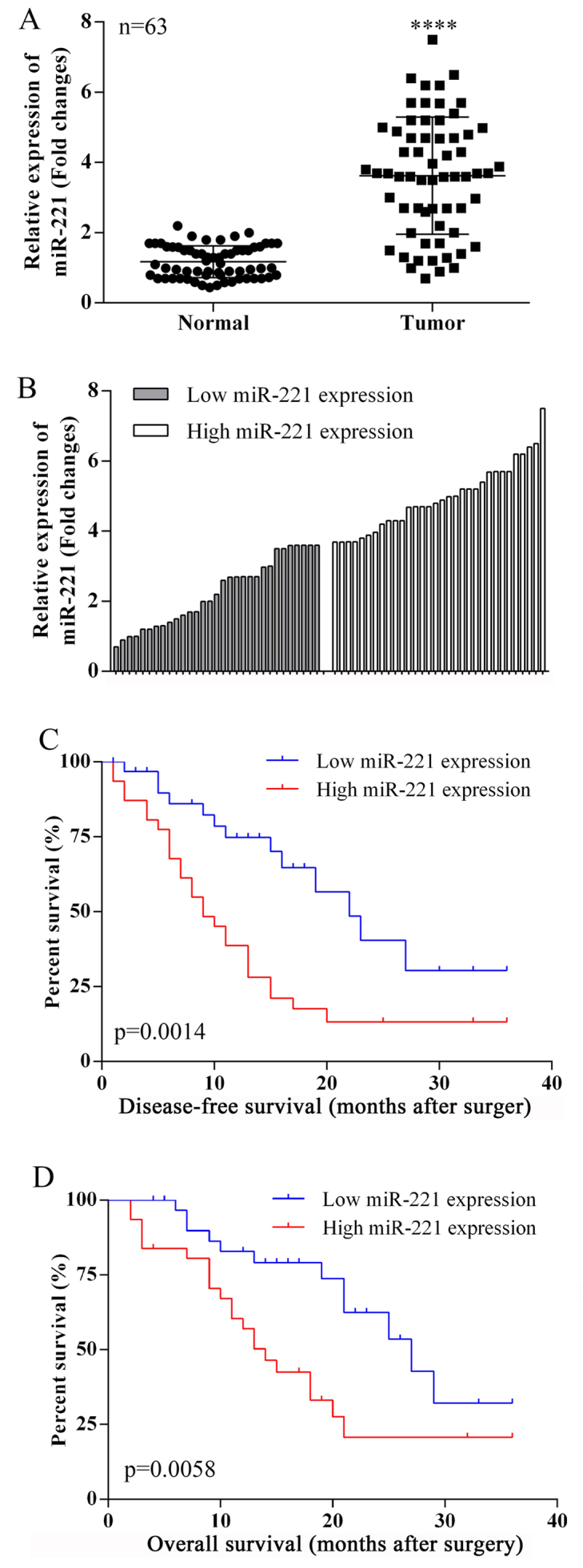
Expression of microRNA-221 (miR-221) in ovarian cancer tissues: clinicopathological correlations. (A) The expression levels of microRNA-221 (miR-221) in ovarian cancer tissues compared with non-tumor tissues (n=63) were detected using quantitative reverse-transcription polymerase chain reaction (qRT-PCR). miR-221 was significantly upregulated in tumor tissues. ****P<0.0001. (B) According to the median ratio of miR-221 expression level (3.68) in tumor tissues, miR-221 expression was classified into two groups: high miR-221 expression group (n=32) and low miR-221 expression group (n=31). (C and D) Kaplan-Meier disease-free survival (DFS) and overall survival (OS) curves according to miR-221 expression level.
miR-221 expression and clinicopathological features in ovarian cancer patients
The clinical pathology findings in 63 ovarian cancer patients studied are shown in Table I. The 63 ovarian cancer patients were classified into two groups based on the median relative miR-221 expression (3.68) in ovarian cancer tissues. Group 1: a low miR-221 expression group (n=31, miR-221 expression ≤ median). Group 2: a high miR-221 expression group (n=32, miR-221 expression ≥ median) (Fig. 1B).
A comparison of the clinicopathological features of the two groups is shown in Table II. The high-miR-221 group (group 2) had a larger tumor size (P=0.0010), deeper depth of tumor invasion (P=0.0003) and higher FIGO stage (P=0.0076) when compared with the low-miR-221 group (group 1). However, the miR-221 expression level was not associated with other parameters such as age (P=0.8052), histologic differentiation (tumor grade) (P=0.9463), lymphatic spread (P=0.4985), or distant metastasis (P=0.6723) (Table II).
Table II.
The relationship between the GAS5 expression and the clinicopathological factors in ovarian cancer patients.
| Clinical parameter | miR-221
|
Chi-squared test P-value | |
|---|---|---|---|
| Group 1 | Group 2 | ||
| Age (years) | 0.8052 | ||
| <55 | 14 | 15 | |
| >55 | 18 | 16 | |
| Size (cm) | 0.0010 | ||
| >5 | 9 | 24 | |
| <5 | 21 | 9 | |
| Histologic differentiation | 0.9463 | ||
| Well | 3 | 2 | |
| Moderate | 11 | 12 | |
| Poor | 14 | 14 | |
| Undifferentiated | 4 | 3 | |
| Invasion depth | 0.0003 | ||
| T1 | 10 | 5 | |
| T2 | 13 | 3 | |
| T3 | 4 | 14 | |
| T4 | 3 | 12 | |
| FIGO stages | 0.0076 | ||
| I | 6 | 2 | |
| II | 15 | 7 | |
| III | 8 | 19 | |
| IV | 1 | 5 | |
| Lymphatic metastasis | 0.4985 | ||
| Yes | 18 | 12 | |
| No | 17 | 16 | |
| Distant metastasis | 0.6723 | ||
| Yes | 3 | 5 | |
| No | 25 | 30 | |
miR-221 expression and patient survival
Disease-free survival (DFS) and overall survival (OS) curves were plotted according to miR-221 expression levels, using the Kaplan-Meier analysis and the log-rank test, respectively. The results are presented in Fig. 1C and 1D. Patients with high miR-221 expression levels had a reduced DFS (P=0.0014) and OS (P=0.0058). For the OS, 3 years of overall accumulative survival rates of ovarian cancer patients with low and high miR-221 expression level were 38.57 and 20.68%, respectively. High expression of miR-221 was associated with a greater OS time for ovarian cancer patients (median OS, 14 months) compared with patients with low miR-221 expression (median OS, 27 months). The 3-year disease-free survival rates for high and low expression of miR-221 patients were 35.21 and 18.54%, respectively. The median survival time for patients with low miR-221 expression was 22 months, and 9 months for patients with high miR-221 expression.
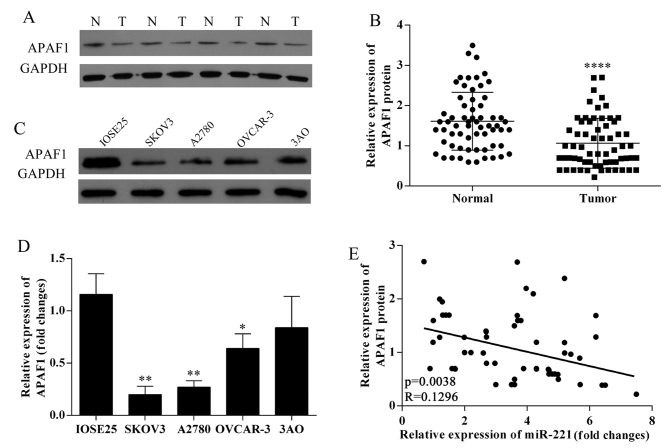
Decreased APAF1 protein expression in ovarian cancer tissues; negative correlation with miR-221 expression
The results of western blotting in ovarian cancer tissues and ovarian cancer lines SKOV3, OVCAR-3, A2780, 3AO and human ovarian surface epithelium cells showed that APAF1 protein expression was downregulated in ovarian cancer tissues (Fig. 2A and B). APAF1 protein expression was down-regulated in ovarian cancer cell lines (Fig. 2C). The expression of APAF1 in SKOV3, OVCAR-3, A2780, 3AO and human ovarian surface epithelium cells were detected using qRT-PCR (Fig. 2D); SKOV3 and A2780 cells were chosen for further study. APAF1 protein expression levels were negatively associated with miR-221 expression levels in 63 ovarian cancer tissues (P=0.0038; R=0.1296; Fig. 2E).
Figure 2.
Correlation between microRNA-221 (miR-221) expression level and the apoptosis protease activating factor 1 (APAF1) protein expression in ovarian cancer. (A) Western blot analyzed APAF1 protein expression in ovarian cancer tissues and normal samples. N, normal sample; T, tumor tissues. (B) Quantification of the APAF1 protein bands (O.D. ratio over GAPDH), ****P<0.0001. (C) Western blot analysis of APAF1 protein expression in ovarian cancer cell lines. (D) Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analyzed APAF1 mRNA expression in ovarian cancer cell lines. (E) APAF1 protein expression levels were negatively correlated with microRNA-221 (miR-221) expression levels (P=0.0038, R=0.1296).
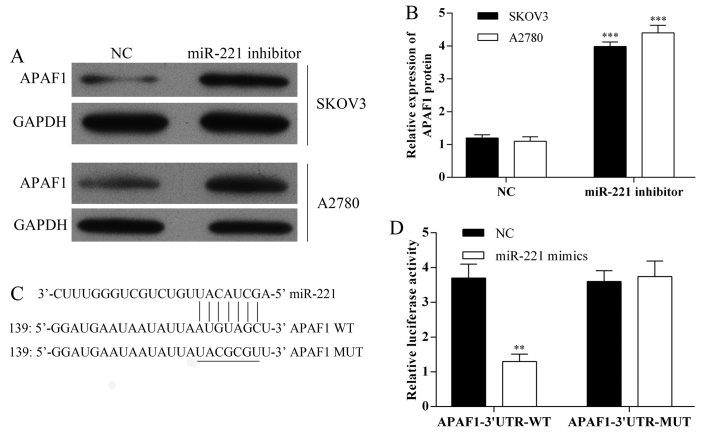
APAF1 as a potential target of miR-221
The expression of APAF1 in SKOV3 and A2780 cells following transfection with miR-221 inhibitor were detected using western blot analysis (Fig. 3A and B). Also, miRNA target predication databases (TargetScan: www.targetscan.org and MICRORNA.ORG: www.microrna.org) were used for computational analysis. miR-221 had one predictive target site in the human APAF1-3′-UTR (Fig. 3C). A Dual-luciferase reporter system was used to determine whether APAF1 was a direct target of miR-221. The results showed that miR-221 mimics downregulated the luciferase activity of the reporter and the luciferase expression of mutant APAF1-3′-UTR was not regulated by miR-221 (Fig. 3D). These results indicated that this site of APAF1-3′-UTR was the regulation site for miR-221.
Figure 3.
The apoptosis protease activating factor 1 (APAF1) gene is a direct target gene of microRNA-221 (miR-221). (A) The expression of apoptosis protease activating factor 1 (APAF1) protein in SKOV3 and A2780 cells, following miR-221 inhibitor transfection, investigated using western blot analysis. (B) Quantification of the APAF1 protein bands (O.D. ratio over GAPDH). ***P<0.001. (C) Sequence alignment of miR-221 and 3′-UTR of APAF1 mRNA using TargetScan (www.targetscan.org) and MICRORNA.ORG (www.microrna.org) algorithms. (D) Cells were co-transfected with miR-221 mimics and a luciferase reporter containing a fragment of the APAF1 3′-UTR harboring either the miR-221 binding site (APAF1-3′-UTR-WT) or a mutant (APAF1-3′-UTR-MUT). The assay showed that luciferase activity in the APAF1-3′-UTR-WT group was significantly decreased but was increased when compared with the luciferase activity of the mutant groups. **P<0.01.
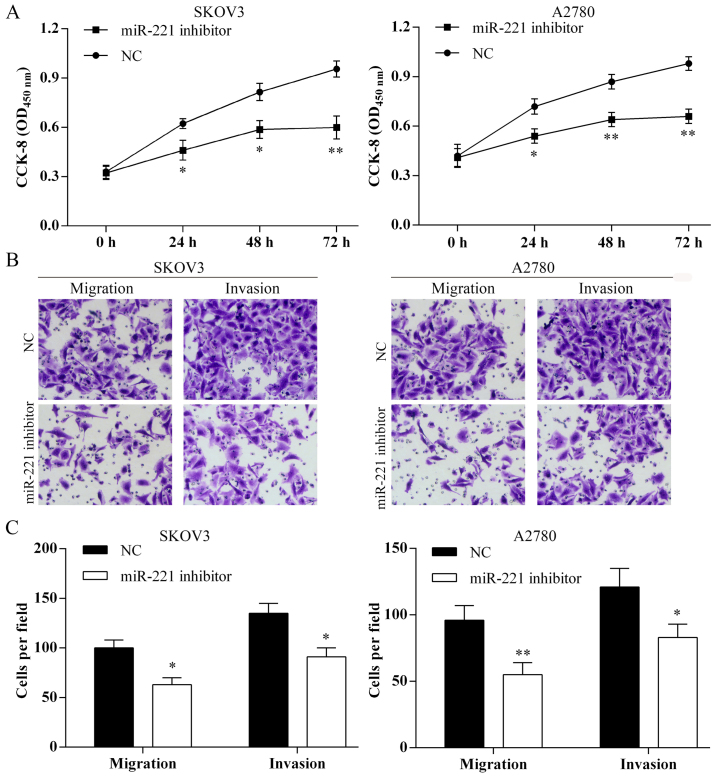
Transfection with miR-221 inhibitor and suppression of ovarian cancer cell proliferation, migration and invasion in vitro
The proliferation of SKOV3 and A2780 cells were suppressed following miR-221 inhibitor transfection (Fig. 4A). Using the Transwell assay the cell migration and invasion of SKOV3 and A2780 were inhibited following miR-221 inhibitor transfection (Fig. 4B and C).
Figure 4.
Transfection of microRNA-221 (miR-221) inhibitor suppressed ovarian cancer cell proliferation, migration and invasion. (A) Cell Counting kit-8 (CCK-8) assay results showed that the ovarian cancer cell proliferation was inhibited by miR-221 inhibitor transfection. *P<0.05; **P<0.01. (B) Cell migration and invasion were detected using Transwell assay (magnifications, ×40). Images of migration and invasion of each cell group are presented. (C) The average migration and invasion cell number per field among different experimental groups. Three independent experiments were performed. *P<0.05; **P<0.01.
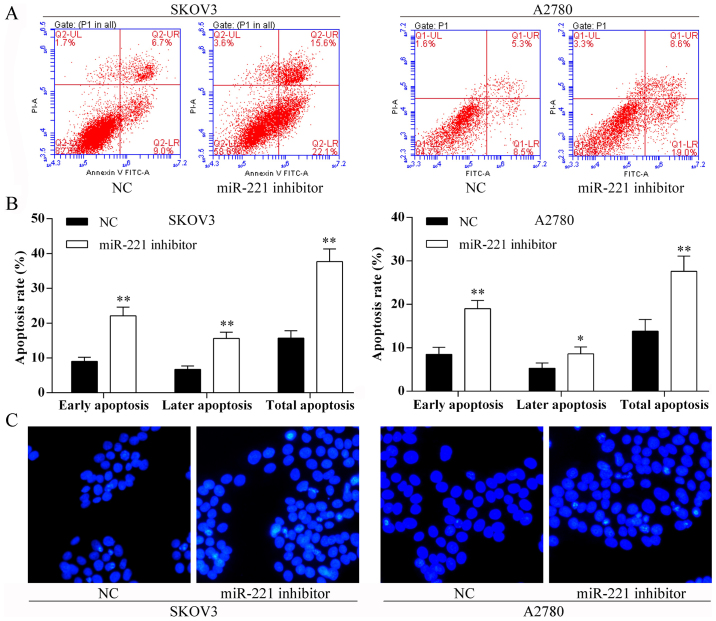
Transfection with miR-221 inhibitor and ovarian cancer cell apoptosis in vitro
Using the Annexin V-FITC/PI staining and Hoechst staining methods, the SKOV3 and A2780 cell apoptosis rates were induced after miR-221 inhibitor transfection (Fig. 5).
Figure 5.
Transfection of microRNA-221 (miR-221) promotes cell apoptosis. (A) Flow cytometric analysis of ovarian cancer cell apoptosis rate following miR-221 inhibitor transfection. LL, viable cells; LR, early apoptotic cells; UR, late apoptotic cells; UL, necrotic cells. (B) The percentage of early apoptotic cells, late apoptotic cells, and total apoptotic cells of each group. *P<0.05; **P<0.01. (C) Hoechst nuclear staining (blue) assay for SKOV3 and A2780 cells with miR-221 inhibitor transfection cell apoptosis.
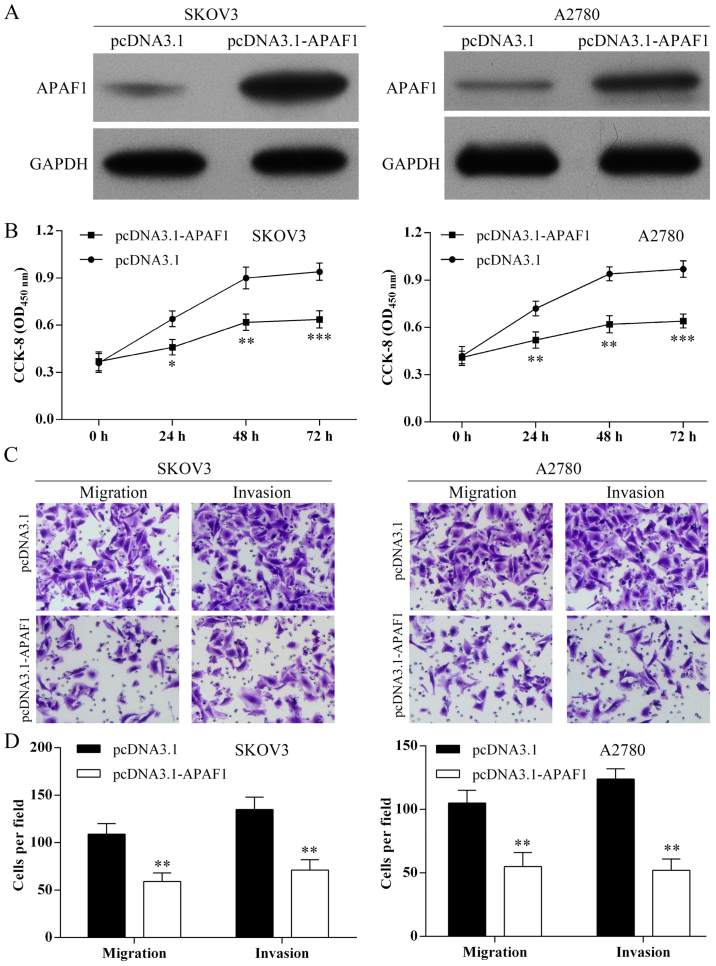
Overexpression of APAF1 and suppression of ovarian cancer cell proliferation, migration and invasion in vitro
As shown in Fig. 6A, APAF1 was overexpressed in SKOV3 and A2780 cells following pcDNA3.1-APAF1 transfection. Using the CCK-8 assay, overexpression of APAF1 inhibited ovarian cancer cell proliferation (Fig. 6B). Using Transwell assay, overexpression of APAF1 suppressed the SKOV3 and A2780 cell migration and invasion (Fig. 6C and D).
Figure 6.
Overexpression of the apoptosis protease activating factor 1 (APAF1) suppresses ovarian cancer cell proliferation, migration and invasion. (A) Western blot analysis was used to detect APAF1 protein expression in ovarian cancer cells after pcDNA301-APAF1 transfection. (B) Cell Counting kit-8 (CCK-8) assay results showed that overexpression of APAF1 inhibited ovarian cancer cell proliferation. ***P<0.001. (C) Cell migration and invasion were detected using Transwell assay (magnifications, ×40). Images of migration and invasion of each cell group are presented. (D) The average migration and invasion cell number per field among different experimental groups. Three independent experiments were performed, *P<0.05; **P<0.01.
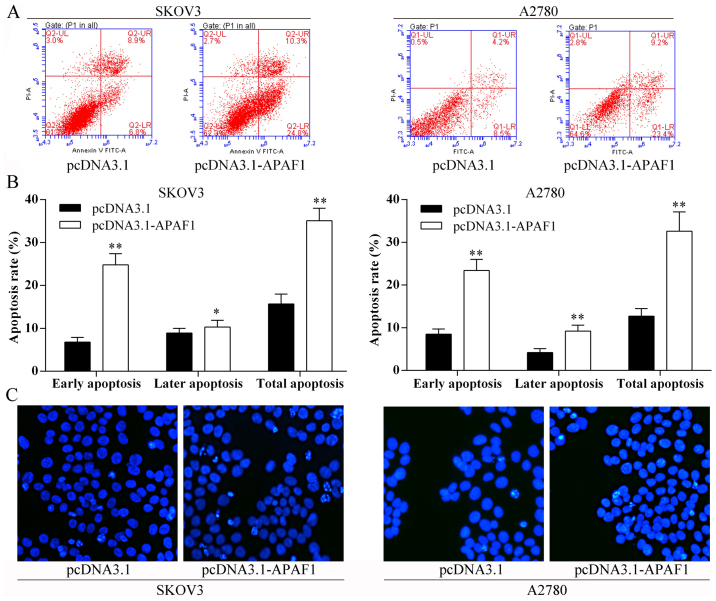
Overexpression of APAF1 and ovarian cancer cell apoptosis in vitro
The Annexin V-FITC/PI staining and Hoechst staining methods showed that the ovarian cancer cell apoptosis rates were induced following overexpression of APAF1 (Fig. 7).
Figure 7.
Overexpression of the apoptosis protease activating factor 1 (APAF1) induced ovarian cancer cell apoptosis. (A) Flow cytometric analysis of ovarian cancer cell apoptosis rate after APAF1 overexpression. LL, viable cells; LR, early apoptotic cells; UR, late apoptotic cells; UL, necrotic cells. (B) The percentage of early apoptotic cells, late apoptotic cells, and total apoptotic cells of each group. *P<0.05; **P<0.01. (C) Hoechst nuclear staining assay for SKOV3 and A2780 cells with pCDNA3.1-APAF-1 transfection cell apoptosis.
Discussion
The present study was designed to investigate the expression of microRNA-221 (miR-221) in ovarian cancer tissues and ovarian carcinoma cell lines and to undertake a preliminary study of the prognostic significance of miR-221 in patients following surgery for ovarian carcinoma. The results showed that the expression of miR-221 was increased in ovarian cancer tissues compared with matched adjacent normal tissue. Increased expression levels of miR-221 were associated with increased ovarian tumor size, increased depth of tumor invasion, a higher tumor stage, reduced patient prognosis and survival. Tumor cell transfection with the miR-221 inhibitor induced apoptosis protease activating factor 1 (APAF1) protein expression and confirmed that APAF1 is a target gene for miR-221. Following miR-221 inhibitor transfection, ovarian cancer cell proliferation, migration and invasion were inhibited in vitro and cell apoptosis was induced in vitro. The findings of the present study indicate that miR-221 may act as an oncogene by regulating tumor cell growth, invasion, migration and apoptosis in ovarian cancer, and that miR-221-APAF1 may represent a new potential diagnosis and therapeutic biomarker in ovarian cancer.
The findings of this study on ovarian cancer cell lines and tissue support the finding of other studies on the role of miR-221 in other types of human cancer (11,12). Also, this study confirmed the findings of other studies on human malignancy that APAF1 expression is associated with inhibition of tumor cell apoptosis (27,31). This study also confirmed that the expression of APAF1 is associated with adverse patient prognosis (32,33). However, this study showed, for the first time that the APAF1 gene is a direct target of miR-221 in human ovarian cancer cells and that the overexpression of APAF1 suppressed ovarian cancer cell proliferation and induced cell apoptosis in vitro.
The present study was preliminary in nature, and so further, larger studies are required to confirm and expand the findings. Although multiple tumor cell lines were studied, the behavior of immortalized tumor cells in vitro cannot replicate that of human cancer cells in vitro. For this reason, human ovarian cancer tissues were studied in parallel. However, the number of human tumor samples was small and confined to those from a single center, we suggest that, for future studies, a larger number of tumor samples should be studied and from multiple centers.
In conclusion, the APAF1 gene was confirmed as a direct target of miR-221 and overexpression of APAF1 suppressed ovarian cancer cell proliferation and induced cell apoptosis in vitro. These findings indicate that miR-221-APAF1 should be studied further as a potential new diagnostic or prognostic biomarker for ovarian cancer.
Acknowledgments
We thank Guangzhou Vipotion Biotechnology Co., Ltd. for the assistance in the vector construction of luciferase reporter assay.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Breuer EK, Murph MM. The role of proteomics in the diagnosis and treatment of women's cancers: current trends in technology and future opportunities. Int J Proteomics. 2011;2011:373584. doi: 10.1155/2011/373584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 6.Meyer SU, Thirion C, Polesskaya A, Bauersachs S, Kaiser S, Krause S, Pfaffl MW. TNF-α and IGF1 modify the microRNA signature in skeletal muscle cell differentiation. Cell Commun Signal. 2015;13:4. doi: 10.1186/s12964-015-0083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian L, Fang YX, Xue JL, Chen JZ. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS One. 2013;8:e75885. doi: 10.1371/journal.pone.0075885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hummel R, Hussey DJ, Haier J. MicroRNAs: Predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 11.Nassirpour R, Mehta PP, Baxi SM, Yin MJ. miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One. 2013;8:e62170. doi: 10.1371/journal.pone.0062170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O'Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41. doi: 10.1126/scisignal.2001538. [DOI] [PubMed] [Google Scholar]
- 13.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita R, Sato M, Kakumu T, Hase T, Yogo N, Maruyama E, Sekido Y, Kondo M, Hasegawa Y. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med. 2015;4:551–564. doi: 10.1002/cam4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berindan-Neagoe I, Monroig PC, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauser B, Zhao Y, Pang X, Ling Z, Myers E, Wang P, Califano J, Gu X. Functions of MiRNA-128 on the regulation of head and neck squamous cell carcinoma growth and apoptosis. PLoS One. 2015;10:e0116321. doi: 10.1371/journal.pone.0116321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 20.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 21.Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Hara AJ, Wang L, Dezube BJ, Harrington WJ, Jr, Damania B, Dittmer DPP. Tumor suppressor microRNAs are underrepresented in primary effusion lymphoma and Kaposi sarcoma. Blood. 2009;113:5938–5941. doi: 10.1182/blood-2008-09-179168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamoto K, Miyoshi K, Murawaki Y. miR-29b, miR-205 and miR-221 enhance chemosensitivity to gemcitabine in HuH28 human cholangiocarcinoma cells. PLoS One. 2013;8:e77623. doi: 10.1371/journal.pone.0077623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/S0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- 25.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/S0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y, Liang W, Hu X, Zhang W, Stetler RA, Vosler P, Cao G, Chen J. Neuroprotection against hypoxic-ischemic brain injury by inhibiting the apoptotic protease activating factor-1 pathway. Stroke. 2010;41:166–172. doi: 10.1161/STROKEAHA.109.561852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yong FL, Wang CW, Roslani AC, Law CW. The involvement of miR-23a/APAF1 regulation axis in colorectal cancer. Int J Mol Sci. 2014;15:11713–11729. doi: 10.3390/ijms150711713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Zio D, Bordi M, Tino E, Lanzuolo C, Ferraro E, Mora E, Ciccosanti F, Fimia GM, Orlando V, Cecconi F. The DNA repair complex Ku70/86 modulates Apaf1 expression upon DNA damage. Cell Death Differ. 2011;18:516–527. doi: 10.1038/cdd.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho CK, Bush JA, Li G. Tissue-specific regulation of Apaf-1 expression by p53. Oncol Rep. 2003;10:1139–1143. [PubMed] [Google Scholar]
- 30.Marsden VS, O'Connor L, O'Reilly LA, Silke J, Metcalf D, Ekert PG, Huang DC, Cecconi F, Kuida K, Tomaselli KJ, et al. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature. 2002;419:634–637. doi: 10.1038/nature01101. [DOI] [PubMed] [Google Scholar]
- 31.Mustika R, Budiyanto A, Nishigori C, Ichihashi M, Ueda M. Decreased expression of Apaf-1 with progression of melanoma. Pigment Cell Res. 2005;18:59–62. doi: 10.1111/j.1600-0749.2004.00205.x. [DOI] [PubMed] [Google Scholar]
- 32.Paik SS, Jang KS, Song YS, Jang SH, Min KW, Han HX, Na W, Lee KH, Choi D, Jang SJ. Reduced expression of Apaf-1 in colorectal adenocarcinoma correlates with tumor progression and aggressive phenotype. Ann Surg Oncol. 2007;14:3453–3459. doi: 10.1245/s10434-007-9541-2. [DOI] [PubMed] [Google Scholar]
- 33.Zlobec I, Minoo P, Baker K, Haegert D, Khetani K, Tornillo L, Terracciano L, Jass JR, Lugli A. Loss of APAF-1 expression is associated with tumour progression and adverse prognosis in colorectal cancer. Eur J Cancer. 2007;43:1101–1107. doi: 10.1016/j.ejca.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 34.Li NF, Broad S, Lu YJ, Yang JS, Watson R, Hagemann T, Wilbanks G, Jacobs I, Balkwill F, Dafou D, et al. Human ovarian surface epithelial cells immortalized with hTERT maintain functional pRb and p53 expression. Cell Prolif. 2007;40:780–794. doi: 10.1111/j.1365-2184.2007.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]