Abstract
The ubiquitin-conjugating enzyme 2C (UBE2C) is the key component in the ubiquitin proteasome system (UPS) by partnering with the anaphase-promoting complex (APC/C). A high UBE2C protein expression level has been reported in various types of human tumors. However, little is known about the precise mechanism by which UBE2C expression is downregulated in gastric cancer. We found in MGC-803 and SGC-7901 gastric cancer cells UBE2C-deficient G2/M phase arrest in the cell cycle and subsequently decreased gastric adenocarcinoma tumorigenesis. In the previous study, we identified Aurora-A (AURKA) as the hub gene of the gastric cancer linkage network based genome-wide association study (eGWAS). Furthermore, knockdown of UBE2C using siRNA markedly reduced the level of phosphorylation AURKA (p-AURKA) via Wnt/β-catenin and PI3K/Akt signaling pathways suppressed the occurrence and development of gastric cancer. Additionally, the expression of E-cadherin was up-regulated and N-cadherin was down-regulated in response to UBE2C knockdown and inhibits epithelial-mesenchymal transition (EMT). Collectively, our data suggest that the activity of AURKA might be regulated by UBE2C through regulating the activity of APC/C. UBE2C may be a new marker in the diagnosis of gastric cancer and may be a potential therapeutic target for the treatment of gastric adenocarcinoma.
Keywords: ubiquitin-conjugating enzyme 2C, PI3K/Akt pathway, Aurora-A, Wnt/β-catenin pathway, epithelial mesenchymal transition
Introduction
Gastric cancer is the fifth most common cancer worldwide, with an estimated 951,000 new cases and 745,000 deaths reported in 2012. Nearly 70% of the cases occurred in less developed regions, with the highest in eastern Asia especially in China (1). A survey of the incidence of malignant tumors and mortality in China (2015) showed that the incidence of gastric cancer was 67.91/million, which was the 2nd highest, just after the lung cancer incidence rate of 73.33/million. In addition, the mortality of 49.80% was also ranked second (2). Gastric cancer remains a common disease that threatens people's health and the median survival time for these patients is only 6–9 months (3). Fortunately, a large number of tumor-suppressor genes and oncogenes have been reported in recent years. However, the molecular mechanisms underlying the development of gastric carcinomas remain poorly understood (4).
Ubiquitination is an important cellular mechanism for the targeted degradation of proteins, and is instrumental for a variety of cellular processes including, but not limited to, cell cycle progression, antigen presentation, transcription, and programmed cell death (5). In eukaryotes, particularly, the ubiquitin proteasome system (UPS) precisely regulates the cell cycle at key checkpoints by targeting cell cycle regulators for proteasome-mediated degradation (6). This regulation requires ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligases (E3) to work in concert to facilitate the ubiquitination of target proteins (7,8).
E2s are classified into four classes and ubiquitin-conjugating enzyme 2C (UBE2C) (also known as UbcH10) is a class III E2 enzyme. The full length UBE2C protein contains 179 amino acids and the first 28 residues comprise an N-terminal extension with various motifs which may contribute to target protein recognition and conjugation (9). UBE2C is located on chromosome 20q13 and has a molecular weight of 19.65 kDa. Interestingly, UBE2C is nearly undetectable in normal tissues, at both the RNA or protein levels. In contrast, increasing evidence indicates that UBE2C protein is overexpressed in many human tumor types, including brain tumors (10), malignant breast carcinomas (11), lung cancer (12,13), anaplastic thyroid carcinomas (14), esophageal adenocarcinomas (15), hepatocellular carcinomas (16) and colorectal cancer (17–19), suggesting that UBE2C may play an important role in tumorigenesis and progression. UBE2C overexpression causes the centrosome duplication instability, and variety of proteins can cause this phenomenon, including the anaphase-promoting complex (APC/C) substrates cyclin B, AURKA and PLK1 (20,21). However, its role in gastric cancer carcinogenesis remains poorly defined.
Compared to conventional genome-wide association studies GWAS revealed associations of single-nucleotide polymorphisms (SNPs), gene expression genome-wide association studies (eGWASs) aimed to discover more common expression and similar to the SNP cell analysis (22–24). In the previous study, we analyzed the 184 differentially expressed genes in gastric cancer from the eGWAS and revealed AURKA is at the core of the gastric cancer linkage network (25). AURKA is a serine/threonine kinase that localizes to spindle poles, and ensures the correct assembly of cellular components during mitosis in normal cells (26). In cancer cells, AURKA overexpression results in the activation of several oncogenic pathways, including those of PI3K/Akt, β-catenin, and NF-κB (27,28). Importantly, we further found that AURKA expression is closely related to UBE2C expression.
In our current report, we demonstrate that gastric cancer malignancies are associated with high levels of UBE2C expression. The knockdown of UBE2C decreases the levels of p-AURKA and inhibits the development of gastric adenocarcinoma by Wnt/β-catenin and PI3K/Akt signaling pathways. AURKA is the center of the network.
Materials and methods
eGWAS analysis
To analyze the molecular signature of gastric cancer, we collected 13 independent datasets from the NCBI Gene Expression Omnibus using the public database eGWAS. The most differentially expressed genes between tumors and non-tumors from each databset were screened, P-values were calculated from the number of positive/negative experiments for each gene, and Fisher's exact test was used to determine significance. The Bonferroni threshold (P=1.0×10−5) indicated that there were 184 gastric cancer susceptibility genes. These 184 genes were analyzed for Gene Ontology (GO) using DAVID online tools, as well as pathway analysis by Medusa. Additionally, we identified the top differentially expressed genes and performed a KEGG pathway analysis using GenMAPP v2.1, calculating the enrichment P-value. Finally, we applied 13 differences of highest expression genes by integrating the experiments, database and literature.
Cell culture
Human gastric cancer cell lines MGC-803 and SGC-7901 were purchased from China Academia Sinica Cell Repository (Shanghai, China). Cells were cultured and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) under standard conditions. Cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Antibodies and other reagents
UBE2C, AURKA, β-catenin, AKT1, p-AKT1, GSK-3β, p-GSK-3β, Slug, Snail and Twist antibodies were purchased from Abcam (Cambridge, UK). E-cadherin, N-cadherin and p-AURKA antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). The GAPDH antibody was purchased from Zhongshan Golden Bridge Biotechnology (Beijing, China). Western blot analysis reagents were purchased from Sigma, PVDF membranes were purchased from Millipore Corp. (Bedford, MA, USA) and RIPA lysis buffer was purchased from Beijing Taike Biotechnology. Lentiviruses containing an UBE2C and AURKA inhibitor sequences (Lenti-si-UBE2C and Lenti-si-AURKA) or negative control (Lenti-NC) were obtained from Shanghai GeneChem Co., Ltd. (Shanghai, China).
Transient transfection
The UBE2C and AURKA silencer siRNAs were synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China). The human gastric cancer cells were seeded and grown in 6-well plates overnight and transfected with negative control siRNA, siRNA against UBE2C, or siRNA against AURKA using X-tremeGENE siRNA transfection reagent (Roche) according to the manufacturer's protocol. The primers for detecting UBE2C forward, 5′-GGATTTCTGCCTTCCCTGAA-3′ and reverse, 5′-GATAGCAGGGCGTGAGGAAC-3′; the primers for detecting AURKA forward, 5′-AAAGAGCAAGCAGCCCCTGC-3′ and reverse, 5′-GAATTCAACCCGTGATATTCTT-3′; and the primers for negative control forward, 5′-CCGGGAAACTGTGGCGTGATGG-3′ and reverse, 5′-AGGTGGAGGAGTGGGTGTCGCTGTT-3′. Briefly, 3 μl of siRNA was added to 50 μl DMEM serum-free medium. In parallel, 3 μl of the transfection reagents was added to 50 μl serum-free DMEM and mixed. Finally, after mixing and incubating the siRNA and X-tremeGENE siRNA transfection reagents separately for 5 min, the reagents were combined, mixed, and placed at room temperature (RT) for 20 min to form the siRNA/transfection complexes. Subsequent experiments were then carried out 48 h after transfection.
Western blot analysis
Protein extracts from siRNA-transfected MGC-803 and SGC-7901 cells were collected, and after samples were adjusted to equal protein concentration and volume, they were subjected to SDS-PAGE on 10% SDS-acrylamide gels. Separated proteins were transferred to PVDF membranes (Millipore Corp.) and then blocked. Antibodies for UBE2C, AURKA, p-AURKA, AKT1, p-AKT1, GSK-3β, p-GSK-3β, and β-catenin were used for western blot analysis, and were incubated at a 1:1,000 dilution, followed by incubation with their respective secondary antibodies (1:2,000 dilution).
Quantitative real-time PCR
Total RNA was extracted from cultured cells using the TRIzol reagent method. Equal amounts (1 μg) of RNA were converted into cDNA using oligo (dT) primer using a PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu, Japan). Real-time PCR analysis involved 2 ng cDNA with 10 μl SYBR Green PCR Master Mix (Applied Biosystems) and 2 μM primers in the ABI StepOne Real-Time PCR System (Applied Biosystems). The PCR primer sequences included those of UBE2C, AURKA, Twist and E-cadherin. The PCR conditions were as follows: 30 cycles of 94°C for 30 sec, 56°C for 30 sec, 72°C for 90 sec, and a final extension at 72°C for 5 min.
Colony formation and cell proliferation assay
Tumor cells transfected with UBE2C siRNAs were cultured at 2,000 cells/well in 6-well plates. The cells were allowed to grow for 14 days, and the medium was changed every 3 days. Upon termination of the experiment, colonies were fixed with paraformaldehyde (4% w/v), stained with crystal violet (0.5% w/v) and counted. For cell proliferation, cells were assayed using the Cell Counting kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's protocol. To determine the effect of UBE2C siRNA on cell proliferation, the tumor cells transfected with UBE2C siRNAs were seeded into each well of a 96-well plate at a density of 1×103 cells in 100 μl of culture medium containing 10% FBS and were grown overnight. All experiments were performed in triplicate. The medium was then replaced with 100 μl fresh medium with 10% CCK-8 reagent, and the cells were incubated for 3.5 h at 37°C. At 0, 24, 48, 72 and 96 h, the absorbance was measured at 450 nm using an EL×800 micro-plate reader (BioTek, USA).
Matrigel invasion and Transwell cell migration assay
The cell invasion capability was assessed using a Matrigel invasion chamber (Becton-Dickenson, Bedford, USA), incubated with 60 μl of 1 mg/ml Matrigel in a 24-well plate (Corning Costar, Cambridge, MA, USA) at 37°C for 1 h to allow gel polymerization. In the cell migration assay, the top chamber was without matrigel. Transfected cells were trypsinized and washed with the appropriate serum-free medium; then, 200 μl of the cell suspension containing 5×104 cells was added to top chamber, followed by the addition of 500 μl of the appropriate medium containing 10% FBS to the well underneath the insert. The cells were incubated at 37°C for 48 h and gently rinsed with phosphate buffered saline (PBS) before, fixing with 4% paraformaldehyde at RT for 15 min. Cells were stained with 0.25% crystal violet for 5 min, rinsed again, and then allowed to dry. The inserts were then viewed under the microscope at ×200 magnification (Olympus Corporation Tokyo, Japan) and the number of cells/field in three random fields were counted.
Wound healing assay
Gastric cancer cells were cultured in 6-well plates overnight. The cell monolayers were scratched in a line across the well using a 200-μm standard pipette tip. The 'wound' was then washed twice with serum-free media to remove cell debris, and the plated cells were incubated in RPMI-1640 medium supplemented with 2.5% FBS (Hyclone). The cell-free wound area was photographed at the indicated times with use of a digital camera connected to an inverted microscope (Nikon TE200; Nikon Corp., Tokyo, Japan). The resulting images were analyzed by the use of Image J software. Wound healing was calculated as the proportion of remaining cell-free area compared with the initial wound area.
Flow cytometric analysis of the cell cycle and apoptosis
The MGC-803 and SGC-7901 cells were transfected with UBE2C siRNA for 48 h, harvested and then fixed with 70% ethanol at -20°C. The fixed cells were then incubated with 50 mg/ml propidium iodide (PI; Sigma, St. Louis, MO, USA), and 1 mg/ml RNAse A for 30 min at RT. The DNA content was then analyzed using a FACSCalibur system (Becton Dickinson). The distribution of cells in each cell cycle phase was determined using ModFit software (Becton Dickinson). For apoptosis analysis, the cells were transfected with si-UBE2C for 48 h and harvested, the phosphatidylserine exposure level was determined by staining the cells with a human Annexin V-FITC kit (MBL, Nagoya, Japan), according to the manufacturer's instructions. At least 1×106 stained cells were analysed by the FACS Calibur for each determination.
Immunofluorescence staining
Twenty-four hours after transfection, cells were plated on glass cover slips and 48 h post-transfection the cover slips were washed extensively in PBS and fixed with 4% paraformaldehyde in PBS. After additional washing, the cells were permeabilized with 1% Triton X-100 in PBS for 5 min. The cover slips were then washed and blocked with 1% BSA for 30 min. Cells were incubated with AURKA primary antibodies overnight at 4°C. The samples were then washed and incubated with species specific secondary rhoda-mine-labeled antibodies (TRITC) in PBS (1:100 dilution) for 2 h. Nuclei were stained with DAPI at RT for 10 min and cover slips were mounted with antifade solution prior to imaging on a confocal microscope (Olympus FV1000S).
Subcutaneous tumor model
Four-week-old immune-deficient nude mice (BALB/c-nu) were purchased from the animal center of the Cancer Institute and Hospital of Chinese Academy of Medical Sciences, bred at the facility of laboratory animals, Tianjin Institute of Biological Engineering. The mice were injected subcutaneously with MGC-803 cells which were stably expressed Lenti-si-UBE2C or Lenti-NC. The cells were suspended with the concentration of 2×106/ml. Mice were monitored daily and all the mice formed tumor subcutaneously. The weight of the mice was measured every 3 days, and the tumor weight was surveyed at the endpoint of the study.
Immunohistochemistry
Immunostaining was performed on paraffin sections of Lenti-si-AURKA tumor specimens and Lenti-NC specimens using the avidin-biotin complex (ABC)-peroxidase method. The sections were incubated with primary antibodies against UBE2C overnight at 4°C. They were then treated with biotinylated secondary antibody (1:100) for 1 hour at RT, followed by incubation with ABC-peroxidase for another 1 hour. Protein expression was detected by coloration with diaminobenzidine (DAB) buffer. After washing with Tris buffer, the sections were incubated with 3,3′-diaminobenzidine (DAB, 30 mg dissolved in 100 ml PBS) for 5 min, rinsed in water and counterstained with hematoxylin.
Statistical analysis
The significance of Kaplan-Meier statistics was tested using the log-rank test. Multivariate analyses were performed using the multivariate Cox regression model. SPSS 16.0 (SPSS, Chicago, IL, USA) was used for all of the calculations. All of the data are represented by the mean ± SD. The statistical significance was set at P<0.05.
Results
eGWAS identifies UBE2C and AURKA as functional genes for gastric cancer
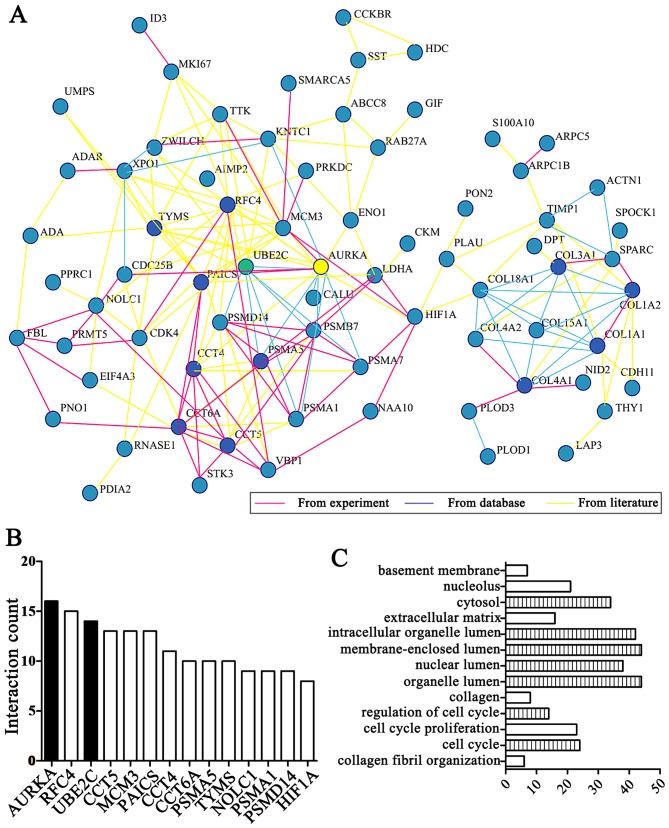
After analyzing 13 independent microarray experiments from the eGWAS in gastric cancer and 679 samples from public repositories, we took 184 genes which repeated differential expression shared the highest ranking. Among them, by integrating the experiments, database and literature, we focused on 13 genes ranking behind AURKA (Fig. 1A and B) and the results demonstrated that AURKA is a hub gene based on linkage analysis. In addition, we found 'cell cycle' functions were the most implicated of the UBE2C and AURKA genes in enrichment of GO terms by DAVID online tool analysis (Fig. 1C).
Figure 1.
UBE2C and AURKA genes were analyzed by eGWAS. (A) AURKA (yellow) is the hub gene by integrating the experiments, database and literature, and UBE2C (green) shows strong linkage. (B) AURKA, RFC4 and UBE2C were markedly differentially expressed genes from network analyzed. (C) GO analysis demonstrates that the differentially expressed genes are significantly enriched in the cell cycle and collagen and UBE2C is significantly increased during the cell cycle and in the organelle lumen (the Bonferroni threshold, P<1.0×10−5). UBE2C, ubiquitin-conjugating enzyme 2C; Aurora-A (AURKA); eGWAS, genome-wide association study; GO, Gene Ontology.
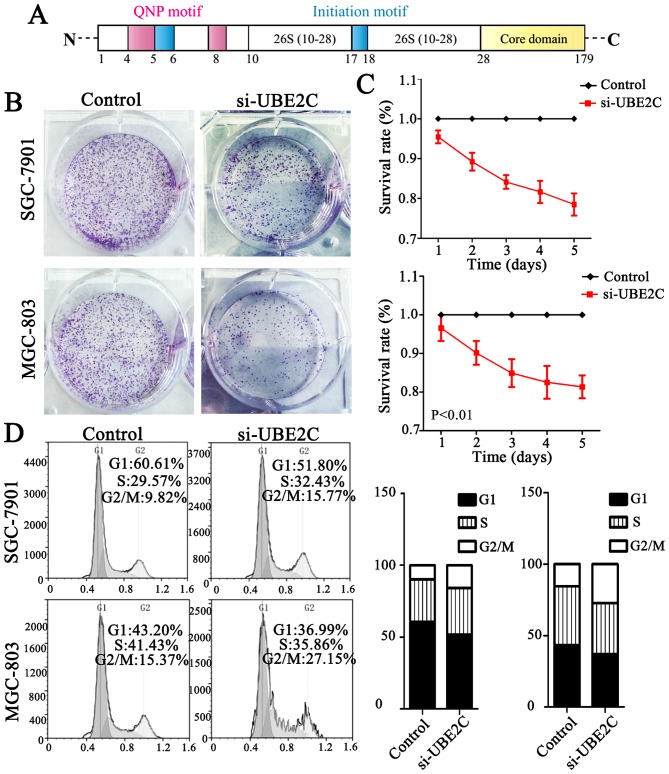
Importantly, UBE2C was markedly differentially expressed second to AURKA in gastric cancer compared with normal gastric mucosa. UBE2C is located on chromosome 20q13 and belongs to the E2 gene family and the first 28 residues comprise an N-terminal extension with various motifs, and the remaining residues form the core domain (Fig. 2A). Taken together UBE2C is frequently overexpressed in gastric cancer and is associated with a worse overall survival.
Figure 2.
UBE2C knockdown inhibits tumor formation and changes cell cycle distribution in MGC-803 and SGC-7901 cells. (A) The structure and motifs of UBE2C. Pink: QNP motif; red: initiation motif. (B) Colony formation assay for assessing the cell proliferation of si-UBE2C-treated cells. (C) Growth curves of MGC-803 and SGC-7901 cells before and after UBE2C gene knockdown. Data are expressed as the mean ± SD of at least three independent experiments, when compared to the control cell group (P<0.01). (D) Cell cycle distribution as a percentage of the total population. Data were obtained by flow cytometry and presented as the mean (n=3). Black bars represent the percentages of the fraction in G1, white bars represent the percentage of the fraction in G2/M, and the gray bars represent the percentage of fraction in S phase. UBE2C, ubiquitin-conjugating enzyme 2C.
Knockdown of UBE2C causes G2/M phase arrest in the cell cycle and suppresses tumor formation
UBE2C is highly expressed in gastric cancer, but whether the downregulation of UBE2C represses tumorigenesis is unkown. Therefore, we knocked down UBE2C using siRNA in MGC803 and SGC7901 cells, and examined the effects on colony formation, cell proliferation and cell cycle distribution. As is shown in Fig. 2B, MGC803 and SGC7901 cells transfected with si-UBE2C exhibited a significant reduction in colony formation after 2 weeks (40.00%), compared with the control group (56.33%). Furthermore, the CCK-8 analysis showed that the survival rate of cells with si-UBE2C treatment was inhibited by 30% (Fig. 2C). In addition, the percentage of SGC7901 and MGC803 cells in the G2/M phase was 9.82 and 15.37% for the control cells, whereas the cells treated with si-UBE2C was significantly increased to 15.77 and 27.15% respectively (Fig. 2D). The extent of siRNA-mediated cell death was analysed in these cell types using an Annexin V assay. The results (Fig. 3) revealed that 22.4 and 38.0% of the UBE2C siRNA-treated SGC7901 and MGC803 cells were Annexin V positive, whereas only 11.1 and 18.9% of the cells treated with control siRNAs showed this positivity at 48 h post-transfection. These results suggested that the suppression of UBE2C expression inhibited the proliferation and induced apoptosis of gastric adenocarcinoma cells in vitro.
Figure 3.
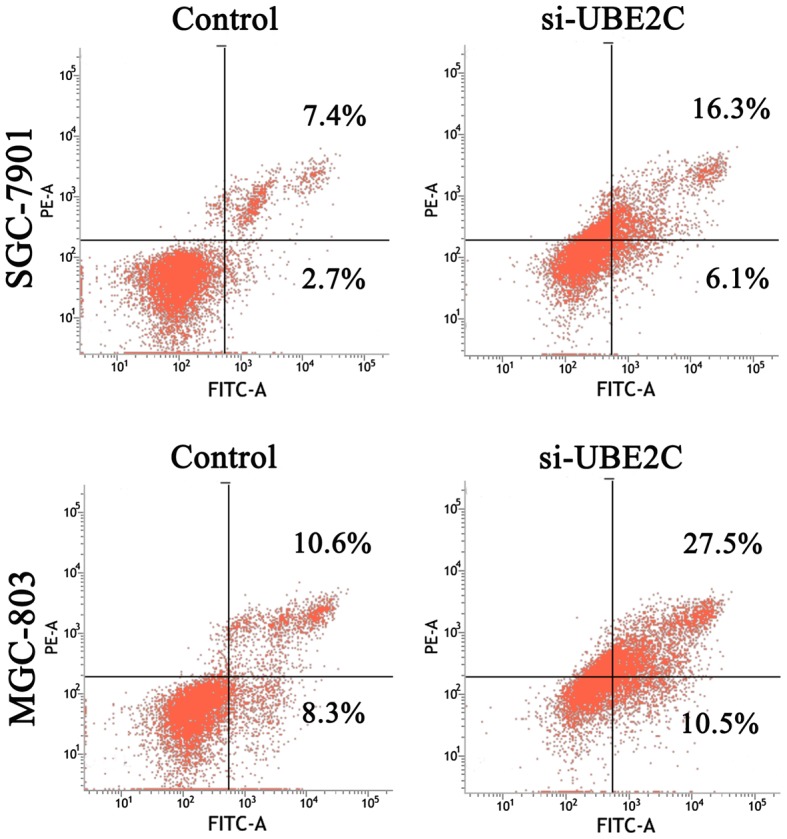
Flow cytometric analysis of the cell apoptosis in MGC-803 and SGC-7901 gastric cancer cells. Compared with the NC-treated control cells, the si-UBE2C-treated cells induced apoptosis of gastric adenocarcinoma cells in vitro. UBE2C, ubiquitin-conjugating enzyme 2C.
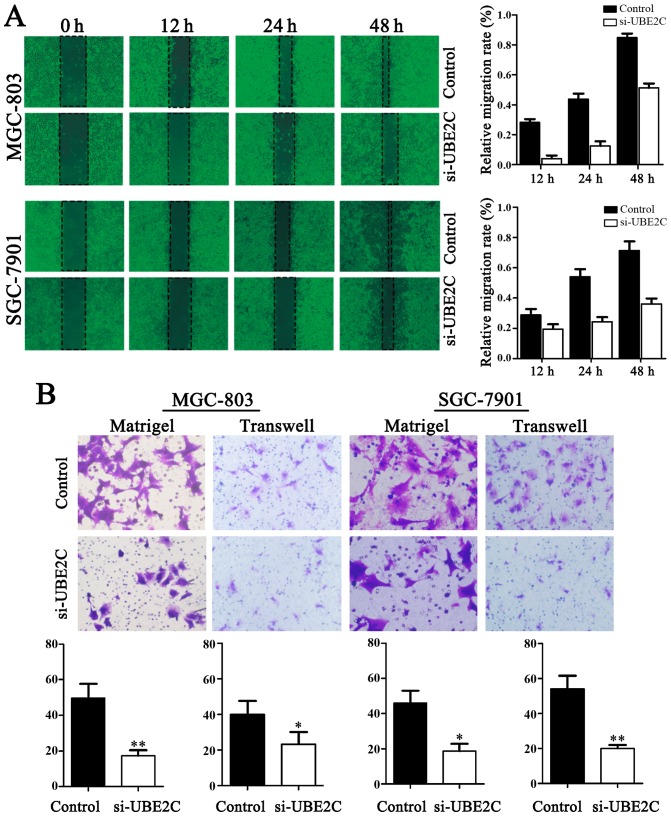
Downregulation of UBE2C inhibits the migration and invasion of gastric adenocarcinoma cells
The migration of cells to the wound area was analyzed at 12, 24 and 48 h after injury, and the results revealed that the knockdown of UBE2C led to a marked inhibition of wound healing, compared with the negative control. Specifically, the relative migration rates of si-UBE2C in MGC803 and SGC7901 cells were 19.60, 32.53 and 37.25%, as well as 2.16, 10.87 and 52.17% after 12, 24 and 48 h, respectively. Conversely, in the negative control the migration rates were 28.57, 53.06 and 71.43%, as well as 27.08, 43.75 and 85.42%, respectively. As shown in Fig. 4, the number of cells invading through the matrigel in MGC803 si-UBE2C-group was significantly decreased (20.7±4.1), compared to control (52.1±2.2). The invasion activity was inhibited in si-UBE2C group (18.7±1.2), compared to the control (49.3±4.1). Collectively, these data indicate that UBE2C play a role in gastric adenocarcinoma cell migration and invasion in vitro.
Figure 4.
Knockdown of UBE2C inhibits the migration and invasion of gastric MGC-803 and SGC-7901 cells. (A) Classic wound healing assay for assessing the migration of si-UBE2C-treated cells. (B) Transwell assays for assessing the invasion of the si-UBE2C-treated cells (P<0.05). UBE2C, ubiquitin-conjugating enzyme 2C.
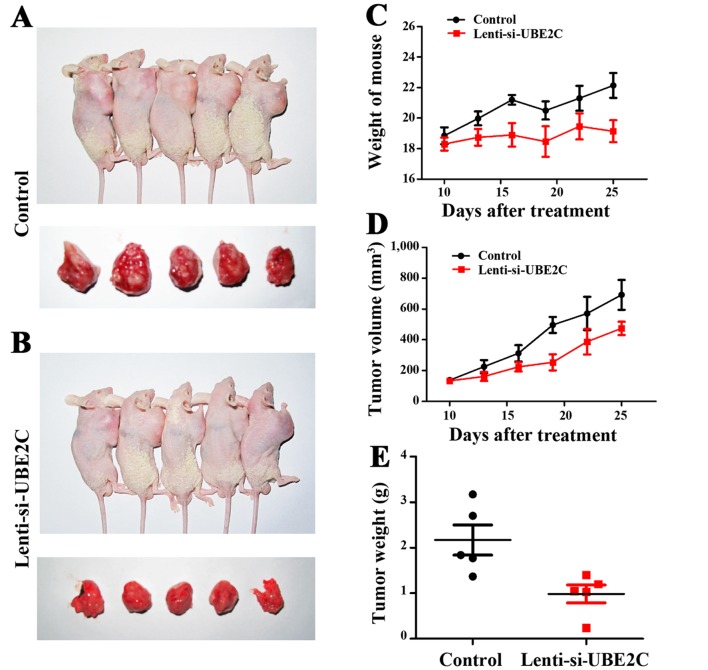
Inhibition of UBE2C suppresses gastric cancer growth in xenograft models
UBE2C suppresses gastric cancer cell migration and invasion in vitro. To verify the role of UBE2C in vivo, the subcutaneous gastric carcinoma model using MGC-803 cells as described previously were established (Fig. 5A and B). The mouse and tumor weights were measured, and the tumor volume was measured using calipers to measure the tumor length and width (Fig. 5C and D). Compared with the Lenti-NC-treated MGC-803 cells, the Lenti-si-UBE2C-treated tumor was suppressed significantly (Fig. 5E). The results confirmed that UBE2C play a role in gastric cancer growth in vivo.
Figure 5.
Treatment with Lenti-si-UBE2C suppresses gastric cancer growth in vivo. (A and B) Mouse and tumor samples from the MGC-803/Control and MGC-803/Lenti-si-AURKA groups. (C–E) The mouse body weights, tumor growth and weight were evaluated using the in vivo proliferation assay, repectively. UBE2C, ubiquitin-conjugating enzyme 2C; AURKA, Aurora-A.
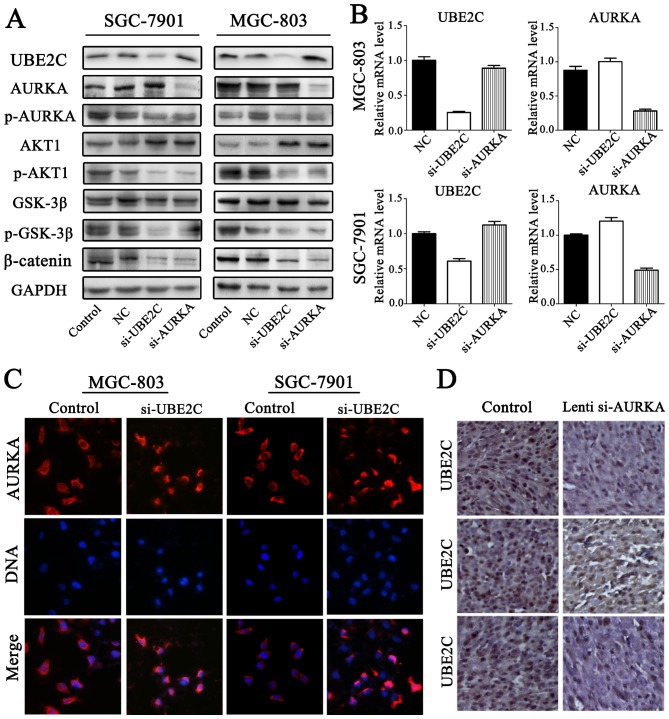
UBE2C deficiency inhibits p-AURKA expression in gastric adenocarcinoma
To accurately determine AURKA and UBE2C expression in gastric adenocarcinoma, the expression levels of p-AURKA were determined through western blot analysis (Fig. 6A and B). The results confirmed that p-AURKA expression was significantly decreased in si-UBE2C treatment group. Furthermore, the levels of AURKA were increased through qt-PCR and western blot analysis. Immunofluorescent microscope analysis suggested that si-UBE2C treatment in MGC803 and SGC7901 cell lines resulted in the increased expression of AURKA in the cytoplasm (Fig. 6C). This was determined by the observation of a significantly greater fluorescent signal intensity in the cytoplasm compared with the control groups, suggesting that the expression of AURKA was negatively correlated with UBE2C. However, immunostaining analysis revealed the UBE2C and AURKA protein expression in gastric cancer, and the expression levels of UBE2C were slightly less in Lenti-si-AURKA treatment compared with control groups (Fig. 6D). The results confirmed that si-UBE2C inhibits the expression of p-AURKA in contrast to inducing AURKA inactivation.
Figure 6.
UBE2C deficiency inhibits the expression of p-AURKA. (A) Protein levels of AURKA and p-AURKA were evaluated by western blot analysis in the si-UBE2C-treated gastric cell lines. (B) mRNA levels of AURKA expression in the indicated cell lines evaluated by quantitative PCR. (C) A large number of AURKA and UBE2C cells were observed in the cytoplasm; the level of AURKA was increased in si-UBE2C-treated cells. (D) Conversely, UBE2C expression was decreased slightly in Lenti-si-AURKA tumors in nude mice. UBE2C, ubiquitin-conjugating enzyme 2C; p-AURKA, phosphorylation AURKA; AURKA, Aurora-A.
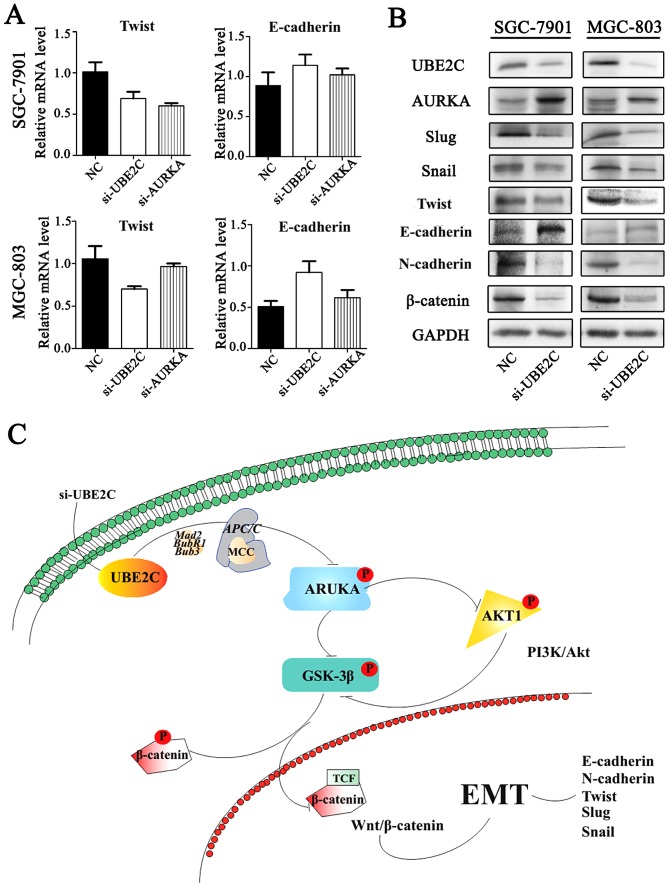
Inhibition of UBE2C represses epithelial-mesenchymal tran- sition (EMT) through Wnt/β-catenin and PI3K/Akt signaling pathways
Our previous studies have shown that AURKA induces EMT through the Wnt/β-catenin and PI3K/Akt signaling pathways. AURKA activity was inhibited by si-UBE2C, it was hypothesized that the loss of UBE2C might also inhibit EMT. To examine this hypothesis, the expression of E-cadherin, N-cadherin and their interrelated genes were analyzed following si-UBE2C treatment. The qt-PCR analysis revealed that the knockdown of UBE2C in MGC803 and SGC7901 cells resulted in increased expression of the epithelial marker E-cadherin and decreased expression of the mesenchymal marker Twist (Fig. 7A). Consistent with these results, western blot analysis results demonstrated a lower protein expression of UBE2C, Twist, Snail, Slug and N-cadherin, whereas E-cadherin was increased (Fig. 7B); these results were consistent with the PCR results.
Figure 7.
UBE2C induces EMT via Wnt/β-catenin and PI3K/Akt signaling pathways. (A) qt-PCR assay for assessing the levels of Twist and E-cadherin in gastric cells after si-UBE2C-treatment. (B) Western blot analysis assay for the expression of EMT-related factors in si-UBE2C-treated cells. (C) si-UBE2C suppresses the Wnt/β-catenin and PI3K/Akt signaling pathways through inhibition of p-AURKA. In particular, si-UBE2C inhibits the phosphorylation of AKT1 and GSK-3β as well as the EMT process. UBE2C, ubiquitin-conjugating enzyme 2C; EMT, epithelial-mesenchymal transition; p-AURKA, phosphorylation AURKA.
Discussion
In our gastric cancer eGWAS, we analyzed 184 genes with differential expression sharing the highest ranking in gastric cancer from independent datasets. The results indicated that AURKA was a hub gene based on linkage analysis (25). Considering previous studies, we hypothesized that there may exist some genes that are co-activated with AURKA in gastric cancer. We used the DAVID online tools to analyze the ontology of the 13 genes and identified UBE2C as being significantly involved in similar processes with AURKA and it is overexpressed in several primary tumors.
Much effort has been spent on the study of the overexpression of UBE2C biological mechanisms in gastric cancer (29,30). This E2 protein catalyzes the ubiquitination and degradation of cyclins A and B, and acts together with the E3 ligase of the APC/C to participate in the regulation of the spindle assembly checkpoint (31). Furthermore, several previous studies have demonstrated that UBE2C can release Mad2, BubR1, Bub3 and other mitotic complexes (MCC) to control the activity of APC/C to induce tumorigenesis (7,32,33). However, little is known about the precise mechanism by which UBE2C expression is downregulated. The first 28 residues of UBE2C comprise an N-terminal extension with various motifs, and the remaining residues form the core domain. The core domain may promote cancer cell proliferation and may play an oncogenic role in gastric cancer (9). The knockdown of UBE2C expression by siRNA resulted in a significant loss of cell proliferation and viability in gastric adenocarcinoma cells. In the current study, we showed that knockdown of UBE2C could reduce gastric cancer tumori-genesis.
In our present analyses, we also investigated the possible link between UBE2C and AURKA in the proliferation of gastric cancer cells. UBE2C may insulate CDC20 from MCC by a mechanism not involving ubiquitin and inhibit the activity of APC/C, thereby effecting the levels of p-AURKA (7,32). The results from our studies have demonstrated that the knockdown of UBE2C decreases the level of p-GSK-3β, p-AKT1, β-catenin and p-AURKA. In addition, knockdown of AURKA by Lenti-siRNA led to a partial reduction in UBE2C expression. Moreover, we demonstrated that AURKA and UBE2C regulate the Wnt/β-catenin and PI3K/Akt signaling pathways in gastric adenocarcinoma. Collectively, UBE2C may play a role in regulating AURKA activity.
Furthermore, we discovered that UBE2C interacts with AURKA, which prevents EMT. Therefore, upon downregulation of UBE2C expression with siRNA, there may be multiple mechanisms that control Wnt/β-catenin and PI3K/Akt signaling pathways, which led to inhibition of EMT (34,35). The suppression of p-AKT1 prevented the inhibitory phosphorylation of p-GSK3β (36), thereby maintaining the degradation of β-catenin. In addition, the downregulation of UBE2C was able to suppress the expression of N-cadherin (37) and increase the expression of E-cadherin (38). Similarly, the expression of Slug (39), Twist and Snail (40) were also decreased, which are also crucial genes in the development of the embryo and stimulate the process of EMT.
In summary, our data suggest that UBE2C may be a promising target for gastric cancer. Although no specific UBE2C inhibitors are currently available for clinical use, proteasome inhibitors form a novel class of chemotherapeutic agents that lead to cell cycle arrest and cell death. Tumor cells are more susceptible to proteasome inhibition due to their rapid division and disordered regulatory pathways. The mechanism of UBE2C in gastric cancer needs further study, and the commissural inhibition of UBE2C and AURKA may be a potential therapy for the treatment of gastric adenocarci-noma. We hope this study can help other researchers to further understand the role of UBE2C.
Acknowledgments
This research was supported by funds from the National High Technology Research and Development Program 863 (2014AA021102 and 2016YFC0902502) and China National Natural Scientific Fund (81372703 and 81172356). We would like to thank the members of the Laboratory of Neuro-Oncology, Tianjin Neurological Institute for their technical assistance.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamura G. Genetic and epigenetic alterations of tumor suppressor and tumor-related genes in gastric cancer. Histol Histopathol. 2002;17:323–329. doi: 10.14670/HH-17.323. [DOI] [PubMed] [Google Scholar]
- 5.Zhong JL, Huang CZ. Ubiquitin proteasome system research in gastrointestinal cancer. World J Gastrointest Oncol. 2016;8:198–206. doi: 10.4251/wjgo.v8.i2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao Z, Zhang H, Cowell J. Ubiquitin-conjugating enzyme UBE2C: Molecular biology, role in tumorigenesis, and potential as a biomarker. Tumour Biol. 2012;33:723–730. doi: 10.1007/s13277-011-0291-1. [DOI] [PubMed] [Google Scholar]
- 7.Xie C, Powell C, Yao M, Wu J, Dong Q. Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int J Biochem Cell Biol. 2014;47:113–117. doi: 10.1016/j.biocel.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Wagner KW, Sapinoso LM, El-Rifai W, Frierson HF, Butz N, Mestan J, Hofmann F, Deveraux QL, Hampton GM. Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene. 2004;23:6621–6629. doi: 10.1038/sj.onc.1207861. [DOI] [PubMed] [Google Scholar]
- 9.Wing SS, Jain P. Molecular cloning, expression and characterization of a ubiquitin conjugation enzyme (E217kB) highly expressed in rat testis. Biochem J. 1995;305:125–132. doi: 10.1042/bj3050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L, Huang CG, Lu YC, Luo C, Hu GH, Liu HM, Chen JX, Han HX. Expression of ubiquitin-conjugating enzyme E2C/UbcH10 in astrocytic tumors. Brain Res. 2008;1201:161–166. doi: 10.1016/j.brainres.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Chou CP, Huang NC, Jhuang SJ, Pan HB, Peng NJ, Cheng JT, Chen CF, Chen JJ, Chang TH. Ubiquitin-conjugating enzyme UBE2C is highly expressed in breast microcalcification lesions. PLoS One. 2014;9:e93934. doi: 10.1371/journal.pone.0093934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrotta I, Bruno L, Maltese L, Russo E, Donato A, Donato G. Immunohistochemical analysis of the ubiquitin-conjugating enzyme UbcH10 in lung cancer: A useful tool for diagnosis and therapy. J Histochem Cytochem. 2012;60:359–365. doi: 10.1369/0022155412439717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallante P, Malapelle U, Berlingieri MT, Bellevicine C, Sepe R, Federico A, Rocco D, Galgani M, Chiariotti L, Sanchez-Cespedes M, et al. UbcH10 overexpression in human lung carcinomas and its correlation with EGFR and p53 mutational status. Eur J Cancer. 2013;49:1117–1126. doi: 10.1016/j.ejca.2012.09.033. [DOI] [PubMed] [Google Scholar]
- 14.Pallante P, Berlingieri MT, Troncone G, Kruhoffer M, Orntoft TF, Viglietto G, Caleo A, Migliaccio I, Decaussin-Petrucci M, Santoro M, et al. UbcH10 overexpression may represent a marker of anaplastic thyroid carcinomas. Br J Cancer. 2005;93:464–471. doi: 10.1038/sj.bjc.6602721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin J, Raoof DA, Wang Z, Lin MY, Thomas DG, Greenson JK, Giordano TJ, Orringer MB, Chang AC, Beer DG, et al. Expression and effect of inhibition of the ubiquitin-conjugating enzyme E2C on esophageal adenocarcinoma. Neoplasia. 2006;8:1062–1071. doi: 10.1593/neo.05832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ieta K, Ojima E, Tanaka F, Nakamura Y, Haraguchi N, Mimori K, Inoue H, Kuwano H, Mori M. Identification of overexpressed genes in hepatocellular carcinoma, with special reference to ubiquitin-conjugating enzyme E2C gene expression. Int J Cancer. 2007;121:33–38. doi: 10.1002/ijc.22605. [DOI] [PubMed] [Google Scholar]
- 17.Bavi P, Uddin S, Ahmed M, Jehan Z, Bu R, Abubaker J, Sultana M, Al-Sanea N, Abduljabbar A, Ashari LH, et al. Bortezomib stabilizes mitotic cyclins and prevents cell cycle progression via inhibition of UBE2C in colorectal carcinoma. Am J Pathol. 2011;178:2109–2120. doi: 10.1016/j.ajpath.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li SZ, Song Y, Zhang HH, Jin BX, Liu Y, Liu WB, Zhang XD, Du RL. UbcH10 overexpression increases carcinogenesis and blocks ALLN susceptibility in colorectal cancer. Sci Rep. 2014;4:6910. doi: 10.1038/srep06910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Chen Y, Hu C, Jing H, Cao Y, Liu X. Association of clinicopathological features with UbcH10 expression in colorectal cancer. J Cancer Res Clin Oncol. 2010;136:419–426. doi: 10.1007/s00432-009-0672-7. [DOI] [PubMed] [Google Scholar]
- 20.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 22.Hu N, Wang Z, Song X, Wei L, Kim BS, Freedman ND, Baek J, Burdette L, Chang J, Chung C, et al. Genome-wide association study of gastric adenocarcinoma in Asia: A comparison of associations between cardia and non-cardia tumours. Gut. 2016;65:1611–1618. doi: 10.1136/gutjnl-2015-309340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh S, Oh S. Epidemiological and genome-wide association study of gastritis or gastric ulcer in korean populations. Genomics Inform. 2014;12:127–133. doi: 10.5808/GI.2014.12.3.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saeki N, Ono H, Sakamoto H, Yoshida T. Genetic factors related to gastric cancer susceptibility identified using a genome-wide association study. Cancer Sci. 2013;104:1–8. doi: 10.1111/cas.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Li Z, Song Y, Wang R, Han L, Wang Q, Jiang K, Kang C, Zhang Q. AURKA induces EMT by regulating histone modification through Wnt/β-catenin and PI3K/Akt signaling pathway in gastric cancer. Oncotarget. 2016;7:33152–33164. doi: 10.18632/oncotarget.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahankali M, Henkels KM, Gomez-Cambronero J, Speranza F. A non-mitotic role for Aurora kinase A as a direct activator of cell migration upon interaction with PLD, FAK and Src. J Cell Sci. 2015;128:516–526. doi: 10.1242/jcs.157339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Do TV, Xiao F, Bickel LE, Klein-Szanto AJ, Pathak HB, Hua X, Howe C, O'Brien SW, Maglaty M, Ecsedy JA, et al. Aurora kinase A mediates epithelial ovarian cancer cell migration and adhesion. Oncogene. 2014;33:539–549. doi: 10.1038/onc.2012.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong T, Zhong Y, Kong J, Dong L, Song Y, Fu M, Liu Z, Wang M, Guo L, Lu S, et al. Overexpression of Aurora-A contributes to malignant development of human esophageal squamous cell carcinoma. Clin Cancer Res. 2004;21:7304–7310. doi: 10.1158/1078-0432.CCR-04-0806. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Han T, Wei G, Wang Y. Inhibition of microRNA-17/20a suppresses cell proliferation in gastric cancer by modulating UBE2C expression. Oncol Rep. 2015;33:2529–2536. doi: 10.3892/or.2015.3835. [DOI] [PubMed] [Google Scholar]
- 30.Shuliang S, Lei C, Guangwu J, Changjie L. Involvement of ubiquitin-conjugating enzyme E2C in proliferation and invasion of prostate carcinoma cells. Oncol Res. 2013;21:121–127. doi: 10.3727/096504013X13832473329953. [DOI] [PubMed] [Google Scholar]
- 31.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 32.Alfieri C, Chang L, Zhang Z, Yang J, Maslen S, Skehel M, Barford D. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature. 2016;536:431–436. doi: 10.1038/nature19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garvanska DH, Larsen MS, Nilsson J. Synergistic inhibition of the APC/C by the removal of APC15 in HCT116 cells lacking UBE2C. Biol Open. 2016;5:1441–1448. doi: 10.1242/bio.020842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasiljevic A, Champier J, Figarella-Branger D, Wierinckx A, Jouvet A, Fevre-Montange M. Molecular characterization of central neurocytomas: Potential markers for tumor typing and progression. Neuropathology. 2013;33:149–161. doi: 10.1111/j.1440-1789.2012.01338.x. [DOI] [PubMed] [Google Scholar]
- 35.Gheldof A, Berx G. Cadherins and epithelial-to-mesenchymal transition. Prog Mol Biol Transl Sci. 2013;116:317–336. doi: 10.1016/B978-0-12-394311-8.00014-5. [DOI] [PubMed] [Google Scholar]
- 36.Dar AA, Belkhiri A, El-Rifai W. The aurora kinase A regulates GSK-3β in gastric cancer cells. Oncogene. 2009;28:866–875. doi: 10.1038/onc.2008.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radice GL. N-cadherin-mediated adhesion and signaling from development to disease: Lessons from mice. Prog Mol Biol Transl Sci. 2013;116:263–289. doi: 10.1016/B978-0-12-394311-8.00012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JY, Kong G. Roles and epigenetic regulation of epithelial-mesenchymal transition and its transcription factors in cancer initiation and progression. Cellular and molecular life sciences. Cell Mol Life Sci. 2016;73:4643–4660. doi: 10.1007/s00018-016-2313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips S, Kuperwasser C. SLUG: Critical regulator of epithelial cell identity in breast development and cancer. Cell Adhes Migr. 2014;8:578–587. doi: 10.4161/19336918.2014.972740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang P, Hu P, Shen H, Yu J, Liu Q, Du J. Prognostic role of Twist or Snail in various carcinomas: A systematic review and meta-analysis. Eur J Clin Invest. 2014;44:1072–1094. doi: 10.1111/eci.12343. [DOI] [PubMed] [Google Scholar]