Abstract
The “deep” proteome has been accessible by mass spectrometry for some time. However, the number of proteins identified in cells of the same type has plateaued at ∼8000–10 000 without ID transfer from reference proteomes/data. Moreover, limited sequence coverage hampers the discrimination of protein isoforms when using trypsin as standard protease. Multienzyme approaches appear to improve sequence coverage and subsequent isoform discrimination. Here we expanded proteome and protein sequence coverage in MCF-7 breast cancer cells to an as yet unmatched depth by employing a workflow that addresses current limitations in deep proteome analysis in multiple stages: We used (i) gel-aided sample preparation (GASP) and combined trypsin/elastase digests to increase peptide orthogonality, (ii) concatenated high-pH prefractionation, and (iii) CHarge Ordered Parallel Ion aNalysis (CHOPIN), available on an Orbitrap Fusion (Lumos) mass spectrometer, to achieve 57% median protein sequence coverage in 13 728 protein groups (8949 Unigene IDs) in a single cell line. CHOPIN allows the use of both detectors in the Orbitrap on predefined precursor types that optimizes parallel ion processing, leading to the identification of a total of 179 549 unique peptides covering the deep proteome in unprecedented detail.
Keywords: deep proteome, LC−MS/MS, sequence coverage, isoform profiling, protein sequence coverage
Introduction
Human primary cells and cell lines are believed to express between 8000 and ∼11 000 gene products dependent on their differentiation state.1−3 Modern proteomic workflows are now able to cover deep cellular proteomes through prefractionation and multienzyme digestion strategies.4,5 The identification of over 8000 cellular proteins is now readily achievable. However, most proteins are detected with only partial sequence coverage, and their level of completeness is biased toward the most abundant (“high content”) proteins.6 Improvements in protein sequence coverage of deep proteomes allow increasingly comprehensive interrogation of protein isoforms, post-translational modifications, amino acid substitutions, deletions, and insertions, all of which represent prime objectives in the future development of proteome research.
Despite the advent of high-speed mass spectrometers,7,8 prefractionation of biological samples is still necessary to overcome the dynamic range of protein abundance and to grant the mass spectrometer enough time for comprehensive sampling. For instance, ion exchange chromatography (strong cation exchange (SCX)9−11 and strong anion exchange (SAX)12,13), isoelectric focusing of peptides,14−16 and high-pH reversed-phase chromatography17−19 have been used with great success to identify an increasing number of proteins in tissues,20,21 cells,22 and other biological samples.23,24 In addition, complementary digestion using proteases with alternative cleavage specificities can increase protein sequence coverage in deep proteome analyses.5,25−27 Interestingly, the fragmentation/detection modes also deliver complementary data to increase peptide identification rates.28,29 However, with each additional variant for sample preparation and data acquisition, the analytical burden is multiplied. In addition to limitations in analyte resolving power and dynamic range, the observed ultradeep/high-sequence-coverage proteome appears to stagnate at the depth of ∼9000 protein groups in a single type of cells when no peptide identifications are transferred from reference proteomes1,30 or super conditions (i.e., “Super-SILAC”31−34), even when current state-of-the-art instrumentation is employed.
The Orbitrap Fusion and its successor, the Orbitrap Fusion Lumos, update the proven LTQ-Orbitrap dual-detector family of instruments35,36 with a view to closing this gap. This combination of a linear ion trap with an Orbitrap mass detector has been iteratively improved through previous generations (Orbitrap Classic/XL, Orbitrap Velos/Elite) to tailor the specific capabilities of each detector for the different requirements in speed, sensitivity, and resolution for precursor (MS1) and fragment ion (MS2) scans and offers different fragmentation types (CID, HCD, and ETD) to generate complementary fragment information,37,38 particularly for modified peptides.39,40 Changes in instrument design, in particular, the addition of a quadrupole element, allowed parallelization of ion isolation/accumulation and detection during the instrument duty cycle in Q-Exactive models,41 thereby increasing speed and shortening the duty cycle at the cost of the presence of the secondary detector (linear ion trap). In the Orbitrap Fusion/Lumos, the two strategies of using a quadrupole for ion isolation and a linear ion trap for fragment spectra acquisition have been combined, which further enhanced parallel data acquisition.7
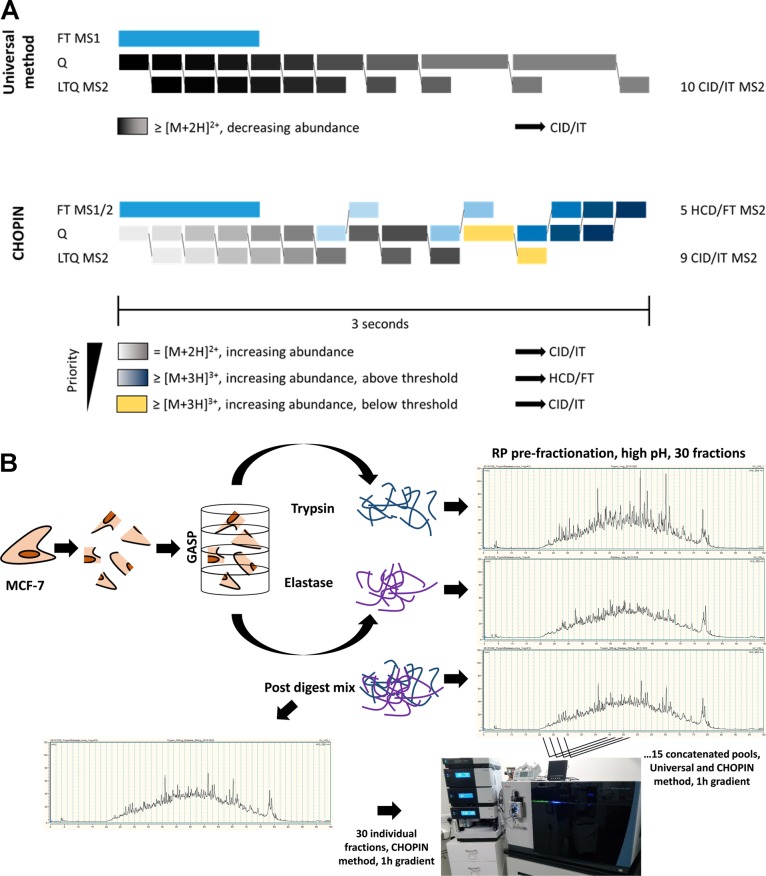
The parallelization capabilities of the Orbitrap Fusion/Lumos are highlighted in the “Universal Method”, which was developed by Thermo Fisher to maximize peptide detection irrespective of sample abundance and complexity.42 Essentially, the instrument is programmed to use longer MS2 acquisition times on low abundant peptides if (i) insufficient novel precursors have been detected and (ii) the duty cycle has not reached a set length. Additionally, the instrument uses the quadrupole, C-trap, Orbitrap, and linear ion trap elements in parallel to maximize usage of each module of the instrument and minimize idle time (Figure 1A). This universal approach may not be as effective as methods specifically optimized for particular samples. However, it has been shown to perform well for the analysis of various sample types and is accessible to all users as a predefined method in the vendor software.36
Figure 1.
Comprehensive cell proteome coverage by prefractionation and CHOPIN MS analysis workflow. (A) Mass spectrometry acquisition methods demonstrating the dynamic segmentation of analytical channels for MS1 FT (Orbitrap), Q (Quadrupole), and MS2 LTQ (Linear Ion Trap) that were designed for the Universal (upper panel) and CHarge Ordered Parallel Ion aNalysis (CHOPIN) method (lower panel). The Universal Method makes use of the parallel acquisition of MS1 scan in the Orbitrap, while peptide fragments are scanned in the LTQ, ordered by decreasing precursor intensity. Additional parallelization is achieved by concurrent MS2 scans and isolation of the following precursor. Precursor ion accumulation is allowed to proceed for up to 250 ms if no previously unselected precursor is found. CHOPIN adds another level of parallelization by triaging intense and highly charged ions to be analyzed by an Orbitrap MS2 scan, while low abundant precursor ions are prioritized for the more sensitive MS2 scan in the linear ion trap. CHOPIN and the data analysis is further described in Supporting Information. (B) Methodological workflow for the analysis of the MCF-7 breast cancer cell line deep proteome. MCF-7 cell extracts were digested with either trypsin or elastase, and peptide mixtures were separated by high-pH reversed-phase (RP) HPLC to collect 30 fractions that were pooled in a concatenated fashion to 15 fractions. Also, tryptic and elastase digest was mixed and prefractionated as above (“Post Digest Mix”, PDM), followed by concatenation or distinct fraction analysis. Each fraction was subsequently analyzed by LC–MS/MS using both the Universal and CHOPIN acquisition methods. Detailed results for each individual experiment are shown in the Supporting Information. (Orbitrap Fusion Lumos photo by RF).
Using these new technical advancements in MS technology in combination with sample prefractionation and high and broad specificity proteolysis, we demonstrate unprecedented coverage of the ultradeep proteome of a breast cancer cell line, thereby providing further insights into global protein sequence coverage, the presence of isoforms, and the PTM landscape.
Methods
Tissue Culture and Cell Lysis
The MCF-7 breast cancer cell line was cultured in DMEM medium (Sigma, no. D6546) supplemented with 10% FCS, 1% penicillin, 1% streptomycin, and 1% glutamine at 37 °C (5% CO2). Five T175 tissue culture flasks of confluent MCF-7 cells were harvested using a trypsin solution (Sigma, no. T3924), washed two times in PBS, and stored at −80 °C until use. The frozen cells were lysed on ice for 30 min in 5 mL of RIPA lysis buffer (Thermo Pierce, no. 89901) supplemented with 4% SDS, 6 M urea, 2 M thiourea, 100 mM DTT, protease, and phosphatase inhibitors (Roche nos. 11836170001 and 04906837001). The lysate was sonicated twice for 1 min (5 s on, 10 s off, repeated four times). After the addition of 1250 units of benzonase (Sigma, no. E1014), the lysate was incubated on ice for 20 min and centrifuged at 21 000 g for 20 min at 4 °C and the pellet discarded. Because of the presence of SDS and DTT in the sample, protein content was estimated by SDS-PAGE and Coomassie staining.
Sample Preparation and Fractionation
Approximately 5 mg of protein was digested using the GASP method.43 In brief, the lysate was mixed with 30% acrylamide, polymerized, and shredded. The gel slurry was fixed in methanol/acetic acid/water (50/40/10%) and washed twice with alternating 6 M urea and 100% acetonitrile to remove SDS. 50 mM ammonium bicarbonate was added to the gel. The gel slurry was split equally into two by volume for digestion by separate enzymes. 100% acetonitrile was added to dehydrate the gel and was removed prior to the addition of 50 μg of trypsin (Promega, no. V5111) or 50 μg of elastase (Worthington Biochemical, no. LS006365). The samples were incubated at 37 °C overnight and further processed as according to the original GASP method to extract peptides from the shredded gel pieces. The samples were desalted on C18 solid-phase extraction cartridges (Sep-Pak plus, Waters) and resuspended in 2% acetonitrile 0.1% formic acid and peptide concentration determined using a peptide quantitation kit (Thermo Pierce, no. 23275).
Off-line high-pH reverse-phase prefractionation was performed on 800 μg of digested material using the loading pump of a Dionex Ultimate 3000 HPLC with an automated fraction collector and a XBridge BEH C18 XP column (3 × 150 mm, 2.5 μm pore size, Waters no. 186006710) over a 100 min gradient using basic pH reverse-phase buffers (A: water, pH 10 with ammonium hydroxide; B: 90% acetonitrile, pH 10 with ammonium hydroxide). The gradient consisted of a 12 min wash with 1% B, then increasing to 35% B over 60 min, with a further increase to 95% B in 8 min, followed by a 10 min wash at 95% B and a 10 min re-equilibration at 1% B, all at a flow rate of 200 μL/min with fractions collected every 2 min throughout the run. 100 μL of the fractions was dried and resuspended in 20 μL of 2% acetonitrile/0.1% formic acid for analysis by LC–MS/MS. Fractions were loaded on the LC–MS/MS following the concatenation scheme shown in Figure 1B with adjusted sample volumes to analyze ∼1 μg on column.
Mass Spectrometry Analysis Methods
Peptide fractions were analyzed by nano-UPLC–MS/MS using a Dionex Ultimate 3000 nano-UPLC with EASY-Spray column (75 μm × 500 mm, 2 μm particle size, Thermo Scientific) with a 60 min gradient of 0.1% formic acid in 5% DMSO to 0.1% formic acid to 35% acetonitrile in 5% DMSO. MS data were acquired with an Orbitrap Fusion7 Lumos instrument using the methods described below. A comprehensive description of the method can be found in the Supporting Information in addition to method transcripts and Xcalibur (Tune v. 2.0.1258.14) methods files.
Universal Method
The Universal method has been developed by Eliuk et al.42 to maximize peptide identification without method optimization for different sample complexities and abundances. In principle, it allows a long ion accumulation time for low abundance precursors with parallel usage of quadrupole, collision cell, and both Orbitrap (FT) and ion trap (IT) detectors (summarized in Figure 1A).
MS scans were acquired at a resolution of 120 000 between 400 and 1500 m/z and an AGC target of 4.0E5. MS/MS spectra were acquired in the linear ion trap (rapid scan mode) after collision-induced dissociation (CID) fragmentation at a collision energy of 35% and an AGC target of 4.0E3 for up to 250 ms, employing a maximal duty cycle of 3 s, prioritizing the most intense ions and injecting ions for all available parallelizable time. Selected precursor masses were excluded for 30 s.
CHOPIN
CHarge Ordered Parallel Ion aNalysis (CHOPIN) employs selection criteria to channel ions to the best suited detector based on precursor ion properties (Figure 1A). The hallmark of CHOPIN is the simultaneous use of both mass detectors for peptide fragment spectra acquisition, which allows the generation of additional MS/MS scans in the Orbitrap at no cost of duty cycle time. Because only high abundant precursors with higher charge states are analyzed in the Orbitrap after high collision energy dissociation (HCD) fragmentation, the success rate of these scans is very high. At the same time, the higher sensitivity of the ion trap is used to analyze low abundant precursor ions. Details and further description of the method used here have been exported into text format and are available in the Supporting Information.
In brief, MS scans were acquired as above. For precursor selection, we prioritized the least abundant signals. Doubly charged ions were scheduled for CID/IT analysis with the same parameters applied as above. Charge states 3–7 with precursor intensity >500 000, however, were scheduled for analysis by a fast HCD/FT scan of maximal 40 ms (15 000 resolution). The remaining charge-state 3–7 ions with intensity <500 000 were scheduled for analysis by CID/IT, as described above. Selected precursor masses were excluded for 12 s, as the gain in MS/MS scan events allows repeated scans of the same precursor across the chromatographic peak without risking undersampling.
Elastase Digests
The elastase digested samples have been analyzed with divergent parameters to address the occurrence of singly charged peptide ions. In the CHOPIN method we added a fourth scan event for singly charged precursor ions to be scanned with a HCD/FT scan, increased collision energy (32% instead of 25%), and a longer injection time (100 ms instead of 40 ms).
However, the “no enzyme” database searches benefit from high mass accuracy MS/MS spectra,44 so we modified the Universal Method to replace the low mass accuracy CID/IT scans for MS/MS data acquisition for 2 HCD/FT scan types recognizing singly charged and multiple charged ions. Because the resulting method does not exactly conform to the parameters of the Universal method anymore, we refer to results obtained with this method as “Universal/FT” and highlight the difference where appropriate. Full details about the method have been exported into text format and are available in the Supporting Information.
Data Analysis
The general workflow of sample processing and identification of MS/MS spectra is shown in Figure S1. CHOPIN produces raw files containing HCD/FT and CID/IT spectra. To allow searching the data in PEAKS,45 we separated both spectra types into separate MGF files by Proteome Discoverer (V. 2.0) using the top 10 (HCD/FT) and top 15 (CID/IT) peaks in every 100 m/z window. CID/IT spectra derived from CHOPIN or Universal Method were then searched in Peaks 7.5 using the default target decoy approach46 with 20 ppm mass error tolerance for the precursor and 0.5 Da for fragment masses while HCD/FT spectra were searched with a 0.05 Da mass tolerance for fragment masses. The selection of a 20 ppm mass accuracy tolerance allowed the inclusion of correctly identified peptides for which the 13C isotope peak was wrongly assigned as monoisotopic precursor mass. These identifications will show as deamidated peptides with a larger mass error. The mass error distribution of deamidated peptides is visualized in Figure S5, showing the population of truly deamidated peptides and wrongly assigned precursor masses.
We allowed up to four missed cleavage sites and no nonspecific cleavage for tryptic samples and set propionamide as fixed cysteine modification and variable modification on lysine and N-termini as well as Deamidation (N,Q) and Oxidation (M) and maximal 1 variable modification per peptide in the de novo and database searches (three variable PTMs for PTM search nodes47). The database used was in all cases the UniProt48 Reference (UPR) Homo sapiens database (retrieved 15.10.2014). The elastase digest and Post Digest Mix data were searched with no enzyme specificity. Peptide false discovery rate (FDR) was adjusted to 1% and proteins grouped according the parsimony principle described by Nesvizhskii and Aebersold.49 Subsequently, the protein identification score threshold was adjusted to achieve a protein FDR of ∼1%. The score thresholds for peptide and protein FDRs as well as identification metrics are shown in Table 1. Because HCD/FT and CID/IT spectra had to be searched individually to appreciate the different fragmentation types and mass accuracies, the results were combined post-search. The result combination includes the following major steps: (i) Read all of the PSMs identified from two sample files, including the ones from both target and decoy databases. (ii) Tune the PSM scores accordingly to make sure the scores of PSMs from different samples are normalized identically. More specifically, the PSM score thresholds at 1% FDR of both samples were calculated; then. using one of the thresholds as the base score, the PSM scores in the other sample were shifted according to the difference between the two score thresholds. (iii) Put all PSMs together and carry out the protein inference algorithm for protein grouping. (iv) Recalculate protein scores and coverage rates. The same procedure was applied to generate single or accumulating results from the prefractionated sample sets.
Table 1. Summary of Identification Metrics Using CHarge Ordered Parallel Ion aNalysis (CHOPIN) and Universal Method on Tryptic (T), Elastase (E), and Post Digest Mix (PDM) Samplesa.
| peptide score threshold @1%FDR | PSMs | MSMS scans | effective peptide FDR @score threshold | protein score threshold @1%FDR | effective protein FDR @score threshold | protein groups @1% FDR | proteins (unique and razor) | |
|---|---|---|---|---|---|---|---|---|
| CHOPIN (T) | 21.1 | 307318 | 582030 | 0.924 | 57 | 1.052 | 8745 | 13019 |
| Universal (T) | 20.2 | 226291 | 539916 | 0.943 | 57 | 1.001 | 8692 | 12770 |
| CHOPIN (E) | 19.8 | 170960 | 660060 | 0.701 | 85 | 1.069 | 4951 | 5521 |
| Universal/FT (E) | 16.4 | 171529 | 349164 | 0.617 | 60 | 0.99 | 5143 | 6866 |
| CHOPIN (PDM) | 14 | 284347 | 714354 | 0.891 | 31 | 0.977 | 7958 | 11974 |
| Universal (PDM) | 14.9 | 192500 | 671699 | 0.84 | 32 | 1.003 | 7517 | 11371 |
| CHOPIN (PDM), unlinked fractions | 14 | 433723 | 1160032 | 0.999 | 37 | 0.977 | 9824 | 13000 |
| All data | 10.8 | 2010579 | 4677255 | 0.996 | 64 | 0.987 | 13728 | 14890 |
| trypsin | 10052 | 13320 | ||||||
| elastase | 7038 | 8257 | ||||||
| PDM | 9834 | 12452 | ||||||
Fractions have been combined and data searched in PEAKS.
Data density was visualized by using the Perseus software (v. 1.5.3.0) platform.50
Results and Discussion
Because the Orbitrap Fusion/Lumos instrument is capable of using a complex data-dependent decision tree, we decided to make additional use of the parallelization capabilities of an Orbitrap Fusion Lumos and developed a data-dependent acquisition method that would use elements of the Universal Method and add in additional MS2 scans for the idling Orbitrap detector. To maximize spectral quality/success rate and detector usage efficiency, we streamlined the ions to the detector that is best suited for their specific properties. Low abundant precursors with a charge state of 2 would be fragmented with CID, and their fragment spectrum was acquired in the more sensitive linear ion trap (CID/IT), while highly abundant precursors with a charge state of >2 would be fragmented using HCD and their fragment spectrum acquired in the Orbitrap (HCD/FT). In addition, higher charged precursors with an abundance below the HCD/FT selection threshold would be acquired with the same detection parameters as doubly charged ions (CID/IT). Consequently, CHOPIN results in hybrid data, containing both spectra types in a single raw file. The duty cycle of this CHarge Ordered Parallel Ion aNalysis (CHOPIN) is depicted in Figure 1A.
To evaluate if CHOPIN would allow the acquisition of more high-quality MS2 spectra in complex samples, we prepared a total cell lysate of MCF-7 cells in the presence of 4% SDS, 6 M urea, 2 M thiourea, 100 mM DTT and sonicated the lysate to maximize lysis and protein solubilization. We used Gel-Aided Sample Preparation (GASP)43 to allow the use of SDS and urea/thiourea for maximum solubilization of the sample to introduce missed cleavage sites where some lysine residues would react with acrylamide to create overlapping peptides, resulting in increased sequence coverage, and for ease of use. Samples where then digested with either trypsin or elastase.
The individual digests were then prefractionated via high-pH reversed phase chromatography (C18, 30 fractions) and concatenated (15 fraction pools) as described in Figure 1B. In addition, we also mixed elastase and tryptic digest and analyzed concatenated and individual fractions. Each fraction was analyzed with CHOPIN and the Universal Method on a 1 h gradient resulting in six data sets of 15 × 1 h LC–MS/MS analyses (trypsin, elastase, Post Digest Mix, each acquired with CHOPIN and Universal Method) and one data set with 30 × 1 h LC–MS/MS analyses (Post Digest Mix, individual fractions, CHOPIN method).
To evaluate how different search algorithms handle data acquired with CHOPIN and the Universal Method, the whole tryptic data set was reprocessed with PEAKS, Mascot,51 Andromeda/MaxQuant,52,53 and SEQUEST54 (Table S6). Additionally, we addressed robustness and reproducibility by analyzing one tryptic fraction in technical triplicates with CHOPIN and Universal Method (Figure S7). In summary, we obtained comparable results with all used search engines, with PEAKS benefiting slightly from its ability to detect post-translational modifications in an unbiased fashion. Overall, we achieved significantly better identification rates and more peptide spectrum matches employing CHOPIN. The results are summarized and discussed in greater detail in the Supporting Information.
CHOPIN Improves Duty Cycle Usage and Success Rate of MS/MS Identification
One duty cycle of the Universal and CHOPIN methods in the tryptic experiment was extracted (Table S1) to exemplify the working principle of the two data acquisition methods under comparable conditions (similar RT, base peak, and base peak intensity). Here the Universal Method results in a Top35 scan event (1 precursor scan followed by 35 MS2 scans) in a 3 s duty cycle. The accumulated injection time for the 35 precursors is 1.8 s and the total MS2 scan time is 2.14 s. Given a 3 s duty cycle the Universal Method gains 0.94 s through parallel handling of MS2 injection and scan. Employing CHOPIN resulted in a Top42 scan event, of which 29 precursors were scanned with CID/IT and 13 were scanned by HCD/FT. Here the accumulated injection time is similar to the Universal Method with 1.79 s; however, because of parallel acquisition of MS2 scan in the Orbitrap and linear ion trap, the instrument spends 2.75 s on MS2 scans, adding up to a total of 4.54 s in a duty cycle of 3 s. The additional level of parallelization by using both detectors for MS2 scans in the same duty cycle gained 2.54 s through parallel handling. In summary, using CHOPIN we gained seven MS2 scans and 0.6 s MS2 scan time over the Universal Method in the exemplified duty cycle.
Because we use HCD/FT for abundant precursors in CHOPIN, the resulting MS2 scans can be expected to have a high success rate. Also, previously scanned intense precursors are moved to the autoexclusion list, effectively precluding them from being selected for a CID/IT scan and therefore improving detector usage efficiency. Consequently, the more sensitive linear ion trap can spend time on less abundant precursors. We plotted the peptide score distribution of the accumulated results of the trypsin digest (Figure 2A, other digests see Figure S2) as a function of peptide mass and identification numbers (density gradient) for each scan type in Chopin (HCD/FT and CID/IT) and for the CID/IT scans using the Universal Method. We observed overall higher scores for the HCD/FT scan mode across the mass range with 32% of all identified spectra (31 066/97 731) yielding a score of 80 or higher. In contrast, only 86 out of 188 037 (0.05%) CID/IT identifications scored in the same range. Using the CID/IT-based Universal Method, only 899 identifications achieved a score of >80, clearly indicating a significantly lower spectrum quality in addition to overall lower identification numbers.
Figure 2.
CHOPIN enhances MS/MS interpretation rates. (A) The density plot shows the number of identifications over precursor mass and peptide score (−10lgP) to demonstrate the gain of spectra quality for peptides by HCD/FT detection (Chopin HCD/FT) in a tryptic digest. The Chopin CID/IT spectra show a similar score distribution compared to peptides identified with the Universal Method. However, the combined data of the CHOPIN result show a clear improvement in the number of identified peptides and confidence. Density plots for the Elastase digest and the Post Digest Mix are shown in Figure S2. (B) Improvements on the peptide level are carried through to ID confidence on the protein group level, especially in the trypsin and Post Digest Mix samples. Because of the inclusion of singly charged precursors, the benefit in the elastase-digested samples is limited to high-confidence identifications.
We observed similar frequencies for low-scoring proteins in the tryptic fractions after Universal and CHOPIN data acquisition, with some benefit for the Universal Method for low-to-medium protein scores (100–200). Interestingly, CHOPIN resulted in considerably more high scoring proteins. For the elastase digest we observed a different score distribution, especially when viewed in context with overall identification numbers (compare Figure 2B and Table S3). While we identified more peptides in the elastase digest with the modified Universal Method (higher success rate of high mass accuracy HCD/FT MS/MS spectra, see Methods section), we needed to use a high protein score threshold to achieve 1% protein FDR (see Table 1). This can be explained by the inclusion of short peptides, frequently generated with a single charge, in the precursor selection algorithm, driving protein FDR. For future use of CHOPIN in elastase digests, we would recommend the addition of a precursor mass threshold to exclude singly charged, short peptides. The benefit of CHOPIN is seen most clearly in the Post Digest Mix, where CHOPIN’s improved duty cycle handles the increased sample complexity and mixed enzyme precursor profile more efficiently (Table 1).
We also compared the proteins and peptides identified with the different acquisition methods by scan types (CID/IT, HCD/FT) for the three experiments. As expected, we can observe a very high success rate for the HCD scans using CHOPIN data acquisition. Interestingly the success rate for CID/IT using CHOPIN is also higher than the success rate using the Universal Method and CID/IT, demonstrating that the CID/IT scan mode is better suited for doubly charged ions than unrestricted use in the Universal Method. In addition to acquiring more spectra due to improved parallelization, CHOPIN increases the spectra quality, yielding a better success rate (Figure S3 and Table S3).
CHOPIN Improves Protein Sequence Coverage
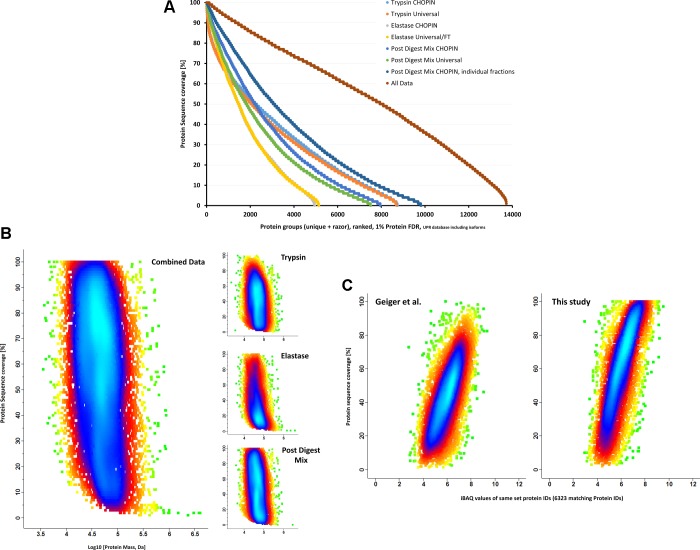
High protein sequence coverage of the deep proteome is key to detecting post-translational modifications in an unbiased way and the discrimination of protein isoforms. Multiple studies have shown to increase proteome sequence coverage by different approaches such as multienzyme proteolysis and extensive prefractionation or combinations thereof. Figure 3A shows the detected protein sequence coverage using the different here employed analysis strategies (trypsin, elastase, and Post Digest Mix, after high-pH fractionation using CHOPIN and Universal Method) and a combined result on the protein level. Data acquisition with CHOPIN consistently resulted in higher sequence coverage than the Universal Method, although the number of detected protein groups does not necessarily increase when a single protease is used (Figure S8). The limitations of tryptic digestion become obvious when the number of proteins with very high sequence coverage is compared with elastase or even the combined digests; only a small number of protein groups are detected with more than 90% sequence coverage: 123, compared with the far greater number from the Post Digest Mixture of trypsin and elastase proteolyzate: 771 (327 protein groups for the Elastase digest and 1462 protein groups for the complete data set).
Figure 3.
Improved global protein sequence coverage using the CHOPIN workflow. (A) Protein sequence coverages observed with different analytical strategies illustrate the benefit of the methods used to improve protein sequence coverage and protein grouping as the number of identified protein groups could be increased significantly. The median protein sequence coverage of 13 728 protein groups (leading protein) was 57%, with 7935 protein groups being identified with more than 50% coverage. (B) Plotting sequence coverage of the combined data (leading protein per group) over molecular protein mass shows a distribution plume similar to a tornado (“Tornado plot”). Interestingly, the density of data points is relatively uniform across protein mass while showing highest density at 70–80% coverage, indicating a similar abundance for the majority of the proteome, independent of molecular weight. The right panel shows the archived protein sequence coverage in the different digests. Trypsin digests alone cannot generate sequence comprehensive data, while elastase digests can cover proteins better. However, the mixture of tryptic and elastase digest (“PDM”) appears to retain the benefits of both proteases and specifically benefits from the improved duty cycle in CHOPIN due to its extreme complexity (compare Table 1). (C) 6323 proteins and corresponding iBAQ values65 could be matched to previously published deep proteome data in MCF-7 cells by Geiger et al.1 The median sequence coverage for the same set proteins could be improved from 43 to 61%.
With the increased sequence coverage generated by CHOPIN and orthogonal digests with trypsin and elastase, more protein isoforms can be distinguished from their canonical variants. This leads to the identification of 13 728 protein groups representing 8949 genes in the combined data. In our database searches (UniProt Reference Homo sapiens database48 containing a total of 85 889 human proteins and isoforms, retrieved 15/10/2014) we used parsimony-based protein inference, as described by Nesvizhskii and Aebersold,49 to report the minimal number of proteins that can be observed with unique and razor peptides. We plotted the sequence coverage of the leading protein of all detected protein groups over their molecular weight (Figure 3B) to illustrate if there is any bias in coverage regarding protein size. The Tornado-shaped plume shows a higher density of data points in the low coverage (0–20%) part of the graph, but we can observe a more even distribution across the plume up to 100%. 7935 protein groups were observed with a sequence coverage for the leading protein of >50% in the merged data (median coverage = 57%). Instead of median sequence coverage this metric can be used to better reflect not only the depth at which a proteome is reported but also the comprehensiveness as it takes “one-hit-wonders” out of the equation.
The unbiased search for peptide modifications by the PEAKS PTM search engine47 allowed for the detection of up to 485 different modifications due to a de novo sequence tag mapping before the database search. In the combined data we discovered a total of 206 different peptide modifications on a total of 193 548 sites (Figure S6). About half of the modifications can be explained by sample processing and plausible artifacts, resulting in a total of 91 modification types on 81 905 sites that can be classified as biological post-translational modifications (Tab. S3).
Because the broad cleavage specificity does not allow us to estimate relative protein abundances within a sample, we retrieved iBAQ values for the MCF-7 proteome from Geiger et al.1 to see if we can cover even low abundant proteins more comprehensively than before (Figure 3C). Here we plotted the protein sequence coverage of proteins common in both data sets over the corresponding iBAQ value retrieved from Geiger et al.1 As expected, highly abundant proteins can be observed with higher protein sequence coverage. However, in our data set the median sequence coverage of the same set proteins could be increased from 42.9 (left panel) to 61% (right panel), with a large proportion of proteins covered with >90% (53 vs 1461 protein groups). This result indicates a step toward complete sequence coverage detection, independent of protein abundance.
Application of Elastase in Total Proteome Digests
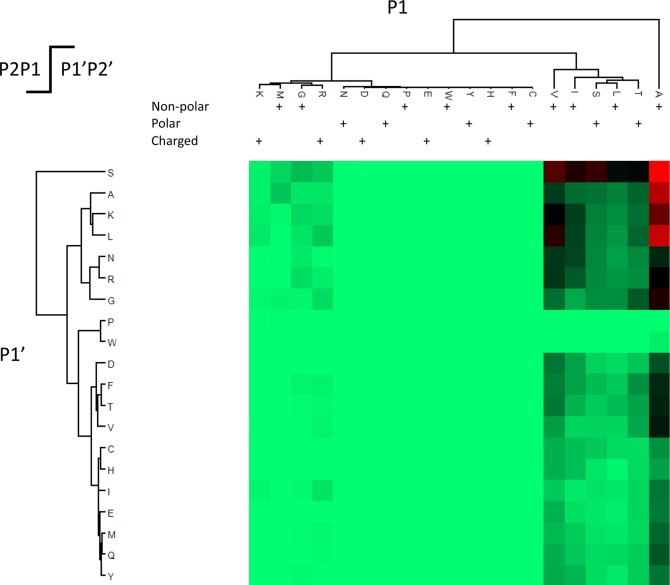
Elastase is often used to increase protein sequence coverage for noncomplex protein samples due to its broad cleavage specificity.55 While unspecific proteases such as Proteinase K have been used in the past on membrane proteins56 and to analyze interpeptide cross-links,57 the data analysis still represents a major challenge as cleavage specificity significantly reduces the computational effort for peptide identification. Recent sequence tag58 or de-novo-based46,59,60 methods for peptide identification can benefit from the detection of sequence information prior to the application of precursor mass and cleavage specificity to reduce the search space and achieve similar result characteristics as standard search algorithms. In this study, for the first time, we used elastase on total cell extracts to supplement for classical multienzyme approaches5,25,61,62 to increase depth and sequence coverage of the MCF-7 proteome. Interestingly, by examining such a complex data set, we refined the distinct cleavage pattern for elastase,55 as shown in Figure 4. We noted that the vast majority of cleavages (86.77%) occur at specifically A, V, I, T, L, and S as P1. Additional 10.3% of cleavages were observed following R, G, M, and K as P1. The identity of P1′ was less relevant with the exception of proline and tryptophan effectively inhibiting cleavage. Taken together, we can conclude that elastase does have a high but broad specificity toward the amino acids A, V, I, T, L, S, R, G, M, K, in the P1 position with a total of 97.7%. Clearly, the ability of elastase to skip multiple cleavage sites generates a peptide population that is highly orthogonal to trypsin-generated peptides and therefore complements a tryptic digest.
Figure 4.
Comprehensive elastase cleavage profile analysis reveals preference toward small aliphatic amino acids. This study demonstrates the feasibility of using elastase as orthogonal protease to trypsin with the potential to replace the classical, narrow specificity multienzyme approach. We detected similar specificity as Rietschel et al.55 based now on 129 677 observed cleavages. 86.77% of cleavages were specific to A, V, I, T, L, and S as P1. However, additional 10.3% of cleavages were detected on R, G, M, and K as P1, indicating a broad but high cleavage specificity of elastase.
On the basis of 129 677 identified peptides in the elastase data, we were able to add peptide IDs orthogonal to the trypsin-derived identifications. In combination, these data allowed the differentiation of protein isoforms that are often inseparable using standard digestion methods. This is further improved due to the randomly introduced missed cleavage sites after tryptic digestion by using the GASP sample preparation methods due to lysine alkylation. As a result, we created peptide populations, which are able to distinguish subtle sequence differences between protein isoforms. This can be demonstrated by comparing the number of identified proteins with the number of identified protein groups (Table 1) in the same workflow. The difference between identified proteins (13 019 for CHOPIN, trypsin) and protein groups (8745 for CHOPIN, trypsin) indicates a high number of protein groups with multiple protein entries. In the combined data both numbers are relatively similar (14 890 proteins vs 13 728 protein groups), indicating most protein groups contained one protein instead multiple products of the same gene.
Even though peptide identification is significantly improved using the de-novo-based search algorithm in PEAKS, an elastase digested cell extract provides a challenge for false-positive estimation due to the presence of short ambiguous peptide sequences. Instead of defining a minimal peptide length we choose the more conservative option to increase the protein score threshold to achieve 1% protein FDR (compare Table 1), which effectively results in the necessity of up to five peptides (unique or razor) being identified with a peptide FDR of 1% for a protein hit in the CHOPIN elastase data when all of the peptide scores for this protein hit are low. Consequently, only 16 out of 13 728 protein groups in the complete data set are identified with only a single (high scoring) peptide. The percentage of isoforms in the here-identified protein groups (27.5%) is very similar to the percentage of isoforms in the database used (25.09%), giving us an indication that protein parsimony is not overly optimistic when isoforms can be distinguished into separate protein groups. Moreover, as shown for the trypsin digestion data sets, the limit of detection of proteins in a whole-cell lysate is determined by the absolute sensitivity of the workflow and to a lesser extent by the data acquisition method if undersampling is avoided (Figure S8). However, CHOPIN could be used to significantly increase the sequence coverage of the proteins detected, which is very beneficial for protein metrics, especially if combined with broad specificity digestion protocols.
Our data also raises questions with regards to protein isoform identification. The unified modeling of both FDR and protein grouping in large data sets is an ongoing debate.63,64 Existing models may well lead to inflated protein group counts from high-coverage data set, particularly with the advent of de-novo-based search tools and of broad specificity proteolysis allowing differentiation between isoforms with almost identical sequences. While standard protein parsimony can be applied for protein grouping and single peptide hits can be virtually excluded as demonstrated here, further advances in the detection of protein isoforms (and PTMs) will likely require new FDR models to minimize false-positives. In the data reported here, the number of protein groups identified when all data are combined with a unified FDR model is considerably higher than achieved by any of the method/digest mix combinations separately (Figure 3A). While the combined data arguably justify these numbers in terms of greater sequence coverage, we report the “All data” total with the above considerations in mind (see also the Supporting Information).
Conclusions
We have developed CHarge Ordered Parallel Ion aNalysis to improve the duty cycle of an Orbitrap Fusion (Lumos) by using both detectors in parallel for MS/MS spectra acquisition in a way that favors spectral quality according to the properties of the peptide precursor. Our results show that this leads to an expanded proteome coverage when combined with a broad specificity digestion approach.
In addition, our study also highlights challenges that lie ahead for future developments in proteome research in the coming years. The analysis of data using different mass detectors with distinct mass errors and fragmentation modes has proved to be beneficial for the identification of the deep high-coverage proteome but also presents a major obstacle in the form of the quantity and variety of data generated by modern hybrid instruments. Available search tools need to adapt to such type of complex MS data to allow combined analysis and more sophisticated statistical evaluation. Second-generation search tools incorporating de novo algorithms allow the unbiased detection of hundreds of different modifications on tens of thousands of sites, even in existing data. As the deep proteome becomes more readily accessible, the focus must move to achieving high protein sequence coverage. Detection of proteins, their isoforms, and PTMs in a comprehensive and unbiased way is crucial to an expanded understanding of the proteome.
Acknowledgments
We would like to thank members of the Kessler lab for insightful discussions. R.F. and B.M.K. were supported by the Kennedy Trust Fund. P.D.C. was supported by a John Fell Fund 133/075 and Welcome Trust grant 105605/Z/14/Z to B.M.K. All mass spectrometry data files associated with this manuscript have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE data repository identifier PXD003977 (https://www.ebi.ac.uk/pride/archive/projects/PXD003977/files).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00915.
Figure S1. Data analysis strategy for the deconvolution of CID/IT HCD/FT hybrid data generated by CHOPIN. Figure S2. Score distribution of identified peptides in Elastase and Post digested mix samples with Universal and CHOPIN method. Figure S3. MS/MS analysis statistics using CHOPIN or the Universal method. Figure S4. Accumulated peptide and protein identifications after high pH pre-fractionation. Figure S5. Mass error distribution of peptides (tryptic digest) identified with and without modifications. Figure S6. Detected modification landscape in MCF-7 cells. Figure S7. Reproducibility of CHOPIN and Universal methods assessed using multiple search engines. Figure S8. FDR modelling demonstrates enhanced peptide, but not protein group identifications by CHOPIN. Table S1. Comparative examples of a typical duty cycle using the CHOPIN and Universal methods. Table S2. Example of data acquisition statistics for one of the tryptic fraction using Universal or CHOPIN method. (PDF)
Table S3. Detected PTM types and classification. (XLSX)
Table S4. Sequence coverage of detected protein groups. (XLSX)
Table S5. Peptide identifications. (ZIP)
Method transcripts for CHOPIN. (PDF)
Method transcript for Universal. (PDF)
Method transcript for CHOPIN Elastase.(PDF)
Method transcript for Universal/FT. (PDF)
Method files for use in Xcalibur for CHOPIN, CHOPIN Elastase, Universal, and Universal/FT. (ZIP)
Author Contributions
∥ S.D. and P.D.C. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Geiger T.; Wehner A.; Schaab C.; Cox J.; Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 2012, 11 (3), M111 014050. 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornburg D.; Drepper C.; Butter F.; Meissner F.; Sendtner M.; Mann M. Deep proteomic evaluation of primary and cell line motoneuron disease models delineates major differences in neuronal characteristics. Mol. Cell. Proteomics 2014, 13 (12), 3410–20. 10.1074/mcp.M113.037291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M.; Schlegl J.; Hahne H.; Moghaddas Gholami A.; Lieberenz M.; Savitski M. M.; Ziegler E.; Butzmann L.; Gessulat S.; Marx H.; Mathieson T.; Lemeer S.; Schnatbaum K.; Reimer U.; Wenschuh H.; Mollenhauer M.; Slotta-Huspenina J.; Boese J. H.; Bantscheff M.; Gerstmair A.; Faerber F.; Kuster B. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509 (7502), 582–7. 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- de Godoy L. M.; Olsen J. V.; Cox J.; Nielsen M. L.; Hubner N. C.; Frohlich F.; Walther T. C.; Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 2008, 455 (7217), 1251–4. 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Guo X.; Trudgian D. C.; Lemoff A.; Yadavalli S.; Mirzaei H. Confetti: a multiprotease map of the HeLa proteome for comprehensive proteomics. Mol. Cell. Proteomics 2014, 13 (6), 1573–84. 10.1074/mcp.M113.035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev R. A. The challenge of the proteome dynamic range and its implications for in-depth proteomics. Proteomics 2013, 13 (5), 723–6. 10.1002/pmic.201200451. [DOI] [PubMed] [Google Scholar]
- Hebert A. S.; Richards A. L.; Bailey D. J.; Ulbrich A.; Coughlin E. E.; Westphall M. S.; Coon J. J. The one hour yeast proteome. Mol. Cell. Proteomics 2014, 13 (1), 339–47. 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltema R. A.; Hauschild J. P.; Lange O.; Hornburg D.; Denisov E.; Damoc E.; Kuehn A.; Makarov A.; Mann M. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol. Cell. Proteomics 2014, 13 (12), 3698–708. 10.1074/mcp.M114.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. P.; Wolters D.; Yates J. R. 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001, 19 (3), 242–7. 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Moczulska K. E.; Pichler P.; Schutzbier M.; Schleiffer A.; Rumpel S.; Mechtler K. Deep and precise quantification of the mouse synaptosomal proteome reveals substantial remodeling during postnatal maturation. J. Proteome Res. 2014, 13 (10), 4310–24. 10.1021/pr500456t. [DOI] [PubMed] [Google Scholar]
- Marino F.; Cristobal A.; Binai N. A.; Bache N.; Heck A. J.; Mohammed S. Mohammed, S., Characterization and usage of the EASY-spray technology as part of an online 2D SCX-RP ultra-high pressure system. Analyst 2014, 139 (24), 6520–8. 10.1039/C4AN01568A. [DOI] [PubMed] [Google Scholar]
- Ritorto M. S.; Cook K.; Tyagi K.; Pedrioli P. G.; Trost M. Hydrophilic strong anion exchange (hSAX) chromatography for highly orthogonal peptide separation of complex proteomes. J. Proteome Res. 2013, 12 (6), 2449–57. 10.1021/pr301011r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J. R.; Zougman A.; Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 2009, 8 (12), 5674–8. 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- Michel P. E.; Reymond F.; Arnaud I. L.; Josserand J.; Girault H. H.; Rossier J. S. Protein fractionation in a multicompartment device using Off-Gel isoelectric focusing. Electrophoresis 2003, 24 (1–2), 3–11. 10.1002/elps.200390030. [DOI] [PubMed] [Google Scholar]
- Horth P.; Miller C. A.; Preckel T.; Wenz C. Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol. Cell. Proteomics 2006, 5 (10), 1968–74. 10.1074/mcp.T600037-MCP200. [DOI] [PubMed] [Google Scholar]
- Magdeldin S.; Yamamoto K.; Yoshida Y.; Xu B.; Zhang Y.; Fujinaka H.; Yaoita E.; Yates J. R. 3rd; Yamamoto T. Deep proteome mapping of mouse kidney based on OFFGel prefractionation reveals remarkable protein post- translational modifications. J. Proteome Res. 2014, 13 (3), 1636–46. 10.1021/pr401122m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilar M.; Olivova P.; Daly A. E.; Gebler J. C. Orthogonality of separation in two-dimensional liquid chromatography. Anal. Chem. 2005, 77 (19), 6426–34. 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Yang F.; Gritsenko M. A.; Wang Y.; Clauss T.; Liu T.; Shen Y.; Monroe M. E.; Lopez-Ferrer D.; Reno T.; Moore R. J.; Klemke R. L.; Camp D. G. 2nd; Smith R. D. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 2011, 11 (10), 2019–26. 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelstrup C. D.; Jersie-Christensen R. R.; Batth T. S.; Arrey T. N.; Kuehn A.; Kellmann M.; Olsen J. V. Rapid and deep proteomes by faster sequencing on a benchtop quadrupole ultra-high-field Orbitrap mass spectrometer. J. Proteome Res. 2014, 13 (12), 6187–95. 10.1021/pr500985w. [DOI] [PubMed] [Google Scholar]
- Wisniewski J. R.; Dus K.; Mann M. Proteomic workflow for analysis of archival formalin-fixed and paraffin-embedded clinical samples to a depth of 10 000 proteins. Proteomics: Clin. Appl. 2013, 7 (3–4), 225–33. 10.1002/prca.201200046. [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Albrechtsen R.; Kronqvist P.; Cox J.; Mann M.; Geiger T. Proteomic maps of breast cancer subtypes. Nat. Commun. 2016, 7, 10259. 10.1038/ncomms10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N.; Wisniewski J. R.; Geiger T.; Cox J.; Kircher M.; Kelso J.; Paabo S.; Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011, 7, 548. 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncarevic S.; Lossner C.; Kuhn K.; Prinz T.; Pike I.; Zucht H. D. In-depth profiling of the peripheral blood mononuclear cells proteome for clinical blood proteomics. Int. J. Proteomics 2014, 2014, 1–9. 10.1155/2014/129259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleijerveld O. B.; Wijten P.; Cappadona S.; McClellan E. A.; Polat A. N.; Raijmakers R.; Sels J. W.; Colle L.; Grasso S.; van den Toorn H. W.; van Breukelen B.; Stubbs A.; Pasterkamp G.; Heck A. J.; Hoefer I. E.; Scholten A. Deep proteome profiling of circulating granulocytes reveals bactericidal/permeability-increasing protein as a biomarker for severe atherosclerotic coronary stenosis. J. Proteome Res. 2012, 11 (11), 5235–44. 10.1021/pr3004375. [DOI] [PubMed] [Google Scholar]
- Wisniewski J. R.; Mann M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal. Chem. 2012, 84 (6), 2631–7. 10.1021/ac300006b. [DOI] [PubMed] [Google Scholar]
- Hosp F.; Scheltema R. A.; Eberl H. C.; Kulak N. A.; Keilhauer E. C.; Mayr K.; Mann M. A Double-Barrel Liquid Chromatography-Tandem Mass Spectrometry (LC–MS/MS) System to Quantify 96 Interactomes per Day. Mol. Cell. Proteomics 2015, 14 (7), 2030–41. 10.1074/mcp.O115.049460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. G.; Kim S.; Maltby D. A.; Ghassemian M.; Bandeira N.; Komives E. A. Expanding proteome coverage with orthogonal-specificity alpha-lytic proteases. Mol. Cell. Proteomics 2014, 13 (3), 823–35. 10.1074/mcp.M113.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M. L.; Savitski M. M.; Zubarev R. A. Improving protein identification using complementary fragmentation techniques in fourier transform mass spectrometry. Mol. Cell. Proteomics 2005, 4 (6), 835–45. 10.1074/mcp.T400022-MCP200. [DOI] [PubMed] [Google Scholar]
- Frese C. K.; Altelaar A. F.; van den Toorn H.; Nolting D.; Griep-Raming J.; Heck A. J.; Mohammed S. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 2012, 84 (22), 9668–73. 10.1021/ac3025366. [DOI] [PubMed] [Google Scholar]
- Geyer P. E.; Kulak N. A.; Pichler G.; Holdt L. M.; Teupser D.; Mann M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst 2016, 2 (3), 185–95. 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- Geiger T.; Cox J.; Ostasiewicz P.; Wisniewski J. R.; Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 2010, 7 (5), 383–5. 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- Deeb S. J.; D’Souza R. C.; Cox J.; Schmidt-Supprian M.; Mann M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell. Proteomics 2012, 11 (5), 77–89. 10.1074/mcp.M111.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N.; Kulak N. A.; Cox J.; Neuhauser N.; Mayr K.; Hoerning O.; Vorm O.; Mann M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell. Proteomics 2012, 11 (3), M111 013722. 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T.; Velic A.; Macek B.; Lundberg E.; Kampf C.; Nagaraj N.; Uhlen M.; Cox J.; Mann M. Initial quantitative proteomic map of 28 mouse tissues using the SILAC mouse. Mol. Cell. Proteomics 2013, 12 (6), 1709–22. 10.1074/mcp.M112.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov A.; Denisov E.; Lange O.; Horning S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 2006, 17 (7), 977–82. 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Eliuk S.; Makarov A. Evolution of Orbitrap Mass Spectrometry Instrumentation. Annu. Rev. Anal. Chem. 2015, 8, 61–80. 10.1146/annurev-anchem-071114-040325. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Tolic N.; Xie F.; Zhao R.; Purvine S. O.; Schepmoes A. A.; Moore R. J.; Anderson G. A.; Smith R. D. Effectiveness of CID, HCD, and ETD with FT MS/MS for degradomic-peptidomic analysis: comparison of peptide identification methods. J. Proteome Res. 2011, 10 (9), 3929–43. 10.1021/pr200052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C.; Li J.; Shen S.; Sheng Q.; Shyr Y.; Qu J. Performance Investigation of Proteomic Identification by HCD/CID Fragmentations in Combination with High/Low-Resolution Detectors on a Tribrid, High-Field Orbitrap Instrument. PLoS One 2016, 11 (7), e0160160. 10.1371/journal.pone.0160160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrychowski M. P.; Huttlin E. L.; Haas W.; Sowa M. E.; Rad R.; Gygi S. P. Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Mol. Cell. Proteomics 2011, 10 (12), M111 009910. 10.1074/mcp.M111.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese C. K.; Altelaar A. F.; Hennrich M. L.; Nolting D.; Zeller M.; Griep-Raming J.; Heck A. J.; Mohammed S. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 2011, 10 (5), 2377–88. 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- Michalski A.; Damoc E.; Hauschild J. P.; Lange O.; Wieghaus A.; Makarov A.; Nagaraj N.; Cox J.; Mann M.; Horning S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 2011, 10 (9), M111 011015. 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliuk S.; Remes P.; Scheffler K.; Senko M.; Zabrouskov V.. A “Universal” Data-dependent Mass Spectrometry Method That Eliminates Time-Consuming Method Optimization for Achieving Maximal Identifications from Each Sample. Thermo Fisher Scientific 2016, http://www.thermoscientific.com/content/dam/tfs/ATG/CMD/cmd-documents/sci-res/posters/ms/events/ismc2014/PN-49014-Universal-MS-Method-PN49014-EN.pdf. [Google Scholar]
- Fischer R.; Kessler B. M. Gel-aided sample preparation (GASP)--a simplified method for gel-assisted proteomic sample generation from protein extracts and intact cells. Proteomics 2015, 15 (7), 1224–9. 10.1002/pmic.201400436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev R.; Mann M. On the proper use of mass accuracy in proteomics. Mol. Cell. Proteomics 2007, 6 (3), 377–81. 10.1074/mcp.M600380-MCP200. [DOI] [PubMed] [Google Scholar]
- Ma B.; Zhang K.; Hendrie C.; Liang C.; Li M.; Doherty-Kirby A.; Lajoie G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17 (20), 2337–42. 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Xin L.; Shan B.; Chen W.; Xie M.; Yuen D.; Zhang W.; Zhang Z.; Lajoie G. A.; Ma B. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 2012, 11 (4), M111 010587. 10.1074/mcp.M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; He L.; Xin L.; Shan B.; Ma B. PeaksPTM: Mass spectrometry-based identification of peptides with unspecified modifications. J. Proteome Res. 2011, 10 (7), 2930–6. 10.1021/pr200153k. [DOI] [PubMed] [Google Scholar]
- UniProt: a hub for protein information. Nucleic Acids Res. 2015, 43 (D1), D204–12. 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I.; Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 2005, 4 (10), 1419–40. 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Sinitcyn P.; Carlson A.; Hein M. Y.; Geiger T.; Mann M.; Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13 (9), 731–40. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Perkins D. N.; Pappin D. J.; Creasy D. M.; Cottrell J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20 (18), 3551–67. . [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11 (12), 2301–2319. 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Cox J.; Neuhauser N.; Michalski A.; Scheltema R. A.; Olsen J. V.; Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10 (4), 1794–805. 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Eng J. K.; McCormack A. L.; Yates J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5 (11), 976–89. 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Rietschel B.; Arrey T. N.; Meyer B.; Bornemann S.; Schuerken M.; Karas M.; Poetsch A. Elastase digests: new ammunition for shotgun membrane proteomics. Mol. Cell. Proteomics 2009, 8 (5), 1029–43. 10.1074/mcp.M800223-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C.; MacCoss M. J.; Howell K. E.; Yates J. R. 3rd A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol. 2003, 21 (5), 532–8. 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- Petrotchenko E. V.; Serpa J. J.; Hardie D. B.; Berjanskii M.; Suriyamongkol B. P.; Wishart D. S.; Borchers C. H. Use of proteinase K nonspecific digestion for selective and comprehensive identification of interpeptide cross-links: application to prion proteins. Mol. Cell. Proteomics 2012, 11 (7), M111 013524. 10.1074/mcp.M111.013524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov I. V.; Seymour S. L.; Patel A. A.; Loboda A.; Tang W. H.; Keating S. P.; Hunter C. L.; Nuwaysir L. M.; Schaeffer D. A. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 2007, 6 (9), 1638–55. 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Bern M.; Cai Y.; Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal. Chem. 2007, 79 (4), 1393–400. 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- Ma B. Novor: real-time peptide de novo sequencing software. J. Am. Soc. Mass Spectrom. 2015, 26 (11), 1885–94. 10.1007/s13361-015-1204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary G.; Wu S. L.; Shieh P.; Hancock W. S. Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J. Proteome Res. 2003, 2 (1), 59–67. 10.1021/pr025557n. [DOI] [PubMed] [Google Scholar]
- Wisniewski J. R.; Rakus D. Multi-enzyme digestion FASP and the ’Total Protein Approach’-based absolute quantification of the Escherichia coli proteome. J. Proteomics 2014, 109, 322–31. 10.1016/j.jprot.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Reiter L.; Claassen M.; Schrimpf S. P.; Jovanovic M.; Schmidt A.; Buhmann J. M.; Hengartner M. O.; Aebersold R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics 2009, 8 (11), 2405–17. 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski M. M.; Wilhelm M.; Hahne H.; Kuster B.; Bantscheff M. A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Mol. Cell. Proteomics 2015, 14 (9), 2394–404. 10.1074/mcp.M114.046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B.; Busse D.; Li N.; Dittmar G.; Schuchhardt J.; Wolf J.; Chen W.; Selbach M. Global quantification of mammalian gene expression control. Nature 2011, 473 (7347), 337–42. 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.