Abstract
Background
Polybasic helical peptides, such as peptide p5, bind human amyloid extracts and synthetic amyloid fibrils. When radiolabeled, peptide p5 has been shown to specifically bind amyloid in vivo thereby allowing imaging of the disease. Structural requirements for heparin and amyloid binding have been studied using analogues of p5 that modify helicity and chirality.
Methods
Peptide-ligand interactions were studied using CD spectroscopy and solution-phase binding assays with radiolabeled p5 analogues. The interaction of a subset of peptides was further studied by using molecular dynamics simulations.
Results
Disruption of the peptide helical structure reduced peptide binding to heparin and human amyloid extracts. The all-D enantiomer and the β-sheet-structured peptide bound all substrates as well as, or better than, p5. The interaction of helical and β-sheet structured peptides with Aβ fibrils was modeled and shown to involve both ionic and non-ionic interactions.
Conclusions
The α-helical secondary structure of peptide p5 is important for heparin and amyloid binding; however, helicity is not an absolute requirement as evidenced by the superior reactivity of a β-sheet peptide. The differential binding of the peptides with heparin and amyloid fibrils suggests that these molecular interactions are different. The all-D enantiomer of p5 and the β-sheet peptide are candidates for amyloid targeting reagents in vivo.
General Significance
Efficient binding of polybasic peptides with amyloid is dependent on the linearity of charge spacing in the context of an α-helical secondary structure. Peptides with an α-helix or β-sheet propensity and with similar alignment of basic residues is optimal.
Abbreviations: TFE, trifluoroethanol; CD, circular dichroism; SPR, surface plasmon resonance; AL, light chain associated amyloid; ATTR, transthyretin-associated amyloid; AA, inflammation-associated amyloidosis; IAPP, islet amyloid-associated polypeptide
Keywords: Polybasic peptides, Amyloid binding, Heparin, Amino acid spacing, CD, Molecular dynamics
Highlights
-
•
The secondary structure of basic peptides influences binding to amyloid deposits.
-
•
Both helical and sheet-structured peptides effectively bind amyloid.
-
•
Structural complementarity enhances electrostatic peptide interactions with amyloid.
-
•
All d-amino acid peptides bind amyloid similar to the l-enantiomeric form.
1. Introduction
Amyloid is a complex pathology in which proteinaceous fibrils, composed of proteins or peptides with a cross-β-sheet secondary structure, deposit in organs and tissues in association with cell-derived hypersulfated heparan sulfate proteoglycans, serum amyloid P component (SAP) and other accessory molecules [1], [2]. The presence of extracellular fibrils, heparan sulfate proteoglycan and SAP are pathognomonic of amyloid. The deposition of amyloid is associated with a growing number of diseases including Alzheimer's disease, type 2 diabetes and plasma cell dyscrasias, and it is also a consequence of aging [3]. Amyloid accumulates in organs and tissues leading to architectural damage, toxicity and ultimately dysfunction [4]. Fibrils, like heparan sulfate (HS) glycosaminoglycans and heparin, are ionic polymers composed of repeating subunits. Additionally, the HS found associated with amyloid is structurally and electrochemically distinct from that found expressed ubiquitously in healthy tissues – it is hypersulfated akin to heparin [5], [6], [7]. These unique biochemical features, the abundance of polyelectrolytic fibrils and hypersulfated HS, serve as specific biomarkers for pathologic amyloid that can be targeted by using polybasic peptides [8], [9], [10].
The heparin-binding peptide, p5, is a 31-residue synthetic reagent that contains a heptad repeat of amino acids (-KAQKAQA-), and it has been shown to specifically bind inflammation-associated AA amyloid and synthetic amyloid fibrils in vitro [9], [11]. Furthermore, when radiolabeled, p5 can be used to specifically detect AA amyloid deposits in vivo in a murine model demonstrating little or no reactivity with normal tissue [12], [13]. Additionally, peptide p5 binds synthetic amyloid fibrils composed of amyloidogenic immunoglobulin variable domain proteins that, in contrast to tissue amyloid, lack HS proteoglycans [11]. The reactivity of peptide p5 with amyloid fibrils, purified human amyloid extracts, and heparin is dependent upon ionic interactions with negatively charged determinants on the glycosaminoglycans and acidic amino acid sidechains in the fibril polymer [10], [14]. Peptide p5+14, a polybasic reagent based on the structure of p5 but with an increased net charge, offers new promise as an amyloid-imaging agent for detecting systemic amyloidosis in patients [15].
Efficient binding of peptides to heparin, and presumably other ionic polymers such as amyloid fibrils and amyloid-associated HS, is critically dependent upon the presence of basic amino acids, lysine or arginine (and protonated histidine), and their spatial orientation as governed by the secondary structure of the peptide [16], [17]. Synthetic polybasic peptides that can adopt a helical secondary structure, presenting the basic side chains aligned along one face of the peptide, are optimal for binding to heparin [17] and for the specific reactivity with AA tissue amyloid in mice [9]. Indeed, the secondary structure of the peptide and the spacing of the basic amino acids is of more importance than the total number of basic amino acids or the net charge of the peptide [9].
To further probe the structural requirements for efficient amyloid binding by synthetic polybasic peptides and explore novel structural variants, we have generated analogues of peptide p5 that: (i) disrupt the helical secondary structure by using glycine residues that favor a disordered secondary structure or by inserting three proline residues spaced along the peptide; (ii) have a propensity for β-sheet structure by using valine and threonine residues to separate the positively charged amino acids, and; (iii) potentially exhibit greater in vivo stability by using all d-amino acids. Structural analyzes and binding studies were used to compare the binding of these variants with amyloid related substrates and heparin quantitatively with the goal of defining properties that might be manipulated to optimize specific amyloid targeting.
2. Materials and methods
2.1. Peptides and proteins
Peptides p5, p5(D), p5(sheet), p5(coil), and p5(Pro3) (Table 1) were purchased from Keck Laboratories (New Haven, CT) or Anaspec (Fremont, CA) as ~70% pure preparations and further purified by RP-HPLC (Biologic DuoFlow; BioRad, Hercules, CA) by using a Zorbax™ 300SB-C3 solid phase (Agilent, Santa Clara, CA) with a linear gradient of 1–51% acetonitrile in water with 0.05% v/v trifluoroacetic acid as the mobile phase (flow rate of 4 mL/min). Fractions of 1.8 mL-volume were collected, pooled, and the purity and integrity of the peptides verified by mass spectrometry [18]. The Aβ(1−40) peptide and human islet amyloid polypeptide (IAPP) were purchased from Anaspec as >90% pure preparations and were used without further purification.
Table 1.
Peptide primary structure and physical data.
| Peptide | Mw | AA | % Helixa | Primary structure |
|---|---|---|---|---|
| p5 | 3257.7 | 31 | 2.60 | GGGYS KAQKA QAKQA KQAQK AQKAQ AKQAK Q |
| p5(D) | 3257.7 | 31 | nd | [GGGYS KAQKA QAKQA KQAQK AQKAQ AKQAK Q]D |
| p5(sheet) | 2106.6 | 18 | 0.05 | VYKVK TKVKT KVKTK VKT |
| p5(coil) | 2491.7 | 31 | 0.02 | GGGYS KGGKG GGKGG KGGGK GGKGG GKGGK G |
| p5(Pro3) | 3221.7 | 31 | 0.23 | GGGYS KAQKA PAKQA KQPQK AQKAP AKQAK Q |
Helix content predicted of peptide, based on amino acid sequence, using Agadir (http://agadir.crg.es/) with parameters of; pH=7; temperature 298 K, and; an ionic strength of 0.15 [29]. nd, not determined.
Recombinant Vλ6 protein Wil was prepared from a periplasmic extract of transformed E. coli and purified as previously described [19].
2.2. Human AL and ATTR extract preparation
Purified human amyloid tissue extracts were prepared using autopsy-derived tissues from patients with light chain – (AL) or transthyretin-associated (ATTR) amyloidosis using the water flotation method as described by Pras et al. [20] without modification. Purified amyloid material isolated in the water wash, and amyloid rich pellet, was collected and stored lyophilized at RT until used.
2.3. Preparation of synthetic amyloid fibrils
To prepare synthetic amyloid fibrils from rVλ6Wil, a 1 mL-volume containing ~1 mg/mL of monomer in phosphate-buffered saline (PBS), 0.01% w/v NaN3, pH7.5, was filtered through 0.2 µm pore-sized filter, added to a 15 mL conical polypropylene tube (BD BioSciences, Bedford, MA) and shaken at a 45° angle at 225 rpm for 3–5 d at 37 °C until the reaction mixture became opaque [19]. For Aβ(1−40) and IAPP fibrils, filtered 1 mL volumes of 0.2 mg/mL peptide in PBS with 0.01% w/v NaN3, pH7.5, were placed at 37 °C without shaking for 5−10 d. The presence of amyloid fibrils was confirmed by using a thioflavin T (ThT; Sigma-Aldrich, St. Louis, MO) fluorescence emission assay. Briefly, a suspension of fibrils at 50 µg/ mL was prepared and 100 µL (5 µg) of the fibril suspension was added to each of three wells on a 96-well microplate. Thirty µL of PBS was added to each of the wells prior to the addition of 10 µL of 300 µM ThT (Sigma-Aldrich, St. Louis, MO). A set of triplicate wells containing PBS and ThT only was used as a background control. The ThT fluorescence emission (490 nm, excitation at 450 nm) was measured using a fluorescence plate reader (Victor 1420 multilabel counter, Perkin Elmer, Wellesley, MA) and corrected by subtraction of mean background fluorescence. Fibril preparations were aliquoted into single use volumes and stored at −80 °C.
2.4. Preparation of murine liver homogenates
The livers from 2 mice with inflammation-associated (AA) amyloidosis or 2 healthy (WT) mice were harvested at necropsy. Approximately 0.5 g of each AA or WT liver was removed, pooled and a 10-fold volume of PBS added. Serine protease inhibitors, leupeptin and phenylmethylsulfonyl fluoride (PMSF), were added each at 100 µg/mL. The solution was mixed vigorously (3×10 s bursts) with a Polytron at setting 6 (Kinematica Inc., Bohemia, NY) and then centrifuged at 4000×g for 10 min. The supernatant was discarded and the pellet resuspended in a 10-fold volume of PBS with 0.05% tween-20% and 0.05% NaN3. The solution was mixed vigorously again by using the Polytron and the resulting suspension stored at 4 °C until used in pull down assays described below.
2.5. Surface plasmon resonance
The binding of p5-related peptides to rVλ6Wil fibrils was measured using a Biacore X surface plasmon resonance instrument (GE Healthcare, Pittsburgh, PA) and all reagents were obtained from GE Healthcare.
Fibrils were attached to CM-5 chips using the amino-coupling method supplied with the instrument software. Briefly, chips were activated by injection (35 µL) of a mixture of N-ethyl-N′-(diethylaminopropyl) carbodiimide/ N-hydroxysuccinimide (EDC/NHS) at a flow rate of 5 µL/min. Immediately thereafter, 35 µL of rVλ6Wil fibrils freshly sonicated for 10 s with a microprobe and Tekmar sonic disruptor, diluted to 100 µg/mL in pH 4.5 NaOAc buffer, was injected. After the coupling reaction, the remaining active groups on the chip were blocked by injection of 35 µL of 1 M ethanolamine-HCl pH 8.5. Non-fibrillar rVλ6Wil was coupled to the Fc-2 channel and served as a control for non-specific binding to the protein. A sensorgram was initiated on each chip in HBS-EP buffer at 10 µL per min. An initial regeneration step consisting of a 5 µL injection of glycine-buffered 1 M NaCl, pH 1.5, was performed and the baseline allowed to re-equilibrate before beginning data collection. Test peptides, p5, p5(D), p5(sheet), p5(coil), and p5(Pro3) were diluted from stock solutions to 300 nM in HBS-EP buffer (0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20) immediately before injection. The peptides were injected (50 µL) and the data collected for 5 min followed by a 200 s delayed wash cycle. The chip was subjected to a regeneration step before the next test injection. Binding and washout data were extracted from the sensorgram, aligned, and analyzed with the BIAevaluation software (Ver. 3) by fitting to a two-state binding model with conformational change [A+B=AB=AB*] or a 1:1 Langmuir isotherm [A+B=AB]. In the latter, the association rate is fit to R=Req(1-e-(kaC+kd)(t-t0))+RI, where Req=( kaC/kaC + kd) Rmax and further where, ka and kd are the association and dissociation rate constants, respectively; Rmax, maximum analyte binding capacity; C, concentration of analyte (M); t0 , injection start time; RI, bulk refractive index contribution. The Langmuir dissociation model equation being R=R0e-kd(t-t0)+Offset, where, R0, is the response at the start of the fitted data; and Offset, is the response at infinite time. .
2.6. Circular dichroism (CD) spectroscopy
Circular dichroism spectra of peptides p5, p5(D), p5(sheet), p5(coil), and p5(Pro3) (0.05 mg/mL in PBS) were acquired using a dual detector DSM 1000 CD instrument (Olis Inc., Bogart, Georgia) with a peptide sample volume of 2.9 mL and a 1 cm cuvette path length. Data were collected in triplicate over the 190 nm −250 nm wavelength range with 1 nm increments. Secondary structure transitions were induced by addition of 2,2,2 trifluoroethanol (TFE) up to 40% by volume or porcine low molecular weight heparin (Enoxaparin sodium – Sanofi, Bridgewater, NJ) up to 1.5 mg/mL (w/v) final concentration. Spectra were corrected for background by subtraction of appropriate buffer control spectrum. Mean residue ellipticity [θ] was calculated according to, [θ]=θ*(MW/no. AA)/(10*conc.*l), where: θ is ellipticity (millidegrees); MW is the molecular weight of the peptide; no. AA, is the number of amino acid residues; conc. is the peptide concentration (mg/mL), and; l, is the cuvette path length (cm).
2.7. Peptide radiolabeling for pulldown assays
In preparation for in vitro pull-down assays, ~50 µg each of peptides p5, p5(D), p5(sheet), p5(coil) and p5(Pro3) was radioiodinated with ~1 mCi of iodine-125 (125I – Perkin Elmer, Waltham, MA) using 20 µg chloramine T as the oxidizing agent [21]. After quenching the reaction with 20 µg sodium metabisulfite, the radiolabeled peptides were diluted into 0.1% sterile gelatin in PBS and free radioiodine removed by size exclusion chromatography on a 5-mL Sephadex G-25 (PD10; GE Healthcare) solid phase, equilibrated with 0.1% gelatin/PBS. Fractions of 200 µL-volume were collected, and those containing the maximal radioactivity (indicative of 125I-labeled peptide) were pooled and the product's radiochemical purity was established by SDS polyacrylamide gel electrophoresis (PAGE) analyzed by phosphor imaging (Cyclone Storage Phosphor System, Perkin Elmer, Shelton, CT).
2.8. Peptide-substrate binding studies – Pulldown assay
Fifty µg of ALκ, ALλ or ATTR amyloid extract, or 25 µg of synthetic fibrils composed of rVλ6Wil Aβ(1−40) or IAPP, suspended in 200 µL of PBS with 0.05% tween-20 (PBST) were centrifuged in a 0.5 mL microfuge tube at 21,000×g for 10 min. The supernatant was discarded and pellet resuspended in 200 µL of either PBS or phosphate-buffered 1.5 M NaCl, both supplemented with 0.05% tween-20. Ten microliters of a 1:100 dilution of 125I-labeled peptide (~100,000 counts per minute [CPM]; ~5 ng peptide) stock was added to the suspension of fibrils or amyloid extract. The sample was mixed by rotation at RT for 1 h. Samples were then centrifuged twice at 15,000×g for 10 min. Supernatants and pellets were separated after each step and the radioactivity in each was measured using a Cobra II gamma counter (Perkin Elmer) with a 1 min acquisition. The percentage of 125I-labeled peptide bound to pellet was determined according to: Bound peptide=[Pellet CPM/(Pellet CPM+Supernatant CPM)]×100.
For binding to liver homogenates, 25 µL (~5 mg) of tissue was centrifuged at 21,000×g for 5 min. The supernatant was discarded, and the pellet resuspended in 200 µL PBST and the assay performed as described above.
2.9. Competition pulldown assays
Competition pulldown assays were performed, as described above using rVλ6Wil fibrils as the substrate. In each case, 125I-labeled peptide (p5, p5(D), p5(sheet), p5(coil), and p5(Pro3)) was mixed with a 1000-fold molar excess of unlabeled p5 and added to the fibrils. Bound peptide was measured as described above. The assays were performed in triplicate and the data presented as the mean of the % maximum value (bound peptide in the absence of competitor) ±95% confidence intervals.
2.10. Computer modeling of peptide binding to amyloid fibrils
To obtain initial structures for molecular dynamics (MD) simulations, the ZDOCK server (http://zdock.umassmed.edu/) was used to generate 100 theoretical models of the interaction of peptides p5, p5(sheet) and p5(coil) with amyloid fibrils composed of Aβ(17-42) (PDB # 2BEG) [22]. ZDOCK searched all possible binding modes in the translational and rotational space between the peptide and fibril and evaluated each pose using an energy-based scoring function. The lowest energy peptide and Aβ(17-42) fibril complex was first solvated in a TIP3P water box and then subjected to 10 ns of molecular dynamics (MD) simulation using NAMD 2.9 [23] with the CHARMM C36 force field [24]. In the simulation, the van der Waals interaction was smoothly turned off between 8.5 and 10 Å using a switching function. Long-range electrostatic interactions were treated using the Particle-Mesh Ewald (PME) method [25] with a 1.0 Å grid spacing. The time step for integration was 1 fs. Langevin dynamics was used to maintain a constant temperature at 310 K, while the Nosé-Hoover Langevin-piston algorithm was used to maintain a constant pressure at 1 bar.
The molecular mechanics-Poisson Boltzmann/surface area (MM-PB/SA) method [26], [27] was used to compute the binding free energies of the peptide with the Aβ(17−42) fibril. The total binding energy ΔGbind (app) was defined as ΔGbind (app)=Gcomplex−Greceptor−Gligand. Each free energy term consisted of the gas phase molecular mechanics energy (ΔEgas), the solvation free energy (ΔGsol), and the vibrational entropy contributions (TΔS). ΔGsol was estimated from the Poisson-Boltzmann (PB) theory and solvent accessible surface area (SASA) calculations which yielded ΔGpolar and ΔGnonpolar. In the PB, energies were evaluated at 0.1 M NaCl concentrations. A surface tension coefficient (γ) of 0.0072 kcal/(mol Å2) was used to calculate the nonpolar solvation free energy contribution. Due to its prohibitive computational cost and the inherent difficulty in determining accurate absolute entropy for large protein-peptide complex systems, the vibrational entropy contribution was not included in our calculation. Egas and ΔGsol were computed for 1000 snapshots extracted evenly from the last 2 ns of the MD trajectory.
2.11. Statistical methods
Binding data for fibrils, extracts and tissue homogenates (Fig. 5) were compared between each respective peptide and peptide p5 using one-way ANOVA. A Bonferroni correction was employed to account for experiment-wise error rates associated with testing multiple hypotheses concurrently. Statistical significance was assumed at a corrected alpha (α) value of 0.001. In Fig. 6, the 95% confidence intervals were calculated for the ratio values (proportions) using the QuickCalcs Graphpad software. Analyses were performed using Prism ver. 6.07 (Graphpad Inc., La Jolla, CA).
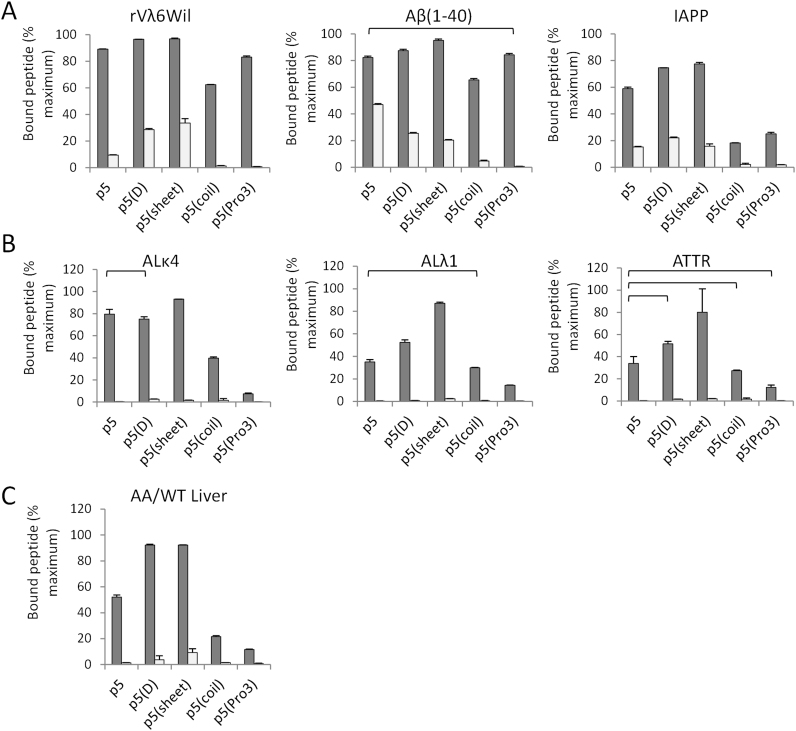
Fig. 5.
Analysis of 125I-labeled peptide binding to amyloid-related substrates using a pulldown assay. Binding of 125I-labeled peptides to synthetic fibrils (A), human AL and ATTR amyloid extracts (B), or liver homogenates from WT and amyloidotic mice (C) was measured in PBS (dark bars) and 1 M NaCl (white bars). Statistical analysis of the binding data in PBS was performed using a Bonferroni multiple comparison ANOVA (p<0.01). Brackets indicate which comparisons were NOT significantly different from the p5 value. All other comparisons with p5 were deemed significant.
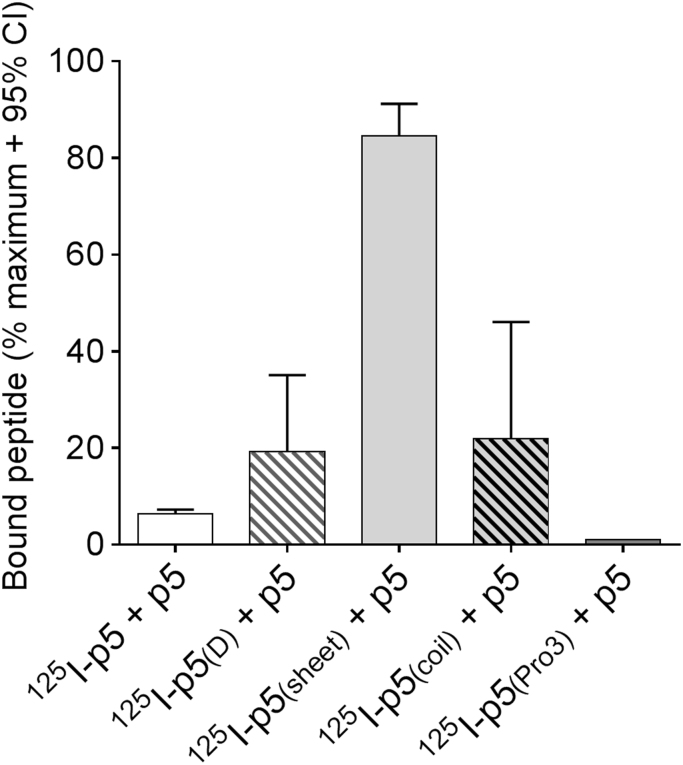
Fig. 6.
Binding of 125I-labeled peptide to rVλ6Wil fibrils in the presence of excess p5 using a completion pulldown assay. Radiolabeled peptides (~5 ng) were incubated with 25 µg of synthetic rVλ6Wil fibrils in the presence of 1000 M excess of peptide p5. The mean % bound peptide (relative to peptide in the absence of competitor; n=2 replicates) with 95% confidence interval is plotted.
3. Results
3.1. Peptide structure prediction
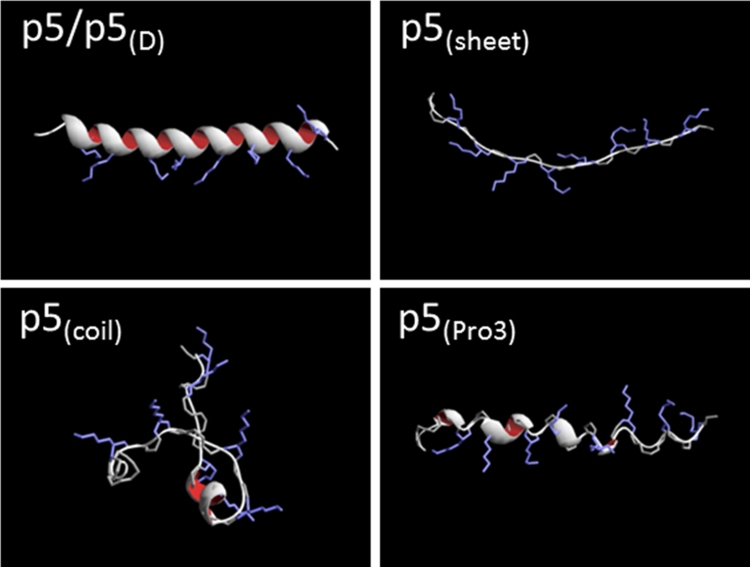
The helical content of peptide p5 and modified derivatives, p5(sheet), p5(coil), and p5(Pro3) in PBS was predicted using the helix/coil transition theory for monomeric peptides, in the Agadir algorithm (Table 1; [28], [29]). In the absence of ligand, the helicity of all peptides was predicted to be low with peptide p5 having ~2.6% helix and the other peptides <0.3% helix. Secondary structure models for each peptide were generated using the iTASSER software package (Fig. 1; [30], [31]). Peptide p5 was predicted to adopt a helical configuration (Fig. 1). Although iTASSER cannot accurately predict the structure of d-amino acid peptides, given that peptide p5(D) has the same amino acid sequence but synthesized with d-amino acids, we posited that it would adopt a similar helix with but with dextro-chirality. In contrast to p5, p5(sheet) was found to adopt an extended sheet motif; whereas, p5(coil) appeared unstructured and p5(Pro3) favored a helical motif but the introduction of prolines residues hindered extensive coil formation (Fig. 1).
Fig. 1.
Predicted secondary structure of peptides used in this study. Peptide p5 was predicted to be α-helical whereas p5(sheet) was an extended sheet. Peptide p5(coil) and p5(Pro3) were random coils or disrupted helices. Structures were generated using the online prediction program, I-TASSER, and rendered using DeepView/Swiss-PDBViewer v4.0.4 (Swiss Institute of Bioinformatics).
3.2. Surface plasmon resonance
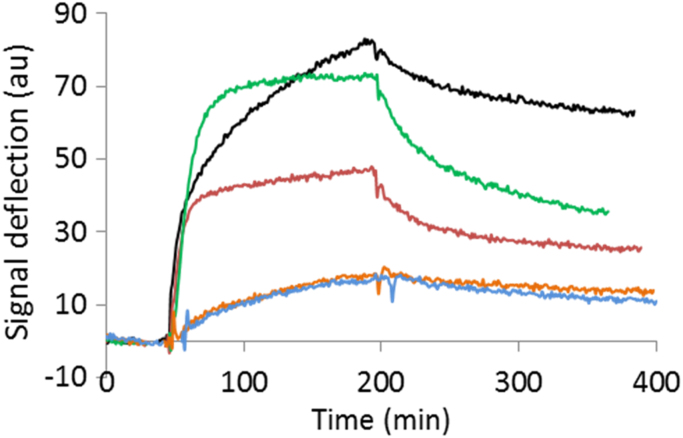
To compare the binding kinetics of each peptide with amyloid fibrils, a natural ligand of peptide p5, we used surface plasmon resonance with synthetic AL fibrils composed of rVλ6Wil protein as the substrate (Fig. 2). The data were fit to a simple Langmuir 1:1 (Table 2) and a more complex, two-state binding (Table 3) equation. Peptides p5 and p5(sheet) bound equivalently with similarly high response unit values (RU); although, p5 was predicted to be ~10% higher than p5(sheet) based on both fitting models. The estimated KD values for p5 and p5(sheet) at 4–15 nM and 10–25 nM, respectively, depending on the fitting model, were similar. Additionally, peptide p5(D) had a predicted KD for rVλ6Wil fibrils of 5–22 nM, but the estimated maximum binding to the fibrils (RUmax) was ~50% less than that of p5 or p5(sheet). Peptides p5(coil) and p5(pro3) had the lowest binding values, ~75% lower RUmax as compared to p5 (Table 2). The KD for these two peptides was the lowest, estimated to be 20–70 nM. The extent of binding of each peptide to rVλ6Wil fibrils, in terms of RU, indicated that:
Fig. 2.
The binding of p5 and p5 peptide analogues to synthetic AL-related light chain fibrils by using surface plasmon resonance. Peptide binding to rVλ6Wil fibrils was monitored using 300 nM of analyte in HBS-EP buffer, pH 7.4. Peptides p5 (black), p5(sheet) (green) and p5(D) (red) bound the fibrils in greater amounts than p5(coil) (orange) and p5(Pro3) (blue). Sensorgrams were analyzed using both a simple Langmuir model and a two-state binding model.
Table 2.
Kinetic SPR parameters for the binding of 300 nM peptide to synthetic rVλ6Wil fibrils using a Langmuir isotherm.
| Peptide | ka (1/Ms) | kd (1/s) | Rmax (RU) | KD (M) | Chi2 |
|---|---|---|---|---|---|
| p5 | 1.2E+05 | 4.4E−04 | 71.5 | 3.7E−09 | 11.3 |
| p5(D) | 3.6E+05 | 1.9E−03 | 36.5 | 5.4E−09 | 1.36 |
| p5(sheet) | 3.4E+05 | 3.4E−03 | 63.0 | 1.0E−08 | 1.32 |
| p5(coil) | 7.7E+04 | 1.4E−03 | 18.9 | 1.8E−08 | 0.61 |
| p5(Pro3) | 8.3E+04 | 2.1E−03 | 17.6 | 2.5E−08 | 0.29 |
Table 3.
Kinetic SPR parameters for the binding of 300 nM peptide to synthetic rVλ6Wil fibrils using a two-state binding model.
| Peptide | ka1 (1/Ms) | kd1 (1/s) | ka2 (1/s) | kd2 (1/s) | Rmax (RU) | KD (M) | Chi2 |
|---|---|---|---|---|---|---|---|
| p5 | 4.8E+05 | 0.203 | 0.0315 | 1.1E−03 | 97.6 | 1.5E−08 | 0.57 |
| p5(D) | 5.5E+05 | 0.052 | 7.4E−03 | 1.7E−03 | 63.4 | 2.2E−08 | 0.27 |
| p5(sheet) | 4.2E+05 | 0.026 | 7.0E−03 | 2.8E−03 | 89.7 | 2.5E−08 | 0.34 |
| p5(coil) | 7.5E+04 | 0.001 | 7.9E−06 | 1.5E−05 | 19.0 | 3.4E−08 | 0.61 |
| p5(Pro3) | 1.2E+05 | 0.045 | 0.0122 | 2.4E−03 | 31.5 | 7.1E−08 | 0.23 |
Both fitting equations reported herein yielded low χ2 values as an indication of a good fit to the model (Table 2, Table 3). However, the more complex two-state model had consistently lower χ2 values. This finding indicated that a conformational change in the peptide may be occurring upon binding to the fibril ligand.
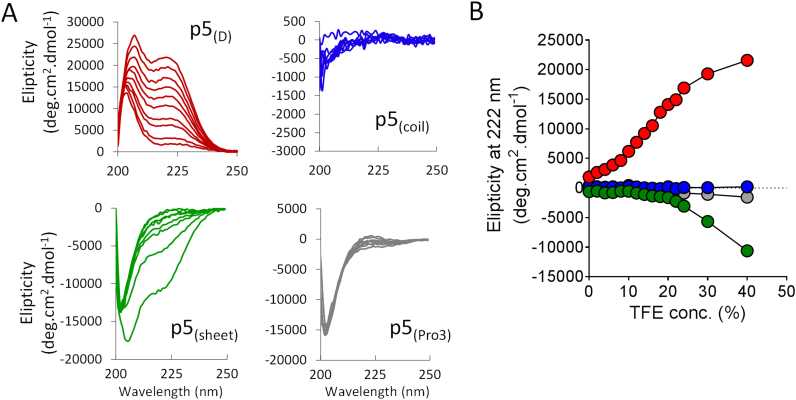
3.3. CD analysis of peptides in TFE
To study the secondary structure of the peptides and structural transitions that may occur, we initially performed CD spectroscopy in physiological saline and following addition of increasing concentrations of TFE, which induces secondary structure in peptides (Fig. 3). The structure of peptide p5(D) in PBS, pH 7.5 was characterized by a maxima at ~200 nm and a week maxima at 222 nm (Fig. 3A). Similar, albeit l-amino acid spectra, were observed for p5(sheet), p5(coil) and p5(Pro3), with minima at ~200 nm and 222 nm (Fig. 3A). Upon addition of TFE to a final concentration of 40%, the spectra of p5(D) and p5(sheet) with a shift in the 200 nm maxima/minima toward 207 nm and a pronounced relative increase in the intensity at 222 nm; however, the p5(coil) and p5(Pro3) were unaltered (Fig. 3A). Addition of TFE induced a helical secondary structure in peptide p5(D) with a transition midpoint of ~16% TFE (Fig. 3B, red). In contrast, the structural transition of p5(sheet) required greater concentrations of TFE, as evidenced by the shift at 222 nm, which did not occur until 20% TFE and was not complete at 40% TFE (Fig. 3B, green).
Fig. 3.
Structural analysis of peptides, by circular dichroism, in response to increasing concentrations of TFE. A) CD spectra of peptides p5(D), p5(sheet), p5(coil), and p5(Pro3) in the presence of increasing concentrations of TFE up to 40% v/v. B) TFE-induced structural transitions of p5(D) (red) and p5(sheet) green are evidenced by changes in the molar elipticity at 222 nm for each peptide. Peptides p5(Pro3) (gray) and p5(coil) (blue) were not structured in the presence of up to 4040% TFE.
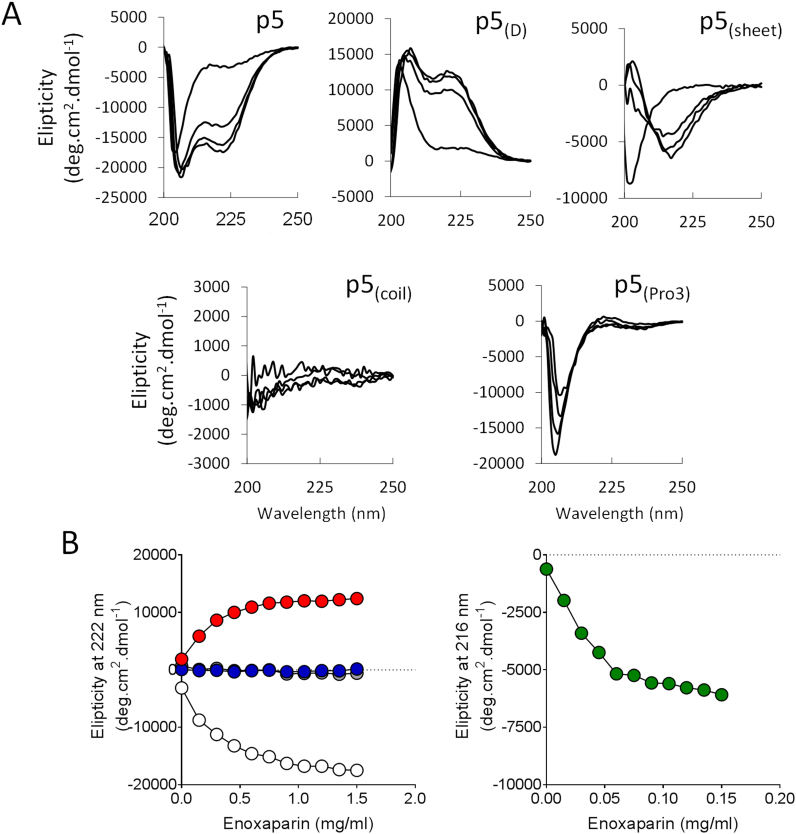
3.4. CD analysis of peptides in the presence of enoxaparin
To study further the structural transitions of the peptides upon interaction with a ligand, we used a second natural substrate, low molecular weight heparin (enoxaparin), in the CD analysis (Fig. 4). The effect of increasing concentrations of enoxaparin on peptides p5 and p5(D) was a transition to a helical motif with double minima observed at 207 nm and 222 nm for p5 and chirally-equivalent maxima for p5(D) (Fig. 4A). The midpoint for the transition was estimated to be ~0.5 mg/mL (~100 µM) enoxaparin (Fig. 4B). In contrast, enoxaparin was a more potent agent for p5(sheet), which adopted a sheet conformation with a maxima at 202 nm and a minima at 216 nm upon addition of only 0.15 mg/mL of enoxaparin (Fig. 4B, green). The midpoint of the structural transition, based on the 216 nm shift in CD spectra, was estimated to be ~0.03 mg/mL (~7 µM) enoxaparin (Fig. 4B, green). Enoxaparin addition to peptides p5(coil) or p5(Pro3) had no effect upon the secondary structure (Fig. 4). The propensity for heparin binding, based upon the midpoint of the structural transition varied for the peptide series according to:
Fig. 4.
Structural analysis of peptides in response to increasing concentrations of low molecular weight heparin (Enoxaparin). (A) Structural changes evidenced in circular dichroism spectra of peptides in the presence of increasing concentrations of enoxaparin up to 1.5 mg/mL. For peptide p5(sheet), enoxaparin was added up to 0.15 mg/mL. (B) Helicity was induced in peptides p5 (white) and p5(D) (red). However, peptides p5(Pro3) (gray) and p5(coil) (blue) were less structured and peptide p5(sheet)(green) adopted a β-sheet configuration, as evidenced by changes in molar elipticity at 222 nm and 216 nm, respectively.
3.5. Binding of peptide to amyloid fibrils, extracts and tissue homogenates
We next studied the interaction of the peptides, when radioiodinated, with synthetic amyloid fibrils or human amyloid extracts either in PBS or 1 M NaCl, as well as murine, amyloid-laden liver homogenates in PBS, using a pulldown assay (Fig. 5, Table 4). In the pulldown assay, 125I-labeled peptides all bound rVλ6Wil, Aβ(1-40) and IAPP synthetic fibrils in PBS (Fig. 5A, dark bars); however, p5(coil) and p5(Pro3) were generally less effective as compare to peptide p5, especially when IAPP fibrils were used as the substrate. Binding to synthetic fibrils was greatly, but variably, diminished in the presence of 1 M NaCl (Fig. 5A, light bars). Peptides p5(D) and p5(sheet) bound all three fibril types in a similar manner to peptide p5 (Fig. 5A). The ratio of binding for each peptide in PBS to that in 1 M NaCl was calculated as a measure of the relative affinity of the peptide for the substrate, assuming the reaction was dominated by electrostatic interactions (Table 4). The calculated binding ratios for 125I-p5, 125I-p5(D) and 125I-p5(sheet) were greater than 125I-p5(coil) and 125I-p5(Pro3) for all amyloid fibrils evaluated (Table 4).
Table 4.
Relative efficiency of peptide binding to synthetic amyloid fibrils and human amyloid extracts based on the ratio of 1 M/0.15 M pulldown data.
| Substrate | 125I-p5 | 125I-p5(D) | 125I-p5(sheet) | 125I-p5(coil) | 125I-p5(Pro3) |
|---|---|---|---|---|---|
| rVλ6Wil fibril | 0.11 | 0.30 | 0.35 | 0.02 | 0.01 |
| Aβ(1–40) fibril | 0.57 | 0.29 | 0.21 | 0.07 | 0.01 |
| IAPP fibril | 0.26 | 0.30 | 0.20 | 0.12 | 0.07 |
| ALκ4 extract | 0.004 | 0.035 | 0.017 | 0.040 | 0.030 |
| ALλ1 extract | 0.013 | 0.015 | 0.028 | 0.021 | 0.022 |
| ATTR extract | 0.008 | 0.032 | 0.026 | 0.059 | 0.018 |
Binding to ALκ, ALλ and ATTR human amyloid extracts was decreased for all peptides relative to the synthetic fibrils (Fig. 5B). Notably, 125I-p5(sheet) exhibited the greatest binding to all substrates (97%, 87%, and 80% bound) with 125I-p5(coil) and 125I-p5(Pro3) having the lowest extract binding (7%, 14%, and 12% for 125I-p5(Pro3)). Binding of all peptides to amyloid extracts in a milieu of 1 M NaCl was significantly reduced to <3% (Fig. 5B).
The peptide interactions with AA amyloid-laden (dark bars) and WT (light bars) murine liver tissue homogenates were obtained from an experimental model of systemic spontaneous amyloidosis. The data indicated that peptides 125I-p5(D) (92%) and 125I-p5(sheet) (92%) bound the amyloid in greater amounts as compared to peptide 125I-p5 (52%; Fig. 5C). The reactivity of 125I-p5(coil) and 125I-p5(Pro3) was much lower, 21% and 11%, respectively, relative to the other peptides (Fig. 5C). The binding to amyloid-free, WT liver, was significantly lower than the AA-liver homogenate at 9%, the maximal value, observed for 125I-p5(sheet) (Fig. 5C).
3.6. Competition binding assays
To further interrogate the binding of the peptides with amyloid fibrils, we examined whether the binding of radioiodinated peptides p5(D), p5(sheet), p5(coil) and p5(Pro3) could be inhibited by an excess of peptide p5 (Fig. 6). In the homologous reaction, a 1000-fold molar excess of p5 inhibited the binding of ~5 ng of 125I-p5 to 25 µg of rVλ6Wil fibrils by ~95% in the pulldown assay (Fig. 6). Inhibition of 125I-labeled p5(D), p5(coil) and p5(Pro3) binding to rVλ6Wil fibrils was similarly reduced by 80%, 80% and 99%, respectively, in the presence of peptide p5. In contrast, a 1000-fold molar excess of p5 reduced the binding of 125I-p5(sheet) to fibrils by only15%.
3.7. Molecular dynamics simulations
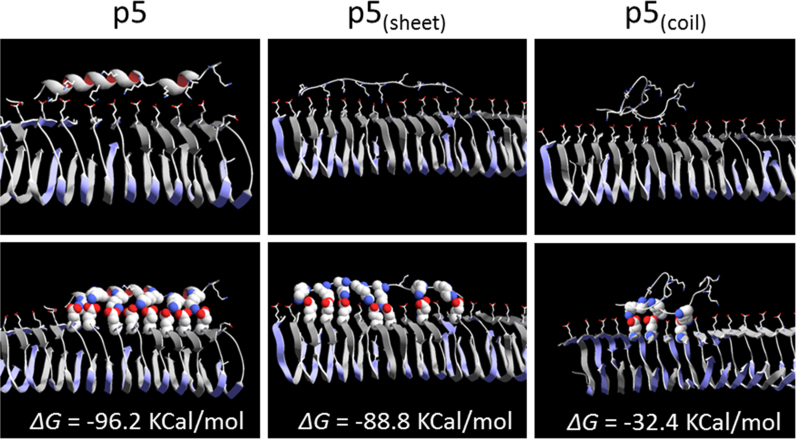
To provide further insight into the binding of p5(sheet) and p5(coil) to fibrils, the interactions with Aβ fibrils were modeled using molecular dynamics simulations and compared to that of p5 (Fig. 7; Table 5). The computer modeling and simulations revealed that peptide p5 and p5(sheet) bound to the longitudinal face of the fibril and that the interaction was dominated by multivalent electrostatic interactions between the lysine side chains and, in this case, Glu21 in the Aβ peptide (Fig. 7). The molecular mechanics combined with the Poisson–Boltzmann and surface area (MM/PBSA) method is commonly used to estimate the binding free energy of two molecules. However, it contains several crude approximations, for example, the lack of conformational entropy and contribution from water molecules in the binding site. Therefore, the MM/PBSA method is only semi-quantitative. Moreover, due to the enthalpy-entropy compensation, the MM/PBSA results (only the enthalpy part) can deviate significantly from the true binding free energies, so they are not accurate for calculating absolute Kd values. Instead, the MM/PBSA approach is very useful in predicting the trend of relative binding in a series of compounds. For these reasons, we report our values as “apparent” ΔG. Although the interaction was dominated by an apparent ΔGPB,elec (~50 kcal/mol, for both peptides), van der Waals interactions (~33 kcal/mol) were also significant (Table 5). The apparent binding free energy for p5 and p5(sheet), −96 and −89k kcal/mol, respectively, was greater than that for the binding of p5(coil) to the fibrils (−32 kcal/mol). The modeling data for p5(coil) indicated that only three salt bridges were formed between the fibril and this peptide (Fig. 7).
Fig. 7.
Molecular dynamics simulation of peptide p5, p5(sheet) and p5(coil) binding to Aβ(17-42) amyloid-fibrils. Images of peptide p5, p5(sheet) and p5(coil) to the Aβ(17-42) fibril (PDB # 2BEG) were taken from molecular dynamics trajectories equilibrated after more than 100 ns each. The upper panels show stick rendering of the interaction. Lower panels show images with space-filled interacting side chains on the surface of the fibril. Images were rendered using DeepView/Swiss-PDBViewer v4.0.4. The estimated free energy of binding (ΔG) is shown for each modeled interaction.
Table 5.
Thermodynamic parameters estimated from molecular dynamic simulations for peptide p5, p5(sheet) and p5(coil) binding to Aβ(17-42) fibrils computed using the MM-PB/SA method.
| ΔEVDWa | ΔGPB, elec | ΔGnonpolar | ΔGbind (app) | |
|---|---|---|---|---|
| p5 | −35.38±6.19 | −53.50±5.18 | −7.31±0.40 | −96.20 |
| p5(sheet) | −31.26±9.67 | −49.10±8.55 | −8.41±0.72 | −88.77 |
| p5(coil) | −26.87±7.13 | −0.72±3.94 | −4.78±0.63 | −32.37 |
Where: ΔEVDW is the van der Waals contribution; ΔGnonpolar is the nonpolar component of the solvation free energy estimated by surface area, and; ΔGPB, elec is the electrostatic component of the apparent total binding energy, ΔGbind (app).
4. Discussion
4.1. Heparin-reactive peptides as specific amyloid targeting agents
Amyloid diseases are complex and heterogeneous. There are no non-invasive methods to detect and quantify these deposits in the USA. We have identified synthetic peptides capable of binding two of the major components of all tissue amyloid deposits, namely the proteinaceous amyloid fibrils and the associated hypersulfated heparan sulfate proteoglycans [9], [11], [15] for use for imaging and therapy. Both constituents are ionic polymers composed of repeating elemental units - amyloidogenic precursor proteins, and acidic disaccharides, respectively. The prototypic peptide initially identified as exhibiting specific amyloid binding was designated p5, a synthetic, polybasic peptide initially develop in silico as a heparin binding reagent [32]. To further probe the structural properties required for amyloid reactivity of polybasic peptides, we generated an all-D enantiomer of p5, a peptide with a propensity for β-sheet formation and, finally, analogues with disrupted helical secondary structure. Each of these structural changes will cause subsequent changes in the distribution of cationic amino acid sidechains. We hypothesized that these data will guide novel amyloid-reactive peptide design with enhanced targeting capabilities.
4.2. Binding of p5 analogues with disrupted helices – the importance of ligand charge
Heparin binds basic peptides and induces a secondary structure in the latter that can be readily detected by using CD spectroscopy [16], [32], [33]. The binding of peptides to heparin is strongly influenced by the propensity to adopt an α-helical structure creating a favorable spatial orientation of the basic amino acids in the peptide, generally a linear array [17]. We have previously demonstrated that the specific binding of heparin-reactive peptides to amyloid in vivo is similarly governed by helicity and charge spacing. Peptide p5 binds heparin with a KD of 0.64 µM [17], and reacts with AA amyloid-laden tissue with an estimated high affinity binding site with a KD of ~0.5 µM [10].
Substituting glycine (p5(coil)) or proline amino acids (p5(Pro3)) into the p5 peptide sequence resulted in a decrease in helical propensity as predicted by both the Agadir (Table 1) algorithm and iTASSER methods (Fig. 1). The former uses helix/coil transition theory, and the latter generates structures based on sequence alignment and threading using homologous sequences found in the x-ray crystal structure database. In contrast to p5, these substitutions rendered peptides p5(coil) and p5(Pro3) incapable of adopting a helical motif in PBS in the presence of TFE or following addition of the ligand, enoxaparin. However, both p5(coil) and p5(Pro3) remained capable of binding synthetic amyloid fibrils composed of rVλ6Wil (AL amyloid) or Aβ(1-40) (Aβ amyloid). Indeed, there was no significant difference in the binding of p5 and p5(Pro3) to Aβ(1-40) fibrils (Fig., 5A). This dichotomy supports our hypothesis that p5-like basic peptides bind to sulfated glycosaminoglycans and amyloid fibrils in two distinct ways.
This may be due to differences in the nature of the substrates, heparin or HS, and synthetic amyloid. For example, synthetic amyloid fibrils composed of rVλ6Wil and Aβ(1-40) are not independent linear arrays. Rather, synthetic fibrils are complex aggregated matrices, i.e. a lattice, with surface charge density that is dependent on the orientation of the proteins comprising the fibrils and the amino acid side chains exposed on the fibril surface. In some regards, the surface presented by amyloid fibrils used in this study may be approximated by a rigid 2-dimensional charged surface array or a synthetic phospholipid membrane [34]. The planar charge distribution on the amyloid surface may allow non-helical, protean peptides such as p5(coil) and p5(Pro3) to bind synthetic rVλ6Wil and Aβ(1-40) fibrils, possibly by interacting with charges of multiple fibrils. In contrast, the inability of p5(coil) and p5(Pro3) to adopt a helical structure significantly hinders the binding to the presumably monodisperse, linear, enoxaparin. Interestingly, peptides p5(coil) and p5(sheet) did not bind well to IAPP fibrils (Fig. 5), as compared to p5. The IAPP peptide differs from rVλ6Wil and Aβ(1-40) sequence because it has no acidic amino acids – only an unprotected carboxylate at the C-terminal providing negative charge for interaction with the peptides. Therefore, the IAPP fibril “lattice” will have a much reduced surface charge density, relative to the other fibrils, which may not support efficient binding of the two helix-disrupted peptides. These data demonstrate the importance of the surface charge density presented by the amyloid fibrils on peptide binding.
4.3. Binding of peptides to amyloid is dominated by electrostatic interactions
The binding of all p5-related peptides to synthetic amyloid fibrils and amyloid extracts was decreased by the addition of 1 M NaCl indicative of an electrostatically-driven interaction (Fig. 5). Human amyloid extracts differ from synthetic fibrils due to the presence of accessory molecules. The presence of HS proteoglycans can serve as an additional target for the peptides. However, constituents such as apolipoproteins and serum amyloid P component may block potential binding sites and hinder peptide binding. Relative to synthetic fibrils, the interaction of all the peptides was markedly reduced when human AL or ATTR extracts were used as a substrate, likely due to this enhanced complexity. The notable exception was p5(sheet), which bound the amyloid extracts as efficiently as synthetic fibrils. The binding of all peptides with amyloid extract was completely inhibited by 1 M NaCl. This contrasts starkly with the effect of NaCl on the binding with pure synthetic fibrils, which was not completely inhibited. This suggests that peptide amyloid interactions are completely dominated by electrostatic interactions and may preferentially involve the HS moieties. In contrast, amyloid fibril-peptide interactions likely involve both electrostatic and as well as non-ionic interactions. This is supported by molecular dynamics simulations which the apparent values calculated indicated that ΔEVDW and ΔGnonpolar account for ~50% of the estimated binding energy (ΔGbind) of the p5 and p5(sheet) interaction with Aβ(17-42) fibrils and essentially all the energy for the weak association of p5(coil) with the synthetic fibril model (Table 5). Thus, we conclude that, whereas the binding of peptides with heparin are dominated by electrostatic interactions [14], [17], [33], [35], reactivity with synthetic amyloid fibrils also involves a significant non-electrostatic component.
4.4. d-amino acid p5 binds amyloid substrate effectively in vitro
Incorporation of d-amino acids into synthetic peptides of biomedical interest prevents proteolysis in vivo, enhances circulating half-life and prevents deiodination of tyrosine amino acids by dehalogenases in the liver and kidneys [36], [37], [38]. Therefore, an all-D enantiomer of peptide p5 (p5(D)) may yield an enhanced amyloid imaging agent. Peptide p5(D) bound heparin with induction of an α-helix and, like p5, reacted with synthetic fibrils and amyloid extracts with similar, or greater efficacy, with the exception of Aβ(1-40) fibrils (Table 4). The binding site of p5(D) on rVλ6Wil fibrils is predicted to be the same as for p5, based on competition studies, which showed that p5 competed equally for binding of 125I-p5 or 125I-p5(D). Therefore, the all-D enantiomer of p5, when radioiodinated, may image amyloid in vivo as well as p5 but with enhanced stability as compared to p5.
4.5. The β-sheet peptide configuration binds amyloid effectively in vitro
The binding of p5 with heparin involves a linear alignment of lysine sidechains along a helical structure that favors multivalent interaction with the sulfate moieties presented by the heparin structure. To further assess the requirement for helicity in amyloid binding peptides, we designed p5(sheet) using valine and threonine residues as spacers between the lysine residues (Table 1). This sequence should have a propensity to adopt β-sheet [39]. The p5(sheet) peptide bound heparin more efficiently than peptide p5. Additionally, p5(sheet) bound all amyloid-related substrates significantly better than p5. When peptides adopt a helical secondary structure with a –KAKAKA-repeat, the binding to heparin yields a KD of 62.5 µM (Wang, BBA 2009), which is 1000-fold weaker than the heptad motif in peptide p5. In contrast, our data suggest that the p5(sheet) peptide with a –KVKTK- repeat motif binds heparin with a 10-fold greater affinity than p5 (see Fig. 4). These data suggest that the sheet conformation, with alternating lysine residues, results in a linear array of charged sidechains that can effectively interact with linear heparin and HS moieties as well as the charged planar array presented by synthetic fibrils. Molecular dynamic simulations of the interaction of p5(sheet) with a prototype Aβ(17-40) amyloid fibril structure demonstrated multivalent electrostatic interactions between the lysine side chains of the peptide and the aspartate side chains that were spaced regularly along the longitudinal axis of the fibril, due to the repeating Aβ elements (Fig. 7). Indeed, despite the difference in charged amino acid spacing found in p5 and p5(sheet), both peptides were predicted to interact with juxtaposing aspartate moieties, based on the simulation. The modeling data are somewhat counter-intuitive since we have demonstrated in vitro that peptide p5(sheet) likely occupies sites on amyloid fibrils that are distinct from the p5 binding site, as its reactivity was not effectively inhibited by an excess of p5 (Fig. 6). By way of reconciling the modeling and binding studies, it seems likely that p5(sheet) may have a higher affinity for the fibrils and, therefore, competes well for the binding of p5. Alternatively, in the presence of an excess of bound p5, the p5(sheet) may bind at alternative sites on the fibrils, something that cannot be accessed by simulations. Given the β-sheet propensity of p5(sheet), this peptide may be able to intercalate into the amyloid fibrils, which are composed of precursor proteins with a β-sheet conformation. Regardless, p5(sheet) is a novel heparin-reactive peptide that can be further studied for its utility in amyloid-targeting.
5. Summary
Amyloid-reactive peptides have great potential as specific amyloid-targeting agents that can be adapted for imaging the pathology and, potentially, as therapeutics. Peptide p5 is the prototypic heparin-reactive, synthetic, α-helical reagent capable of very specific amyloid binding in vivo. The helical secondary structure of p5 is important for the binding to heparin and amyloid extracts. An all-D enantiomer of p5, which may be more stable in vivo, exhibited enhanced amyloid reactivity. In addition, the p5(sheet) peptide, with a propensity for β-sheet formation in the presence of ligand, displayed enhanced reactivity to heparin and significantly better amyloid binding, as compared to p5. Our data suggest that polybasic peptides with a β-sheet propensity afford a new class of reagents with enhanced binding properties for multiple, biomedically relevant needs, i.e. the neutralization of small molecular weight heparin and as agents for imaging and treating amyloid in patients with systemic amyloidosis.
Acknowledgments
This work was supported by PHS grant R01DK079984 from The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), as well as funds from the Molecular Imaging and Translational Research Program and the Department of Medicine at UTMCK.
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.bbrep.2016.08.007.
Appendix A. Transparency document
Supplementary material
.
References
- 1.Blancas-Mejia L.M., Ramirez-Alvarado M. Systemic amyloidosis. Annu. Rev. Biochem. 2013 doi: 10.1146/annurev-biochem-072611-130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wechalekar A.D., Gillmore J.D., Hawkins P.N. Systemic amyloidosis. Lancet. 2015 doi: 10.1016/S0140-6736(15)01274-X. [DOI] [PubMed] [Google Scholar]
- 3.Pinney J.H., Hawkins P.N. Amyloidosis. Ann. Clin. Biochem. 2012;49:229–241. doi: 10.1258/acb.2011.011225. [DOI] [PubMed] [Google Scholar]
- 4.McWilliams-Koeppen H.P., Foster J.S., Hackenbrack N., Ramirez-Alvarado M., Donohoe D., Williams A., Macy S., Wooliver C., Wortham D., Morrell-Falvey J., Foster C.M., Kennel S.J., Wall J.S. Light chain amyloid fibrils cause metabolic dysfunction in human cardiomyocytes. PloS One. 2015;10:e0137716. doi: 10.1371/journal.pone.0137716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ancsin J.B. Amyloidogenesis: historical and modern observations point to heparan sulfate proteoglycans as a major culprit, amyloid: the international journal of experimental and clinical investigation: the official journal of the International. Soc. Amyloidosis. 2003;10:67–79. doi: 10.3109/13506120309041728. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl B., Lindahl U. Amyloid-specific heparan sulfate from human liver and spleen. J. Biol. Chem. 1997;272:26091–26094. doi: 10.1074/jbc.272.42.26091. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., Li J.P. Heparan sulfate proteoglycans in amyloidosis. Prog. Mol. Biol. Transl. Sci. 2010;93:309–334. doi: 10.1016/S1877-1173(10)93013-5. [DOI] [PubMed] [Google Scholar]
- 8.Smits N.C., Kurup S., Rops A.L., ten Dam G.B., Massuger L.F., Hafmans T., Turnbull J.E., Spillmann D., Li J.P., Kennel S.J., Wall J.S., Shworak N.W., Dekhuijzen P.N., van der Vlag J., van Kuppevelt T.H. The heparan sulfate motif (GlcNS6S-IdoA2S)3, common in heparin, has a strict topography and is involved in cell behavior and disease. J. Biol. Chem. 2010;285:41143–41151. doi: 10.1074/jbc.M110.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wall J.S., Richey T., Stuckey A., Donnell R., Macy S., Martin E.B., Williams A., Higuchi K., Kennel S.J. In vivo molecular imaging of peripheral amyloidosis using heparin-binding peptides. Proc. Natl. Acad. Sci. USA. 2011;108:E586–E594. doi: 10.1073/pnas.1103247108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wall J.S., Williams A., Richey T., Stuckey A., Huang Y., Wooliver C., Macy S., Heidel E., Gupta N., Lee A., Rader B., Martin E.B., Kennel S.J. A binding-site barrier affects imaging efficiency of high affinity amyloid-reactive peptide radiotracers in vivo. PloS One. 2013;8:e66181. doi: 10.1371/journal.pone.0066181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin E.B., Williams A., Heidel E., Macy S., Kennel S.J., Wall J.S. Peptide p5 binds both heparinase-sensitive glycosaminoglycans and fibrils in patient-derived AL amyloid extracts. Biochem. Biophys. Res. Commun. 2013;436:85–89. doi: 10.1016/j.bbrc.2013.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin E.B., Kennel S.J., Richey T., Wooliver C., Osborne D., Williams A., Stuckey A., Wall J.S. Dynamic PET and SPECT imaging with radioiodinated, amyloid-reactive peptide p5 in mice: a positive role for peptide dehalogenation. Peptides. 2014;60C:63–70. doi: 10.1016/j.peptides.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wall J.S., Richey T., Williams A., Stuckey A., Osborne D., Martin E., Kennel S.J. Comparative analysis of peptide p5 and serum amyloid P component for imaging AA amyloid in mice using dual-isotope SPECT. Mol. Imaging Biol.: Off. Publ. Acad. Mol. Imaging. 2012;14:402–407. doi: 10.1007/s11307-011-0524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wall J.S., Solomon A., Kennel S.J. Development and evaluation of agents for targeting visceral amyloid. Tijdschr. voor Nucleaire Geneeskd. 2011;33:807–814. [PMC free article] [PubMed] [Google Scholar]
- 15.Wall J.S., Martin E.B., Richey T., Stuckey A.C., Macy S., Wooliver C., Williams A., Foster J.S., McWilliams-Koeppen P., Uberbacher E., Cheng X., Kennel S.J. Preclinical validation of the heparin-reactive peptide p5+14 as a molecular imaging agent for visceral amyloidosis. Molecules. 2015;20:7657–7682. doi: 10.3390/molecules20057657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nitz M., Rullo A., Ding M.X. Heparin dependent coiled-coil formation. Chembiochem: A Eur. J. Chem. Biol. 2008;9:1545–1548. doi: 10.1002/cbic.200800056. [DOI] [PubMed] [Google Scholar]
- 17.Rullo A., Nitz M. Importance of the spatial display of charged residues in heparin-peptide interactions. Biopolymers. 2010;93:290–298. doi: 10.1002/bip.21339. [DOI] [PubMed] [Google Scholar]
- 18.Martin E.B., Williams A., Richey T., Stuckey A., Heidel R.E., Kennel S.J., Wall J.S. Comparative evaluation of p5+14 with SAP and peptide p5 by dual-energy SPECT imaging of mice with AA amyloidosis. Sci. Rep. 2016;6:22695. doi: 10.1038/srep22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wall J., Schell M., Murphy C., Hrncic R., Stevens F.J., Solomon A. Thermodynamic instability of human lambda 6 light chains: correlation with fibrillogenicity. Biochemistry. 1999;38:14101–14108. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 20.Pras M., Schubert M., Zucker-Franklin D., Rimon A., Franklin E.C. The characterization of soluble amyloid prepared in water. J. Clin. Investig. 1968;47:924–933. doi: 10.1172/JCI105784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wall J.S., Paulus M.J., GLeason S., Gregor J., Solomon A., Kennel S.J. Micro-imaging of amyloid in mice. Methods Enzymol. 2006;412:161–182. doi: 10.1016/S0076-6879(06)12011-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luhrs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Dobeli H., Schubert D., Riek R. 3D structure of alzheimer’s amyloid-beta(1-42) fibrils. Proc. Natl. Acad. Sci. USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F.T., Mattos C., Michnick S., Ngo T., Nguyen D.T., Prodhom B., Reiher W.E., Roux B., Schlenkrich M., Smith J.C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 25.Darden T., York D., Pedersen L. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98 [Google Scholar]
- 26.Kollman P., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D., Cheatham Tr. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 27.Chen Q., Buolamwini J.K., Smith J.C., Li A., Xu Q., Cheng X., Wei D. Impact of resistance mutations on inhibitor binding to HIV-1 integrase. J. Chem. Inf. Model. 2013;53:3297–3307. doi: 10.1021/ci400537n. [DOI] [PubMed] [Google Scholar]
- 28.Munoz V., Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Biol. 1994;1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- 29.Munoz V., Serrano L. Development of the multiple sequence approximation within the AGADIR model of alpha-helix formation: comparison with Zimm–Bragg and Lifson–Roig formalisms. Biopolymers. 1997;41:495–509. doi: 10.1002/(SICI)1097-0282(19970415)41:5<495::AID-BIP2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinforma. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayaraman G., Wu C.W., Liu Y.J., Chien K.Y., Fang J.C., Lyu P.C. Binding of a de novo designed peptide to specific glycosaminoglycans. FEBS Lett. 2000;482:154–158. doi: 10.1016/s0014-5793(00)01964-5. [DOI] [PubMed] [Google Scholar]
- 33.Wang J., Rabenstein D.L. Interaction of heparin with two synthetic peptides that neutralize the anticoagulant activity of heparin. Biochemistry. 2006;45:15740–15747. doi: 10.1021/bi061346a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Tal N., Honig B., Peitzsch R.M., Denisov G., McLaughlin S. Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys. J. 1996;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guglier S., Hricovini M., Raman R., Polito L., Torri G., Casu B., Sasisekharan R., Guerrini M. Minimum FGF2 binding structural requirements of heparin and heparan sulfate oligosaccharides as determined by NMR spectroscopy. Biochemistry. 2008;47:13862–13869. doi: 10.1021/bi801007p. [DOI] [PubMed] [Google Scholar]
- 36.Araujo R.L., Carvalho D.P. Bioenergetic impact of tissue-specific regulation of iodothyronine deiodinases during nutritional imbalance. J. Bioenerg. Biomembr. 2011;43:59–65. doi: 10.1007/s10863-011-9327-x. [DOI] [PubMed] [Google Scholar]
- 37.Leonard J.L., Ekenbarger D.M., Frank S.J., Farwell A.P., Koehrle J. Localization of type I iodothyronine 5′-deiodinase to the basolateral plasma membrane in renal cortical epithelial cells. J. Biol. Chem. 1991;266:11262–11269. [PubMed] [Google Scholar]
- 38.Leonard J.L., Visser T.J., Leonard D.M. Characterization of the subunit structure of the catalytically active type I iodothyronine deiodinase. J. Biol. Chem. 2001;276:2600–2607. doi: 10.1074/jbc.M006973200. [DOI] [PubMed] [Google Scholar]
- 39.Pace C.N., Scholtz J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material