Abstract
Rationale: Emphysema has considerable variability in the severity and distribution of parenchymal destruction throughout the lungs. Upper lobe–predominant emphysema has emerged as an important predictor of response to lung volume reduction surgery. Yet, aside from alpha-1 antitrypsin deficiency, the genetic determinants of emphysema distribution remain largely unknown.
Objectives: To identify the genetic influences of emphysema distribution in non–alpha-1 antitrypsin–deficient smokers.
Methods: A total of 11,532 subjects with complete genotype and computed tomography densitometry data in the COPDGene (Genetic Epidemiology of Chronic Obstructive Pulmonary Disease [COPD]; non-Hispanic white and African American), ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints), and GenKOLS (Genetics of Chronic Obstructive Lung Disease) studies were analyzed. Two computed tomography scan emphysema distribution measures (difference between upper-third and lower-third emphysema; ratio of upper-third to lower-third emphysema) were tested for genetic associations in all study subjects. Separate analyses in each study population were followed by a fixed effect metaanalysis. Single-nucleotide polymorphism–, gene-, and pathway-based approaches were used. In silico functional evaluation was also performed.
Measurements and Main Results: We identified five loci associated with emphysema distribution at genome-wide significance. These loci included two previously reported associations with COPD susceptibility (4q31 near HHIP and 15q25 near CHRNA5) and three new associations near SOWAHB, TRAPPC9, and KIAA1462. Gene set analysis and in silico functional evaluation revealed pathways and cell types that may potentially contribute to the pathogenesis of emphysema distribution.
Conclusions: This multicohort genome-wide association study identified new genomic loci associated with differential emphysematous destruction throughout the lungs. These findings may point to new biologic pathways on which to expand diagnostic and therapeutic approaches in chronic obstructive pulmonary disease.
Clinical trial registered with www.clinicaltrials.gov (NCT 00608764).
Keywords: genetics, emphysema, emphysema distribution, chronic obstructive pulmonary disease
At a Glance Commentary
Scientific Knowledge on the Subject
Chronic obstructive pulmonary disease (COPD), the leading cause of chronic respiratory morbidity and mortality worldwide, is a complex disease characterized by phenotypic and genotypic heterogeneity. Emphysematous lung destruction is frequently observed in subjects with COPD, but there is considerable variability in the severity and regional distribution of this destruction throughout the lungs. The genetic determinants of emphysema distribution remain largely unknown.
What This Study Adds to the Field
In four cohorts of current and former smokers without alpha-1 antitrypsin deficiency, our study identifies genome-wide significant associations with emphysema distribution near HHIP, CHRNA5, SOWAHB, TRAPPC9, and KIAA1462. Two loci (4q31 near HHIP and 15q25 near CHRNA5) have been previously associated with COPD susceptibility and total amount of emphysema, and three loci are newly identified (4q13 near SOWAHB, 8q24 near TRAPPC9, and 10p12 near KIAA1462). This study also reveals pathways that may potentially contribute to the pathogenesis of emphysema distribution.
Emphysema, irreversible destruction of lung parenchyma, is frequently observed in patients with chronic obstructive pulmonary disease (COPD), but there is considerable variability in its severity and regional distribution throughout the lung. This regional distribution has emerged as a clinically important phenotype capturing specific features about COPD subtypes (1). The pattern of emphysema distribution influences lung function impairment and gas exchange abnormalities (2–9). The NETT (National Emphysema Treatment Trial), a large multicenter randomized study comparing lung volume reduction surgery with medical treatment, has shown that upper lobe emphysema is an important predictor of the response to lung volume reduction surgery and the survival rate of patients with severe COPD (10, 11). Similarly, less invasive methods of lung volume reduction, such as bronchoscopically placed endobronchial valves and lung volume reduction coils, have been shown to improve clinical outcomes in subjects with upper lobe–predominant emphysema (12–14). However, in a randomized control trial evaluating the efficacy of a γ-selective retinoid agonist in the treatment of emphysema, patients with lower lung emphysema deteriorated faster on placebo and seemed to respond better to the treatment compared with those with predominantly upper lobe disease (15).
The regional heterogeneity of emphysematous destruction is not fully explained by differences in smoking behavior or demographic factors, and the determinants of upper versus lower lobe emphysema predominance remain largely unknown. Alpha-1 antitrypsin deficiency is often associated with basilar emphysema, whereas polymorphisms in the MMP9 gene and xenobiotic enzyme genes (GSTP1 and EPHX1) in non–alpha-1 antitrypsin–deficient smokers have been associated with apical emphysema predominance in candidate gene studies (8, 16, 17). These findings suggest the presence of genetic influences on emphysema distribution patterns. To investigate this hypothesis and identify genome-wide significant genetic markers, we performed a genome-wide association study (GWAS) of emphysema distribution in four large population-based cohorts of current and former cigarette smokers. Understanding the genetic influences on emphysema distribution may provide further insight into COPD susceptibility, disease pathophysiology, and treatment strategies. Some of these results have been previously reported as an abstract (18).
Methods
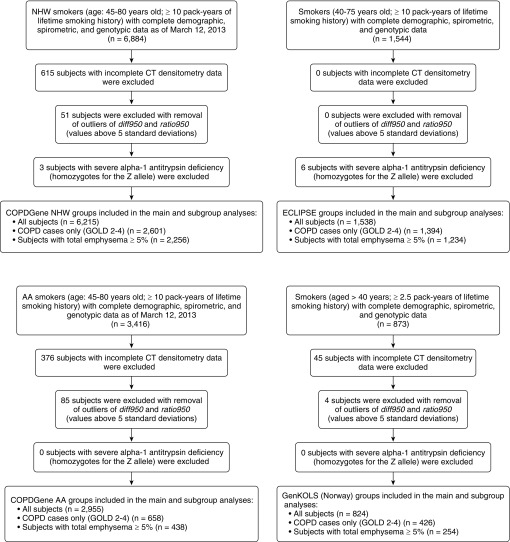
We analyzed 11,532 non–alpha-1 antitrypsin–deficient current and former smokers with complete genotype and computed tomography (CT) densitometry data from four cohorts: (1) COPDGene (Genetic Epidemiology of COPD) study non-Hispanic white (COPDGene NHW) subjects, (2) COPDGene African American (COPDGene AA) subjects, (3) GenKOLS (Genetics of Chronic Obstructive Lung Disease), and (4) ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints) study (Figure 1). Detailed descriptions including study populations, genotyping quality control, and genotyping imputation have been previously published (19).
Figure 1.
Participant flow diagrams of the studied cohorts: COPDGene (Genetic Epidemiology of COPD) NHW (non-Hispanic white subjects) (top left), COPDGene AA (African Americans) (bottom left), ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints) (top right), and GenKOLS (Genetics of Chronic Obstructive Lung Disease) (bottom right). COPD = chronic obstructive pulmonary disease; CT = computed tomography; GOLD = Global Initiative for Chronic Obstructive Lung Disease.
Emphysema percentage (CT densitometry low-attenuation area less than −950 Hounsfield units [%LAA950]) was measured in each lung third at end-expiration using Slicer software (www.slicer.org) (20). From these measurements, we constructed two measures of emphysema distribution for each study participant: the difference between upper-third and lower-third emphysema (diff950), and the ratio of upper-third to lower-third emphysema (ratio950). A rank-based inverse normal transformation was applied to both phenotypes to reduce the impact of outliers and deviations from normality. Furthermore, diff950 and ratio950 values above 5 SDs were considered outliers and were removed from the analyses. The correlation coefficients between diff950 and ratio950 range between 0.79 and 0.91 over the four studied populations. diff950 is less strongly correlated with total emphysema compared with ratio950 in all four study populations (0.03–0.34 vs. 0.21–0.42). These findings suggest distinct information may be captured on emphysema distribution using both diff950 and ratio950. Although commonly used as binary variables (with prespecified but not well-agreed-on cutoffs) to categorize the emphysema distribution of patients and study participants, we performed the categorization only to demonstrate the prevalence and the characteristics of the extremes of these phenotypes in our cohorts and we used diff950 and ratio950 as continuous traits for the genetic analyses because continuous variables are generally more informative than categorical ones.
Heritabilities of diff950 and ratio950 in COPDGene NHW subjects were assessed using Genome-wide Complex Trait Analysis (version 1.21) (21) and likelihood ratio tests. GWAS was then conducted within each population for each of the two emphysema distribution phenotypes under an additive linear regression model using PLINK (version 1.07) (22). Results were adjusted for age, sex, pack-years of smoking, current smoking, scanner type, and principal components of genetic ancestry. Results from all studies were combined into fixed-effect metaanalyses in METAL (version 2010–08–01) (23). Metaanalysis P values less than 5 × 10−8 were considered genome-wide significant.
To determine whether the genome-wide significant results are affected by emphysema severity and/or case-control status, we (1) reran the GWAS with total emphysema added as covariate, (2) performed analyses separately in COPD cases (subjects with COPD grade 2–4 as defined by the Global Initiative for Chronic Obstructive Lung Disease spirometric grading system based on FEV1) (“cases only”) and in subjects with at least 5% total emphysema, (3) assessed whether the top single-nucleotide polymorphisms (SNPs) from the present study also achieved genome-wide significance in previously published GWAS of COPD case-control status and %LAA (19, 24); and (4) evaluated associations with genetic variants previously associated with COPD susceptibility, emphysema, or emphysema distribution (“candidate SNP analysis”) (16, 17, 19, 24–28). P values less than 0.05 were considered as a suggestive association for candidate SNPs.
To explore their potential functional consequences, the lead SNPs from each genome-wide significant region and the SNPs in linkage disequilibrium (LD) with the lead SNPs (r2 ≥ 0.8 in the 1,000 Genomes Phase 1 data) were queried against the HaploReg database (version 3) and the expression quantitative trait loci (eQTL) data in the GTEx consortium (lung and whole blood) and the ECLIPSE study (whole blood and induced sputum) (29–31). The threshold for significance in the eQTL analysis was a false discovery rate (FDR) less than 5%, calculated using the QVALUE package (32).
To identify the functional pathways that are significantly overrepresented in the list of genes inferred from the GWAS, gene- and pathway-based analyses were performed using VEGAS (version 2.0) and MetaCore, respectively (33, 34). Bonferroni correction for 17,640 genes (P < 2.8 × 10−6) was used to account for multiple testing in VEGAS results (33). FDR value less than 5% was used to define a statistically significant association with a given process or pathway.
Additional methods are available in the online supplement.
Results
Baseline Characteristics of Participants
In total, 6,215 subjects from COPDGene NHW, 2,955 subjects from COPDGene AA, 1,538 subjects from ECLIPSE, and 824 subjects from GenKOLS had complete phenotype and genotype data and were included in the analysis. The total sample size across all cohorts was 11,532. The demographic and clinical characteristics of these subjects are shown in Table 1. The median ages at enrollment across the cohorts ranged from 53 to 64, with COPDGene AA subjects slightly younger than the other cohorts. GenKOLS subjects had the lowest median exposure to smoking. FEV1 percent predicted was the highest in COPDGene AA subjects (median, 87 [interquartile range, 71–99]) and the lowest in ECLIPSE (median, 48 [interquartile range, 36–64]).
Table 1.
Baseline Characteristics of Study Subjects
| COPDGene NHW | COPDGene AA | ECLIPSE | GenKOLS (Norway) | |
|---|---|---|---|---|
| Sample size | 6,215 | 2,955 | 1,538 | 824 |
| Age, yr | 62.30 (55.20 to 68.80) | 53.00 (49.20 to 58.20) | 64.00 (58.00 to 69.00) | 59.50 (52.40 to 68.20) |
| Sex, male, n (%) | 3,293 (53) | 1,647 (55.7) | 996 (65) | 481 (58.4) |
| Pack-years | 42.00 (30 to 58.7) | 34.40 (22.5 to 46.55) | 43.00 (30.00 to 58.00) | 22.40 (13.61 to 34.20) |
| FEV1, %predicted | 78.30 (55.85 to 93.20) | 86.60 (71.10 to 98.95) | 48.20 (36.50 to 64.35) | 79.03 (54.75 to 93.09) |
| Total emphysema (%LAA950) | 2.78 (0.84 to 8.74) | 0.93 (0.35 to 2.75) | 14.61 (6.64 to 25.31) | 1.74 (0.47 to 7.84) |
| Emphysema distribution difference | −0.27 (−1.55 to 1.25) | −0.05 (−0.47 to 0.59) | 0.72 (−3.60 to 12.20) | −0.14 (−1.22 to 0.80) |
| Emphysema distribution ratio | 0.75 (0.41 to 1.44) | 0.85 (0.45 to 1.80) | 1.15 (0.64 to 2.11) | 0.78 (0.35 to 1.63) |
| Emphysema predominance, n | ||||
| Lower lobe predominance | 2,036 | 851 | 265 | 295 |
| Diffuse emphysema | 3,116 | 1,448 | 872 | 371 |
| Upper lobe predominance | 1,063 | 656 | 401 | 158 |
| GOLD grade, n | ||||
| PRISm | 649 | 476 | 0 | 0 |
| 0 | 2,382 | 1,658 | 144 | 398 |
| 1 | 583 | 163 | 0 | 0 |
| 2 | 1,354 | 416 | 569 | 250 |
| 3 | 838 | 183 | 634 | 124 |
| 4 | 409 | 59 | 191 | 52 |
| COPD cases (COPD GOLD 2–4) | 2,601 | 658 | 1,394 | 426 |
| Subjects with total emphysema > 5% | 2,256 | 438 | 1,234 | 254 |
Definition of abbreviations: AA = African American; COPD = chronic obstructive pulmonary disease; COPDGene = Genetic Epidemiology of COPD; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; GenKOLS = Genetics of Chronic Obstructive Lung Disease; GOLD = Global Initiative for Chronic Obstructive Lung Disease; NHW = non-Hispanic white; PRISm = Preserved Ratio Impaired Spirometry; %LAA950 = percentage of lung voxels with attenuation on computed tomography densitometry lower than −950 Hounsfield units.
Continuous variables are expressed as median (interquartile range).
Emphysema distribution difference is the difference between upper-third and lower-third emphysema. Emphysema distribution ratio is the ratio between upper-third and lower-third emphysema. Emphysema predominance was categorized based on the ratio between upper-third and lower-third emphysema: Subjects with emphysema ratio less than 0.5 (“lower lobe–predominant group”), subjects with emphysema ratio between 0.5 and 2 (“diffuse emphysema group”), and subjects with emphysema ratio greater than 2 (“upper lobe–predominant group”) in each of the studied cohorts.
The ratio of upper lung to lower lung emphysema has traditionally been used to define subjects with upper lobe– versus lower lobe–predominant emphysema. We therefore compared the radiologic, demographic, clinical, and spirometric characteristics of groups of subjects parsed with a focus on the extremes of upper and lower lobe emphysema distributions: those with ratio950 values less than 0.5 (“lower lobe–predominant group”), those with ratio950 values between 0.5 and 2, and those with ratio950 values greater than 2 (“upper lobe–predominant group”) in each of the studied cohorts. Compared with subjects with lower lobe–predominant emphysema, those with upper lobe–predominant emphysema have higher pack-year histories of smoking, higher Modified Medical Research Council dyspnea scores, and lower FEV1 values and 6-minute-walk distance (see Table E1 in the online supplement). These differences in characteristics are less pronounced in COPD cases and the emphysema greater than 5% subgroups (see Tables E2 and E3).
Heritability
By using the Genome-wide Complex Trait Analysis method in the COPDGene NHW population, we estimated heritability of emphysema distribution phenotypes adjusting for age, sex, pack-years of smoking, current smoking, scanner type, and principal components of genetic ancestry (21, 35). The heritabilities of diff950 and ratio950 were respectively estimated to be 16% (SE, 6.3%; P = 0.005) and 20.5% (SE, 6.4%; P = 0.001), indicating modest but statistically significant heritability of these two emphysema distribution phenotypes.
GWAS: Individual Studies and Metaanalysis
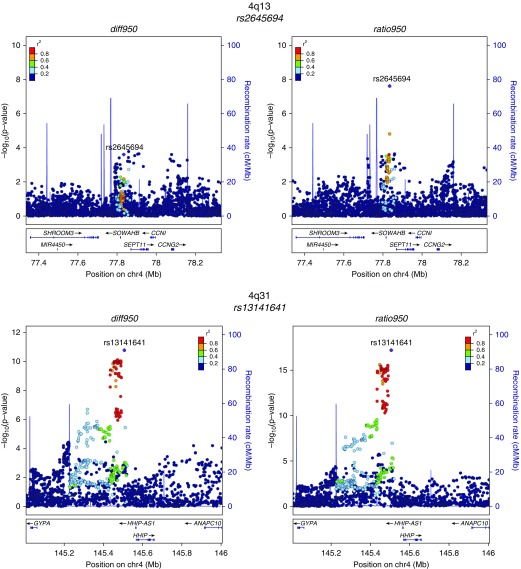
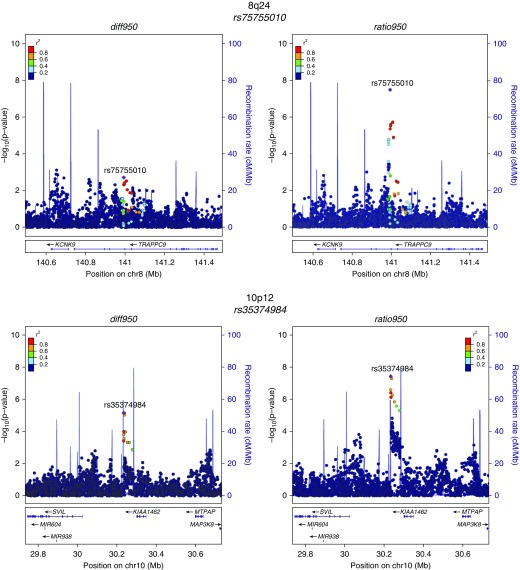
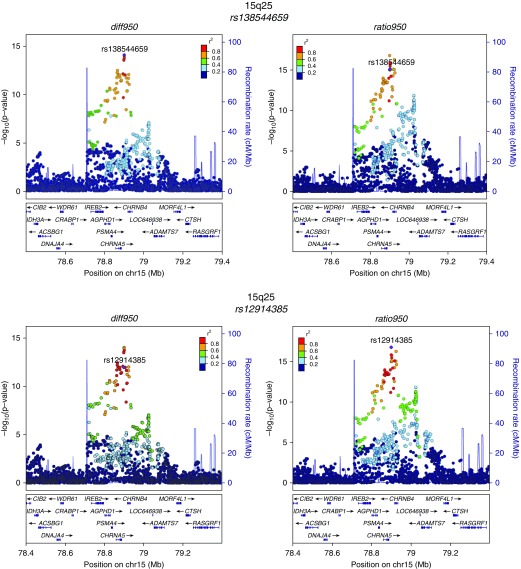
Quantile–quantile plots of the observed against expected P value distributions and genomic control values demonstrated an excess of low P values without systematic inflation in the GWAS test statistics (see Figure E1). Genome-wide significant results from the metaanalysis are shown in Table 2, and their regional association plots are shown in Figure 2. Loci with prior evidence of association with COPD susceptibility (4q31 near HHIP and 15q25 near CHRNA5) had the most significant associations with both emphysema distribution difference and ratio phenotypes. Additional new associations were also identified at genome-wide significance near SOWAHB (4q13), TRAPPC9 (8q24), and KIAA1462 (10p12) for ratio950. One of the SNPs with the strongest association signals was directly genotyped (rs12914385) and five were imputed (rs138544659, rs13141641, rs2645694, rs75755010, rs35374984). However, the imputed SNPs had high imputation quality (imputation r2 ≥ 0.8) in all studied cohorts.
Table 2.
Genome-Wide Significant Associations for Emphysema Distribution Phenotypes
| Metaanalysis | COPDGene NHW (n = 6,215) | COPDGene AA (n = 2,955) | ECLIPSE (n = 1,538) | GenKOLS (n = 824) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Locus | Nearest Gene | Effect Allele | Effect Allele Frequency | Effect Size | P Value | Effect Size (SE) | P Value | Effect Size (SE) | P Value | Effect Size (SE) | P Value | Effect Size (SE) | P Value |
| diff950 | ||||||||||||||
| rs138544659 | 15q25 | CHRNA3/5 | T | 0.70 | −0.12 | 9.87 × 10−15 | −0.12 (0.02) | 1.22 × 10−9 | −0.12 (0.04) | 0.002 | −0.12 (0.04) | 0.003 | −0.12 (0.05) | 0.03 |
| rs13141641 | 4q31 | HHIP | T | 0.63 | 0.09 | 1.76 × 10−11 | 0.10 (0.02) | 6.51 × 10−9 | 0.03 (0.04) | 0.45 | 0.13 (0.03) | 0.0002 | 0.05 (0.05) | 0.24 |
| ratio950 | ||||||||||||||
| rs13141641 | 4q31 | HHIP | T | 0.62 | 0.12 | 6.34 × 10−18 | 0.15 (0.02) | 4.92 × 10−17 | 0.04 (0.04) | 0.34 | 0.11 (0.03) | 0.002 | 0.05 (0.04) | 0.24 |
| rs12914385 | 15q25 | CHRNA3/5 | T | 0.38 | 0.11 | 1.72 × 10−17 | 0.13 (0.02) | 4.36 × 10−14 | 0.07 (0.03) | 0.03 | 0.08 (0.04) | 0.02 | 0.13 (0.04) | 0.003 |
| rs2645694 | 4q13 | SOWAHB | T | 0.30 | 0.08 | 2.42 × 10−8 | 0.08 (0.02) | 1.93 × 10−5 | 0.08 (0.03) | 0.003 | 0.07 (0.04) | 0.1 | 0.07 (0.05) | 0.17 |
| rs75755010 | 8q24 | TRAPPC9 | A | 0.08 | −0.15 | 3.43 × 10−8 | −0.15 (0.04) | 0.001 | −0.19 (0.04) | 4.98 × 10−6 | −0.01 (0.09) | 0.9 | −0.10 (0.10) | 0.31 |
| rs35374984 | 10p12 | KIAA1462 | A | 0.29 | 0.10 | 3.83 × 10−8 | 0.09 (0.03) | 0.001 | 0.10 (0.03) | 0.0004 | 0.12 (0.06) | 0.06 | 0.18 (0.08) | 0.03 |
Definition of abbreviations: AA = African American; COPDGene = Genetic Epidemiology of COPD; diff950 = inverse normally transformed upper-third/lower-third percent emphysema difference; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; GenKOLS = Genetics of Chronic Obstructive Lung Disease; NHW = non-Hispanic white; ratio950 = inverse normally transformed upper-third/lower-third percent emphysema ratio; SNP = single-nucleotide polymorphism.
Results of the individual studies and metaanalysis are shown.
Figure 2.
Regional association plots for metaanalysis genome-wide significant loci. These plots were generated using the LocusZoom tool with hg19/1,000 Genomes Mar 2012 European as the reference panel for non-Hispanic white subjects and hg19/1,000 Genomes Mar 2012 African for African American subjects. chr = chromosome; diff950 = inverse normally transformed difference between upper-third and lower-third emphysema; ratio950 = inverse normally transformed ratio between upper-third and lower-third emphysema.
Rerunning the GWAS with total emphysema as a covariate did not significantly affect the overall results, with some partial attenuation of the association signal at 4q13 and 10p12 (see Table E4). For the regions yielding genome-wide significance in all subjects, we examined the results separately in COPD cases only and in subjects with at least 5% total emphysema. In these analyses, most of the SNPs continued to be either genome-wide significant or nominally significant in the subgroups compared with the total sample, suggesting an effect beyond case-control status and emphysema severity (Table 3). However, it is important to note that although the overall P values for the SNPs in 4q13 and 8q24 remained significant in the various subgroups for ratio950, the associations with the diff950 were partially attenuated in the “cases only” and “subjects with total emphysema greater than or equal to 5%” subgroups. We also queried previously published GWAS of COPD case-control status and GWAS of %LAA from our group for the top SNPs from this present emphysema distribution study (19, 24). The P values in Table E5 indicate that for all loci where there is a significant COPD association, the COPD risk allele is associated with upper lobe predominance. In addition, only 4q13 and 8q24 loci are not significantly associated with case-control status or with % LAA. Taken together, these findings suggest that some of the signals may be mediated by case-control status and/or emphysema severity. Conditioning on the genome-wide significant SNPs from the metaanalysis significantly attenuated all of the genome-wide significant association signals (see Figure E2).
Table 3.
Analyses in COPD Cases Only and Only in Subjects with More Than 5% Total Emphysema
| diff950 Metaanalysis | ratio950 Metaanalysis | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All Subjects (n = 11,532) | Cases (n = 5,079) | Subjects with Total Emphysema ≥5% (n = 4,182) | All Subjects (n = 11,532) | Cases (n = 5,079) | Subjects with Total Emphysema ≥5% (n = 4,182) | ||||||||||
| SNP | Locus | Nearest Gene | Effect Allele | Effect Size | P Value | Effect Size | P Value | Effect Size | P Value | Effect Size | P Value | Effect Size | P Value | Effect Size | P Value |
| rs138544659 | 15q25 | CHRNA3/5 | T | −0.12 | 9.87 × 10−15 | −0.16 | 1.19 × 10−9 | −0.18 | 1.21 × 10−7 | −0.13 | 8.04 × 10−16 | −0.16 | 1.9 × 10−12 | −0.12 | 2.72 × 10−8 |
| rs13141641 | 4q31 | HHIP | T | 0.09 | 1.76 × 10−11 | 0.14 | 2.61 × 10−9 | 0.15 | 1.09 × 10−7 | 0.12 | 6.34 × 10−18 | 0.15 | 1.59 × 10−12 | 0.11 | 3.01 × 10−8 |
| rs12914385 | 15q25 | CHRNA3/5 | T | 0.10 | 1.13 × 10−12 | 0.13 | 6.5 × 10−8 | 0.12 | 4 × 10−5 | 0.11 | 1.72 × 10−17 | 0.14 | 8.86 × 10−12 | 0.08 | 2.29 × 10−5 |
| rs2645694 | 4q13 | SOWAHB | T | 0.05 | 0.0002 | 0.06 | 0.02 | 0.06 | 0.06 | 0.08 | 2.42 × 10−8 | 0.02 | 0.001 | 0.04 | 0.06 |
| rs75755010 | 8q24 | TRAPPC9 | A | −0.08 | 0.002 | −0.08 | 0.16 | −0.08 | 0.26 | −0.15 | 3.43 × 10−8 | −0.14 | 0.001 | −0.08 | 0.08 |
| rs35374984 | 10p12 | KIAA1462 | A | 0.08 | 7.11 × 10−6 | 0.09 | 0.01 | 0.12 | 0.01 | 0.10 | 3.83 × 10−8 | 0.10 | 0.001 | 0.08 | 0.004 |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; diff950 = inverse normally transformed difference between upper-third and lower-third emphysema; ratio950 = inverse normally transformed ratio between upper-third and lower-third emphysema; SNP = single-nucleotide polymorphism.
The SNPs included are the ones that reached genome-wide significance in all subjects. The numbers reported are the effect sizes and P values of the metaanalyses performed in the corresponding subgroup.
Association Analysis of Candidate SNPs
In the analysis of previously reported COPD and/or emphysema distribution variants, a total of 22 variants were considered (see Table E6). We extracted the P values of these SNPs from the metaanalyses of diff950 and ratio950. Eight of the SNPs that were previously associated with emphysema, lung function decline, or COPD susceptibility were also nominally associated with emphysema distribution near TGFB2, HHIP, RIN3, SERPINA10, IREB2/CHRNA3/5, AGPHD, MYO1D, and CYP2A6/ADCK4. Among the variants previously associated with emphysema distribution, only rs7221059 near MGAT5B was nominally significant in our study (P = 0.02).
Enhancer Enrichment in Top GWAS Loci
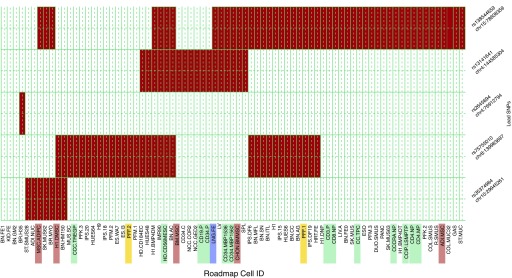
Previous studies have implicated genetic control of gene expression as a key functional mechanism for most complex disease GWAS associations (36). Publicly available data from the ENCODE and Roadmap Epigenomics projects provide an unprecedented opportunity to link genetic variation with experimental regulatory data from a wide array of cell lines. To determine the overlap of the genome-wide significant loci found in this study with enhancer regions in cell lines from the ENCODE and the Epigenomics Roadmap Projects, we queried the HaploReg database for the metaanalyses lead SNPs and all SNPs in LD with these lead SNPs (r2 ≥ 0.8 in the 1,000 Genomes data). As illustrated in Figure 3 and Table E7, a substantial number of these SNPs are located within annotated enhancer regions in several cell lines, notably fetal lung, fibroblasts, and lymphoblastoid cell lines.
Figure 3.
Heatmap of Roadmap enhancer enrichment. Cell type–specific enhancer activity was measured for each of five loci representing the six metaanalysis lead single-nucleotide polymorphisms (SNPs; two of the lead SNPs, rs138544659 and rs12914385, were in linkage disequilibrium and counted as one locus). Enhancer activity in cell types from the ENCODE and Roadmap projects were evaluated for relevant SNPs at each locus and compiled into a matrix with cell types along the columns and loci along the rows. Cell-type descriptions were searched for key words related to immune, endothelial, and lung cells to identify the relevant lines and label their text in the heatmap. Immune cell-type labels are highlighted in green, lung cell types in blue, fibroblasts in yellow, and mesenchymal stem cells in red. No endothelial cell types displayed enhancer activity at the six loci. chr = chromosome.
eQTL Results
To further explore the potential functional consequences of individual loci described in this study, we searched for evidence of functional impact using existing data sources. We queried the lung and whole-blood dataset in the GTEx consortium plus the whole-blood and induced sputum data from the ECLIPSE study for genome-wide significant associations from the metaanalysis and their high LD SNPs (r2 ≥ 0.8) looking to identify loci that are simultaneously and significantly associated with emphysema distribution and the expression of nearby genes (29, 30, 37). None of the loci described in this study were a cis eQTL in lung and/or whole blood in GTEx. In the ECLIPSE data, only the 15q25 locus was found to harbor cis eQTL SNPs near CHRNA3, IREB2, and PSMA4 genes at an FDR less than 5%. These identified eQTL SNPs were observed in both whole-blood and induced sputum samples.
Gene-based and Pathway Analyses
We then performed gene-based association analysis for 17,640 genes using VEGAS version 2. The results are listed in Table E8. Twelve genes withstood Bonferroni correction (P < 2.8 × 10−6): eight genes from 15q25 region (CHRNA3, CHRNA5, CHRNB4, HYKK/AGPHD1, IREB2, LOC646938, PSMA4, ADAMTS7), one gene on chromosome 2 (ATAD2B), one gene on chromosome 11 (FAR1), one gene on chromosome 13 (PCDH9), and one gene on chromosome 19 (GLTSCR1). All genes with a P value less than 0.01 from the VEGAS analysis were selected for MetaCore analyses (1,117 genes for diff950 and 1,243 genes for ratio950). At FDR values less than 5%, the top-ranked gene groups suggested enrichment for several pathways of interest, including regulation of apoptosis, transcription (transcription factor Tubby signaling pathways), immune response (classical, alternative and lectin-induced complement pathways; FcεRI pathway; CRTH2, LTBR1, and C5a signaling), cell adhesion, cytoskeleton remodeling (transforming growth factor [TGF], WNT, and Rho GTPase signaling), and development (adiponectin signaling; TGF-β–dependent induction of EMT via RhoA, PI3K, and ILK; S1P1 signaling pathway) (Table 4).
Table 4.
Top Significantly Enriched Pathways for Emphysema Distribution Phenotypes Provided by Metacore Pathway Analysis
| Pathway | FDR | Number of Pathway Genes in the Data | Total Number of Genes in the Pathway |
|---|---|---|---|
| Upper-third/lower-third percent emphysema difference | |||
| NETosis: neutrophil extracellular trap–associated cell death | 0.013 | 7 | 31 |
| Transcription: transcription factor Tubby signaling pathways | 0.026 | 5 | 17 |
| SLE genetic marker-specific pathways in antigen-presenting cells | 0.026 | 10 | 84 |
| Development: adiponectin signaling | 0.031 | 7 | 45 |
| Cytoskeleton remodeling: TGF, WNT, and cytoskeletal remodeling | 0.031 | 11 | 111 |
| Signal transduction: activin A signaling regulation | 0.031 | 6 | 33 |
| Upper-third/lower-third percent emphysema ratio | |||
| Immune response: alternative complement pathway | 2.38 × 10−8 | 16 | 53 |
| Immune response: lectin-induced complement pathway | 6.32 × 10−7 | 14 | 50 |
| Immune response: classical complement pathway | 9.77 × 10−7 | 14 | 53 |
| Immune response: role of the membrane attack complex in cell survival | 0.005 | 8 | 35 |
| Chemotaxis: leukocyte chemotaxis | 0.011 | 11 | 75 |
| Nociception: nociceptin receptor signaling | 0.011 | 11 | 76 |
| Immune response: FcεRI pathway | 0.014 | 9 | 55 |
| Aberrant B-Raf signaling in melanoma progression | 0.014 | 9 | 55 |
| Immune response: CRTH2 signaling in T-helper 2 cells | 0.014 | 8 | 44 |
| Development: TGF-β–dependent induction of EMT via RhoA, PI3K, and ILK | 0.017 | 8 | 46 |
| Immune response: LTBR1 signaling | 0.02 | 9 | 60 |
| Immune response: C5a signaling | 0.025 | 8 | 50 |
| Neurophysiologic process: constitutive and regulated NMDA receptor trafficking | 0.025 | 9 | 63 |
| Transcription: sirtuin6 regulation and functions | 0.026 | 9 | 64 |
| Breast cancer, general schema | 0.032 | 7 | 41 |
| Transport: ACM3 in salivary glands | 0.034 | 7 | 42 |
| Development: S1P1 signaling pathway | 0.043 | 7 | 44 |
| Cell adhesion: IL-8–dependent cell migration and adhesion | 0.046 | 6 | 33 |
| Cytoskeleton remodeling: regulation of actin cytoskeleton by Rho GTPases | 0.049 | 5 | 23 |
Definition of abbreviations: EMT = epithelial–mesenchymal transition; FDR = false discovery rate; SLE = systemic lupus erythematosus; TGF = transforming growth factor.
FDR values less than 0.05 indicate a statistically significant association with a given process or pathway.
Discussion
This multicohort GWAS of emphysema distribution identified five independent loci achieving genome-wide significance. Two loci (4q31 near HHIP and 15q25 near CHRNA5) have been previously associated with COPD susceptibility and total amount of emphysema, and three loci are newly identified (4q13 near SOWAHB, 8q24 near TRAPPC9, and 10p12 near KIAA1462). In addition, of 22 previously identified loci tested for association, nine loci were nominally associated with emphysema distribution. In silico functional evaluation revealed strong enrichment of enhancer regions among the top GWAS loci and indicated that regulation of gene expression in specific cell types (particularly lung fibroblasts and lymphoblastoid cell lines) may be a key functional mechanism for genetic factors influencing emphysema distribution. Gene set analyses provided insights into biologic mechanisms that may be relevant to the pathogenesis of emphysema distribution, including complement pathways, immune response, cytoskeleton remodeling, cell adhesion, and chemotaxis.
Ito and colleagues (17) found that the MMP9 gene was associated with upper lobe–predominant emphysema. DeMeo and colleagues (16) found variants in genes encoding xenobiotic enzymes (GSTP1 and EPHX1) as differentially associated with distributional features of emphysema. None of these genes (MMP9, GSTP1, EPHX1) replicated in our study. In a GWAS of percent emphysema on CT in the Multi-Ethnic Study of Atherosclerosis of COPD (MESA), Manichaikul and colleagues (25) assessed lung fields of cardiac CT scans of 7,914 participants with relatively normal spirometry (FEV1/FVC, 0.69–0.82) and had no genome-wide significant associations for upper-lower lobe emphysema ratio across race/ethnic groups using these cardiac CT images. However, genome-wide significance was reached among Hispanics (rs10411619 near MAN2B) and among Chinese (rs7698250 near DHX15 and rs7221059 near MGAT5B) (25). The cohorts we analyzed in our study did not include Hispanic and Chinese subjects. Only rs7221059 was nominally significant in our candidate SNP analysis. In our study, given that we are unsure which measure of emphysema distribution best captures genetic features, we opted to analyze both the difference and ratio between upper-third and lower-third emphysema. The presence of known COPD susceptibility loci (15q25 and 4q31) among the top signals for emphysema distribution supports the validity of these metrics. The associations of these loci with emphysema distribution were not purely driven by total emphysema or COPD status, as supported by analyses adjusting for emphysema or performed only in COPD subjects.
The chromosome 4 association with emphysema distribution (rs2645694) lies near the SOWAHB gene. This gene belongs to the ankyrin repeat domain family. Ankyrin repeats mediate protein-protein interactions in very diverse families of proteins. However, no phenotype, disease or trait is known to be directly associated with the SOWAHB gene. The SHROOM3 gene, 110-kb upstream from SOWAHB, was previously shown to facilitate canonical TGF-β1 signaling, increase α1 collagen expression, and potentiate interstitial renal allograft fibrosis (38). Although SHROOM3 has not previously been associated with COPD, prior genetic studies have demonstrated an association of gene polymorphisms of the TGF-β superfamily with COPD (39, 40).
The SNP on chromosome 8q24 lies near the TRAPPC9 gene. TRAPPC9 encodes a protein that plays a role in nuclear factor-κB signaling. Nuclear factor-κB activation has been implicated in disease pathogenesis in rodent models of COPD (41). In addition, increased markers of nuclear factor-κB pathway activity have been demonstrated in sputum macrophages during COPD exacerbations and in bronchial biopsies of patients with stable COPD (42, 43).
The KIAA1462 gene on chromosome 10p12 was previously associated with coronary artery disease and its gene product is a component of endothelial cell-cell junctions (44, 45). Damaged pulmonary endothelial integrity was observed in rats exposed to second-hand smoke and in lung tissues of subjects with COPD (46). Another interesting gene downstream of KIAA1462 is the SVIL gene, the product of which was shown to act as a high-affinity link between the actin cytoskeleton and the membrane (47). The role of actin and cytoskeletal processes in emphysema is less well established, but cytoskeletal-associated pathways containing genes, such as ACTN1 and DBN1, have been previously associated with immune cell activation that plays a significant role in COPD pathogenesis (48, 49).
Among the significant genes from the gene-based analysis, ADAMTS7 in 15q25 and PCDH9 in 13q21 are of particular interest. In a mouse model of cigarette smoke–induced bronchitis and emphysema, IREB2 was characterized as at least one of the functional genes in the 15q25 region (50). Recent eQTL analyses have also implicated CHRNA3 and PSMA4, located in close proximity to IREB2, as strong candidate genes for COPD susceptibility (30). Our present study proposes ADAMTS7 as yet another potential causal gene in this 15q25 region. This gene has not been previously associated with lung diseases. However, polymorphisms in the ADAM33 gene, another member of the ADAMTS family, have been shown to be related to an accelerated decline in FEV1, airway hyperresponsiveness, and airway inflammation (51, 52). PCDH9 is a protocadherin belonging to the cadherin superfamily of adhesion molecules. In a GWAS of lung function decline in adults with and without asthma, two SNPs near PCDH9 (rs17077331 and rs17077335) were associated with FEV1/FVC decrease in patients without asthma (53).
Current theories concerning disease pathogenesis in COPD and emphysema include an imbalance between protease and antiprotease activity, induced apoptosis of alveolar cells through deregulation of pathways involved in oxidative stress, chronic inflammation, innate and adaptive immune responses, and aberrant mechanical stress-induced tissue remodeling leading to destruction of the extracellular matrix in the lung (48, 54–57). The precise reasons for the upper lobe predominance in non–alpha-1 antitrypsin–deficient smokers with emphysema are unclear, but it has been attributed to regional differences in perfusion, transit time of leukocytes, clearance of deposited dust, mechanical stress, and pleural pressure (5, 16, 17, 58).
The biologic relevance of our findings is supported by strong enrichment in known COPD and emphysema functional pathways. This similarity suggests that regional differences in emphysema severity share some of the processes that occur in general COPD pathogenesis and progression. However, our study also revealed biologic pathways that may be specifically involved in and are unique to the pathogenesis of emphysema distribution per se. It corroborates and extends the observations from Campbell and colleagues’ (59) gene expression profile of smokers’ lung tissue samples from regions within the same lung with varying amounts of emphysematous destruction. Taken together, these observations indicate that although they share common genes and biologic processes, emphysema distribution and general emphysema and COPD may also have distinct genetic signatures and biologic underpinnings. Our metaanalysis is the largest GWAS to date (n = 11,532) reporting associations for emphysema distribution in smokers. It is also the first analyzing two quantitative emphysema distribution phenotypes (diff950 and ratio950) that quantify correlated but complementary aspects of emphysema distribution. These imaging phenotypes were generated from standardized chest CT scans across a wide range of COPD disease severity, as compared with cardiac CT scans of subjects with only mild COPD in the study by Manichaikul and coworkers (25). Furthermore, in our study, additional in silico functional analyses were performed to determine the downstream effects of the identified SNPs and detect key biologic pathways potentially involved in the pathogenesis of emphysema distribution.
This study highlights new associations with distributional features of emphysema but has the following limitations. First, our analysis included cohorts with different imaging protocols, proportions of disease severity, and racial groups. We handled this heterogeneity by performing GWAS separately within each cohort and by adjusting for the scanner type and total emphysema. In addition, we conducted secondary analyses that showed that the overall results were, for the most part, not impacted by the affection status and scanner types. Second, we analyzed emphysema distribution using upper and lower lung thirds. However, several other imaging biomarkers have also been described in the literature for tracking the regional heterogeneity of emphysema distribution and need to be explored, such as lobar, upper/lower lung halves, core-rind, and centrilobular/panlobular/paraseptal distributions (60). Third, we performed inverse normal transformation of the studied phenotypes given their highly skewed distributions and the concern of violating the assumption of normally distributed residuals. This is a conservative approach because inverse normal transformation carries the risk of reducing statistical power (61). Fourth, for this analysis, we did not adjust for severity and types of airway disease. The state of the airway assessment by CT is suboptimal because the critical sites of airflow obstruction (1- to 2-mm-diameter airways) are below the resolution of the CT scanner. Fifth, the new GWAS signals from this metaanalysis were not replicated in a separate cohort. However, we did find consistent evidence for association across the COPDGene, ECLIPSE, and GenKOLS cohorts. Sixth, although publicly available cell line regulatory data are extensive and provide an unprecedented opportunity to link genetic variation with experimental regulatory data from a wide array of cell lines, they are not comprehensive and important emphysema distribution-related cell types may not be present in ENCODE and Roadmap data resources. Moreover, available data on variability of regulatory annotation in specific cell types in various conditions are limited. Lastly, while pathway analyses were able to detect interesting biologic pathways that may be involved in disease pathogenesis, such analyses are only hypothesis-generating and require functional confirmation.
In summary, this multicohort GWAS of emphysema distribution provides evidence for a role of common genetic variants in contributing to the patterns of smoking-related emphysema distribution. It confirms established associations for COPD GWAS loci, suggests new associations, and provides important clues to new biologic pathways on which to expand diagnostic and therapeutic approaches in COPD. Further validation of these associations in larger populations is required along with functional investigations to identify a role for the SNPs and genes highlighted in this study.
Footnotes
Supported by NHLBI grants R01HL089897, R01HL089856, R01HL124233, R01HL126596, R01HL113264, K01HL125858, P01105339, P01HL114501, and 2T32HL07427-32. The COPDGene study (NCT00608764) is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, GlaxoSmithKline, Siemens, and Sunovion. The Norway GenKOLS (Genetics of Chronic Obstructive Lung Disease, GSK code RES11080) and ECLIPSE (NCT00292552; GSK code SCO104960) studies were funded by GlaxoSmithKline. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or the National Institutes of Health.
Author Contributions: D.L.D. had full access to all of the data in the study, takes responsibility for the integrity of the data and the accuracy of the data analysis, had authority over manuscript preparation, and the decision to submit the manuscript for publication. Study concept and design, A.B., P.J.C., E.K.S., and D.L.D. Acquisition, analysis, or interpretation of data, A.B., S.M.L., M.H.C., C.P.H., R.P.B., G.R.W., E.H.-S., P.B., A.G., N.M.L., T.H.B., H.O.C., J.D.C., E.K.S., P.J.C., and D.L.D. Drafting of the manuscript, A.B., P.J.C., and D.L.D. Critical revision of the manuscript for important intellectual content, all authors. Statistical analysis, A.B., N.M.L., P.J.C., and D.L.D. Obtained funding, J.D.C. and E.K.S. Study supervision, all authors.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201605-0997OC on September 26, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Castaldi PJ, Dy J, Ross J, Chang Y, Washko GR, Curran-Everett D, Williams A, Lynch DA, Make BJ, Crapo JD, et al. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax. 2014;69:415–422. doi: 10.1136/thoraxjnl-2013-203601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aziz ZA, Wells AU, Desai SR, Ellis SM, Walker AE, MacDonald S, Hansell DM. Functional impairment in emphysema: contribution of airway abnormalities and distribution of parenchymal disease. AJR Am J Roentgenol. 2005;185:1509–1515. doi: 10.2214/AJR.04.1578. [DOI] [PubMed] [Google Scholar]
- 3.Chae EJ, Seo JB, Song JW, Kim N, Park BW, Lee YK, Oh YM, Lee SD, Lim SY. Slope of emphysema index: an objective descriptor of regional heterogeneity of emphysema and an independent determinant of pulmonary function. AJR Am J Roentgenol. 2010;194:W248–W255. doi: 10.2214/AJR.09.2672. [DOI] [PubMed] [Google Scholar]
- 4.Gietema HA, Zanen P, Schilham A, van Ginneken B, van Klaveren RJ, Prokop M, Lammers JW. Distribution of emphysema in heavy smokers: impact on pulmonary function. Respir Med. 2010;104:76–82. doi: 10.1016/j.rmed.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Gurney JW. Cross-sectional physiology of the lung. Radiology. 1991;178:1–10. doi: 10.1148/radiology.178.1.1984285. [DOI] [PubMed] [Google Scholar]
- 6.Mair G, Miller JJ, McAllister D, Maclay J, Connell M, Murchison JT, MacNee W. Computed tomographic emphysema distribution: relationship to clinical features in a cohort of smokers. Eur Respir J. 2009;33:536–542. doi: 10.1183/09031936.00111808. [DOI] [PubMed] [Google Scholar]
- 7.Nakano Y, Sakai H, Muro S, Hirai T, Oku Y, Nishimura K, Mishima M. Comparison of low attenuation areas on computed tomographic scans between inner and outer segments of the lung in patients with chronic obstructive pulmonary disease: incidence and contribution to lung function. Thorax. 1999;54:384–389. doi: 10.1136/thx.54.5.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parr DG, Stoel BC, Stolk J, Stockley RA. Pattern of emphysema distribution in alpha1-antitrypsin deficiency influences lung function impairment. Am J Respir Crit Care Med. 2004;170:1172–1178. doi: 10.1164/rccm.200406-761OC. [DOI] [PubMed] [Google Scholar]
- 9.Saitoh T, Koba H, Shijubo N, Tanaka H, Sugaya F. Lobar distribution of emphysema in computed tomographic densitometric analysis. Invest Radiol. 2000;35:235–243. doi: 10.1097/00004424-200004000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE National Emphysema Treatment Trial Research Group. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348:2059–2073. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 11.Martinez FJ, Foster G, Curtis JL, Criner G, Weinmann G, Fishman A, DeCamp MM, Benditt J, Sciurba F, Make B, et al. NETT Research Group. Predictors of mortality in patients with emphysema and severe airflow obstruction. Am J Respir Crit Care Med. 2006;173:1326–1334. doi: 10.1164/rccm.200510-1677OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venuta F, Anile M, Diso D, Carillo C, De Giacomo T, D’Andrilli A, Fraioli F, Rendina EA, Coloni GF. Long-term follow-up after bronchoscopic lung volume reduction in patients with emphysema. Eur Respir J. 2012;39:1084–1089. doi: 10.1183/09031936.00071311. [DOI] [PubMed] [Google Scholar]
- 13.Deslée G, Mal H, Dutau H, Bourdin A, Vergnon JM, Pison C, Kessler R, Jounieaux V, Thiberville L, Leroy S, et al. REVOLENS Study Group. Lung volume reduction coil treatment vs usual care in patients with severe emphysema: the REVOLENS Randomized Clinical Trial. JAMA. 2016;315:175–184. doi: 10.1001/jama.2015.17821. [DOI] [PubMed] [Google Scholar]
- 14.Sciurba FC, Chandra D, Bon J. Bronchoscopic lung volume reduction in COPD: lessons in implementing clinically based precision medicine. JAMA. 2016;315:139–141. doi: 10.1001/jama.2015.17714. [DOI] [PubMed] [Google Scholar]
- 15.Jones P. Tesra (treatment of emphysema with selective retinoid agonist) study results. Am J Respir Crit Care Med. 2011;183:A6418. [Google Scholar]
- 16.DeMeo DL, Hersh CP, Hoffman EA, Litonjua AA, Lazarus R, Sparrow D, Benditt JO, Criner G, Make B, Martinez FJ, et al. Genetic determinants of emphysema distribution in the national emphysema treatment trial. Am J Respir Crit Care Med. 2007;176:42–48. doi: 10.1164/rccm.200612-1797OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito I, Nagai S, Handa T, Muro S, Hirai T, Tsukino M, Mishima M. Matrix metalloproteinase-9 promoter polymorphism associated with upper lung dominant emphysema. Am J Respir Crit Care Med. 2005;172:1378–1382. doi: 10.1164/rccm.200506-953OC. [DOI] [PubMed] [Google Scholar]
- 18.El Boueiz A, Lutz SM, Bowler RP, Cho MH, McDonald ML, Laird NM, Beaty TH, Crapo JD, Silverman EK, Castaldi PJ, et al. Genome-wide association study identifies novel genetic determinants of emphysema distribution patterns [abstract]. Presented at the annual meeting for the American Society of Human Genetics. October 18-22, 2014, San Diego, CA [Google Scholar]
- 19.Cho MH, Castaldi PJ, Hersh CP, Hobbs BD, Barr RG, Tal-Singer R, Bakke P, Gulsvik A, San José Estépar R, Van Beek EJ, et al. NETT Genetics, ECLIPSE, and COPDGene Investigators. A genome-wide association study of emphysema and airway quantitative imaging phenotypes. Am J Respir Crit Care Med. 2015;192:559–569. doi: 10.1164/rccm.201501-0148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coxson HO, Rogers RM, Whittall KP, D’yachkova Y, Paré PD, Sciurba FC, Hogg JC. A quantification of the lung surface area in emphysema using computed tomography. Am J Respir Crit Care Med. 1999;159:851–856. doi: 10.1164/ajrccm.159.3.9805067. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, DeMeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manichaikul A, Hoffman EA, Smolonska J, Gao W, Cho MH, Baumhauer H, Budoff M, Austin JH, Washko GR, Carr JJ, et al. Genome-wide study of percent emphysema on computed tomography in the general population. The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilk JB, Shrine NR, Loehr LR, Zhao JH, Manichaikul A, Lopez LM, Smith AV, Heckbert SR, Smolonska J, Tang W, et al. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am J Respir Crit Care Med. 2012;186:622–632. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong X, Cho MH, Anderson W, Coxson HO, Muller N, Washko G, Hoffman EA, Bakke P, Gulsvik A, Lomas DA, et al. ECLIPSE Study NETT Investigators. Genome-wide association study identifies BICD1 as a susceptibility gene for emphysema. Am J Respir Crit Care Med. 2011;183:43–49. doi: 10.1164/rccm.201004-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castaldi PJ, Cho MH, San José Estépar R, McDonald ML, Laird N, Beaty TH, Washko G, Crapo JD, Silverman EK COPDGene Investigators. Genome-wide association identifies regulatory Loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190:399–409. doi: 10.1164/rccm.201403-0569OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castaldi PJ, Cho MH, Zhou X, Qiu W, Mcgeachie M, Celli B, Bakke P, Gulsvik A, Lomas DA, Crapo JD, et al. Genetic control of gene expression at novel and established chronic obstructive pulmonary disease loci. Hum Mol Genet. 2015;24:1200–1210. doi: 10.1093/hmg/ddu525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, et al. AMFS Investigators. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra A, Macgregor S. VEGAS2: software for more flexible gene-based testing. Twin Res Hum Genet. 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- 35.Zhou JJ, Cho MH, Castaldi PJ, Hersh CP, Silverman EK, Laird NM. Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am J Respir Crit Care Med. 2013;188:941–947. doi: 10.1164/rccm.201302-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu W, Cho MH, Riley JH, Anderson WH, Singh D, Bakke P, Gulsvik A, Litonjua AA, Lomas DA, Crapo JD, et al. ECLIPSE Investigators. Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS One. 2011;6:e24395. doi: 10.1371/journal.pone.0024395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menon MC, Chuang PY, Li Z, Wei C, Zhang W, Luan Y, Yi Z, Xiong H, Woytovich C, Greene I, et al. Intronic locus determines SHROOM3 expression and potentiates renal allograft fibrosis. J Clin Invest. 2015;125:208–221. doi: 10.1172/JCI76902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warburton D, Shi W, Xu B. TGF-β-Smad3 signaling in emphysema and pulmonary fibrosis: an epigenetic aberration of normal development? Am J Physiol Lung Cell Mol Physiol. 2013;304:L83–L85. doi: 10.1152/ajplung.00258.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Königshoff M, Kneidinger N, Eickelberg O. TGF-beta signaling in COPD: deciphering genetic and cellular susceptibilities for future therapeutic regimen. Swiss Med Wkly. 2009;139:554–563. doi: 10.4414/smw.2009.12528. [DOI] [PubMed] [Google Scholar]
- 41.Marwick JA, Kirkham PA, Stevenson CS, Danahay H, Giddings J, Butler K, Donaldson K, Macnee W, Rahman I. Cigarette smoke alters chromatin remodeling and induces proinflammatory genes in rat lungs. Am J Respir Cell Mol Biol. 2004;31:633–642. doi: 10.1165/rcmb.2004-0006OC. [DOI] [PubMed] [Google Scholar]
- 42.Caramori G, Romagnoli M, Casolari P, Bellettato C, Casoni G, Boschetto P, Chung KF, Barnes PJ, Adcock IM, Ciaccia A, et al. Nuclear localisation of p65 in sputum macrophages but not in sputum neutrophils during COPD exacerbations. Thorax. 2003;58:348–351. doi: 10.1136/thorax.58.4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Stefano A, Caramori G, Oates T, Capelli A, Lusuardi M, Gnemmi I, Ioli F, Chung KF, Donner CF, Barnes PJ, et al. Increased expression of nuclear factor-kappaB in bronchial biopsies from smokers and patients with COPD. Eur Respir J. 2002;20:556–563. doi: 10.1183/09031936.02.00272002. [DOI] [PubMed] [Google Scholar]
- 44.Erdmann J, Willenborg C, Nahrstaedt J, Preuss M, König IR, Baumert J, Linsel-Nitschke P, Gieger C, Tennstedt S, Belcredi P, et al. Genome-wide association study identifies a new locus for coronary artery disease on chromosome 10p11.23. Eur Heart J. 2011;32:158–168. doi: 10.1093/eurheartj/ehq405. [DOI] [PubMed] [Google Scholar]
- 45.Akashi M, Higashi T, Masuda S, Komori T, Furuse M. A coronary artery disease-associated gene product, JCAD/KIAA1462, is a novel component of endothelial cell-cell junctions. Biochem Biophys Res Commun. 2011;413:224–229. doi: 10.1016/j.bbrc.2011.08.073. [DOI] [PubMed] [Google Scholar]
- 46.Kratzer A, Chu HW, Salys J, Moumen Z, Leberl M, Bowler R, Cool C, Zamora M, Taraseviciene-Stewart L. Endothelial cell adhesion molecule CD146: implications for its role in the pathogenesis of COPD. J Pathol. 2013;230:388–398. doi: 10.1002/path.4197. [DOI] [PubMed] [Google Scholar]
- 47.Hasegawa H, Hyodo T, Asano E, Ito S, Maeda M, Kuribayashi H, Natsume A, Wakabayashi T, Hamaguchi M, Senga T. The role of PLK1-phosphorylated SVIL in myosin II activation and cytokinetic furrowing. J Cell Sci. 2013;126:3627–3637. doi: 10.1242/jcs.124818. [DOI] [PubMed] [Google Scholar]
- 48.van der Strate BW, Postma DS, Brandsma CA, Melgert BN, Luinge MA, Geerlings M, Hylkema MN, van den Berg A, Timens W, Kerstjens HA. Cigarette smoke-induced emphysema: a role for the B cell? Am J Respir Crit Care Med. 2006;173:751–758. doi: 10.1164/rccm.200504-594OC. [DOI] [PubMed] [Google Scholar]
- 49.Gordón-Alonso M, Sala-Valdés M, Rocha-Perugini V, Pérez-Hernández D, López-Martín S, Ursa A, Alvarez S, Kolesnikova TV, Vázquez J, Sánchez-Madrid F, et al. EWI-2 association with α-actinin regulates T cell immune synapses and HIV viral infection. J Immunol. 2012;189:689–700. doi: 10.4049/jimmunol.1103708. [DOI] [PubMed] [Google Scholar]
- 50.Cloonan SM, Glass K, Laucho-Contreras ME, Bhashyam AR, Cervo M, Pabón MA, Konrad C, Polverino F, Siempos II, Perez E, et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat Med. 2016;22:163–174. doi: 10.1038/nm.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sadeghnejad A, Ohar JA, Zheng SL, Sterling DA, Hawkins GA, Meyers DA, Bleecker ER. Adam33 polymorphisms are associated with COPD and lung function in long-term tobacco smokers. Respir Res. 2009;10:21. doi: 10.1186/1465-9921-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gosman MM, Boezen HM, van Diemen CC, Snoeck-Stroband JB, Lapperre TS, Hiemstra PS, Ten Hacken NH, Stolk J, Postma DS. A disintegrin and metalloprotease 33 and chronic obstructive pulmonary disease pathophysiology. Thorax. 2007;62:242–247. doi: 10.1136/thx.2006.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imboden M, Bouzigon E, Curjuric I, Ramasamy A, Kumar A, Hancock DB, Wilk JB, Vonk JM, Thun GA, Siroux V, et al. Genome-wide association study of lung function decline in adults with and without asthma. J Allergy Clin Immunol. 2012;129:1218–1228. doi: 10.1016/j.jaci.2012.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park JW, Ryter SW, Choi AM. Functional significance of apoptosis in chronic obstructive pulmonary disease. COPD. 2007;4:347–353. doi: 10.1080/15412550701603775. [DOI] [PubMed] [Google Scholar]
- 55.Postma DS, Timens W. Remodeling in asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3:434–439. doi: 10.1513/pats.200601-006AW. [DOI] [PubMed] [Google Scholar]
- 56.Suki B, Sato S, Parameswaran H, Szabari MV, Takahashi A, Bartolák-Suki E. Emphysema and mechanical stress-induced lung remodeling. Physiology (Bethesda) 2013;28:404–413. doi: 10.1152/physiol.00041.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marc MM, Kristan SS, Rozman A, Kern I, Flezar M, Kosnik M, Korosec P. Complement factor C5a in acute exacerbation of chronic obstructive pulmonary disease. Scand J Immunol. 2010;71:386–391. doi: 10.1111/j.1365-3083.2010.02385.x. [DOI] [PubMed] [Google Scholar]
- 58.West JB. Distribution of mechanical stress in the lung, a possible factor in localisation of pulmonary disease. Lancet. 1971;1:839–841. doi: 10.1016/s0140-6736(71)91501-7. [DOI] [PubMed] [Google Scholar]
- 59.Campbell JD, McDonough JE, Zeskind JE, Hackett TL, Pechkovsky DV, Brandsma CA, Suzuki M, Gosselink JV, Liu G, Alekseyev YO, et al. A gene expression signature of emphysema-related lung destruction and its reversal by the tripeptide GHK. Genome Med. 2012;4:67. doi: 10.1186/gm367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Owsijewitsch M, Ley-Zaporozhan J, Kuhnigk JM, Kopp-Schneider A, Eberhardt R, Eichinger M, Heussel CP, Kauczor HU, Ley S. Quantitative emphysema distribution in anatomic and non-anatomic lung regions. COPD. 2015;12:257–266. doi: 10.3109/15412555.2014.933950. [DOI] [PubMed] [Google Scholar]
- 61.Beasley TM, Erickson S, Allison DB. Rank-based inverse normal transformations are increasingly used, but are they merited? Behav Genet. 2009;39:580–595. doi: 10.1007/s10519-009-9281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]