Abstract
Mesenchymal stem cells (MSCs) have been reported to participate in the formation of supportive tumor stroma. The abilities of proliferation and invasion of human epithelial ovarian cancer (EOC) cells were significantly enhanced when indirectly cocultured with human omental adipose-derived MSCs (O-ADSCs) in vitro. However, the underlying mechanisms remain poorly understood. In this study, EOC cells were cultured with conditioned medium (CM) from O-ADSCs (O-ADSC), and the effect of O-ADSC CM on the proteomic profile of EOC cells was assessed by two-dimensional gel electrophoresis (2-DE), followed by liquid chromatography and tandem mass spectrometry. The 2-DE assays revealed a global increase in protein expression in the EOC cells treated with CM. Nine proteins were identified from 11 selected protein spots with differential expression after treatment with CM from O-ADSCs. All the nine proteins have been linked to carcinoma and apoptosis, and the migration ability of tumor cells can be regulated by these proteins. Moreover, the upregulation of prohibitin and serine/arginine-rich splicing factor 1 in EOC cells treated with CM was further confirmed by quantitative real-time polymerase chain reaction. These results suggest that O-ADSCs affect the proteomic profile of EOC cells via paracrine mechanism in favor of EOC progression.
Keywords: ovarian cancer, mesenchymal stromal cells, mesenchymal stem cells, omentum, proteomic
Introduction
Ovarian cancer is the most lethal gynecologic malignancy and the fifth leading cause of cancer deaths among females. The curative rate is >90% in patients with ovary-confined cancers. However, early symptoms of ovarian cancer are often confused with irritable bowel or premenstrual syndrome. Most patients are diagnosed at a late stage with dissemination of the tumor beyond the ovaries.1
The omentum is one of the most common sites of ovarian cancer metastasis. However, the reason why the omentum is susceptible to ovarian cancer metastasis is yet unknown. Omental tumor microenvironment has been reported to promote ovarian cancer cell adhesion, invasion, and proliferation. Human omental adipose-derived mesenchymal stem cells (O-ADSCs) are a population of multipotent mesenchymal stem cells (MSCs) contained in the omentum tissue that show tropism toward tumorigenic sites and participate in the formation of supportive tumor stroma. O-ADSCs could engraft in ovarian cancer stroma and contribute to the formation of a hospitable environment for cancer cells.2,3 Furthermore, O-ADSCs promoted pericyte coverage of blood vessels to enhance tumor vascularization and induced chemotherapy resistance by regulating metabolism via nitric oxide pathways in ovarian cancer.4,5 MSCs are known to interact with cancer cells through not only direct cell-to-cell contact but also the secretion of paracrine factors. Recent research showed that conditioned medium (CM) from MSCs significantly increased resistance to ciprofloxacin in urinary tract cancer cell lines.6 In ovarian cancer, MSC secretions were able to confer chemoresistance to carboplatin through the inhibition of effector caspase activation and apoptosis blockade.7 Despite intensive investigation, the influence of MSCs on tumor progression remains unclear. Several studies suggested that MSCs promoted tumor growth, while others reported contradicting results.8–12 To explore the impact of O-ADSCs on human epithelial ovarian cancer (EOC) development, we previously showed that O-ADSCs significantly promoted the proliferation and invasion of EOC cells in an indirect coculture system.13 However, the underlying mechanisms remain poorly understood. The aim of this study was therefore to demonstrate the proteomic alterations in ovarian cancer cells resulting from treatment with CM from O-ADSCs and explore how O-ADSCs contribute to ovarian cancer progression.
Materials and methods
Reagent and apparatus
Dulbecco’s Modified Eagle’s Medium (DMEM) was purchased from Gibco (Waltham, MA, USA), and fetal bovine serum (FBS) was also purchased from Gibco. Urea, CHAPS zwitterionic detergent (CHAPS), dithiothreitol (DTT), immobilized pH gradient (IPG) buffer, Tris-HCl, and IPG strips (17 cm, pH 4–7, linear) were purchased from Bio-Rad (Hercules, CA, USA). Collagenase (Type I), protease inhibitory cocktail, and thiourea were purchased from Sigma-Aldrich (St Louis, MO, USA). Sodium dodecyl sulfate (SDS), glycerol, Tris base, agarose, glycine, and Coomassie R-250 were purchased from Biosharp (Hefei, China). Iodoacetamide was purchased from TCI (Portland, OR, USA). Bromophenol blue was purchased from Fisher Scientific (Pittsburgh, PA, USA). TRIzol reagent and reverse transcription kit were purchased from Takara (Shiga, Japan). Fast SYBR Green Master Mix was purchased from Toyobo (Osaka, Japan).
Isolation and culture of O-ADSCs from omentum tissue
Omentum tissue was obtained from cancer-free female donors diagnosed with uterine fibroids, who underwent abdominal myomectomy. This study protocol was approved by the ethics committee of Tongji Medical College, Huazhong University of Science and Technology (IORG number IORG 0003571), and all donors provided written informed consent. The omentum specimen was placed in DMEM supplemented with 10% FBS for transport to the laboratory after collection. In a laminar hood, the tissue was washed with sterile phosphate-buffered saline (PBS) for several times, and then, it was finely minced. The minced tissue was placed into a new sterile 15 mL conical tube containing 10 mL DMEM and 1 mg/mL collagenase (Type I). The tissue was incubated for 1 h at 37°C with gentle shaking to dissociate the cells. After inactivation of collagenase by adding one volume of DMEM containing 10% FBS, the cells were collected by centrifugation for 10 min at 300× g, and the cell pellet was resuspended in PBS and passed through a 100 μm filter. The collected cells were centrifuged again. Cell pellet was resuspended in complete culture medium (DMEM) containing 10% FBS. Isolated cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
Collection of CM from O-ADSCs
O-ADSCs (5×105) were seeded in 100 mm diameter Petri dishes in DMEM containing 10% FBS until reaching 80% confluence as the culture medium was changed every other day. After the last change of medium, the cells were further cultured in fresh complete culture medium for additional 48 h and the medium was collected as CM from O-ADSCs, which thus contained DMEM, FBS, and secreted factors from O-ADSCs. O-ADSC CM was centrifuged for 5 min at 300× g and, finally, 0.22 μm filtered and stored at −20°C. Before use, O-ADSC CM was thawed and, then, used to treat EOC cell lines for experiment.
Cancer cells culture
Three human EOC cell lines, SKOV3, A2780, and HO-8910, were used in this study. SKOV3 and A2780 were purchased from China Center for Type Culture Collection (CCTCC; Wuhan, China). HO-8910 was purchased from Shanghai Fuxiang Biotechnology Co, Ltd. (Shanghai, China). All of these cell lines were cultured in DMEM supplemented with 10% FBS at 37°C in a humidified atmosphere of 5% CO2.
Treatment of cancer cells with O-ADSC CM
Cancer cells were seeded on six-well plates at a density of 1×105 cells/well. After 24 h preincubation, standard culture medium was exchanged for O-ADSC CM. For analyzing the effect of O-ADSC CM on the proteomic profile of EOC cells, SKOV3 was cultured for 7 days. For further confirmation of protein with differential expression in EOC cells treated with O-ADSC CM, cells were cultured for 0.5–1, 2, 4, and 6–8 days. EOC cells were cultured in DMEM containing 10% FBS as control.
Two-dimensional gel electrophoresis (2-DE)
Cells were lysed with cold rehydration mix (7 M urea, 2 M thiourea, 4% CHAPS, 65 mM DTT, and 0.2% IPG buffer) including 1 mM protease inhibitory cocktail for 10 min and then disrupted by ultrasound (VCX 130; Sonics, Newtown, CT, USA) for 10 cycles (on for 5 s and off for 10 s) at the amplitude of 35% on ice followed by centrifuging at 16,060× g at 4°C (Labofuge 400R; Heraeus, Hanau, Germany) for 70 min, and finally, the supernatants were collected. Protein concentration was measured by Bradford assay (Bio-Rad) according to the manufacturer’s protocol.
2-DE was performed using a PROTEAN® IEF Cell apparatus (Bio-Rad) according to the manufacturer’s instructions. For isoelectric focusing (IEF), a sample containing 2,500 μg protein was added to the above rehydration mix to make a final volume of 500 μL. The rehydration was performed with IPG strips (17 cm, pH 4–7, linear) for 14 h at 17°C. The IEF was run according to the following procedure: 350 V linear for 1 h, 750 V rapid for 1.5 h, 1,000 V rapid for 1.5 h, 10,000 V linear for 5 h, followed by 10,000 V rapid for 60,000 V h. After completion of IEF, the strips were conditioned in an equilibration buffer (7 M urea, 20% SDS, 20% glycerol, and 0.375 M Tris-HCl) containing 2% DTT for 13 min with gentle shaking and, then, transferred to an equilibration buffer containing 2.5% iodoacetamide for another 13 min with gentle shaking. For the second-dimension protein separation, the strips were placed in a gel cassette. The top reservoir contained the sealant solution (25 mM Tris base, 192 mM agarose, 0.1% SDS, and 0.001% bromophenol blue) and the bottom reservoir contained Tris-glycine buffer (25 mM Tris base, 192 mM glycine, 0.1% SDS, and pH 8.3). The strips were run at a constant voltage setting: 80 V for 1 h, then 120 V until the bromophenol dye front migrated to the lower end of the gels. These experiments were repeated three times.
Gel imaging and analysis
After electrophoresis, the gels were stained by a modified Coomassie R-250 method, as previously described.10 Then, the gels were imaged using a scanner (GS-800 Calibrated Densitometer; Bio-Rad) and further analyzed using PDQuest (Version 8.0; Bio-Rad).
Protein analysis by liquid chromatography and tandem mass spectrometry (LC–MS/MS) technology
According to their nearly neutral isoelectric point (pI) and more obvious difference in abundance, spots of interest were manually excised from the 2-DE gels and destained by washing with a mixture of 200 mM NH4HCO3/acetonitrile (1:1). Following reducing with DTT, alkylating with iodoacetamide, and digesting in-gel with trypsin (Promega, Madison, WI, USA), the proteins were degraded into peptides, which were lyophilized and subsequently dissolved in 2% acetonitrile/0.1% formic acid. Each fraction was subjected to analysis by LC–MS/MS using a Nano HPLC (Eksigent, Dublin, CA, USA) coupled to a QSTAR Elite mass spectrometer (Applied Biosystems, Waltham, MA, USA).14
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using the TRIzol reagent, and cDNA was synthesized from 1,000 ng of total RNA using a reverse transcription kit according to the manufacturer’s protocol. The qRT-PCR was performed using an ABI StepOnePlus thermocycler (Applied Biosystems) using the Fast SYBR Green Master Mix. The sequences of the primers are listed in Table 1. Expressions of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TATA box binding protein (TBP), and ribosomal protein, large, P0 (RPLP0) were used for normalization. The reactions for serine/arginine-rich splicing factor 1 (SRSF1), TBP, and GAPDH were performed by two-step thermal cycling method: 95°C for 1 min, 40 PCR cycles at 95°C for 30 s, 59°C for 20 s, followed by generating melting curves to check the specificity of the reactions. The reaction condition for prohibitin (PHB) and RPLP0 was similar, but the annealing temperature was different as follows: PHB (61°C) and RPLP0 (60°C). Each reaction was repeated in triplicate. The results were calculated using the Ct (2−ΔΔCt) method. These experiments were performed in three times using three EOC cell lines (SKOV3, A2780, and HO-8910).
Table 1.
Primer sequences for qRT-PCR
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| PHB | GTCCTTGACACATCTGACCTTCGGG | CAGCAGAGATGATGGCCGCCT |
| SRSF1 | GGAACAACGATTGCCGCATCTA | CTTGAGGTCGGATGTCGCGGATA |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
| TBP | TGCACAGGAGCCAAGAGTGAA | CACATCACAGCTCCCCACCA |
| RPLP0 | TTAAACCCTGAGTGGCAATCC | CCACATTCCCCCGGATATGA |
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PHB, prohibitin; qRT-PCR, quantitative real-time polymerase chain reaction; RPLP0, ribosomal protein, large, P0; SRSF1, serine/arginine-rich splicing factor 1; TBP, TATA box binding protein.
Statistical analysis
Statistical analyses were performed using SPSS 11.0. The results of qRT-PCR were expressed as mean ± SD, and differences between groups were analyzed using the Student’s t-test and one-way analysis of variance as appropriate. All tests were two tailed, and the statistical significance level was set at P<0.05.
Results
O-ADSC CM alters the proteomic profile of human ovarian cancer cells
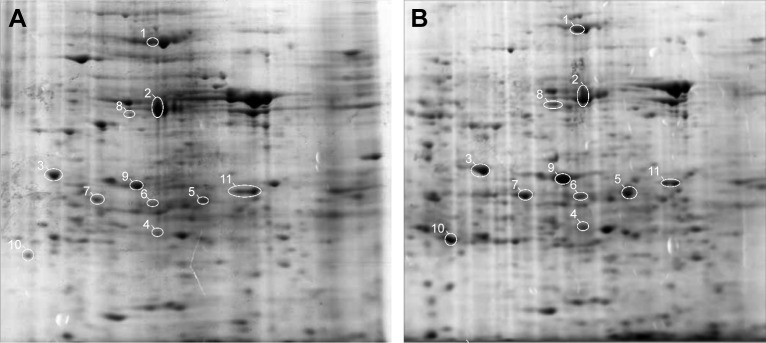
To explore the effects of O-ADSCs on the proteomic profile in EOC, proteomic expression analysis by 2-DE was performed using SKOV3 cells cultured in O-ADSC CM for 7 days. SKOV3 cells cultured in DMEM supplemented with 10% FBS were used as control. The results of gel analysis revealed an adequate reproducibility of assays and a global increase in protein expression in the cells treated with CM (757±60.7 vs 558±15.0 spots, P=0.019). 2-DE analysis revealed a total of 165 spots, which demonstrated a threefold increase or decrease in the protein expression level in the O-ADSC CM-treated SKOV3 cells relative to the control cells. Among these spots, 119 spots exhibited an increased expression and 46 spots showed a decreased expression in SKOV3 cells treated with O-ADSC CM compared to those cultured in DMEM containing 10% FBS (Figure 1).
Figure 1.
Representative two-dimensional electrophoretic images of human epithelial ovarian cancer SKOV3 cells untreated (A) and treated with CM from O-ADSC (B).
Notes: Eleven spots with significant alterations in expression that were subjected to mass spectrometry identification are encircled in white. Among them, spots 1–10 are enlarged and spot 11 becomes smaller after treatment with O-ADSC CM. No magnification.
Abbreviations: CM, conditioned medium; O-ADSC, omental adipose-derived mesenchymal stem cell.
Functional significance of differentially expressed proteins
Eleven spots with differential expression were chosen for protein identification by LC–MS/MS analysis (Figure 1). Finally, the following nine proteins were identified (Table 2): PHB, SRSF1, β-actin (ACTB), tropomyosin 4 (TPM4), Cathepsin D (CTSD), heat shock protein 60 (HSP60), cytokeratin 1 (CK1), pyruvate kinase isozyme M1/M2 (KPYM), and peroxiredoxin 3 (PRDX3). All the proteins have been reported to be relevant in tumor progression, and they may be associated with tumor formation (HSP60, PHB, and SRSF1) and cancer chemoresistance (KPYM, PHB, CTSD, and PRDX3) and used as clinical markers for cancer diagnosis (CK1 and TPM4). We summarized their associated diseases and functions in Table 3.
Table 2.
Characteristics of differentially expressed protein alterations in SKOV3 cells between treated and untreated groups with O-ADSC CM
| Spot number | Accession number | Protein name | Mascot score | MW (Da) | pI | Up/downregulated after treatment with CM | Abbreviation name |
|---|---|---|---|---|---|---|---|
| 1 | JAC06007 | 60 kDa heat shock protein | 4,335 | 61,187 | 6.73 | Upregulated | HSP60 |
| 2 | P60709 | β-Actin | 1,871 | 42,052 | 5.29 | Upregulated | ACTB |
| 3 | NP-01193728 | Pyruvate kinase isozyme M1/M2 | 1,421 | 58,470 | 7.95 | Upregulated | KPYM |
| 4 | CAG46507 | Prohibitin | 1,012 | 29,843 | 5.57 | Upregulated | PHB |
| 5 | NP-008855 | Serine/arginine-rich splicing factor 1 | 501 | 27,842 | 10.37 | Upregulated | SRSF1 |
| 6 | No significant hits to report | ||||||
| 7 | AA14156 | Cathepsin D | 676 | 45,037 | 5.6 | Upregulated | CTSD |
| 8 and 9 | P04624 | Cytokeratin 1 | 631 | 66,170 | 8.15 | Upregulated | CK1 |
| 10 | P30048 | Peroxiredoxin 3 | 436 | 28,017 | 5.77 | Upregulated | PRDX3 |
| 11 | NP-001138632 | Tropomyosin 4 | 667 | 28,619 | 5.19 | Downregulated | TPM4 |
Abbreviations: CM, conditioned medium; MW, molecular weight; O-ADSC, omental adipose-derived mesenchymal stem cell; pI, isoelectric point.
Table 3.
Associated disease and functions of identified proteins
| Spot number | Protein name | Associated disease (reference) | Protein function (reference) |
|---|---|---|---|
| 1 | 60 kDa heat shock protein | Colorectal cancer40 and hepatocellular carcinoma41 | Involved in protein import and correct folding and associated with tumorigenesis40 and underexpression following treatment with chemotherapeutic agent41 |
| 2 | β-Actin | Colorectal neoplasia42 | Always used as denominators of gene expression quantitation, but overexpression in colorectal neoplasia42 |
| 3 | Pyruvate kinase isozyme M1/M2 | Human malignant melanoma43 | Associated in cell apoptosis43 |
| 4 | Prohibitin | Ovarian cancer18 | Stabilization of mitochondrial function,17,29 inhibits the intrinsic apoptotic pathway,18 and required for Ras-induced Raf–MEK–ERK activation and epithelial cell migration16 |
| 5 | Serine/arginine-rich splicing factor 1 | Non-small-cell lung cancer35 and breast cancer36 | A splicing factor as an oncoprotein,34 involved in tumor poor prognosis by promoting survivin stability,35 promotes mammary epithelial cell transformation,36 and enhances Wnt signaling activation20 |
| 7 | Cathepsin D | Pancreatic cancer44 and human cervical cancer45 | Correlates with malignant progression44 and antiapoptotic mediator by inducing autophagy45 |
| 8 and 9 | Cytokeratin 1 | Breast cancer46 and hepatocellular carcinoma44 | Diagnosis marker44,47 |
| 10 | Peroxiredoxin 3 | Hepatocellular carcinoma,48 prostate cancer,49 breast cancer,50 and ovarian cancer51 | Involved in chemoresistance48–50 associated with NF-κB signaling pathway52 |
| 11 | Tropomyosin 4 | Ovarian cancer53 | Diagnosis marker53 |
Abbreviations: MEK, mitogen-activated protein/extracellular signal-regulated kinase kinase; ERK, extracellular signal-regulated kinase; NF-κB, nuclear factor-κB.
O-ADSC CM upregulates the mRNA level of PHB and SRSF1 in human ovarian cancer
Mutational activation of the Ras–Raf–MEK–ERK kinase pathway is frequently observed in a wide range of human cancers, including gynecological cancer, and PHB has been demonstrated to be mandatory for this signaling pathway.15,16 As listed in Table 2, PHB was involved in tumor progression in ovarian cancer.17,18 Resistance to chemotherapy usually arises in ovarian cancer. Major signaling pathways were observed to upregulate in this process, including Wnt/β-catenin, and SRSF1 was required for the synthesis of β-catenin protein.19,20 Recent studies have shown that SRSF1 determined wild-type P53 expression in ovarian cancer.21 Therefore, we further verified the differential expression of PHB and SRSF1 in three EOC cell lines (SKOV3, A2780, and HO-8910) with or without treatment with O-ADSC CM by qRT-PCR.
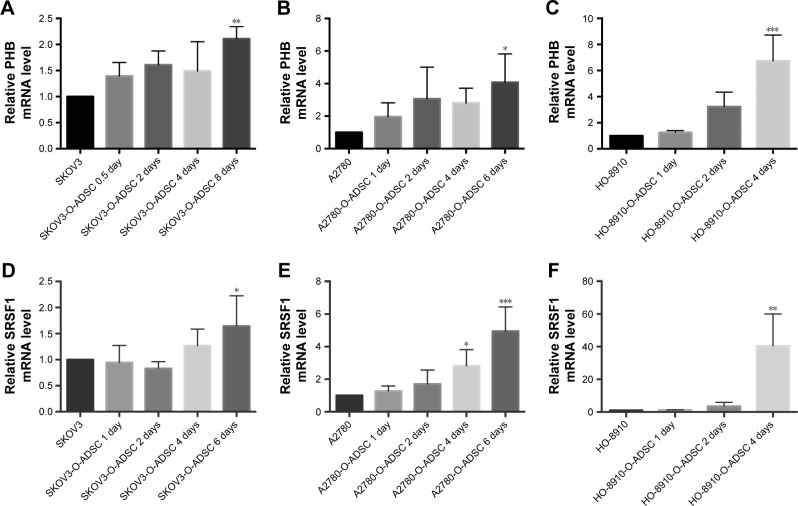
Compared with untreated human EOC cells, PHB mRNA expression was increased in SKOV3 (2.110±0.235 folds, P=0.006), A2780 (3.522±0.956 folds, P=0.015), and HO-8910 (6.748±1.969 folds, P<0.001) cells after treatment with O-ADSC CM for 4–8 days (Figure 2A–C). Similarly, O-ADSC also upregulated the mRNA level of SRSF1 in SKOV3 (1.844±0.44 folds, P=0.016), A2780 (2.825±0.991 folds, P=0.038; 4.952±1.488 folds, P<0.001), and HO-8910 (40.476±19.574 folds, P=0.033) cells treated with O-ADSC CM for 4–6 days (Figure 2D–F).
Figure 2.
O-ADSC CM upregulates the mRNA levels of PHB (A–C) and SRSF1 (D–F) in human epithelial ovarian cancer cell lines SKOV3, A2780, and HO-8910.
Notes: The histograms demonstrate the results of qRT-PCR. *P<0.05, **P<0.01, and ***P<0.001 compared to control cells that were not treated with O-ADSC CM.
Abbreviations: CM, conditioned medium; O-ADSC, omental adipose-derived mesenchymal stem cell; PHB, prohibitin; qRT-PCR, quantitative real-time polymerase chain reaction; SRSF1, serine/arginine-rich splicing factor 1.
Discussion
Given that O-ADSCs represent an important component in the tumor microenvironment promoting ovarian cancer progression, we were interested in the influence of O-ADSCs on the proteomic profile of EOC cells, which can be helpful in understanding the biological mechanisms underlying the protumorigenic role of O-ADSCs. CM, a cell-free bioactive substrate, plays a pivotal role in various pathophysiological processes through paracrine mechanism. Treating tumor cells with CM from MSCs has been used as a useful model in many cancers, including ovarian cancer.22 Therefore, we evaluated the effect of O-ADSCs on the proteomic profile of EOC cells through this model. In this study, we found significant proteomic alterations in EOC cells after O-ADSC CM treatment and identified a serial of proteins with differential expression on the base of differential expression analysis of 2-DE results. Indeed, these proteins have been implicated in regulating a variety of biological behaviors of tumor cells, including cell apoptosis, migration, and chemoresistance. These findings strengthen the evidence supporting the tumor-promoting effect of O-ADSCs even in the absence of direct contact with tumor cell and open a door for further research on the paracrine-based mechanism.
Recently, adipose-derived MSCs (ADSCs) have been shown to release extracellular vesicles (EVs) that might include exosomes, microvesicles, and other entities.23,24 Accumulating evidence indicates that EVs play a critical role in the tumor–stroma cell interaction.25–27 Moreover, a series of paracrine factors secreted by ADSCs, including FGF10, VEGFC, and matrix metalloproteinases (MMPs), have been reported to promote tumor progression through microenvironment network, such as activating Wnt signaling in colon cancer cells.28 These findings support that ADSCs can regulate tumor cell function in the absence of direct cell–cell contact as shown in this study.
We found that PHB and SRSF1 were significantly upregulated in EOC cells cultured with O-ADSC CM. PHB, mainly positioning in mitochondria, plays an important role to maintain mitochondria stabilization.29 In ovarian cancer, PHB has been reported to regulate cancer cell proliferation, apoptosis, and migration, partially through mediating MMP-2 and MMP-9 expressions.17,18 Moreover, in PHB-deficient cells, C-Raf kinase failed to interact with active Ras induced by epidermal growth factor and the adhesion complex proteins cadherin and β-catenin relocalize to the plasma membrane, resulting in stabilizing adherent junctions.16 There is now compelling evidence showing that PHB undergoes phosphorylation in response to insulin, growth factor, and immune stimulation, playing an important role in these signaling pathways.16,30,31 The expressions of MMPs are also regulated by growth factors in ovarian cancer.32 Several studies have demonstrated that MSCs are capable of producing a wide range of growth factors including epidermal growth factor.33 Maybe secreted growth factors are one of the mechanisms involved in elevated PHB level and probably further regulated MMPs in EOC cells treated with O-ADSC CM. SRSF1 is a splicing factor that identified as an oncoprotein.34 SRSF1 is overexpressed in several human cancers and has been shown to promote the proliferation, transformation, and survival of cancer cells by enhancing survivin expression.35,36 In addition, SRSF1 could specially enhance β-catenin accumulation and Wnt signaling activation.20 Moreover, ADSCs regulated the expression of survivin and the activation of Wnt signaling in cancer cells via paracrine mechanism.25,28,37 Thus, the proteomic alterations resulting from O-ADSC CM, together with previous literature, cast light on the paracrine-mediated tumor-promoting activity of O-ADSCs in ovarian cancer.
In our previous study, O-ADSCs were found to be able to exacerbate the proliferation, invasion, and migration of EOC cells.13 Recently, Chen et al28 found that ADSCs could enhance the metastatic capacity of colon cancer cells by inducing epithelial–mesenchymal transition (EMT) and colon cancer cells in turn enhanced the paracrine signaling of ADSCs with unaltered differentiation potential. More importantly, emerging evidence showed that cancer cells could induce ADSCs to exhibit the phenotype of tumor-associated myofibroblasts (TAFs), which provided a favorable environment for tumor progression.38,39 In the present study, O-ADSC CM was used to investigate the effect of normal ADSCs on EOC cells. This experiment modal excludes the effect of direct ADSC–EOC cell contact and the influence of tumor cell on ADSC and, thus, may represent the conditions of an early-stage tumor.
Conclusion
Our findings indicate that O-ADSC CM alters the proteomic profile of EOC cells to promote tumor malignant progression, which shed light on the interaction between O-ADSCs and EOC cells, but the identification of the specific molecules, which are responsible for the observed protumorigenic effect of O-ADSC CM on EOC cells, should be further investigated.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 81272860, 81472443, 81302269, and 81572572).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Nowicka A, Marini FC, Solley TN, et al. Human omental-derived adipose stem cells increase ovarian cancer proliferation, migration, and chemoresistance. PLoS One. 2013;8(12):e81859. doi: 10.1371/journal.pone.0081859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamat P, Schweizer R, Kaenel P, et al. Human adipose-derived mesenchymal stromal cells may promote breast cancer progression and metastatic spread. Plast Reconstr Surg. 2015;136(1):76–84. doi: 10.1097/PRS.0000000000001321. [DOI] [PubMed] [Google Scholar]
- 4.Klopp AH, Zhang Y, Solley T, et al. Omental adipose tissue-derived stromal cells promote vascularization and growth of endometrial tumors. Clin Cancer Res. 2012;18(3):771–782. doi: 10.1158/1078-0432.CCR-11-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salimian Rizi B, Caneba C, Nowicka A, et al. Nitric oxide mediates metabolic coupling of omentum-derived adipose stroma to ovarian and endometrial cancer cells. Cancer Res. 2015;75(2):456–471. doi: 10.1158/0008-5472.CAN-14-1337. [DOI] [PubMed] [Google Scholar]
- 6.Maj M, Bajek A, Nalejska E, et al. Influence of mesenchymal stem cells conditioned media on proliferation of urinary tract cancer cell lines and their sensitivity to ciprofloxacin. J Cell Biochem. 2016 Nov 22; doi: 10.1002/jcb.25794. Epub. [DOI] [PubMed] [Google Scholar]
- 7.Castells M, Milhas D, Gandy C, et al. Microenvironment mesenchymal cells protect ovarian cancer cell lines from apoptosis by inhibiting XIAP inactivation. Cell Death Dis. 2013;4:e887. doi: 10.1038/cddis.2013.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klopp AH, Gupta A, Spaeth E, Andreeff M, Marini F., 3rd Concise review: dissecting a discrepancy in the literature: do mesenchymal stem cells support or suppress tumor growth? Stem Cells. 2011;29(1):11–19. doi: 10.1002/stem.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 10.Ning H, Lei HE, Xu YD, et al. Conversion of adipose-derived stem cells into natural killer-like cells with anti-tumor activities in nude mice. PLoS One. 2014;9(8):e106246. doi: 10.1371/journal.pone.0106246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahara K, Ii M, Inamoto T, et al. Adipose-derived stromal cells inhibit prostate cancer cell proliferation inducing apoptosis. Biochem Biophys Res Commun. 2014;446(4):1102–1107. doi: 10.1016/j.bbrc.2014.03.080. [DOI] [PubMed] [Google Scholar]
- 12.Spaeth EL, Dembinski JL, Sasser AK, et al. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS One. 2009;4(4):e4992. doi: 10.1371/journal.pone.0004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu Y, Tang H, Guo Y, et al. Adipose-derived mesenchymal stem cells promote cell proliferation and invasion of epithelial ovarian cancer. Exp Cell Res. 2015;337(1):16–27. doi: 10.1016/j.yexcr.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Cai J, Huang Z, et al. Proteomic identification of target proteins following Drosha knockdown in cervical cancer. Oncol Rep. 2013;30(5):2229–2237. doi: 10.3892/or.2013.2672. [DOI] [PubMed] [Google Scholar]
- 15.Marampon F, Gravina GL, Popov VM, et al. Close correlation between MEK/ERK and Aurora-B signaling pathways in sustaining tumorigenic potential and radioresistance of gynecological cancer cell lines. Int J Oncol. 2014;44(1):285–294. doi: 10.3892/ijo.2013.2167. [DOI] [PubMed] [Google Scholar]
- 16.Rajalingam K, Wunder C, Brinkmann V, et al. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat Cell Biol. 2005;7(8):837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 17.Gregory-Bass RC, Olatinwo M, Xu W, et al. Prohibitin silencing reverses stabilization of mitochondrial integrity and chemoresistance in ovarian cancer cells by increasing their sensitivity to apoptosis. Int J Cancer. 2008;122(9):1923–1930. doi: 10.1002/ijc.23351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Liao H, Zheng HC, et al. Effect of luteinizing hormone-induced prohibitin and matrix metalloproteinases on ovarian epithelial tumor cell proliferation. Am J Cancer Res. 2015;5(1):114–124. [PMC free article] [PubMed] [Google Scholar]
- 19.Tomao F, Papa A, Strudel M, et al. Investigating molecular profiles of ovarian cancer: an update on cancer stem cells. J Cancer. 2014;5(5):301–310. doi: 10.7150/jca.8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Y, Huang B, Shi Z, et al. SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing beta-catenin biosynthesis. EMBO Mol Med. 2013;5(5):737–750. doi: 10.1002/emmm.201202218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patwardhan GA, Hosain SB, Liu DX, et al. Ceramide modulates pre-mRNA splicing to restore the expression of wild-type tumor suppressor p53 in deletion-mutant cancer cells. Biochim Biophys Acta. 2014;1841(11):1571–1580. doi: 10.1016/j.bbalip.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reza AM, Choi YJ, Yasuda H, Kim JH. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Scientific Reports. 2016;6:38498. doi: 10.1038/srep38498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Shen H, Ma J, Sun L, Wei M. Extracellular Vesicles Derived from Adipose Mesenchymal Stem Cells Regulate the Phenotype of Smooth Muscle Cells to Limit Intimal Hyperplasia. Cardiovascular Drugs and Therapy. 2016;30(2):111–118. doi: 10.1007/s10557-015-6630-5. [DOI] [PubMed] [Google Scholar]
- 24.Lopatina T, Bruno S, Tetta C, Kalinina N, Porta M, Camussi G. Platelet-derived growth factor regulates the secretion of extracellular vesicles by adipose mesenchymal stem cells and enhances their angiogenic potential. Cell Commun Signal. 2014;12:26. doi: 10.1186/1478-811X-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin R, Wang S, Zhao RC. Exosomes from human adipose-derived mesenchymal stem cells promote migration through Wnt signaling pathway in a breast cancer cell model. Mol Cell Biochem. 2013;383(1–2):13–20. doi: 10.1007/s11010-013-1746-z. [DOI] [PubMed] [Google Scholar]
- 26.Silvers CR, Liu YR, Wu CH, Miyamoto H, Messing EM, Lee YF. Identification of extracellular vesicle-borne periostin as a feature of muscle-invasive bladder cancer. Oncotarget. 2016;7(17):23335–23345. doi: 10.18632/oncotarget.8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko SF, Yip HK, Zhen YY, et al. Adipose-Derived Mesenchymal Stem Cell Exosomes Suppress Hepatocellular Carcinoma Growth in a Rat Model: Apparent Diffusion Coefficient, Natural Killer T-Cell Responses, and Histopathological Features. Stem Cells International. 2015;2015:853506. doi: 10.1155/2015/853506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D, Liu S, Ma H, et al. Paracrine factors from adipose-mesenchymal stem cells enhance metastatic capacity through Wnt signaling pathway in a colon cancer cell co-culture model. Cancer Cell Int. 2015;15:42. doi: 10.1186/s12935-015-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen NG, Lu CC, Lin YH, et al. Proteomic approaches to study epigallocatechin gallate-provoked apoptosis of TSGH-8301 human urinary bladder carcinoma cells: roles of AKT and heat shock protein 27-modulated intrinsic apoptotic pathways. Oncol Rep. 2011;26(4):939–947. doi: 10.3892/or.2011.1377. [DOI] [PubMed] [Google Scholar]
- 30.Ande SR, Moulik S, Mishra S. Interaction between O-GlcNAc modification and tyrosine phosphorylation of prohibitin: implication for a novel binary switch. PLoS One. 2009;4(2):e4586. doi: 10.1371/journal.pone.0004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Nath N, Fusaro G, Chellappan S. Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol Cell Biol. 1999;19(11):7447–7460. doi: 10.1128/mcb.19.11.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh S, Basu M, Roy SS. ETS-1 protein regulates vascular endothelial growth factor-induced matrix metalloproteinase-9 and matrix metalloproteinase-13 expression in human ovarian carcinoma cell line SKOV-3. J Biol Chem. 2012;287(18):15001–15015. doi: 10.1074/jbc.M111.284034. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.English K. Mechanisms of mesenchymal stromal cell immunomodulation. Immunol Cell Biol. 2013;91(1):19–26. doi: 10.1038/icb.2012.56. [DOI] [PubMed] [Google Scholar]
- 34.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14(3):185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ezponda T, Pajares MJ, Agorreta J, et al. The oncoprotein SF2/ASF promotes non-small cell lung cancer survival by enhancing survivin expression. Clin Cancer Res. 2010;16(16):4113–4125. doi: 10.1158/1078-0432.CCR-10-0076. [DOI] [PubMed] [Google Scholar]
- 36.Anczukow O, Rosenberg AZ, Akerman M, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19(2):220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Zhang Z, Yao C. Survivin is upregulated in myeloma cell lines cocultured with mesenchymal stem cells. Leuk Res. 2010;34(10):1325–1329. doi: 10.1016/j.leukres.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Cho JA, Park H, Lim EH, Lee KW. Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int J Oncol. 2012;40(1):130–138. doi: 10.3892/ijo.2011.1193. [DOI] [PubMed] [Google Scholar]
- 39.Cho JA, Park H, Lim EH, et al. Exosomes from ovarian cancer cells induce adipose tissue-derived mesenchymal stem cells to acquire the physical and functional characteristics of tumor-supporting myofibroblasts. Gynecol Oncol. 2011;123(2):379–386. doi: 10.1016/j.ygyno.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Stierum R, Gaspari M, Dommels Y, et al. Proteome analysis reveals novel proteins associated with proliferation and differentiation of the colorectal cancer cell line Caco-2. Biochim Biophys Acta. 2003;1650(1–2):73–91. doi: 10.1016/s1570-9639(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Owusu L, Duan W, et al. Anti-metastatic and differential effects on protein expression of epigallocatechin-3-gallate in HCCLM6 hepatocellular carcinoma cells. Int J Mol Med. 2013;32(4):959–964. doi: 10.3892/ijmm.2013.1446. [DOI] [PubMed] [Google Scholar]
- 42.Polley AC, Mulholland F, Pin C, et al. Proteomic analysis reveals field-wide changes in protein expression in the morphologically normal mucosa of patients with colorectal neoplasia. Cancer Res. 2006;66(13):6553–6562. doi: 10.1158/0008-5472.CAN-06-0534. [DOI] [PubMed] [Google Scholar]
- 43.Nawarak J, Huang-Liu R, Kao SH, et al. Proteomics analysis of kojic acid treated A375 human malignant melanoma cells. J Proteome Res. 2008;7(9):3737–3746. doi: 10.1021/pr7008737. [DOI] [PubMed] [Google Scholar]
- 44.Sulpizio S, Franceschini N, Piattelli A, Di Sebastiano P, Innocenti P, Selvaggi F. Cathepsins and pancreatic cancer: the 2012 update. Pancreatology. 2012;12(5):395–401. doi: 10.1016/j.pan.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Hah YS, Noh HS, Ha JH, et al. Cathepsin D inhibits oxidative stress-induced cell death via activation of autophagy in cancer cells. Cancer Lett. 2012;323(2):208–214. doi: 10.1016/j.canlet.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 46.Attallah AM, El-Far M, Omran MM, et al. Circulating levels and clinical implications of epithelial membrane antigen and cytokeratin-1 in women with breast cancer: can their ratio improve the results? Tumour Biol. 2014;35(11):10737–10745. doi: 10.1007/s13277-014-2375-1. [DOI] [PubMed] [Google Scholar]
- 47.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29(1):22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 48.Ismail S, Mayah W, Battia HE, et al. Plasma nuclear factor kappa B and serum peroxiredoxin 3 in early diagnosis of hepatocellular carcinoma. Asian Pac J Cancer Prev. 2015;16(4):1657–1663. doi: 10.7314/apjcp.2015.16.4.1657. [DOI] [PubMed] [Google Scholar]
- 49.Ummanni R, Barreto F, Venz S, et al. Peroxiredoxins 3 and 4 are overexpressed in prostate cancer tissue and affect the proliferation of prostate cancer cells in vitro. J Proteome Res. 2012;11(4):2452–2466. doi: 10.1021/pr201172n. [DOI] [PubMed] [Google Scholar]
- 50.McDonald C, Muhlbauer J, Perlmutter G, Taparra K, Phelan SA. Peroxiredoxin proteins protect MCF-7 breast cancer cells from doxorubicin-induced toxicity. Int J Oncol. 2014;45(1):219–226. doi: 10.3892/ijo.2014.2398. [DOI] [PubMed] [Google Scholar]
- 51.Kalinina EV, Berezov TT, Shtil AA, et al. Expression of peroxiredoxin 1, 2, 3, and 6 genes in cancer cells during drug resistance formation. Bull Exp Biol Med. 2012;153(6):878–881. doi: 10.1007/s10517-012-1849-7. Article in English, Russian. [DOI] [PubMed] [Google Scholar]
- 52.Duan J, Lang Y, Song C, Xiong J, Wang Y, Yan Y. siRNA targeting of PRDX3 enhances cisplatininduced apoptosis in ovarian cancer cells through the suppression of the NFkappaB signaling pathway. Mol Med Rep. 2013;7(5):1688–1694. doi: 10.3892/mmr.2013.1370. [DOI] [PubMed] [Google Scholar]
- 53.Bailey MJ, Shield-Artin KL, Oliva K, Ayhan M, Reisman S, Rice GE. Stage-specific analysis of plasma protein profiles in ovarian cancer: Difference in-gel electrophoresis analysis of pooled clinical samples. J Carcinog. 2013;12:10. doi: 10.4103/1477-3163.114216. [DOI] [PMC free article] [PubMed] [Google Scholar]