Abstract
The androgen receptor (AR) is required for prostate cancer (PCa) survival and progression, and ablation of AR activity is the first line of therapeutic intervention for disseminated disease. While initially effective, recurrent tumors ultimately arise for which there is no durable cure. Despite the dependence of PCa on AR activity throughout the course of disease, delineation of the AR-dependent transcriptional network that governs disease progression remains elusive, and the function of AR in mitotically active cells is not well understood. Analyzing AR activity as a function of cell cycle revealed an unexpected and highly expanded repertoire of AR-regulated gene networks in actively cycling cells. New AR functions segregated into two major clusters: those that are specific to cycling cells and retained throughout the mitotic cell cycle (“Cell Cycle Common”), versus those that were specifically enriched in a subset of cell cycle phases (“Phase Restricted”). Further analyses identified previously unrecognized AR functions in major pathways associated with clinical PCa progression. Illustrating the impact of these unmasked AR-driven pathways, dihydroceramide-desaturase 1 (DEGS1) was identified as an AR regulated gene in mitotically active cells that promoted pro-metastatic phenotypes, and in advanced PCa proved to be highly associated with development of metastases, recurrence after therapeutic intervention, and reduced overall survival. Taken together, these findings delineate AR function in mitotically active tumor cells, thus providing critical insight into the molecular basis by which AR promotes development of lethal PCa and nominate new avenues for therapeutic intervention.
Introduction
Prostate cancer (PCa) is the most commonly diagnosed non-cutaneous malignancy in the USA, and the second most lethal form of cancer (1). While localized disease is readily treatable through surgery and/or radiation, there is no curative intervention for advanced PCa. Advanced PCa typically remains reliant on androgen receptor (AR) signaling throughout disease progression (2, 3). As a hormone nuclear receptor, AR requires androgen (testosterone or dihydrotestestone, DHT) binding for activation. Once active, AR binds DNA and initiates transcriptional programs required for proliferation and survival, however the mechanisms governing these events have remained largely elusive. By contrast, the mechanism by which AR regulates proteases important for prostate function (e.g. Prostate Specific Antigen, PSA, encoded by KLK3), are well known, and PSA is used clinically to monitor tumor development and progression (4). Given the exquisite dependence of PCa on AR function, therapy for disseminated disease suppresses AR signaling, achieved by ablation of circulating ligand (androgen deprivation therapy, ADT), often used in combination with direct AR antagonists (2, 5). While initially efficacious, these regimens ultimately fail and castration resistant prostate cancer (CRPC) develops due to reactivation of the AR signaling axis (3, 6, 7). At present, there remain no durable means to manage recurrent AR activity and treat CRPC; as such developing understanding of AR function and mechanisms to target downstream processes represents a primary focus in the field.
Despite the importance of AR in driving PCa progression, the specific gene networks controlled by AR to induce advanced tumor phenotypes are not well understood. Recent studies revealed a role for AR in regulating PCa cell metabolism particularly as involved in glycolysis and anabolism flux, thereby supporting tumor cell growth (8, 9). Additionally, AR can indirectly modulate cell cycle progression via mTOR-dependent pathways that lead to Cyclin D1 accumulation, consequent activation of cyclin dependent kinase 4 (CDK4) and phosphorylation/inactivation of the RB tumor suppressor, and subsequent G1-S checkpoint transition (10). These observations provide clues to the biochemical framework for how AR promotes tumor cell growth, but the molecular basis for AR-dependent phenotypes associated with cell survival, cell migration, and metastasis remains largely unsolved. It is notable that genome wide analyses of AR activity have been performed in cells synchronized in G0 through steroid deprivation, leaving unresolved the role of AR in mitotically active cells (9, 11).
Studies herein reveal that AR induces differential transcriptional networks in mitotically active cells, unmasking critical new insight into AR function. Molecular dissection demonstrated that networks specific to actively cycling cells can be subdivided into those that are common to all active phases of the cell cycle, versus those that are specifically enriched in distinct cell cycle phases. Networks identified were strongly associated with novel AR cistromes through genome-wide binding analyses, demonstrating the unexpected finding that AR function is significantly expanded in cells committed to the mitotic cell cycle, and that AR function is sensitive to cell cycle position. Functional analyses unveiled novel mechanisms through which AR likely promotes pro-tumorigenic phenotypes including cell migration and tumor metastases. Of particular significance was the discovery of DEGS1, a desaturase involved primarily in the formation of ceramide, as a key target through which AR promotes pro-metastatic phenotypes. Analyses in human tumor samples further identified DEGS1 as strongly associated with disease progression and poor outcome, outperforming current biomarkers of aggressive disease. Taken together, the studies herein demonstrate that AR signaling is widely expanded in mitotically active cells, and promotes pro-tumorigenic networks underpinning cancer progression.
Results
Androgens Induce Distinct Gene Expression Programs in Cycling Cells
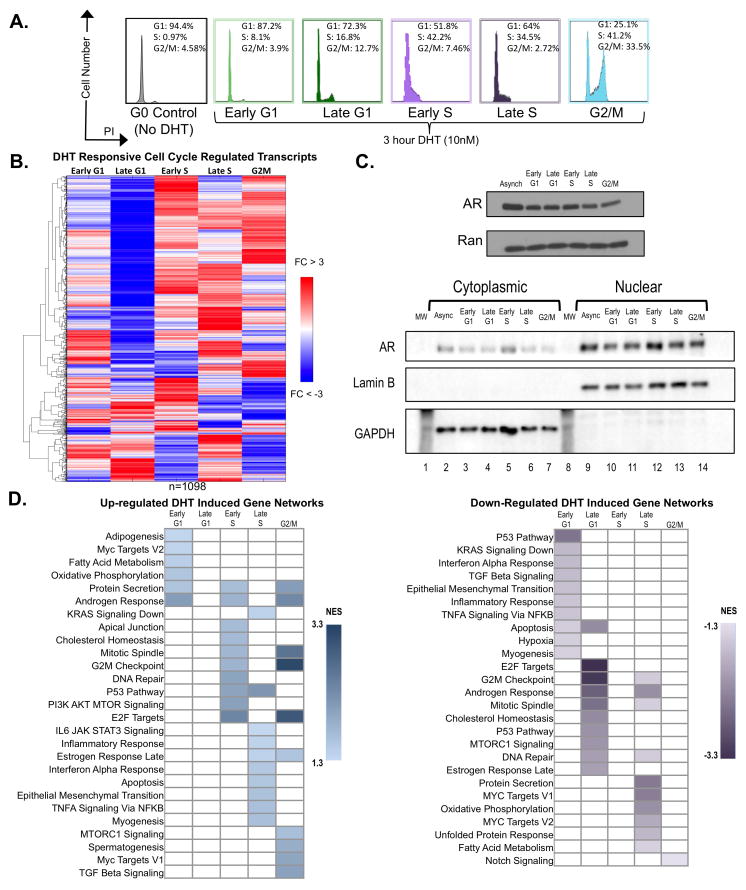
Despite the known ability of AR to promote PCa growth and progression, the molecular basis of AR function remains incompletely understood. As the majority of genome-wide analyses have been performed in cells that have exited the cell cycle via steroid deprivation prior to hormone stimulation, it was posited that AR functions supporting lethal tumor phenotypes may be uncovered by assessing AR activity in mitotically active cells. To explore this, human PCa models were utilized to discretely assess cell cycle-dependent effects of androgen and downstream cellular consequences. A baseline for AR activity in resting cells deprived of androgen was established using hormone therapy sensitive (HTS) PCa cells synchronized in G0 through steroid starvation (Fig 1A). In parallel, cells cultured in steroid replete conditions were enriched for distinct phases of cell cycle and subsequently subjected to a 3-hr pulse of DHT. Cells were enriched prior to DHT stimulation in early G1 (through use of a CDK4 inhibitor, 87.2% G1 phase), late G1 (via CDK2 suppression, 72.3% G1 phase), in early S (by DNA polymerase inactivation, 42.2% S phase), in mid-S phase (through suppression of ribonucleotide reductase, 34.5% S phase), and G2/M (through release from S-phase into nocodazole, 33.5% G2/M). Robust enrichment in desired phases was validated through flow cytometric analyses (Fig 1A). Differentially enriched transcripts were identified through gene expression analyses and compared to unstimulated (G0, steroid deprived) cells using a stringent cutoff for both fold change (FC≥1.5) and false discovery rate (FDR≤10%). After applying filtering parameters, 1098 transcripts demonstrated differential (positive or negative) cell cycle-regulated expression compared to unstimulated cells in G0. To identify androgen-responsive transcriptomes specific to distinct cell cycle phases, genes were clustered using a center-means based approach, considering only expression levels in DHT stimulated samples. Clustering analysis revealed that maximum up-regulation occurred after cell cycle commitment in early S phase and beyond, with maximum down-regulation observed in late G1 phase (Fig 1B). Additionally, changes in gene expression were not due to appreciable changes in AR protein levels (Fig 1C top), or alterations in AR nuclear localization (Fig 1C bottom). Further, in order to examine whether these expression changes could be mimicked through longer DHT stimulation times, genes altered specifically at 24 hour DHT stimulation were examined(12). Intriguingly, less than 50% of genes changed at 24 hours were seen to undergo cell cycle regulated gene expression in the current study, suggesting that while longer DHT stimulation times may identify a subset of these targets, it is not sufficient to identify a majority of genes differentially regulated by cell cycle phase (Fig S1). These observations suggest that AR-cell cycle interplay elicits distinct alterations in AR transcriptome in mitotically active cells, dependent on cell cycle phase.
Figure 1.
Gene set enrichment analysis (GSEA) identified DHT-responsive gene networks demonstrating cell-cycle enrichment specific to actively cycling PCa cells. Briefly, transcripts enriched in individual phases were systematically compared to the remaining datasets, resulting in identification of DHT-responsive gene networks that are up-regulated (Fig 1D left panel, Fig S2, Supplementary Table 1) or down-regulated (Fig 1D right panel, Fig S2, Supplementary Table 1) in a phase restricted manner. As expected, pathways previously identified as androgen regulated (e.g. androgen response and E2F hallmark signatures) (13–15) were up-regulated in early G1, early S, and G2/M phases (Fig 1D left). Both pathways were subsequently down-regulated in late G1 and late S (Fig 1D right), suggesting that previously reported androgen regulated pathways are cell cycle-dependent. Additionally, as AR has been recently shown to play an important role in DNA repair (16, 17), it is notable that hallmark DNA repair and p53 regulated genes were up-regulated by DHT exclusively in S and G2/M phases, suggesting that the capacity of AR to modulate DNA repair is also sensitive to cell cycle position. Finally, pathways exclusively up-regulated in late S phase include epithelial to mesenchymal transition (EMT) and inflammatory signaling, pathways known to promote clinical progression in human disease (18, 19). These observations reveal putative new downstream consequences of AR signaling that provide insight into the means by which this receptor modulates aggressive tumor phenotypes. Mechanistically, these data reveal for the first time that AR signaling elicits distinct transcriptional programs across the cell cycle, and harbors expanded functions in mitotically active cells.
The AR Cistrome is Expanded in Mitotically Active Cells
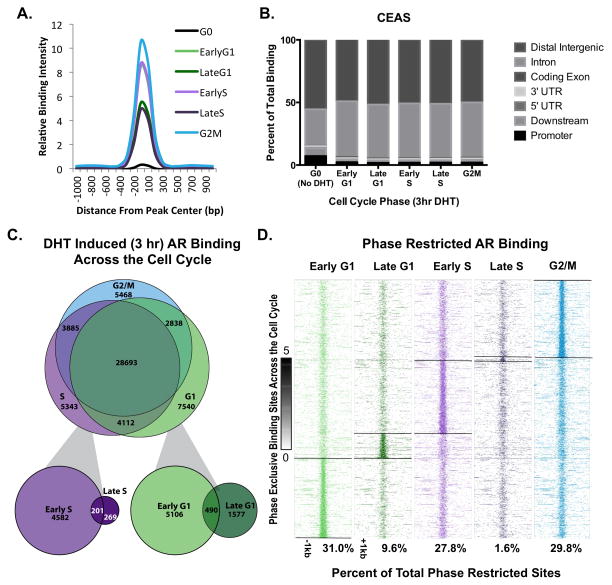
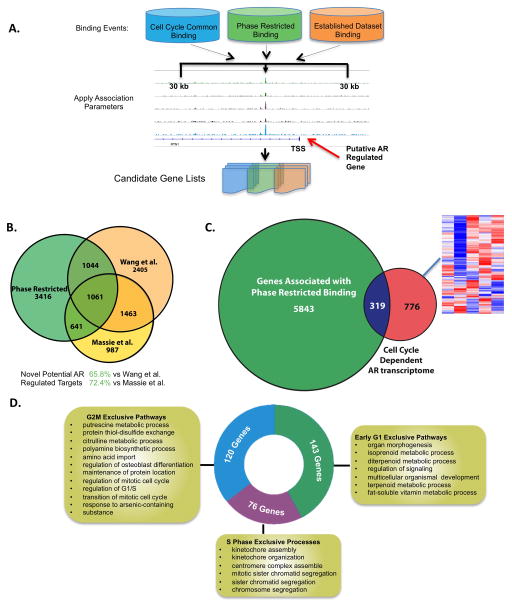
To discern the molecular basis of the observed expanded gene networks controlled by AR in mitotically active cells, parallel studies were performed to assess the cell cycle specific AR cistrome. AR binding sites (ARBSs) were found through chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq), using standard methodology (20). Statistically significant ARBSs within each phase were identified using MACS peak calling algorithm and a rigorous cutoff (p<1e−8). As expected, DHT significantly increased AR binding intensity in all phases when compared to steroid deprived cells; however, there was noted variance in DHT-induced intensity of AR binding amongst the cell cycle phases, with G2/M enriched cells demonstrating the highest intensity, and late G1 and late S exhibiting the weakest intensity (Fig 2A), perhaps reflective of differential chromatin accessibility in early versus late cell cycle. Using the cis-regulatory element annotation system (CEAS, Figure 2B), in unstimulated (G0) cells, AR bound intronic (29.2%) and distal intergenic (55.4%) regions, with a small fraction observed in promoter (0–3000bp from TSS, 7.2%) and downstream (0–3000bp from TTS, 6.3%) regions. Comparatively, AR binding in mitotically active cells occurred most frequently (>95%) in intronic and distal intergenic regions, indicating a shift in preferential binding regions after DHT treatment and cell cycle activation. Additionally, binding events in early G1, early S, and G2/M contained comparable numbers of AR occupied sites (38,620; 41,023; and 40,884 respectively), whereas binding in late G1 and late S was slightly reduced (26,202 and 20,720 respectively, Fig 2C). These data demonstrate that AR exhibits a differential binding profile amongst different phases of the cell cycle, consistent with differential DHT-induced gene networks in mitotically active cells.
Figure 2.
Further analyses of expanded AR binding events in mitotically active cells resulted in identification of two major patterns of AR binding, including occupancy at sites that are specific to the mitotic cell cycle but retained in all phases (deemed cell cycle common, or CCC) counterbalanced by occupancy events that are enriched in specific phases of the mitotic cell cycle (phase restricted binding, or PRB). Comparatively, 28,693 sites were classified as CCC pattern events, and 17,056 meeting the criteria of PRB binding events (Fig 2C,D). In the PRB subclass, early G1, early S, and G2/M phase showed similar numbers of phase-restricted binding events, with late G1 phase exhibiting a marked decrease in AR binding events, and late S phase showing the least evidence of phase-restricted AR occupancy (Fig 2D). These collective observations are consistent with the identification of cell cycle specific AR signaling (Figure 1), and suggest a causative role for AR PRB in the observed phase specific, DHT-dependent transcriptional networks.
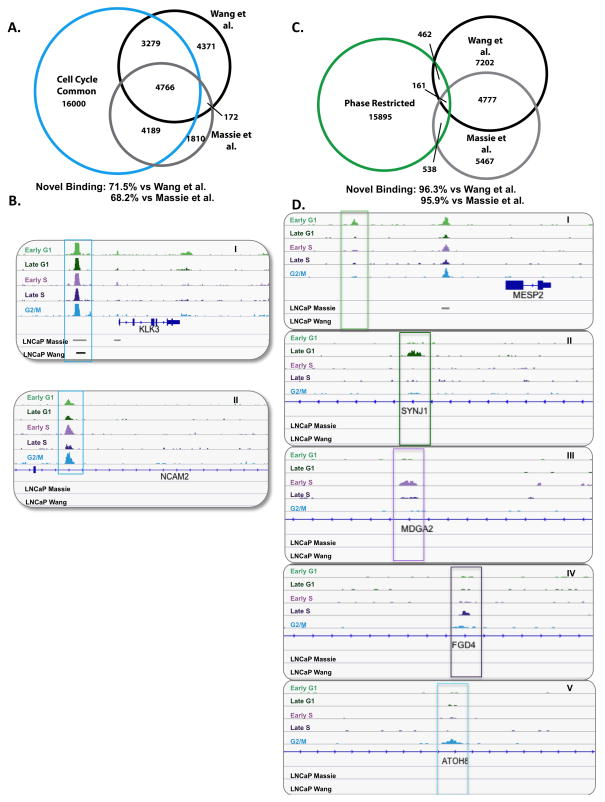
The concept that AR performs unique functions in mitotically active cells was further underscored by comparison with previously reported studies of AR cistromes. As shown, AR binding analyses herein captured the majority of previously reported sites of AR occupancy, wherein cells were arrested in G0 by steroid deprivation prior to DHT stimulation (Fig S3)(9, 11). Comparing CCC binding to all post-G0 phases in mitotically active cells illustrated high confidence overlap with binding reported by Massie et al., with over 80% of ARBSs identified in G0 also exhibiting CCC binding in mitotically active cells. Additionally, 60% of AR occupied sites identified by Wang et al. coincided with CCC binding, with the reduction in overlapped binding likely due to differences in experimental method (ChIP-Chip performed by Wang et al. vs ChIP-Seq performed in the current study, Fig 3A, Fig S3). As expected, binding present in G0 and maintained throughout the cell cycle is exemplified by occupancy at well-characterized AR targets such as KLK3 (PSA) and TMPRSS2 (Fig 3B, panel I, Fig S4). Notably, a significant fraction of CCC AR occupied sites detected in mitotically active cells are novel. With respect to CCC binding, approximately 70% of the total number of ARBSs were previously unreported (Fig 3A), illustrated by AR occupancy seen in Fig 3B, panels I and II. Collectively, these findings indicate that AR binding sites found in G0 are sustained throughout the mitotic cell cycle, and that AR binding capacity and function are significantly expanded in mitotically active cells.
Figure 3.
While the fraction of novel CCC binding is substantial, assessment of PRB sites reveals further insight into AR function in cycling cells. Over 90% of the PRB events were not identified in the G0 enriched datasets (Fig 3C), and represent restricted binding events from each cell cycle phase (Fig 3D). Collectively, these data indicate that while AR function in G0 cells is largely preserved in cycling cells, AR function in mitotically active cells is greatly expanded beyond G0, eliciting patterns that are unique to the post G0-cell cycle as well as those that are restricted to specific cell cycle phases. These findings provide the first evidence that AR is differentially programmed in cycling cells, and yield significant new insight into the mechanisms by which AR likely promotes tumor development and progression.
Evidence for Differential AR Cofactor Requirements Throughout the Cell Cycle
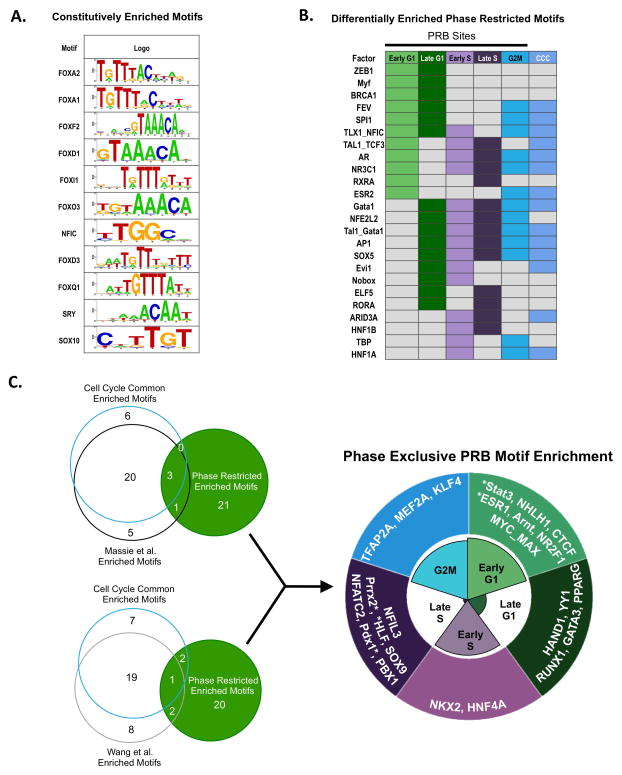
The observations that androgen induces distinct transcriptional networks in a cell cycle-dependent manner (Fig 1), and that AR exhibits an expanded, differential cistrome in mitotically active cells (Fig 2) suggests that cell cycle specific signaling is directly regulated by AR. To probe the mechanisms and consequence of cell cycle dependent AR regulation, motif analyses were performed utilizing PRB events. Enriched motifs were divided into three cohorts: constitutively enriched (significantly represented throughout all phases of PRB; Fig 4A), differentially enriched (enrichment altered throughout two or more PRB phases: Fig 4B), and phase exclusive enriched (motifs present only in single phase indicated; Fig 4C, Fig S5, Supplementary Table 2). Additionally, motifs significantly represented using CCC binding and previously reported (G0 enriched) datasets were generated for comparison to identify cell cycle specific alterations. Interestingly, motifs constitutively enriched in all phases of PRB binding were concurrently enriched in CCC and G0 enriched datasets (Fig 4A). This cohort includes FOXA1, which has been well described as an AR pioneer factor, is required for AR activity, and has been shown to contribute to AR reprogramming in tumor transformation (Fig 4A) (21, 22). Additionally, the nuclear factor I family member NFIC, which interacts with FOXA1 to regulate AR, was enriched in all datasets evaluated (23). The AR binding motif was also enriched within CCC binding, in addition to a number of other previously described AR interactors including GATA and AP-1, consistent with the overlap of CCC motifs and those previously described (21, 24, 25). Interestingly, the AR binding motif was enriched in all PRB events except late G1; while lack of enrichment for the AR motif in G1 could be due to the decrease in overall binding in this phase (Fig 4B), these data suggest potential AR occupancy at non-canonical motifs in late G1. Overall, these data suggest that the requirement for pioneer factor function is retained in all phases of cell cycle, and that AR generally relies on canonical AR binding motifs across the cell cycle. In contrast, novel motif enrichment was observed in AR events that are phase restricted. Over 80% of the PRB enriched motifs (of 25 total) were shown to be novel when compared to those enriched in CCC binding or those identified in previous studies (Fig 4C), suggesting that PRB enriched motifs contribute to differential AR function in active cell cycle. This cohort of motifs was enriched for factors with known roles in metastasis and DNA damage, e.g. ZEB1 and BRCA1 (26, 27), putting forth the concept that differentially enriched, PRB-specific AR cofactors may not only support cell cycle phase specific AR function, but likely contribute to AR-induced tumor phenotypes. Taken together, the high degree of motif overlap between previously published binding studies and CCC binding coupled with the lack of overlap when comparing the phase-restricted enriched motifs suggest a likely role for cell cycle regulated AR cofactors in controlling and facilitating the expanded AR cistrome and regulated gene networks in mitotically active cells.
Figure 4.
Expanded AR Function in Mitotically Active Cells Reveals Link to Lethal Tumor Phenotypes
To investigate functional consequence of PRB events, a “guilty by association” approach was utilized to identify potential targets of phase-restricted AR activity impacted by AR binding. Genes with transcriptional start sites (TSS) within 30kb of ARBSs were identified, as depicted in Fig 5A, and subsequently compared to genes identified using published datasets. As expected, genes associated with CCC binding events overlapped to a high degree with those associated with previous reports (88% of Massie dataset validated, 65% of Wang dataset validated), replicating the overlap seen using binding alone (Fig S6). Importantly, genes associated with PRB nominated a novel AR-regulated gene network, further indicating that the function of AR is expanded in mitotically active cells (Fig 5B). Genes nominated by proximity to PRB events were then compared to the cohort that exhibited differential DHT-induced gene expression (Fig 1B). Strikingly, of the 1098 genes identified as showing phase specific AR expression, 319 contained PRB in putative regulatory regions (Fig 5C). To discern the biological impact of PRB related, androgen induced transcriptional changes, gene ontology (GO) analyses was performed. Due to the low number PRB sites in late G1 and late S phases, ontology analyses were restricted to events in only early G1, early S, and G2/M phases. Genes associated with PRB events in early G1 revealed a significant impact on metabolic processes, whereas early S genes unmasked the influence of AR on mitotic progression, including kinetochore assembly, and genes with PRB events in G2/M show impact on both metabolic processes and cell cycle regulation (Fig 5D). Combined, these data identify critical new transcriptional networks likely to be controlled by phase-restricted AR binding, exposing a 319 gene set with both phase-restricted binding as well as differential expression. These data suggest for the first time that PRB events are linked to androgen-induced phase specific transcriptional responses in cycling cells, thus revealing differential AR function throughout the cell cycle.
Figure 5.
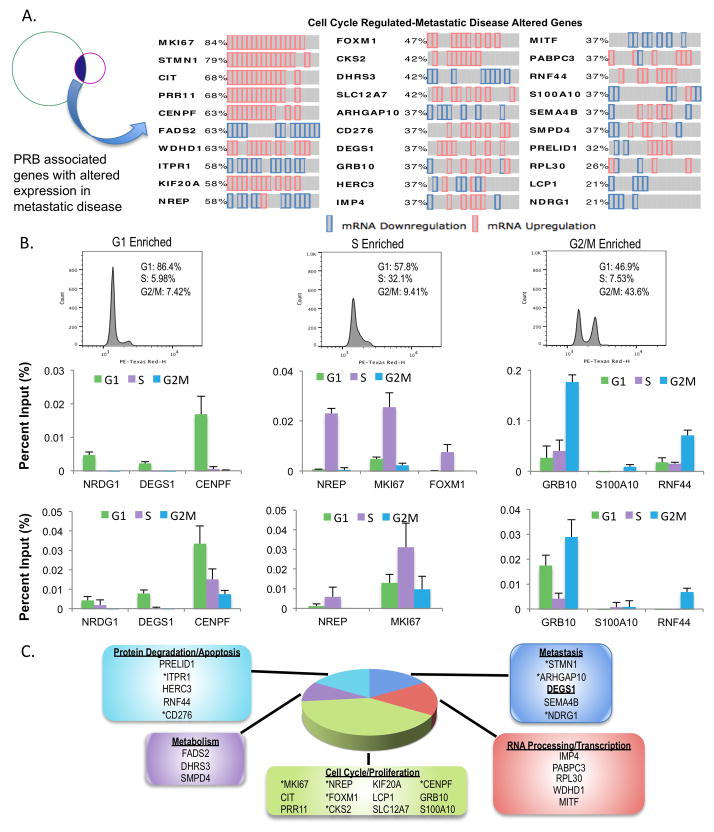
To explore the biological relevance of the newly identified AR-associated cistrome and transcriptome, the 319 gene set (genes exhibiting both PRB and cell cycle regulated gene expression) was examined for expression in primary and metastatic PCa in order to identify targets likely to have clinical implications. Genes with altered expression in over 10% of primary disease were removed, so as to isolate those with changes exclusively after metastatic tumor development, nominating 30 prioritized clinically relevant targets (Fig 6A), of which 19 were up-regulated and 11 down-regulated. Additionally, a sampling of this subset was validated for PRB using an alternate means of synchronization in order to further assess cell cycle regulation (Fig 6B top). Targets were identified that exhibited binding restricted to early G1 (NRDG1, DEGS1, and CENPF), early S (NREP, MKI67, and FOXM1), or G2/M (GRB10, S100A10, RNF44) and validated by ChIP-qPCR in their respective phases (Fig 6B middle), thus strengthening these findings. Further, synchronization in an additional HTS model system was utilized (VCaP, Fig S7), and ChIP-qPCR for these sites exhibited PRB in this distinct model system (Fig 6B bottom). Functional classification for the prioritized 30 gene set showed association with numerous processes associated with lethal malignancies, including metabolism, cell cycle/proliferation, protein degradation/apoptosis, RNA processing/transcription, and metastasis (Fig 6C). A specific emphasis was placed on metastasis, as AR is known to promote metastatic phenotypes (28, 29), but the underlying basis for this function is incompletely understood. Within this group, several newly identified genes under likely AR regulation have been previously implicated in at least some aspect of PCa biology. For example, stathmin-1 (STMN1) is associated with EMT phenotypes (30), whereas NDRG1 has been suggested to modulate metastatic development (31), and ARHGAP10 harbors a known role in cellular proliferation (32).
Figure 6.
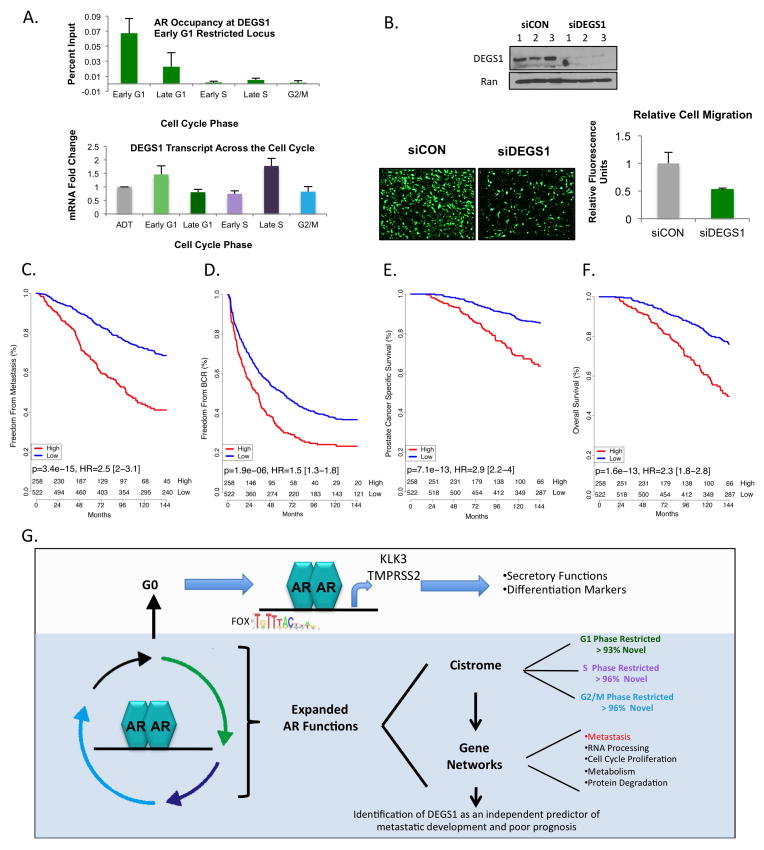
The present study provides an unexpected link of AR to modulation of these factors, strongly suggestive that AR utilizes these pathways to promote clinical tumor progression. Further, semaphorin 4B (SEMA4B), a protein involved in axon extension, was identified in this cohort and has been previously implicated in metastasis of non-small cell lung cancer (33). Further systematic analysis revealed dihydroceramide desaturase 1 (DEGS1), a desaturase invovled in the ceramide synthesis pathway (34), amongst the most clinically significant AR-regulated genes. As shown, validation via ChIP-qPCR demonstrated phase-restricted AR binding to the DEGS1 regulatory region in early G1 (Fig 7A top), with maximum gene expression occurring in late S phase (Fig 7A bottom). Intriguingly, this phase was where expression of genes involved in EMT co-occurred, a process also linked to metastasis. Taken together, prioritization of clinical targets with both PRB and differential expression highlighted a 30 gene set over-represented exclusively in metastatic disease, with DEGS1 being prioritized due to its potential in promoting metastasis.
Figure 7.
Given the likely impact of DEGS1 on PCa tumor behavior, RNAi silencing was utilized to ablate DEGS1 protein levels (Fig 7B top). Notably, DEGS1 depletion significantly impeded migratory capacity (~50%), suggesting that this newly identified AR-regulated target activity promotes pro-metastatic functions (Fig 7B bottom left, with quantification bottom right). Interestingly, while bioactive lipids ceramide-1-phosphate (C1P) and sphingosine-1-phosphate (S1P) have been shown to induce cell proliferation (35, 36), siDEGS1 had no effect on Brdu incorporation, suggesting that DEGS1 does not alter proliferative potential in this disease type (Fig S8). To further assess the putative role of DEGS1 in PCa progression, a large cohort of patients with localized, high risk PCa (n=780) with a median of 11.2 years of clinical follow-up was utilized to determine the clinical relevance of DEGS1. As shown, univariate analyses identified elevated DEGS1 was strongly associated with reduction in freedom from metastasis (Fig 7C, p=3.4e-15) and rapid biochemical recurrence (Fig 7D, p= 1.9e-06), thus nominating DEGS1 as an AR-regulated predictive marker for metastatic progression. Concordantly, DEGS1 was robustly associated with poor prognosis and reduced survival, both with regard to PCa-specific survival (Fig 7E, p=7.1e-13) and overall survival (Fig 7F, p=1.6e-13). Given the lack of predictive biomarkers to identify aggressive PCa, DEGS1 was further examined through multivariate analyses and elevated DEGS1 levels showed significant value as an independent predictor of poor outcomes when compared with Gleason score and PSA (Fig S9). Taken together, these data show for the first time that genes regulated by AR in mitotically active cells are likely drivers of PCa cell progression and metastatic events and unveil DEGS1 as an unexpected AR regulated gene of clinical relevance (Fig 7G).
Discussion
Elucidating the mechanisms by which AR induces lethal tumor phenotypes is critical for the development of more effective therapeutic strategies to manage PCa. Herein, new and unexpected functions of AR were unveiled through profiling AR activity in mitotically active cells. Key findings demonstrate that: i) androgen engages a distinct transcriptional network in post G0, mitotically active cells; ii) the function of AR in actively cycling cells is strikingly expanded during mitotic progression, demonstrating binding events that are specific to mitotically active cells and/or restricted to specific cell cycle phase; iii) a distinct set of transcription factor motifs are enriched dependent on cell cycle phase; iv) genes exhibiting dual cell cycle regulated binding and expression nominate cell cycle specific targets with relevance in late stage disease; v) DEGS1 was identified as a cell cycle specific AR target gene which alters cell migration in PCa; and vi) DEGS1 independently predicts for metastatic development and poor prognosis. Taken together, these data uncover novel functions for AR in mitotically active cells, allowing the identification of targets with clinical relevance in aggressive disease.
The finding that AR functions differentially in cycling cells when compared to resting cells sheds new light on the function of AR activity in PCa, and is consistent with early studies suggesting that AR function may be modified by components of the cell cycle machinery (37). Herein, differential AR activity in the mitotic cell cycle was demonstrated at a genome-wide level, with cell cycle phase altering both AR-chromatin interaction as well as downstream changes corresponding to cell cycle regulated, DHT induced transcriptional output. The newly described AR cistromes in mitotically active cells can be divided into binding maintained throughout the cell cycle in cells committed to mitotisis (cell cycle common, CCC binding) or binding enriched in a specific cell cycle phase (phase restricted binding, PRB). The CCC binding profile notably encompasses the vast majority of previously reported AR binding sites, largely identified in DHT-stimulated cells that were synchronized in G0 (9, 11), through additional AR binding regions were also identified. These binding overlaps include those regulating well-characterized AR target genes (e.g. KLK3 and TMPRS22). Such concordance between previously identified AR cistromes and the CCC binding profile suggests that AR binding events in DHT stimulated, G0 phase cells are largely maintained in actively cycling cells at all phases, and may serve as the “baseline” of AR function. However, this subset comprised only ~30% of the CCC cluster, with nearly 70% of CCC binding events identifying novel AR functions. An even greater expansion of AR activity was revealed by the PRB subset (~95% novel binding events not previously observed in genome-wide mapping studies enriched in G0 phase), further supporting the idea that AR-cell cycle interplay results in novel AR function in cycling cells. PRB-associated genes corresponded to differentially enriched processes, with early G1-phase restricted binding associated with metabolic processes, early S-phase associated binding with chromosomal organization, and G2/M phase restricted binding associated with mitotic regulation. These findings are consistent with previous cellular observations suggesting that AR promotes G1-S transition (38), and provide some mechanistic basis for this long-appreciated function for AR. In addition to cell cycle regulated gene networks, new binding and transcriptional targets of AR were identified that assist in defining the mechanism(s) by which AR promotes androgen responsiveness (13), metabolism (8, 9), DNA damage (16, 17), and cell cycle transitions (14, 38). Further, as a subset of prioritized PRB sites were validated in multiple model systems, and using a variety of synchronization methods, the expansion of the AR cistrome in mitotically active cells is a phenomenon likely to be seen across model systems and cell types. The data herein demonstrate that the AR-dependent transcriptional networks governing these cancer-associated activities are regulated as a consequence of cell cycle position, further underscoring the importance of AR-cell cycle interplay in promoting gene expression networks and suggesting implications for tumors with alterations in cell cycle regulatory networks.
The discovery that AR activity is modulated as a function of cell cycle position, and that these functions of AR appear to be critical for AR-mediated tumor progression, lead to provocative hypotheses as to how AR function may be controlled. Data herein demonstrate that AR binding events in mitotically active cells are enriched for co-occurrence of differential transcription factor motifs (Fig 4B), suggesting that AR may depend on differential cooperating factors to elicit cell cycle-specific transcriptional signaling. Known motifs of cofactors thought to be requisite for AR activity (e.g. “pioneer factors”, such FOX and GATA) (21, 39, 40) associated with AR binding irrespective of cell cycle position, consistent with the importance of these factors for AR activity. However, selective enrichment of putative transcriptional cooperating factors were observed that have been previously linked to malignant phenotypes. Exemplifying this cohort in G1 phase are ZEB1, recently shown to be strongly associated with PCa migration and invasion (26, 41) and BRCA1, which has a well described role in DNA-repair and transcriptional regulation (42, 43). Ongoing studies are directed at interrogating the molecular impact of each motif enrichment and putative cooperating cofactor activity on AR function, both with regard to CCC and PRB events. Such regulation is likely to be complex, as 25 motifs in total were exclusively associated with PRB binding in a single phase of cell cycle. Strikingly, PRB-enriched motifs showed little overlap with CCC-associated motifs, congruent with the expanded cistrome seen in mitotically active cells. Notable within this subset is enrichment of CTCF in early G1-phase, as CTCF is involved in nucleosome positioning and chromosome remodeling (44). CTCF was previously shown to be associated with AR binding (45), but this interaction was contingent upon loss of FOXA1 and subsequent expansion of AR cistrome; thus, the present data bring forth new understanding of CTCF-AR interplay. Additionally, the c-myc motif was exclusively enriched in early G1; this motif was shown to be enriched in AR binding in clinical CRPC samples (46) and is known to be important for PCa survival, up-regulated in PCa, and associated with aggressive disease (47–50). As AR and c-myc are also known to regulate similar metabolic pathways (51), and have been seen to co-regulate target genes (52), the phase specific enrichment provides the context to determine how these two major transcriptional effectors of cancer progression act in concert to promote disease progression. Overall, these analyses implicate cell cycle specific transcriptional regulators as cooperating with AR to promote disease progression, and provide the molecular basis for further delineation of cell cycle specific AR function.
Complementing these findings, multiple components of the cell cycle machinery have been shown to directly interact with and influence AR activity, and may also contribute to cell cycle specific AR function. The early G1 kinase CDK6 enhances AR activity in reporter assays (53), and the CDK4/6 catalytic subunit Cyclin D1 binds and modulates AR activity in a target-selective manner (54–56). Notably, a highly oncogenic variant of Cyclin D1 associated with advanced disease induces mis-regulation of AR and directs expression of gene networks promoting metastatic phenotypes (57). In late G1, Cyclin E can enhance AR transcriptional activity through direct interaction, suggesting an intricate balance between these factors may play a role in the cell cycle regulated activity seen in G1 phase (58). Additionally, the RB/E2F1 axis, which controls G1-S transition has been shown to regulate AR activity. AR expression is directly regulated by E2F1, and RB loss therefore results in enhanced AR expression and output (14). Finally, AR activity in G2/M is intricately modulated, as CDK1 can inhibit AR activity through phosphorylation on serine 308, and the phosphatase Cdc25b enhances AR activity (59, 60). Determining the contribution of these factors in controlling cell cycle specific AR activity will be of high priority, given the advent of preclinical findings assessing CDK-targeting therapeutics in PCa and trials for clinical assessment for CDK4/6 inhibitors. For example, the CDK4/6 inhibitor palbociclib was shown to suppress PCa growth in model systems and to selectively cooperate with AR-directed therapies (61), and clinical evaluation is currently in progress (http://clinicaltrials.gov, NCT02059213). The present study has defined AR activity in response to palbociclib, both with regard to AR binding and downstream transcriptional regulation, and will allow for future studies directed at maximizing AR suppression in the presence of palbociclib.
The discovery of previously unrecognized AR transcriptional targets that promote tumor progression represents a major advance in our understanding of PCa progression. The newly identified AR-regulated genes are linked to diverse processes relevant to cancer progression including metastasis, RNA processing/transcription, cell cycle/proliferation, metabolism, and protein degradation/apoptosis. While each represents a significant advance in understating of AR function, the link to metastasis was particularly striking. Several newly identified AR regulated genes in this class (STMN1, NDRG1, and ARHGAP10/ARHGAP21) have been tentatively linked to metastatic progression, but the mechanisms governing expression were unknown (30, 31). The Rho/Rac signaling pathway was recently identified as a driver in promotion of metastatic phenotypes in PCa, as part of cooperation between AR and DNA-repair factors (29). Here, the Rho GTPase-activating protein ARHGAP10 was found to undergo cell cycle specific regulation and as such will be of further interest to elucidate the extent of the interplay between AR, cell cycle components, and DNA damage factors in controlling metastasis. SEMA4B, a protein involved in inhibition of axonal extension, was also up-regulated in metastatic disease. While a role for SEMA4B in PCa has not yet been described, in models of non-small cell lung cancer (NSCLC) it was shown to inhibit proliferation and cell metastasis (33). Finally, findings herein identified DEGS1 as a novel, robust predictor of metastatic disease and poor outcome in PCa that is regulated by AR. DEGS1 is poorly understood at the molecular level, but is known to be involved in ceramide synthesis, and as shown here, promotes cell migratory phenotypes in PCa. These findings strengthen and complement preliminary preclinical observations in esophageal carcinoma linking DEGS1 to cell migration (34). Analyses herein of human tissues demonstrated that DEGS1 upregulation is tightly linked with a decrease in freedom from biochemical recurrence, freedom from metastasis, and overall survival, underlying importance as an AR-induced effector of lethal disease phenotypes. Strikingly, DEGS1 independently predicted for aggressive disease, on par with currently utilized standards (PSA and Gleason score). As Gleason score is currently the strongest currently utilized predictor of poor outcome, added biomarkers with the ability to predict for disease progression such as DEGS1 are critical. Translational potential is also noted, as a recent study examining the use of a sphingosine kinase inhibitor ABC294640 in PCa was seen to additionally inhibit DEGS activity, suggesting that drugs specifically targeting DEGS1 may be of therapeutic import (62).
Taken together, molecular assessment of AR activity not only reveals differential function across the cell cycle and illuminates new understanding of the mechanisms by which AR promotes disease progression, but underscore the importance of discerning cancer-promoting transcription factor function in mitotically active cells. The present findings yield critical new insight into AR function and nominate downstream effectors of AR activity as potentially impactful therapeutic targets and biomarkers of lethal disease phenotypes.
Experimental Procedures
Cell Cycle Arrest
Cells were plated in hormone replete media 24 hours prior to treatment. Cells were treated 24 hours prior to harvest with (final concentration): 0.5μM PD-0332991 (early G1 phase enrichment), 5μg/mL roscovitine (late G1 phase enrichment), 2μg/mL aphidicolin (early S enrichment), 1mM hydroxyurea (late S enrichment), 2μg/mL aphidicolin for 15 hours, followed by washout and treatment with 50ng/mL nocodozole for 9 hours (G2/M phase enrichment).
Gene Expression Analysis
Cells were arrested as described above and stimulated with 10nM DHT for 3 hours prior to harvest. mRNA was processed and transcriptional analyses performed. Details for full gene expression analyses are available in supplemental materials and methods.
ChIP-Seq
Cells were arrested as described above, with biological triplicate for each condition, and ChIP-Seq performed as previously described(20). Extended details for ChIP-Seq data analyses are available in the supplemental methods.
RNA Interference
Cells were seeded at equal density in hormone replete conditions. Cells were then transfected for 6–8 hours in serum free conditions with either control or siDEGS1 according to manufacturer specifications. Cells were maintained for an additional 72 hours in serum replete media, treated as specified and harvested at indicated time-point. Immunoblot analysis was conducted to confirm knockdown at 72 hours post transfection.
Clinical Analyses
Microarray data on radical prostatectomy samples were obtained from the Mayo Clinic and profiled using the Affymetrix Human Exon 1.0 ST array as described previously (63, 64). Two independent cohorts of patients: Mayo Clinic I (MCI, n=545) (63) and Mayo Clinic II (MCII, n=235) (64), were pooled into the MC cohort (N=780) and evaluated. Microarray data was downloaded from the NCBI Gene Expression Omnibus (GSE46691, GSE62116)
DEGS1 expression was split into high versus low by comparing the top 1/3 of DEGS1 expression versus the bottom 2/3 in each cohort. Kaplan Meier curves for freedom from biochemical recurrence (BCR), freedom from metastasis, prostate cancer specific survival (PCSS), and overall survival (OS), and P-values were generated using the log-rank test. Cox regression was used for both univariate and multivariate analyses. In the multivariate analysis: age was treated as a continuous variable; PSA was grouped into low (<10), intermediate (10–20), and high (>20); Surgical margin status (SMS), seminal vesicle invasion (SVI), extracapsular extension (ECE), and lymph node invasion (LNI) were treated as binary variables; and Gleason score was grouped into low (<7), intermediate (7), or high (8–10). Stratification by cohort was used when performing pooled analysis to account for baseline differences between cohorts (65).
Supplementary Material
Acknowledgments
The authors gratefully thank Mandeep Takhar for her assistance in providing microarray data analysis, K. Knudsen lab members for their input, Bin Fang for assistance in creation of motif diagrams, and E. Schade for graphical support and expertise. This work was supported by grants to WL (R01HG007538 and R01CA193466) and to KEK by grants from the NCI (CA159945, CA176401), P30-CA056036, and in part by a grant to KEK with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Knudsen KE, Penning TM. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol Metab. 2010;21(5):315–24. doi: 10.1016/j.tem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15(15):4792–8. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beekman KW, Hussain M. Hormonal approaches in prostate cancer: application in the contemporary prostate cancer patient. Urol Oncol. 2008;26(4):415–9. doi: 10.1016/j.urolonc.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Knudsen KE, Kelly WK. Outsmarting androgen receptor: creative approaches for targeting aberrant androgen signaling in advanced prostate cancer. Expert Rev Endocrinol Metab. 2011;6(3):483–93. doi: 10.1586/eem.11.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan X, Balk SP. Mechanisms mediating androgen receptor reactivation after castration. Urol Oncol. 2009;27(1):36–41. doi: 10.1016/j.urolonc.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan X, Cai C, Chen S, Chen S, Yu Z, Balk SP. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33(22):2815–25. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barfeld SJ, Itkonen HM, Urbanucci A, Mills IG. Androgen-regulated metabolism and biosynthesis in prostate cancer. Endocr Relat Cancer. 2014;21(4):T57–66. doi: 10.1530/ERC-13-0515. [DOI] [PubMed] [Google Scholar]
- 9.Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30(13):2719–33. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Chen SY, Ross KN, Balk SP. Androgens induce prostate cancer cell proliferation through mammalian target of rapamycin activation and post-transcriptional increases in cyclin D proteins. Cancer Res. 2006;66(15):7783–92. doi: 10.1158/0008-5472.CAN-05-4472. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waltering KK, Helenius MA, Sahu B, Manni V, Linja MJ, Janne OA, et al. Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Cancer Res. 2009;69(20):8141–9. doi: 10.1158/0008-5472.CAN-09-0919. [DOI] [PubMed] [Google Scholar]
- 13.Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A. 2002;99(18):11890–5. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120(12):4478–92. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallik I, Davila M, Tapia T, Schanen B, Chakrabarti R. Androgen regulates Cdc6 transcription through interactions between androgen receptor and E2F transcription factor in prostate cancer cells. Biochim Biophys Acta. 2008;1783(10):1737–44. doi: 10.1016/j.bbamcr.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013;3(11):1254–71. doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3(11):1245–53. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nauseef JT, Henry MD. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat Rev Urol. 2011;8(8):428–39. doi: 10.1038/nrurol.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7(4):256–69. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48(3):240–8. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27(3):380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet. 2015;47(11):1346–51. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabowska MM, Elliott AD, DeGraff DJ, Anderson PD, Anumanthan G, Yamashita H, et al. NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression. Mol Endocrinol. 2014;28(6):949–64. doi: 10.1210/me.2013-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bubulya A, Wise SC, Shen XQ, Burmeister LA, Shemshedini L. c-Jun can mediate androgen receptor-induced transactivation. J Biol Chem. 1996;271(40):24583–9. doi: 10.1074/jbc.271.40.24583. [DOI] [PubMed] [Google Scholar]
- 25.Chen SY, Cai C, Fisher CJ, Zheng Z, Omwancha J, Hsieh CL, et al. c-Jun enhancement of androgen receptor transactivation is associated with prostate cancer cell proliferation. Oncogene. 2006;25(54):7212–23. doi: 10.1038/sj.onc.1209705. [DOI] [PubMed] [Google Scholar]
- 26.Graham TR, Zhau HE, Odero-Marah VA, Osunkoya AO, Kimbro KS, Tighiouart M, et al. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008;68(7):2479–88. doi: 10.1158/0008-5472.CAN-07-2559. [DOI] [PubMed] [Google Scholar]
- 27.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481(7381):287–94. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 28.Augello MA, Den RB, Knudsen KE. AR function in promoting metastatic prostate cancer. Cancer Metastasis Rev. 2014;33(2–3):399–411. doi: 10.1007/s10555-013-9471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodwin JF, Kothari V, Drake JM, Zhao S, Dylgjeri E, Dean JL, et al. DNA-PKcs-Mediated Transcriptional Regulation Drives Prostate Cancer Progression and Metastasis. Cancer Cell. 2015;28(1):97–113. doi: 10.1016/j.ccell.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams K, Ghosh R, Giridhar PV, Gu G, Case T, Belcher SM, et al. Inhibition of stathmin1 accelerates the metastatic process. Cancer Res. 2012;72(20):5407–17. doi: 10.1158/0008-5472.CAN-12-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W, Yue F, Zheng M, Merlot A, Bae DH, Huang M, et al. The proto-oncogene c-Src and its downstream signaling pathways are inhibited by the metastasis suppressor, NDRG1. Oncotarget. 2015;6(11):8851–74. doi: 10.18632/oncotarget.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazarini M, Traina F, Machado-Neto JA, Barcellos KS, Moreira YB, Brandao MM, et al. ARHGAP21 is a RhoGAP for RhoA and RhoC with a role in proliferation and migration of prostate adenocarcinoma cells. Biochim Biophys Acta. 2013;1832(2):365–74. doi: 10.1016/j.bbadis.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Jian H, Zhao Y, Liu B, Lu S. SEMA4B inhibits growth of non-small cell lung cancer in vitro and in vivo. Cell Signal. 2015;27(6):1208–13. doi: 10.1016/j.cellsig.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 34.Zhou W, Ye XL, Sun ZJ, Ji XD, Chen HX, Xie D. Overexpression of degenerative spermatocyte homolog 1 up-regulates the expression of cyclin D1 and enhances metastatic efficiency in esophageal carcinoma Eca109 cells. Mol Carcinog. 2009;48(10):886–94. doi: 10.1002/mc.20533. [DOI] [PubMed] [Google Scholar]
- 35.Chalfant CE, Spiegel S. Sphingosine 1-phosphate and ceramide 1-phosphate: expanding roles in cell signaling. J Cell Sci. 2005;118(Pt 20):4605–12. doi: 10.1242/jcs.02637. [DOI] [PubMed] [Google Scholar]
- 36.Gangoiti P, Granado MH, Wang SW, Kong JY, Steinbrecher UP, Gomez-Munoz A. Ceramide 1-phosphate stimulates macrophage proliferation through activation of the PI3-kinase/PKB, JNK and ERK1/2 pathways. Cell Signal. 2008;20(4):726–36. doi: 10.1016/j.cellsig.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Martinez ED, Danielsen M. Loss of androgen receptor transcriptional activity at the G(1)/S transition. J Biol Chem. 2002;277(33):29719–29. doi: 10.1074/jbc.M112134200. [DOI] [PubMed] [Google Scholar]
- 38.Balk SP, Knudsen KE. AR, the cell cycle, and prostate cancer. Nucl Recept Signal. 2008;6:e001. doi: 10.1621/nrs.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He B, Lanz RB, Fiskus W, Geng C, Yi P, Hartig SM, et al. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc Natl Acad Sci U S A. 2014;111(51):18261–6. doi: 10.1073/pnas.1421415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yun EJ, Baek ST, Xie D, Tseng SF, Dobin T, Hernandez E, et al. DAB2IP regulates cancer stem cell phenotypes through modulating stem cell factor receptor and ZEB1. Oncogene. 2015;34(21):2741–52. doi: 10.1038/onc.2014.215. [DOI] [PubMed] [Google Scholar]
- 42.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25(43):5864–74. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 43.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25(43):5854–63. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 44.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4(7):e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30(19):3962–76. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23(1):35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21(9):1156–67. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palaskas N, Larson SM, Schultz N, Komisopoulou E, Wong J, Rohle D, et al. 18F-fluorodeoxy-glucose positron emission tomography marks MYC-overexpressing human basal-like breast cancers. Cancer Res. 2011;71(15):5164–74. doi: 10.1158/0008-5472.CAN-10-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan L, Peng G, Sahgal N, Fazli L, Gleave M, Zhang Y, et al. Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene. 2015 doi: 10.1038/onc.2015.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barfeld SJ, Fazli L, Persson M, Marjavaara L, Urbanucci A, Kaukoniemi KM, et al. Myc-dependent purine biosynthesis affects nucleolar stress and therapy response in prostate cancer. Oncotarget. 2015;6(14):12587–602. doi: 10.18632/oncotarget.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schrecengost RS, Dean JL, Goodwin JF, Schiewer MJ, Urban MW, Stanek TJ, et al. USP22 regulates oncogenic signaling pathways to drive lethal cancer progression. Cancer Res. 2014;74(1):272–86. doi: 10.1158/0008-5472.CAN-13-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim JT, Mansukhani M, Weinstein IB. Cyclin-dependent kinase 6 associates with the androgen receptor and enhances its transcriptional activity in prostate cancer cells. Proc Natl Acad Sci U S A. 2005;102(14):5156–61. doi: 10.1073/pnas.0501203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burd CJ, Petre CE, Morey LM, Wang Y, Revelo MP, Haiman CA, et al. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci U S A. 2006;103(7):2190–5. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Comstock CE, Augello MA, Schiewer MJ, Karch J, Burd CJ, Ertel A, et al. Cyclin D1 is a selective modifier of androgen-dependent signaling and androgen receptor function. J Biol Chem. 2011;286(10):8117–27. doi: 10.1074/jbc.M110.170720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knudsen KE, Cavenee WK, Arden KC. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 1999;59(10):2297–301. [PubMed] [Google Scholar]
- 57.Augello MA, Berman-Booty LD, Carr R, 3rd, Yoshida A, Dean JL, Schiewer MJ, et al. Consequence of the tumor-associated conversion to cyclin D1b. EMBO Mol Med. 2015;7(5):628–47. doi: 10.15252/emmm.201404242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto A, Hashimoto Y, Kohri K, Ogata E, Kato S, Ikeda K, et al. Cyclin E as a coactivator of the androgen receptor. J Cell Biol. 2000;150(4):873–80. doi: 10.1083/jcb.150.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chua SS, Ma Z, Ngan E, Tsai SY. Cdc25B as a steroid receptor coactivator. Vitam Horm. 2004;68:231–56. doi: 10.1016/S0083-6729(04)68008-3. [DOI] [PubMed] [Google Scholar]
- 60.Koryakina Y, Knudsen KE, Gioeli D. Cell-cycle-dependent regulation of androgen receptor function. Endocr Relat Cancer. 2015;22(2):249–64. doi: 10.1530/ERC-14-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Comstock CE, Augello MA, Goodwin JF, de Leeuw R, Schiewer MJ, Ostrander WF, Jr, et al. Targeting cell cycle and hormone receptor pathways in cancer. Oncogene. 2013;32(48):5481–91. doi: 10.1038/onc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venant H, Rahmaniyan M, Jones EE, Lu P, Lilly MB, Garrett-Mayer E, et al. The Sphingosine Kinase 2 Inhibitor ABC294640 Reduces the Growth of Prostate Cancer Cells and Results in Accumulation of Dihydroceramides In Vitro and In Vivo. Mol Cancer Ther. 2015;14(12):2744–52. doi: 10.1158/1535-7163.MCT-15-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190(6):2047–53. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15(13):1469–80. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.