Abstract
Plants live in biogeochemically diverse soils that harbor extraordinarily diverse microbiota. Plant organs associate intimately with a subset of these microbes; this community’s structure can be altered by soil nutrient content. Plant-associated microbes can compete with the plant and with each other for nutrients; they can also provide traits that increase plant productivity. It is unknown how the plant immune system coordinates microbial recognition with nutritional cues during microbiome assembly. We establish that a genetic network controlling phosphate stress response influences root microbiome community structure, even under non-stress phosphate conditions. We define a molecular mechanism regulating coordination between nutrition and defense in the presence of a synthetic bacterial community. We demonstrate that the master transcriptional regulators of phosphate stress response in Arabidopsis also directly repress defense, consistent with plant prioritization of nutritional stress over defense. Our work will impact efforts to define and deploy useful microbes to enhance plant performance.
Plant organs create distinct physical and chemical environments that are colonized by specific microbial taxa1. These can be modulated by the plant immune system2 and by soil nutrient composition3. Phosphorus is present in the biosphere at high concentrations, but plants can only absorb orthophosphate (Pi), a form also essential for microbial proliferation4, 5 and scarce in soil6. Thus, plants possess adaptive phosphate starvation responses (PSR) to manage low Pi availability that typically occurs in the presence of plant-associated microbes. Common strategies for increasing Pi uptake capacity include rapid extension of lateral roots foraging into topsoil where Pi accumulates7 and establishment of beneficial relationships with some soil microorganisms8, 9. For example, the capacity of a specific mutualistic fungus to colonize Arabidopsis roots is modulated by plant phosphate status implying coordination between the PSR and the immune system8, 10. Descriptions of pathogen exploitation of PSR-immune system coordination are emerging11, 12.
We demonstrate that Arabidopsis mutants with altered phosphate starvation responses (PSR) assemble atypical microbiomes, either in phosphate-replete wild soil, or during in vitro colonization with a synthetic bacterial community (SynCom). This SynCom competes for phosphate with the plant and induces PSR in limiting phosphate. PSR in these conditions requires the master transcriptional regulator PHR1 and its weakly redundant paralog, PHL1. The severely reduced PSR observed in phr1 phl1 mutants is accompanied by transcriptional changes in plant defense leading to enhanced immune function. Negative regulation of immune system components by PHR1 is direct, as measured by target gene promoter occupancy, and functional, as validated by pathology phenotypes. Thus, PHR1 directly activates microbiome-enhanced response to phosphate limitation while repressing microbially-driven plant immune system outputs.
The root microbiome in plants with altered phosphate stress response
We linked PSR to the root microbiome by contrasting the root bacterial community of wild-type Arabidopsis Col-0 with three types of PSR mutants (Fig. 1a, b, Supplementary Text 1; Extended Data Fig. 1; Supplementary Table 1). PSR, historically defined in axenic seedlings and measured by Pi concentration in the plant shoot, is variable across these mutants. In replete Pi and axenic conditions, phr1 plants accumulate less free Pi than wild-type13; pht1;1, pht1;1 pht1;4 and phf1 accumulate very low Pi levels and express constitutive PSR14, 15; and pho2, nla and spx1 spx2 express diverse magnitudes of Pi hyper-accumulation16– 18. We grew plants in a previously characterized wild soil19 that is not overtly phosphate deficient (Extended Data Fig. 2). Generally, the Pi concentration of PSR mutants grown in this wild soil recapitulated those defined in axenic conditions, except for phf1 and nla which displayed the opposite phenotype to that observed in axenic agar, and phr1 which accumulated the same Pi concentration as Col-0 (Fig. 1b). These results suggest that complex chemical conditions, soil microbes, or a combination of these can alter Pi metabolism in these mutants.
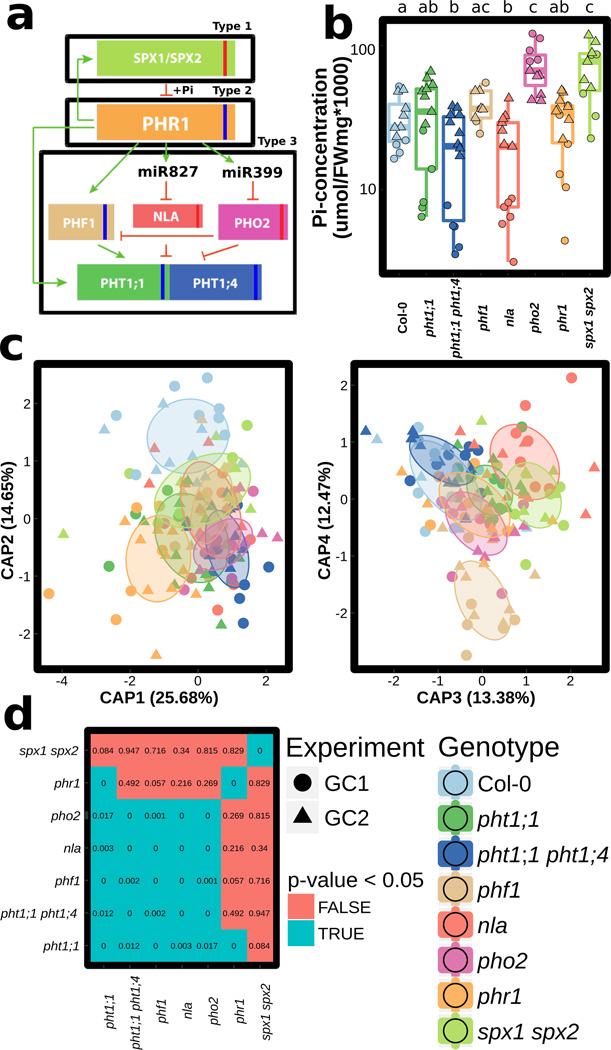
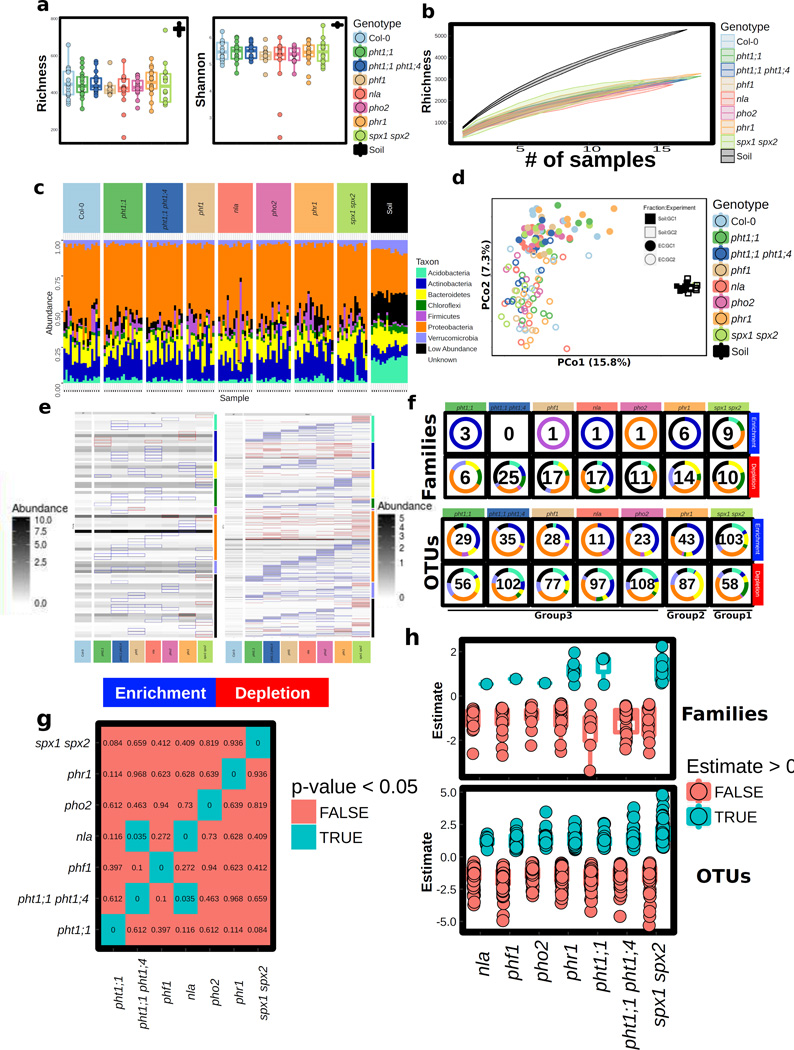
Figure 1. Phosphate Stress Response (PSR) mutants assemble an altered root microbiota.
a, Diagram of PSR regulation in Arabidopsis. Red and blue stripes indicate whether these mutants hyper- or hypo-accumulate Pi, respectively, in axenic, Pi replete conditions. The master PSR regulator PHR1 is a Myb-CC family transcription factor13 bound under phosphate replete conditions by the negative regulators SPX1 and SPX2 in the nucleus18. During PSR, PHR1 is released from SPX and regulates genes whose products include high-affinity phosphate transporters (PHT1;1 and PHT1;4)13. Transporter accumulation at the plasma membrane is controlled by PHF117, while PHO2 and NLA mediate PHT1 degradation16, 17b, Phosphate (Pi) concentration in shoots of plant genotypes (grown in growth chambers, 16-h dark/8-h light regime, 21°C day 18°C night for 7 weeks) in a natural soil. Statistical significance was determined by ANOVA while controlling for experiment (indicated by point shape); genotype grouping is based on a post-hoc Tukey test, and is indicated by letters at the top; genotypes with the same letter are indistinguishable at 95% confidence. Biological replicate numbers are: Col-0 (n=12), pht1:1 (n=13), pht1;1 pht1;4 (n=14), phf1 (n=9), nla (n=13), pho2 (n=11), phr1 (n=14) and spx1 spx2 (n=11) distributed across two independent experiments. c, Constrained ordination of root microbiome composition showing the effect of plant genotype: phr1 separates on the first two axes, spx1 spx2 on the third axis and phf1 on the fourth axis. Ellipses show the parametric smallest area around the mean that contains 50% of the probability mass for each genotype. Biological replicate numbers are: Col-0 (n=17), pht1:1 (n=18), pht1;1 pht1;4 (n=17), phf1 (n=13), nla (n=16), pho2 (n=16), phr1 (n=18) and spx1 spx2 (n=14) distributed across two independent experiments d, Table of p-values from Monte Carlo pairwise comparisons between mutants at the OTU level. A significant p-value (cyan) indicates that two genotypes are more similar than expected by chance.
Bacterial root endophytic (EC) community profiles were consistent with previous studies2, 19. Constrained ordination revealed significant differences between bacterial communities across the Pi accumulation gradient represented by these PSR mutants [5.3 % constrained variance, canonical analysis of principal coordinates (CAP)] (Fig. 1c). Additionally, CAP confirmed that phr1 and spx1 spx2 carried different communities, as evidenced by their separation on the first three ordination axes, and that phf1 was the most affected of Pi-transport mutant (Fig. 1c). Specific bacterial taxa had differential abundances inside the roots of mutant plants compared to wild-type. Mutants from the same PSR type had a similar effect on the root microbiome at a low taxonomic level [97 % identity Operational Taxonomic Unit (OTU)] (Fig. 1d), while they had no overlapping effect at a higher taxonomic level (Family, Extended Data Fig. 1g). This suggests that closely related groups of bacteria have differential colonization patterns on the same host genotypes. Importantly, we found that the enrichment and depletion profiles were better explained by PSR mutant signaling type rather than the mutant’s capacity for Pi accumulation: all of the Pi-transport-related mutants had a similar effect on the root microbiome, and the antagonistic PSR regulators phr1 and spx1 spx2 each exhibited unique patterns (Fig. 1a, d and Extended Data Fig. 1f, g). Our results indicate that PSR components influence root microbiome composition in plants grown in a phosphate-replete wild soil, leading to alteration of the abundance of specific microbes across diverse levels of Pi accumulation representing diverse magnitudes of PSR.
Phosphate starvation response in a microcosm reconstitution
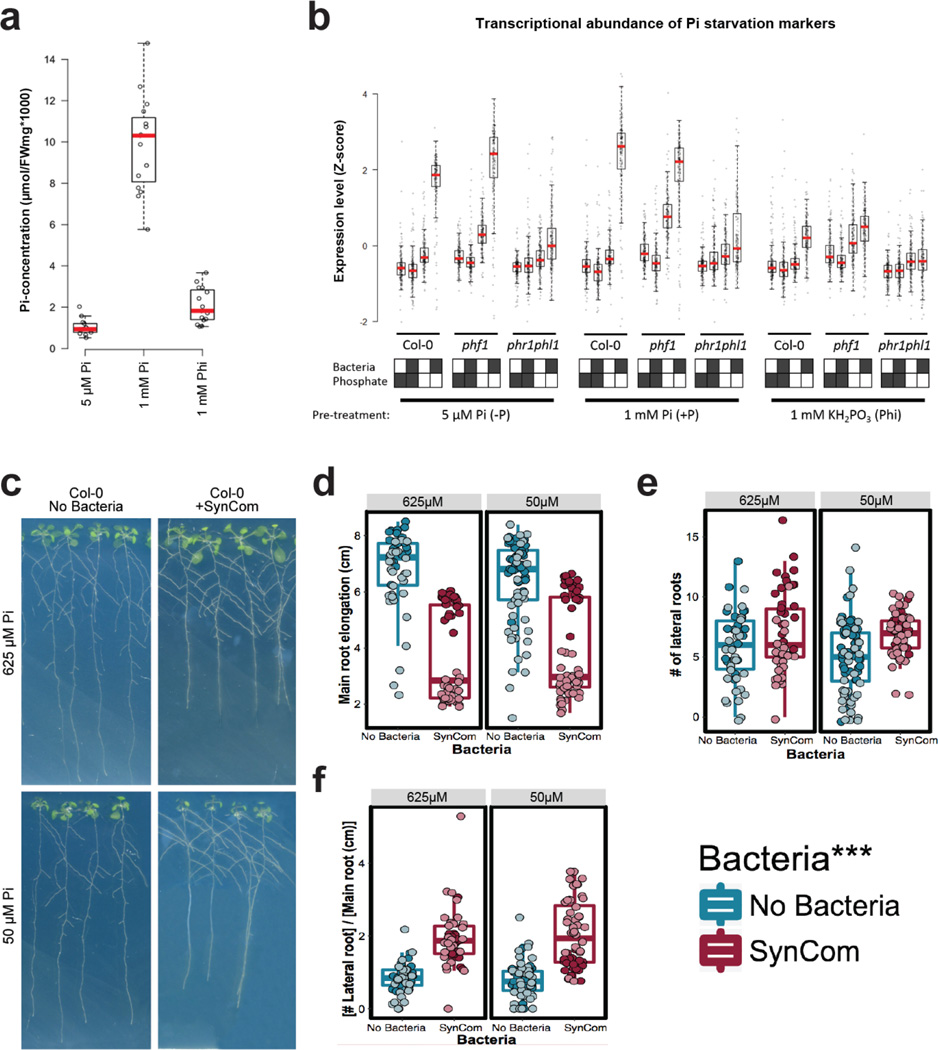
Our observations in a wild soil suggested complex interplay between PSR and the presence of a microbial community. Thus, we deployed a tractable but complex bacterial synthetic community (SynCom) of 35 taxonomically diverse, genome-sequenced bacteria isolated from the roots of Brassicaceae (nearly all from Arabidopsis) and two wild soils. This SynCom approximates the phylum level distribution observed in wild-type root endophytic compartments (Extended Data Fig. 3, Supplementary Table 1, Supplementary Table 2). We inoculated seedlings of Col-0, phf1 and the double mutant phr1 phl1 (a redundant paralogue of phr113) grown on agar plates in low or high Pi (Supplementary Text 2). Twelve days later, we noted that the SynCom had a negative effect on shoot Pi accumulation of Col-0 plants grown on low Pi, but not on plants grown on replete phosphate (Fig. 2a). As expected, both PSR mutants accumulated less Pi than Col-0; the SynCom did not rescue this defect. Thus, in this microcosm, plant-associated microbes drive a context-dependent competition with the plant for Pi.
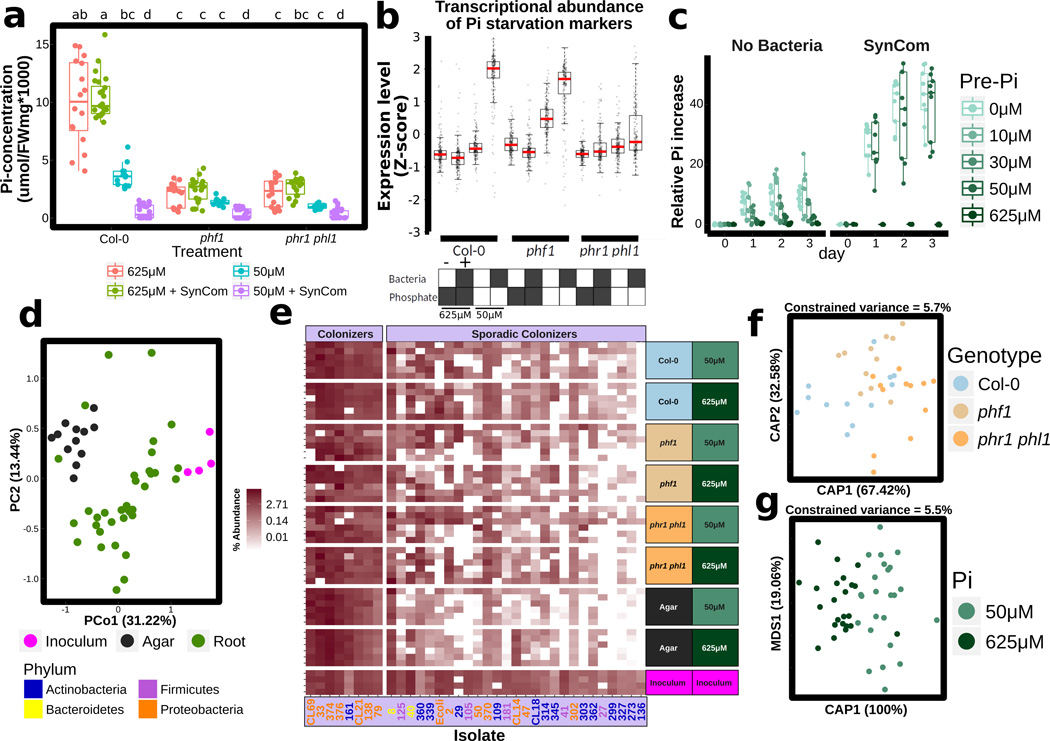
Figure 2. A bacterial Synthetic Community (SynCom) differentially colonizes PSR mutants.
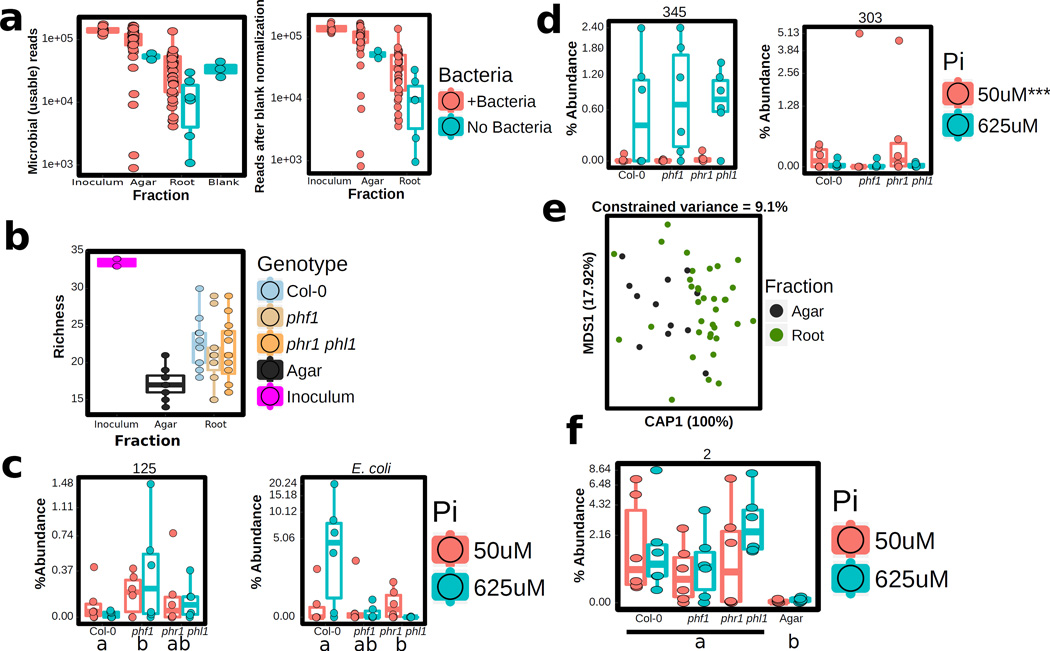
a, Pi concentration in shoots of plants grown on different Pi regimens with or without the SynCom. Plants were germinated on Johnson medium containing 0.5 % sucrose, supplemented with 1 mM Pi for 7 d in a vertical position, then transferred to 50 µM Pi or 625 µM Pi media (without sucrose) alone or with the SynCom at 105 c.f.u/mL, for another 12 d. Biological replicates numbers are: Col-0 (n=16 (625 µM Pi), 24 (625 µM Pi + SynCom), 12 (50 µM Pi), 24 (50 µM Pi + SynCom)), phf1 (n=16, 18, 12, 24) and phr1 phl1 (n=16, 18, 12, 24) from three independent experiments. Statistical significance was determined via ANOVA while controlling for experiment, and the letters indicate the results of a post-hoc Tukey test. Groups of samples that share at least one letter are statistically indistinguishable. b, Expression levels of 193 core PSR genes. The RPKM expression values of these genes were z-score transformed and used to generate box and whiskers plots that show the distribution of the expression values of this gene set. Boxes at bottom indicate presence/absence of SynCom and Pi at the concentration indicated. This labeling is maintained throughout. Data is the average of 4 biological replicates. c, Functional activation of PSR by the SynCom. Plants were grown on five different Pi levels (0 µM Pi, 10 µM Pi, 30 µM Pi, 50 µM Pi and 625 µM Pi) without the SynCom (left) and on three different Pi levels (0 µM Pi, 50 µM Pi and 625 µM Pi) with the SynCom (right). Plants were then transferred to full (1 mM Pi) condition to evaluate the capacity of the plants for Pi accumulation over time (Methods). Shoots were harvested every 24 h for 3 days and Pi concentration was measured. Pi increase represents the normalized difference in Pi content at day n with respect to the Pi content on the day of transfer to 1 mM Pi. Shoots with SynCom-activated PSR accumulated approximately 20–40 times more Pi than non-inoculated shoots. Absolute Pi concentration values are available in Supplementary Table 4. For all Pi concentrations and SynCom treatments n=6 at day 0, and n=9 at all other time points, distributed across two independent experiments. d, PCoA of SynCom experiments showing that Agar and Root samples are different from starting inoculum. Biological replicate numbers are: Inoculum (n=4), Agar (n=12) and Root (n=35) across two independent experiments. e, Heatmap showing percent abundances of SynCom isolates (columns) in all samples (rows). Strain name colors correspond to Phylum (bottom left). Within each block, samples are sorted by experiment. For each combination of genotype and Pi level, there are n=6 biological replicates evenly distributed across two independent experiments, except for Inoculum for which there are n=4 technical replicates evenly distributed across two independent experiments. f, Constrained ordination showing the effect of plant genotype and g, media Pi concentration effect on the root communities. The proportion of total variance explained (constrained) by each variable is indicated on top of each plot; for g, remaining unconstrained ordination was subjected to multi-dimensional scaling (MDS); the first MDS axis (MDS1) is shown. For f and g, biological replicate numbers are: Col-0 (n=12), phf1 (n=11), phr1 phl1 (n=12), 50uM Pi (n=24) and 625uM Pi (n=23) distributed across two independent experiments.
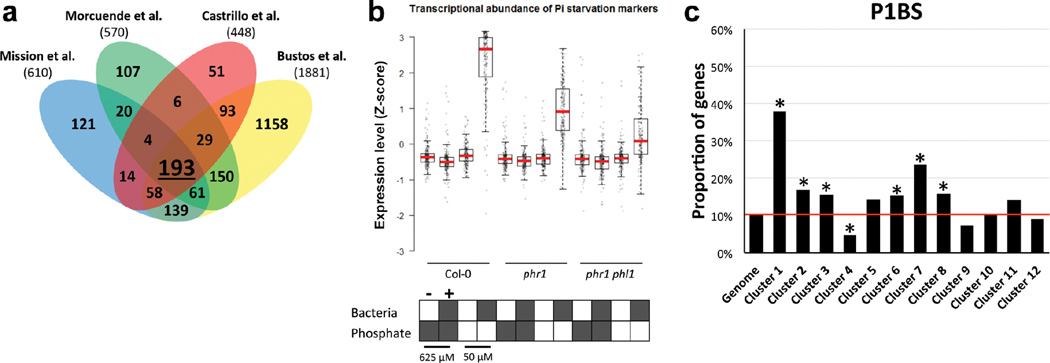
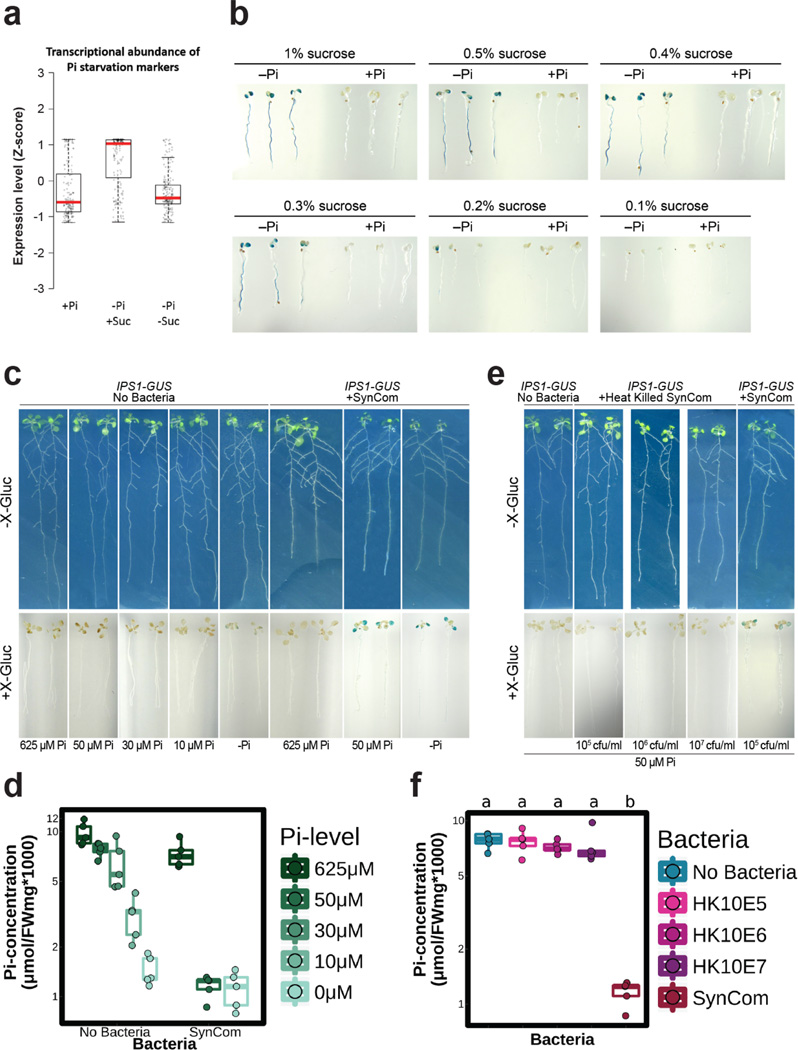
We sought to establish whether PSR was activated by the SynCom. We generated a literature-based core set of 193 PSR transcriptional markers and explored their expression in transcriptomic experiments (Extended Data Fig. 4a, b, Supplementary Table 3). In axenic low Pi conditions, only the constitutive Pi-stressed mutant phf1 exhibited induction of these PSR markers. By contrast, Col-0 plants expressed only a marginal induction of PSR markers compared to those plants grown at high Pi (Fig. 2b). This is explained by the purposeful absence of sucrose, a key component for the PSR induction in vitro20 (Supplementary Text 2; Extended Data Fig. 5) that cannot be used in combination with bacterial SynCom colonization protocols. Remarkably, the SynCom greatly enhanced the canonical transcriptional response to Pi starvation in Col-0 (Fig. 2b); this was dependent on PHR1 and PHL1 (Fig. 2b; Extended Data Fig. 4b). Various controls validated these conclusions (Supplementary Text 2; Extended Data Fig. 4–Fig. 6). Importantly, shoots of plants pre-colonized with SynCom on 0 or 50 µM Pi, but not on 650 µM Pi, accumulated 20–40 times more Pi than shoots from similarly treated non-colonized plants when subsequently transferred to full Pi conditions in the absence of additional bacteria (Fig. 2c and Supplementary Table 4). This demonstrates functional PSR activation by the SynCom. We thus propose that the transcriptional response to low Pi induced by our SynCom reflects an integral microbial element of normal PSR in complex biotic environments.
We evaluated agar- and root-associated microbiomes of plants grown with the SynCom (Supplementary Text 3; Fig. 2d, e; Extended Data Fig. 7e, f; Supplementary Table 5). In line with results from plants grown in wild soil, we found that PSR mutants failed to assemble a wild-type SynCom microbiome (Fig. 2f). Some strains were differentially abundant across PSR mutants phf1 and phr1 phl1 (Fig. 2e, f; Extended Data Fig. 7c), Pi concentration (Fig. 2g; Extended Data Fig. 7d), or sample fraction (Extended Data Fig. 7b, e, f). These results established a microcosm reconstitution system to study plant PSR under chronic competition with plant-associated microbes and allowed us to confirm that the tested PSR mutants influence root microbiome membership.
Coordination between phosphate stress response and immune system output
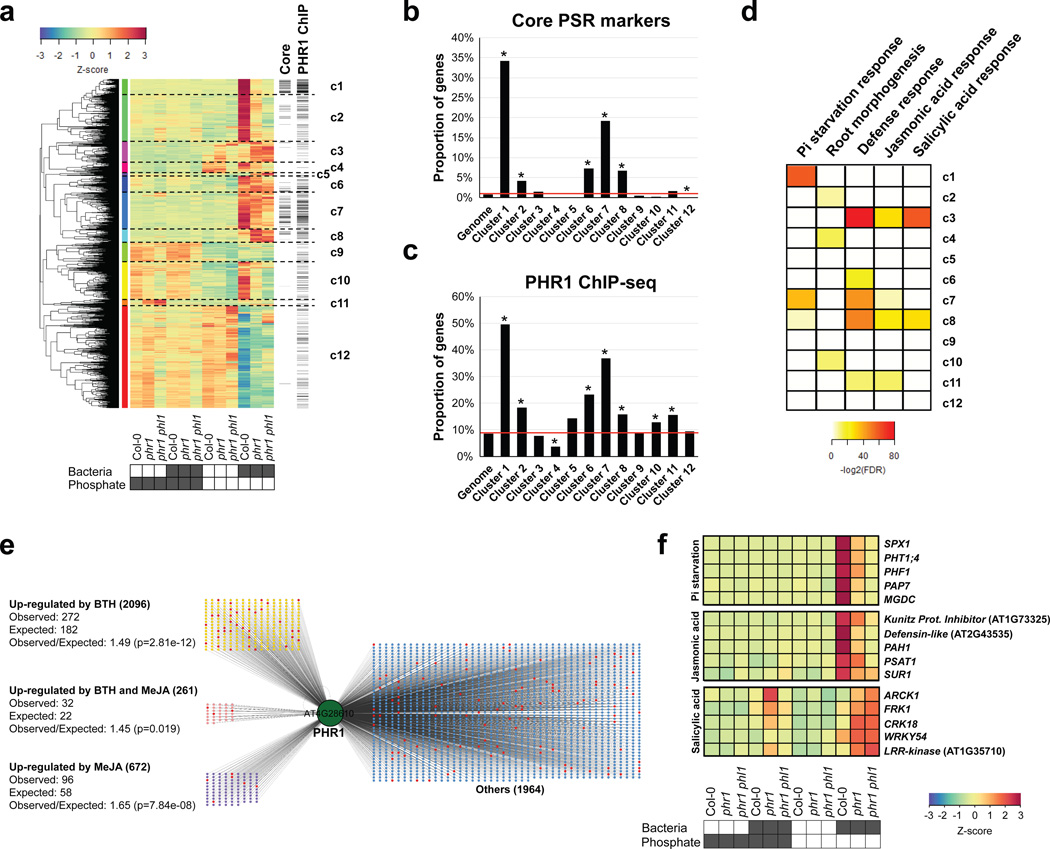
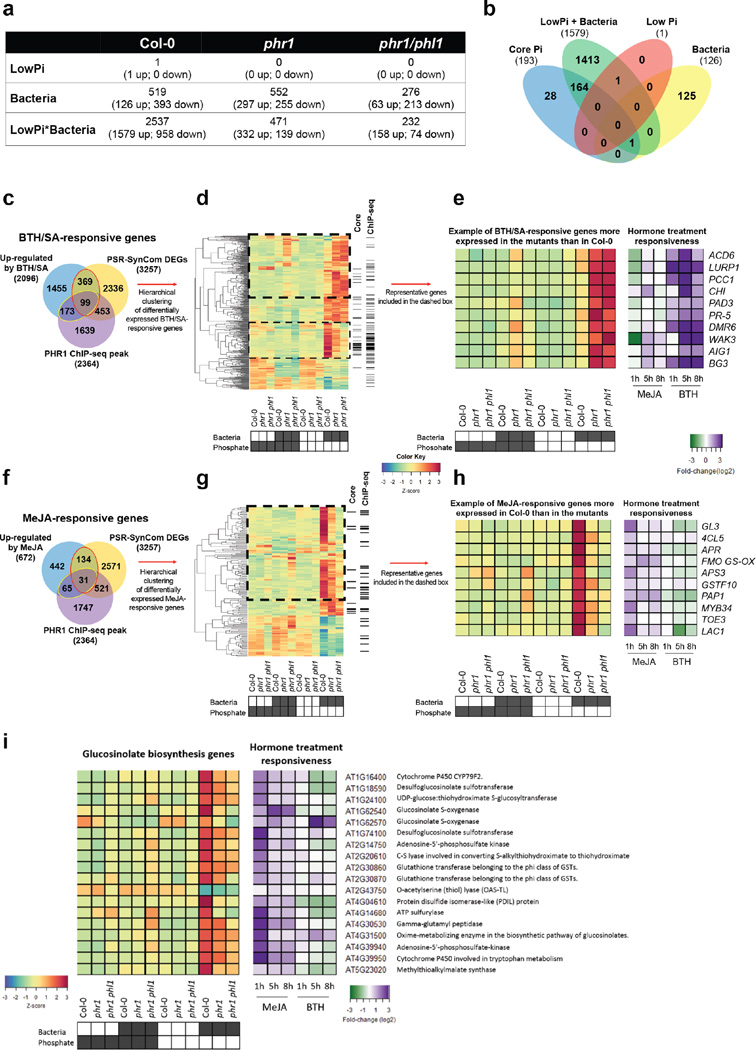
We noted that phr1 phl1 and phf1 differentially activated transcriptional PSR in the presence of our SynCom (Fig. 2b). Therefore, we investigated the transcriptomes of plants growing in the SynCom to understand how these microbes activate PHR1-dependent PSR. We identified differentially expressed genes (DEGs) that responded to either low Pi, presence of the SynCom, or the combination of both (hereafter PSR-SynCom DEGs) (Supplementary Text 4; Extended Data Fig. 8a, b; Supplementary Table 6). Hierarchical clustering (Fig. 3a, Supplementary Table 7) revealed gene sets (c1, c2, c7 and c10) that were more strongly activated in Col-0 than in phr1 or phr1 phl1. These clusters contained most of the core PSR markers regulated by PHR1 (Fig. 3b). They were also enriched in PHR1 direct targets identified in an independent ChIP-seq experiment (Fig. 3c, Supplementary Table 8), PHR1 promoter binding motifs (Extended Data Fig. 4c), and genes involved in biological processes related to PSR (Fig. 3d and Supplementary Table 9). PHR1 surprisingly contributed to transcriptional regulation of plant immunity. Five of the twelve clusters (Fig. 3a; c3, c6, c7, c8 and c11) were enriched in genes related to plant immune system output; four of these were over-represented for jasmonic acid (JA) and/or salicylic acid (SA) pathway markers (Fig. 3d, c3, c7, c8, and c11; Supplementary Table 9) and three of these four were enriched for PHR1 direct targets (Fig. 3c). SA and JA are plant hormone regulators of immunity and at least SA modulates Arabidopsis root microbiome composition2.
Figure 3. PHR1 mediates interaction of the PSR and plant immune system outputs.
a, Hierarchical clustering of 3257 genes that were differentially expressed in the RNA-seq experiment. Plants were germinated on Johnson medium containing 0.5 % sucrose supplemented with 1 mM Pi for 7 d, then transferred to 50 µM Pi or 625 µM Pi media (without sucrose) alone or with the Synthetic Community at 105 c.f.u/mL, for another 12 d (plates vertical). Columns on the right indicate genes that are core PSR markers (‘core’ lane) or had a PHR1 binding peak (‘PHR1 ChIP’ lane). b, Proportion of PSR marker genes per cluster. c, Proportion of PHR1 direct targets genes per cluster. The red line in b and c denotes the proportion of genes in the whole Arabidopsis genome that contain the analyzed feature. Asterisk denotes significant enrichment or depletion (p ≤ 0.05; hypergeometric test). d, Summary of the Gene Ontology enrichment analysis for each of the twelve clusters. The enrichment significance is shown as -log2(FDR). White means no enrichment. The complete results are in Supplementary Table 9. e, The set of genes bound by PHR1 (At4g28610) in ChIP-seq experiments is enriched in genes that are up-regulated by BTH/SA and/or MeJA. Red nodes are core PSR marker genes. f, Example of genes bound by PHR1 and differentially expressed in our experiment. PSR marker genes (top) and JA response (middle) are more expressed in wild-type plants, whereas SA-responsive genes (bottom) exhibit higher transcript levels in phr1 and phr1 phl1. The heatmaps show the average measurement of ten biological replicates for Col-0 and phr1 and six for phr1 phl1. The color key (blue to red) related to a, and f, represents gene expression as Z-scores.
To further explore PHR1 function in the regulation of plant immunity, we generated transcriptomic time course data for treatment-matched Col-0 seedlings following application of Methyl Jasmonate (MeJA) or the SA analogue Benzothiadiazole (BTH; Supplementary Table 10). We found a considerable over-representation of SA- and JA-activated genes among the PSR-SynCom DEGs (468 versus 251 predicted for SA and 165 vs. 80 predicted for JA; p<0.0001, hypergeometric test) (Extended Data Fig. 8c–h, Supplementary Table 7). A large proportion of SA-responsive genes were more strongly expressed in phr1 and phr1 phl1 than in Col-0; these were strongly enriched for classical SA-dependent defense genes (Extended Data Fig. 8d, e). A second group of SA-responsive genes that were less expressed in phr1 and phr1 phl1 than in Col-0 lacked classical SA-dependent defense genes and were weakly enriched for genes likely contributing to PSR (Extended Data Fig. 8d). By contrast, most JA-responsive genes exhibited lower expression in phr1 and phr1 phl1 (Extended Data Fig. 8g, h), including a subset of 18 of 46 genes known or predicted to mediate biosynthesis of defense-related glucosinolates (Extended Data Fig. 8i)21. This agrees with the recent observation that phr1 exhibited decreased glucosinolate levels during Pi starvation22. Analyses of SA- and JA- up-regulated genes revealed enrichment of direct PHR1 targets (Fig. 3e), consistent with the converse observation that some PHR1-regulated clusters enriched in direct targets were also enriched in defense genes (Fig. 3c, d). Many of the SA- and JA- responsive genes were PSR-SynCom DEGs (Fig. 3f; Extended Data Fig. 8c–h, Supplementary Table 7). Thus, PHR1 directly regulates an unexpected proportion of the plant immune system during PSR triggered by our SynCom.
PHR1 integrates plant immune system output and phosphate stress response
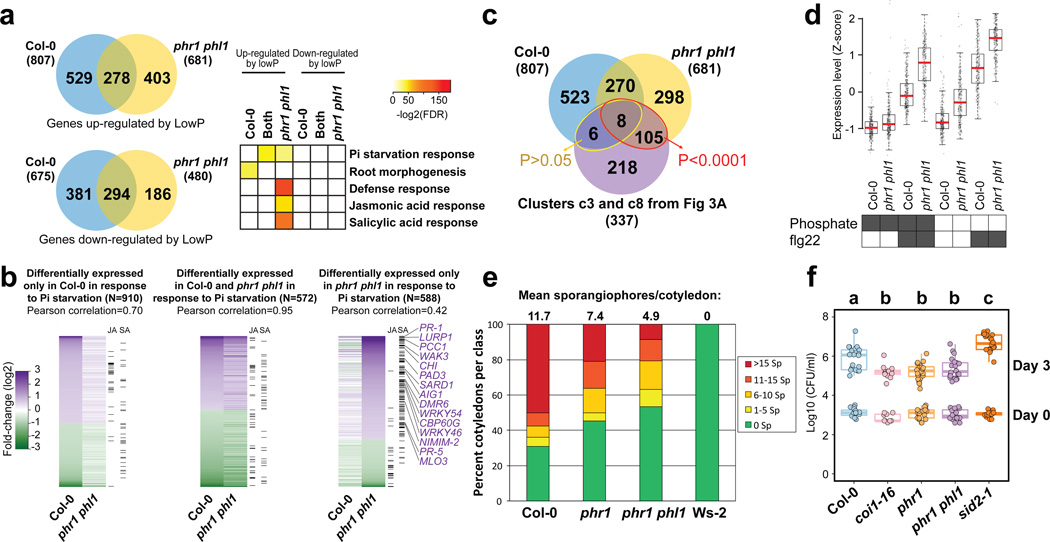
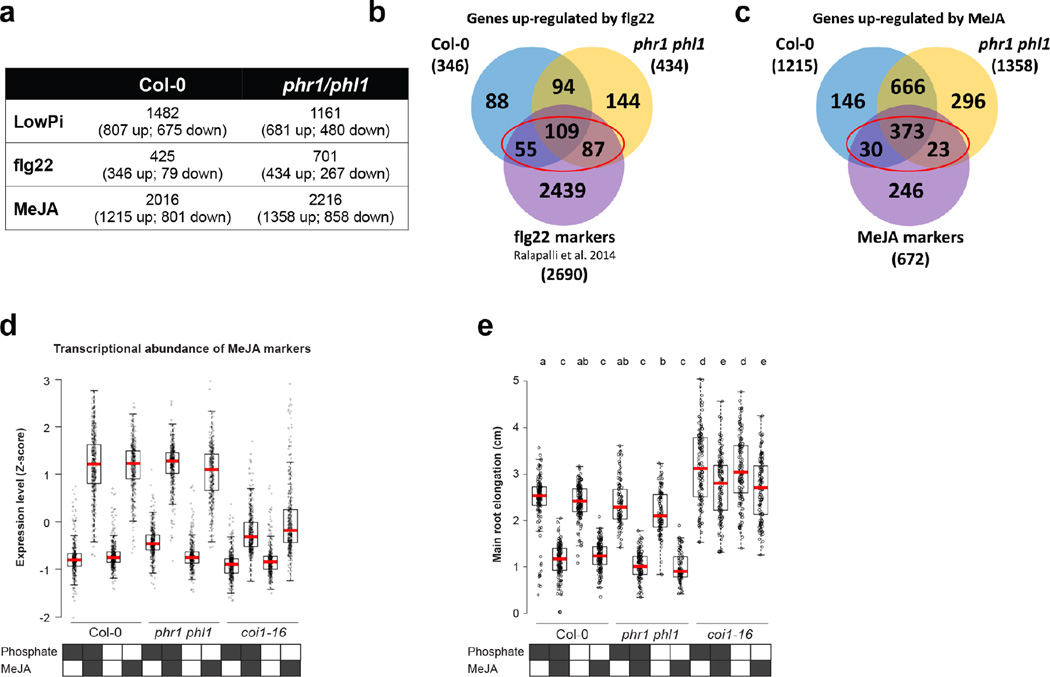
We tested whether PHR1 also controls the expression of plant defense genes under conditions typically used to study PSR (axenic growth, sucrose present, no microbiota involved). We performed RNA-seq in response to low Pi in these conditions and identified 1482 DEGs in Col-0 and 1161 DEGs in phr1 phl1 (Fig. 4a, b; Extended Data Fig. 9, Supplementary Table 11). A significant number of our BTH/SA-activated genes were also up-regulated in phr1 phl1, but not in Col-0 in response to low Pi (Fig. 4a, b; Supplementary Table 12). A large number of these overlapped with the defense genes induced in phr1 phl1 by our SymCom (Fig. 4c; red ellipse, 113/337 = 33%; clusters c3 and c8 from Fig. 3a). At least 14/113 are direct PHR1 targets (Supplementary Table 12).
Figure 4. Loss of PHR1 activity results in enhanced activation of plant immunity.
a, Venn diagram (left) showing the overlap between genes up-regulated and down-regulated in Col-0 and phr1 phl1 in response to phosphate starvation. Gene ontology enrichment (right) analyses indicate that defense-related genes are up-regulated exclusively in phr1 phl1. The complete enrichment results are shown in Supplementary Table 14. Color key (white to red) represents the gene ontology enrichment significance shown as -log2(FDR). White means no enrichment. b, Fold-change of genes differentially expressed in Col-0, phr1 phl1 or in both genotypes in response to phosphate starvation. Columns on the right indicate whether each gene is also up-regulated by MeJA or BTH/SA. Arabidopsis plants were germinated on Johnson medium (1 % sucrose) containing 1 mM Pi for 7 d in a vertical position and then transferred to the same medium containing 1 % sucrose either alone or supplemented with 1 mM Pi for 12 d. c, Venn diagram showing the overlap among genes up-regulated in Col-0 and phr1 phl1 during a typical PSR (from a) and the defense genes up-regulated in phr1 phl1 in response to the SynCom (from Fig. 3a; clusters c3 and c8). The red ellipse indicates 113 defense genes that were up-regulated in phr1 phl1 during classical PSR and during PSR triggered by the SynCom; yellow ellipse indicates the 14 genes up-regulated genes under the same conditions. p-values refer to enrichment results using hypergeometric tests. dphr1 phl1 exhibits enhanced transcriptional activation of 251 genes differentially expressed following chronic flg22 exposure. Averaged from six biological replicates. ephr1 exhibits enhanced disease resistance to the biotrophic oomycete pathogen Hyaloperonospora arabidopsidis isolate Noco2. Infection classes were defined by the number of asexual sporangiophores (Sp) per cotyledon and displayed as a color gradient from green (more resistant) to red (more susceptible); the mean number of sporangiophores per cotyledon is noted above each bar. Col-0 and Ws-2 represent susceptible and resistant controls, respectively. More than 100 cotyledons counted per genotype; the experiment was performed at least five times with similar results. fphr1 mutants exhibit enhanced disease resistance to the hemibiotrophic bacterial pathogen Pseudomonas syringae DC3000. The coi1-16 (n= 9 (day zero), 13 (day three)) and sid2-1 (n= 16, 20) mutants were controls for resistance and susceptibility, respectively. Col-0 (n=16, 20), phr1 (n=17, 20), phr1 phl1 (n=16, 20) and control plants were inoculated under typical experimental conditions: phosphate replete in non-axenic potting soil (Extended Data Fig. 2). The experiment includes at least 9 biological replicates from three independent experiments. Statistical comparisons among genotypes were one-way ANOVA tests followed by a post-hoc Tukey analysis; genotypes with the same letter above the graph are statistically indistinguishable at 95 % confidence.
To underscore the role of PHR1 in the regulation of response to microbes, we analyzed the transcriptional profile of Col-0 and phr1 phl1 plants exposed to the flagellin peptide flg22. We chose a chronic exposure to flg22 to mimic the condition of plants in contact with a microbiome. We found that phr1 phl1 plants displayed higher expression of flg22-responsive genes23 than Col-0, independent of phosphate status (Supplementary Text 5; Fig. 4d; Extended Data Fig. 9a, b; Supplementary Table 11; Supplementary Table 13). This indicates that PHR1 negatively regulates the immune response triggered by flg22.
We hypothesized that phr1 phl1 would express an altered response to pathogen infection. The phr1 and phr1 phl1 mutants exhibited enhanced disease resistance against both Hyaloperonospora arabidopsidis isolate Noco2, and Pseudomonas syringae DC3000 (Fig. 4e, f). Collectively, these results confirm the role of PHR1 as a direct integrator of PSR and the plant immune system.
Conclusions
Plant responses to phosphate stress are inextricably linked to life in microbe-rich soil. We demonstrate that genes controlling PSR contribute to assembly of a normal root microbiome. Surprisingly, our SynCom enhanced the activity of PHR1, the master regulator of the PSR, in plants grown under limited phosphate. This led to our discovery that PHR1 is a direct regulator of a functionally relevant set of plant immune system genes. Despite being required for the activation of JA-responsive genes during PSR24, we found that PHR1 is unlikely to be a general regulator of this response (Extended Data Fig. 9c–e, Supplementary Table 12), but rather may fine-tune JA response in specific biological contexts.
We demonstrate that PSR and immune system outputs are directly integrated by PHR1 (and, likely, PHL1). We provide a mechanistic explanation for previous disparate observations that PSR and defense regulation are coordinated and implications that PHR1 is the key regulator8, 11, 12, 24. We provide new insight into the intersection of plant nutritional stress response, immune system function, and microbiome assembly and maintenance; systems that must act simultaneously and coordinately in natural and agricultural settings. Our findings will drive investigations aimed at enhancing phosphate use efficiency using microbes.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
METHODS
Census study experimental procedures
For experiments in wild soil, we collected the top-soil (approx. 20 cm) from a site free of pesticide and fertilizer at Mason Farm (MF; North Carolina, USA; +35° 53′ 30.40′′, −79° 1′ 5.37′′) 19. Soil was dried, crushed and sifted to remove debris. To improve drainage, soil was mixed 2:1 volume with autoclaved sand. Square pots (2 × 2 inch square) were filled with the soil mixture and used to grow plants. Soil micronutrient analysis is published by Lundberg, D.S. et al. 19.
All Arabidopsis thaliana mutants used in this study were in the Columbia (Col-0) background (Supplementary Table 16). All seeds were surface-sterilized with 70 % bleach, 0.2 % Tween-20 for 8 minutes, and rinsed 3X with sterile distilled water to eliminate any seed-borne microbes on the seed surface. Seeds were stratified at 4 °C in the dark for 2 days.
To determine the role of phosphate starvation response in controlling microbiome composition, we analyzed five mutants related to the Pi-transport system (pht1;1, pht1;1 pht1;4, phf1, nla, and pho2) and two mutants directly involved in the transcriptional regulation of the Pi-starvation response (phr1 and spx1 spx2). All these genes are expressed in roots 13– 18.
Seeds were germinated in sterile square pots filled with MF soil prepared as described above. We also used pots without plants as “bulk soil” controls. All pots, including controls, were watered from the top with non-sterile distilled water to avoid chlorine and other tap water additives 2 times a week. Plants were grown in growth chambers with a 16-h dark/8-h light regime at 21 °C day 18 °C night for 7 weeks. In all experiments, pots with plants of different genotypes were randomly placed in trays according to true random numbers derived from atmospheric noise; we obtained those numbers from www.random.org. We positioned trays in the growth chamber without paying attention to the pots they contained, and we periodically reshuffled them without paying attention to the pot labels.
Plants and bulk soil controls were harvested and their endophytic compartment (EC) microbial communities isolated as described19. DNA extraction was performed using 96-well format MoBio PowerSoil Kit (MOBIO Laboratories) following the manufacturer’s instruction.
The method of Ames25 was used to determine the phosphate concentration in the shoots of seedlings grown on different Pi regimens and treatments. Main root length elongation was measured using ImageJ software26 and for shoot area and number of lateral roots WinRhizo software27 was used.
Processing of 16S sequencing data
For wild soil experiment 16S sequencing, we processed libraries according to Caporaso, et al.28. Three sets of index primers were used to amplify the V4 (515F-806R) region of the 16S rRNA gene of each sample. In each case, the reverse primer had a unique molecular barcode for each sample28. PCR reactions with ∼20 ng template were performed with 5 Prime Hot Master Mix in triplicate using plates 2, 4 and 5 from the 16S rRNA Amplification Protocol28. PCR blockers mPNA and pPNA29 were used to reduce contamination by plant host plastid and mitochondrial 16S amplicon. The PCR program used was:
Temperature cycling
95 °C for 3 min
35 cycles of
95 °C for 45 seconds
78 °C (PNA) for 30 seconds
50 °C for 60 seconds
72 °C for 90 seconds
4 °C until use
Reactions were purified using AMPure XP magnetic beads and quantified with Quant IT Picogreen. Amplicons were pooled in equal amounts and then diluted to 5.5 pM for sequencing. Samples were sequenced on an Illumina MiSeq machine at UNC, using a 500-cycle V2 chemistry kit. The library was spiked with 25 % PhiX control to increase sequence diversity. The raw data for the wild soil experiments is available in the EBI Sequence Read Archive (accession PRJEB15671).
For SynCom experiment 16S library, we amplified the V3-V4 regions of the bacterial 16S rRNA gene using primers 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). Libraries were created using a modified version of the Lundberg, D.S. et al.29. Basically, the molecule-tagging step was changed to an exponential amplification to account for low DNA yields with the following reaction:
| Reaction | |
| 5 µL | Kapa Enhancer |
| 5 µL | Kapa Buffer A |
| 1.25 µL | 5 µM 338F |
| 1.25 µL | 5 µM 806R |
| 0.375 µL | mixed PNAs (1:1 mix of 100 µM pPNA and 100 µM mPNA) |
| 0.5 µL | Kapa dNTPs |
| 0.2 µL | Kapa Robust Taq |
| 8 µL | dH2O |
| 5 µL | DNA |
Temperature cycling
95 °C for 60 seconds
24 cycles of
95 °C for 15 seconds
78 °C (PNA) for 10 seconds
50 °C for 30 seconds
72 °C for 30 seconds
4 °C until use
Bead cleaning
Following PCR cleanup to remove primer dimers, the PCR product was indexed using the same reaction and 9 cycles of the cycling conditions described in Lundberg, D.S. et al.29. Sequencing was performed at UNC on an Illumina MiSeq instrument using a 600-cycle V3 chemistry kit. The raw data for the SynCom experiments is available in the EBI Sequence Read Archive accession PRJEB15671.
For wild soil census analysis, sequences from each experiment were pre-processed following standard method pipelines from2, 19. Briefly, sequence pairs were merged, quality-filtered and de-multiplexed according to their barcodes. The resulting sequences were then clustered into Operational Taxonomic Unit (OTUs) using UPARSE30 implemented with USEARCH7.1090, at 97 % percent identity. Representative OTU sequences (Supplementary Dataset 1) were taxonomically annotated with the RDP classifier31 trained on the Greengenes database (4/February/2011; Supplemental Dataset 1). We used a custom script (https://github.com/surh/pbi/blob/master/census/1.filter_contaminants.r) to remove organellar OTUs, and OTUs that had no more than a kingdom-level classification, and an OTU count table was generated (Supplementary Table 1, Supplementary Dataset 1).
SynCom sequencing data were processed with MT-Toolbox32. Categorizable reads from MT-Toolbox (i.e. reads with correct primer and primer sequences that successfully merged with their pair) were quality filtered with Sickle by not allowing any window with Q-score under 20, and trimmed from the 5’ end to a final length of 270 bp. The resulting sequences were matched to a reference set of the strains in the SynCom generated from Sanger sequences, the sequence from a contaminant strain (47Yellow) that grew in the plate from strain 47 (Supplementary Table 2) and Arabidopsis organellar sequences. Sequence mapping was done with USEARCH7.1090 with the option –”usearch_global” at a 98 % identity threshold. 90 % of sequences matched an expected isolate, and those sequence mapping results were used to produce an isolate abundance table. The remaining unmapped sequences were clustered into OTUs with the same settings used for the census experiment, the vast majority of those OTUs belonged to the same families as isolates in the SynCom, and were probably unmapped due to PCR and/or sequencing errors. We combined the isolate and OTU count tables into a single master table. The resulting table was processed and analyzed with the code at (https://github.com/surh/pbi/blob/master/syncom/7.syncomP_16S.r). Matches to Arabidopsis organelles were discarded. PCR blanks were included in the sequencing and the average counts per strain observed on those blanks were subtracted from the rest of the samples following33. Extended Data Fig. 7a shows the number of usable reads across samples, and the remaining number after subtracting sterile controls (blanks).
In vitro plant growth conditions
For physiological, transcriptional analysis or pathology experiments, we used phr1, phr1 phl1, phf1, and coi1–16, sid2-1 mutants, which are all in the Col-0 genetic background (Supplementary Table 16). For all physiological and transcriptional analysis in vitro, Arabidopsis seedlings were grown on Johnson medium [KNO3 (0.6 g/L), Ca(NO3)2.4H2O (0.9 g/L), MgSO4.7H2O (0.2 g/L), KCl (3.8 mg/L), H3BO3 (1.5 mg/L), MnSO4.H2O (0.8 mg/L), ZnSO4.7H2O (0.6 mg/L), CuSO4. 5H2O (0.1 mg/L), H2MoO4 (16.1 µg/L), FeSO4.7H2O (1.1 mg/L), Myo-Inositol (0.1 g/L), MES (0.5 g/L), pH 5.6 – 5.7] solidified with 1 % bacto-agar (BD, Difco). Media were supplemented with Pi (KH2PO4) at distinct concentrations depending on the experiment; 1 mM Pi was used for complete medium and approximately 5 µM Pi (traces of Pi in the agar) was the Pi concentration in the medium not supplemented with Pi. Unless otherwise stated, plants were grown in a growth chamber in a 15-h dark/9-h light regime (21 °C day /18 °C night).
For Synthetic Community experiments, plants were germinated on Johnson medium containing 0.5 % sucrose, with 1 mM Pi, 5 µM Pi or supplemented with KH2PO3 (phosphite) at 1 mM for 7 d in a vertical position, then transferred to 50 µM Pi or 625 µM Pi media (without sucrose) alone or with the Synthetic Community at 105 c.f.u/mL, for another 12 d. For the heat-killed SynCom experiments, plants were grown as above. Heat-killed SynComs were obtained by heating different concentrations of bacteria: 105 c.f.u/mL, 106 c.f.u/mL and 107 c.f.u/mL at 95 °C for 2 h in an oven. The whole content of the heat-killed SynCom solutions were added to the media.
For the functional activation of the PSR by the SynCom, plants were germinated on Johnson medium containing 0.5 % sucrose, 1 mM Pi for 7 d in a vertical position, then transferred to 0, 10, 30, 50 and 625 µM Pi alone, or to 0, 50 and 625 µM Pi with the Synthetic Community at 105 c.f.u / mL, for another 12 d. At this point, we harvested our time zero (3 replica per conditions, each replica was 5 shoots harvested across all plates used). The remaining plants were transferred again to 1mM Pi to evaluate the capacity of the plants for Pi accumulation in a time series analysis. We harvested plant shoots every 24 h for 3 days and Pi-concentration was determined. Pi increase was calculated as: (Pi concentration day n – Pi concentration day 0) / Pi-concentration day 0. Relative increase in Pi concentration is plotted in Fig. 2c. Both relative and absolute Pi concentration values are provided in Supplementary Table 4.
We repeated this experiment twice. For the first experiment, we used 6 plates with 10 plants per condition (48 plates and 480 plants in total). We harvested three replicas per time point with 5 shoots each. In all cases, shoots were harvested across all plates used. For the second experiment, we used 11 plates with 10 plants per condition (88 plates and 880 plants). In this case, we harvested 6 replicas for 1, 2 and 3 days after the re-feeding with Pi, and three replicas for time zero. Each replica contains 5 shoots harvested across all the plates used.
For the demonstration that sucrose is required for the induction of PSR in sterile conditions, plants overexpressing the PSR reporter construct IPS1:GUS13 were grown in Johnson medium containing 1 mM Pi or 5 µM Pi supplemented with different concentrations of sucrose. After 12 days, the expression of the reporter constructs IPS1:GUS, highly induced by low Pi, was followed by GUS staining. Plants were grown in a growth chamber in a 15-h light/9-h dark regime (21 °C day /18 °C night).
For the ChIP-seq experiment, phr1 harboring the PromPHR1:PHR1-MYC construct18 and Col-0 seedlings were grown on Johnson medium 1 mM Pi, 1 % sucrose for 7 days and then transferred to a media not supplemented with Pi for another 5 days. Plants were grown in a growth chamber in a 15-h light/9-h dark regime (21 °C day /18 °C night). A total of 2364 genes were identified as regulated by PHR1. The ChIP-seq data will be fully presented in de Lorenzo, et al.34.
For the transcriptional analysis under conditions typically used to study PSR (axenic growth with sucrose present; no microbiota involved), with Methyl Jasmonate (MeJA) and the 22-amino acid flagellin peptide (flg22), plants were germinated on Johnson medium (1 % sucrose) containing 1 mM Pi for 7 d in a vertical position and then transferred to 1 mM Pi and 5 µM Pi media containing 1 % sucrose either alone or supplemented with 10 µM MeJA (Sigma) or 1 µM flg22 (Sigma) for 12 d.
For growth inhibition assays, seedlings were grown on Johnson medium (1 % Sucrose) in 1 mM Pi and 5 µM Pi conditions for 5 d, transferred to 1 mM Pi and 5 µM Pi media supplemented or not with 10 µM MeJA for 5 d. Main root length was then measured using ImageJ software26.
Bacterial isolation and culture
For Synthetic Community experiments, we selected 35 diverse bacterial strains. 32 of them were isolated from roots of Arabidopsis and other Brassicaceae species grown in two wild soils19. Two strains came from Mason Farm unplanted soil19, and Escherichia coli DH5α was included as a control (Supplementary Table 2). More than half (19/35) of the strains belonged to families enriched in the EC of plants grown in Mason Farm soil (Supplementary Table 2)2, 19. The strains were chosen from a larger isolate collection in a way that maximizes SynCom diversity while retaining enough differences in their 16S rRNA gene to allow for easy and unambiguous identification.
A single colony of bacteria to be tested was inoculated in 4 mL of 2XYT medium (16 g/L Tryptone, 10 g/L Yeast Extract, 5 g/L NaCL, ∼5.5 mM Pi) in a test tube. Bacterial cultures were grown while shaking at 28°C overnight. At this point, the Pi concentration was reduced to by dilution to 5 mM Pi average in the supernatants (10 cultures used for the quantification). Cultures were then rinsed with a sterile solution of 10 mM MgCl2 followed by a centrifugation step at 2600 g for 8 min. This process was repeated twice. The concentration of Pi in the supernatant after the first wash with MgCL2 was 0.06 mM Pi and after the second wash it was reduced to 0.005 mM Pi. In the suspension of SynCom member cells in MgCl2, the average concentration of Pi was 0.08 mM. The OD600nm was measured and assuming that 1 OD600nm unit is equal to 109 c.f.u/mL we equalized individual bacterium concentration to a final value of 105 c.f.u/mL of medium. The concentration of Pi in the final SynCom was 0.09 µM Pi. Thus, based on these results, we were not Pi fertilizing the plant by adding the SynCom. Medium was cooled down (to 40–44°C) near the solidification point and then the bacteria mix was added to the medium with agitation. We monitored the pH in the media after adding 1, 5, 10 mL of 10 mM MgCl2 which represents almost ten times the volume we used to add the SynCom. After adding MgCl2 the pH in the media remained stable. We also analyzed the pH after adding the SynCom at 105, 106 and 107 c.f.u/ml of media and found no pH changes. Therefore, we considered that the MES buffer we used was appropriate for this experiment.
To isolate and quantify bacteria from plant roots in the SynCom experiment, plant roots were harvested, and rinsed 3 times with sterile distilled water to remove agar particles and weakly associated microbes. Plant material was then freeze-dried. Root pulverization and DNA extraction was conducted as described above.
To isolate and quantify bacteria from agar samples, a freeze and squeeze protocol was used. Syringes with a square of sterilized miracloth at the bottom were completely packed with agar and kept at −20 °C for a week. Samples were thawed at room temperature and syringes were squeezed gently into 50 mL tubes. Samples were centrifuged at max speed for 20 min and most of the supernatant discarded. The remaining 1–2 mL of supernatant, containing the pellet, was moved into clean microfuge tubes. Samples were centrifuged again, supernatant was removed, and pellets were used for DNA extraction. DNA extraction was performed using 96-well format MoBio PowerSoil Kit (MOBIO Laboratories).
Pathology studies
For oomycete pathology studies, Hyaloperonospora arabidopsidis (Hpa) isolate Noco2 was propagated on the susceptible Arabidopsis ecotype Col-0. Spores of Hpa were suspended in deionized sterile water at a concentration of 5×104 spores/mL. The solution containing spores was spray-inoculated onto 10-d-old seedlings of Arabidopsis grown in fertilized potting soil. Inoculated plants were grown at 21 °C under a 9-h light regime. Asexual sporangiophores were counted 5 d post-inoculation on at least 100 cotyledons for each genotype.
For bacterial pathology studies, Pseudomonas syringae pv. tomato DC3000 was suspended in 10 mM MgCl2 to a final concentration of 105 c.f.u/mL. 35–40-d-old plants of Arabidopsis grown on soil were hand-infiltrated using a needle-less syringe on the abaxial leaf surface. Leaf discs (10 mm diameter) were collected after 1 h and 3 d post inoculation, and bacterial growth was measured as described35.
Genome-wide gene expression analyses
We performed 3 different sets of RNA-seq experiments in this study. (I) The first set (Fig. 2b, Fig. 3 and Extended Data Fig. 4b) evaluated the effect of the SynCom on the phosphate starvation response of Arabidopsis seedlings. In addition to wild-type Col-0 (4 replicates), phf1 (4 replicates) and phr1 phl1 (4 replicates) were included in the experiment shown in Fig. 2b, whereas Col-0 (10 replicates), phr1 (10 replicates) and phr1 phl1 (6 replicates) were used in the experiment shown in Fig. 3 and Extended Data Fig. 4b. (II) The second experiment (Extended Data Fig. 6a, b) is an expansion of the first and was designed to evaluate whether different pre-treatments (1 mM Pi, 5 µM Pi, 1 mM Phosphite [Phi]) influence the phosphate starvation response triggered by the SynCom. We used Col-0 (4 replicates), phf1 (4 replicates) and phr1 phl1 (4 replicates) in this experiment. (III) Finally, the third experiment evaluated the effect of MeJA and flg22 on the phosphate starvation response (Fig. 4 and Extended Data Fig. 9) of Arabidopsis seedlings. The genotypes Col-0 (6 replicates) and phr1 phl1 (6 replicates) were used. The experiments listed above were repeated between two and five independent times and each repetition (defined as “batch” in the generalized linear model, see RNA-seq data analysis, below) included two biological replicates per genotype per condition. Supplementary Table 15 contains the metadata information of all RNA-seq experiments. Raw reads and read counts are available at the NCBI Gene Expression Omnibus under accession number GSE87339.
RNA isolation and RNA-seq library construction
Total RNA was extracted from roots of Arabidopsis according to Logemann, et al.36. Frozen seedlings were pulverized in liquid nitrogen. Samples were homogenized in 400 µl of Z6-buffer; 8 M guanidinium-HCl, 20 mM MES, 20 mM EDTA pH 7.0. Following the addition of 400 µl phenol:chloroform:isoamylalcohol; 25:24:1, samples were vortexed and centrifuged (20000 g, 10 min) for phase separation. The aqueous phase was transferred to a new 1.5 ml tube and 0.05 volumes of 1N acetic acid and 0.7 volumes 96 % ethanol were added. The RNA was precipitated at −20 °C overnight. Following centrifugation, (20000 g, 10 min, 4 °C) the pellet was washed with 200 µl sodium-acetate (pH 5.2) and 70 % ethanol. The RNA was dried, and dissolved in 30 µl of ultrapure water and stored at −80 °C until use.
Illumina-based mRNA-seq libraries were prepared from 1000 ng RNA. Briefly, mRNA was purified from total RNA using Sera-mag oligo(dT) magnetic beads (GE Healthcare Life Sciences) and then fragmented in the presence of divalent cations (Mg2+) at 94 °C for 6 min. The resulting fragmented mRNA was used for first-strand cDNA synthesis using random hexamers and reverse transcriptase, followed by second strand cDNA synthesis using DNA Polymerase I and RNAseH. Double-stranded cDNA was end-repaired using T4 DNA polymerase, T4 polynucleotide kinase and Klenow polymerase. The DNA fragments were then adenylated using Klenow exo-polymerase to allow the ligation of Illumina Truseq HT adapters (D501–D508 and D701–D712). All enzymes were purchased from Enzymatics. Following library preparation, quality control and quantification were performed using a 2100 Bioanalyzer instrument (Agilent) and the Quant-iT PicoGreen dsDNA Reagent (Invitrogen), respectively. Libraries were sequenced using Illumina HiSeq2500 sequencers to generate 50 bp single-end reads.
RNA-seq data analysis
Initial quality assessment of the Illumina RNA-seq reads was performed using the FASTX-Toolkit. Cutadapt37 was used to identify and discard reads containing the Illumina adapter sequence. The resulting high-quality reads were then mapped against the TAIR10 Arabidopsis reference genome using Tophat38, with parameters set to allow only one mismatch and discard any read that mapped to multiple positions in the reference. The Python package HTSeq39 was used to count reads that mapped to each one of the 27,206 nuclear protein-coding genes. Extended Data Fig. 10 shows a summary of the uniquely mapped read counts per library. Raw sequencing data and read counts are available at the NCBI Gene Expression Omnibus accession number GSE87339.
Differential gene expression analyses were performed using the generalized linear model (glm) approach40 implemented in the edgeR package41. This software was specifically developed and optimized to deal with over-dispersed count data, which is produced by RNA-seq. Normalization was performed using the trimmed mean of M-values method (TMM42; function calcNormFactors in edgeR). The glmFit function was used to fit the counts in a negative binomial generalized linear model with a log link function40. For the SynCom experiment (Fig. 3), the model includes the covariates: phosphate content (High or Low), bacteria (present or absent) and batch effect. A term for the interaction between Phosphate and Bacteria was included as represented below:
The model used to analyze the effect of MeJA and flg22 (Fig. 4) included the following covariates: phosphate content (High or Low), MeJA (present or absent), flg22 (present or absent) and batch effect.
In each model, the term “Batch” refers to independent repetitions of the experiment (see the Genome-wide gene expression analyses section). Data from the different genotypes were fitted independently with the same model variables. The Benjamini-Hochberg method (False Discovery Rate; FDR)43 was applied to correct the p-values after performing multiple comparisons. Genes with FDR below or equal to 0.01 and fold-change variation of at least 1.5X were considered differentially expressed.
Transcriptional activation of the phosphate starvation response was studied using a literature-curated set of phosphate starvation marker genes (Extended Data Fig. 4a, Supplementary Table 3). This core set consists of 193 genes that were up-regulated by phosphate starvation stress across four different gene expression experiments13, 44– 46. The RPKM (Reads Per Kilobase of transcript per Million mapped reads) expression values of these 193 genes were z-score transformed and used to generate box and whiskers plots to show the distribution of the expression values of this gene set.
Hierarchical clustering analyses were performed with the heatmap.2 function in R from the gplots package47 using the sets of differentially expressed genes identified in each experiment. Genes were clustered based on the Euclidean distance and with the complete-linkage method. Genes belonging to each cluster were submitted to Gene Ontology (GO) enrichment analyses on the PlantGSEA platform48 in order to identify over-represented biological processes.
Defining markers of the MeJA and SA responses
Genes whose transcription is induced by MeJA (672 genes), BTH/SA (2096 genes) or both hormones (261 genes) were used as markers of the activation of these immune response output sectors in Arabidopsis (Supplementary Table 10)49. These gene sets were defined using two-week old Col-0 seedlings grown on potting soil and sprayed with MeJA (50 µM; Sigma), BTH (300 µM; Actigard 50WG) or a mock solution (0.02 % Silwet, 0.1 % ethanol). Samples were harvested 1 h, 5 h and 8 h after the treatment in two independent experiments. Total RNA was extracted with the RNeasy Plant Mini kit (Qiagen) and then used to prepare Illumina mRNA-seq libraries. The bioinformatics pipeline to generate count tables and the criteria used to define differentially expressed genes between conditions (Hormone treatment vs. Mock treatment) was the same as described above. Raw sequencing data are available at the NCBI Gene Expression Omnibus under the accession number GSE90077.
Statistical analyses
Most statistical analyses were performed in the R statistical environment50 and follow methods previously described2. As described in the following subsections, a number of packages were used, and many were called through AMOR-0.0–1451, which is based on code from Lebeis, et al.2. All scripts and knitr52 output from R scripts are available upon request. Most plots are ggplot253 objects generated with functions in AMOR51. For all linear modeling analyses (ANOVA, ZINB, GLM), terms for batch and biological replicate were included whenever appropriate. Code for both census and SynCom analysis is available at https://github.com/surh/pbi.
For wild soil and SynCom experiments, the number of samples per genotype and treatment was determined based on our previously published work, which showed that seven and five samples are enough to detect differences in wild soils and SynCom experiments, respectively.2, 19. For RNA-seq experiments, we used at least four replicates per condition, which is sufficient for parameter estimation with the edgeR software41.
Alpha and beta diversity were calculated on count tables that were rarefied to 1000 reads. Samples with less than this number of usable reads (i.e. high quality non-organellar reads) were discarded. Alpha diversity (Shannon index, richness) metrics were calculated using the “diversity” function in vegan54, and differences between groups were tested with ANOVA (Extended Data Fig. 1a). Site diversity (Extended Data Fig. 1b) was calculated with the “sitediv” function in AMOR51. Unconstrained ordination was performed with vegan (Bray-Curtis), and Principal Coordinate Analysis (PCoA) was performed with AMOR (Extended Data Fig. 1d)51. Canonical Analysis of Principal Coordinates (CAP) is a form of constrained ordination55 and was performed using the “capscale” function of the vegan package in R54. CAP was performed on the full counts of the EC samples only, using the “Cao” distance. Constraining was done separately on plant genotype while conditioning on sequencing depth and biological replicate. This approach allowed us to focus on the portion of variation that is associated with plant genotype, conditionally, independent of other factors.
For the SynCom experiments, richness was directly calculated in R. Principal Coordinate Analysis was performed with the “PCO” function of AMOR51 using the “Cao” distance which was calculated with vegan54 on an abundance table rarefied to 1500 reads per sample. Canonical Analysis of Principal Coordinates (CAP) was performed using the “capscale” function of the vegan package54 in R. CAP was performed on the full counts of the root samples only, using the “Cao” distance. Constraining was done separately on Fraction, Pi level and plant genotype while conditioning on sequencing depth and the other covariates.
Differentially abundant bacterial taxa across fraction and genotype in the wild soil experiments were identified using the same approach as in Lebeis, et al.2. Briefly, we used a Zero-Inflated Negative Binomial (ZINB) framework that allowed us to test for the effect of specific variables, while both controlling for the other covariates and accounting for the excess of zero entries in the abundance tables. These zero-entries likely represented under-sampling and not true absences. The same analysis was performed at the family and OTU-level on the measurable OTUs (taxa that have an abundance of at least 25 counts in at least five samples)19. Results are in Extended Data Fig. 1e–h, Supplementary Table 1. Extended Data Fig. 1h shows the distribution of significant genotypic effects on bacterial abundances at both taxonomic levels; in both cases the behavior is similar, indicating small and even effects of all genotypes.
For the comparison of enrichment profiles between genotypes, we followed the same Monte-Carlo approach described in Lebeis, et al.2. Briefly we looked at the enrichment/depletion profile of bacterial taxa for each mutant compared to wild-type Col-0, and asked, for each pair of mutants, if they were more similar than expected by chance and assed significance by random permutation. Results are in Fig. 1d, Extended Data Fig. 1g.
To define differentially abundant strains in SynCom experiments, we found that a Negative Binomial GLM approach gave more stable results than the ZINB approach. We used the edgeR package41 to fit a quasi Negative Binomial GLM model with the glmQLFit function, and significance was tested with the glmQLFtest function56. Results of all relevant pairwise comparisons are in Extended Data Fig. 7 and Supplementary Table 5.
For the definition of robust colonizers in synthetic community experiments, we calculated the average relative abundance of E. coli on all root samples and counted, for each strain, how many times it was more abundant than E. coli’s average on the same set of root samples. Then we used a one-sided binomial test to ask if the probability of a given strain to be more abundant than the average E.coli was significantly higher than a coin toss (50%). Strains that passed the test were labeled as robust-colonizers, the rest of the strains were labeled as Sporadic or Non-Colonizers. The results are indicated in Fig. 2e and Supplementary Table 2.
Data and software accessibility
All data generated from this project is publicly available. Raw sequences from soil census and SynCom colonization are available at the EBI Sequence Read Archive under accession PRJEB15671. Count tables, metadata, taxonomic annotations and OTU representative sequences from the Mason Farm census and Syncom experiments are available as Supplementary Datasets 1 and Supplementary Datasets 2 respectively. Custom scripts used for statistical analysis and plotting are available at (https://github.com/surh/pbi). Raw sequences from transcriptomic experiments are available at the NCBI Gene Expression Omnibus under the accession number GSE87339. The corresponding metadata information is provided in Supplementary Table 15. All code is available upon request.
Extended Data
Extended Data Figure 1. The Arabidopsis PSR alters highly specific bacterial taxa abundances.
a, Alpha diversity of bacterial root microbiome in wild-type Col-0, PSR mutants and bulk soil samples. We used ANOVA methods and no statistical differences were detected between plant genotypes after controlling for experiment. b, Additive beta-diversity curves showing how many OTUs are found in bulk soil samples or root endophytic (EC) samples of the same genotype as more samples (pots) are added. The curves show the mean and the 95 % confidence interval calculated from 20 permutations. c, Phylum-level distributions of plant root endophytic communities from different plant genotypes and bulk soil samples. d, Principal Coordinates Analysis based on Bray-Curtis dissimilarity of root and bulk soil bacterial communities showing a large effect of experiment on variation, as expected according to previous studies19. For a-d the number of biological replicates per genotype and soil are: Col-0 (n=17), pht1;1 (n=18), pht1;1 pht1;4 (n=17), phf1 (n=13), nla (n=16), pho2 (n=16), phr1 (n=18), spx1 spx2 (n=14) and Soil (n=17) e, Bacterial taxa that are differentially abundant (DA) between PSR mutants and Col-0. Each row represents a bacterial Family (left) or OTU (right) that shows a significant abundance difference between Col-0 and at least one mutant. The heat-map grey scale shows the mean abundance of the given taxa in the corresponding genotype, and significant enrichments and depletions with respect to Col-0 are indicated with a red or blue rectangle, respectively. Taxa are organized by phylum shown on the right bar colored according to f. f, Doughnut plot showing Family-level (top) and OTUs- level (bottom) differences in endophytic root microbiome compositions between mutants (columns) and Col-0 plants. The number inside each doughnut indicates how many bacterial Families are enriched or depleted in each mutant with respect to Col-0, and the colors in the doughnut show the phylum level distribution of those differential abundances. g, Tables of p-values from Monte Carlo pairwise comparisons between mutants. A significant p-value (cyan) indicates that two genotypes are more similar than expected by chance. Results of Family level comparison are shown. This plot should be compared with the corresponding OTU-level plot in Fig. 1d. h, Distributions of plant genotypic effects on taxonomic abundances at the Family (up) or OTU (down) level. For each genotype, the value of the linear model coefficients for individual OTUs or Families is plotted grouped by their sign. Positive values indicate that a given taxon has increased abundance in a mutant with respect to Col-0, while a negative value represents the inverse pattern. Only coefficients from significant comparisons are shown. The number of taxa (ie. points) on each box and whisker plots is indicated in the corresponding doughnut plot in f.
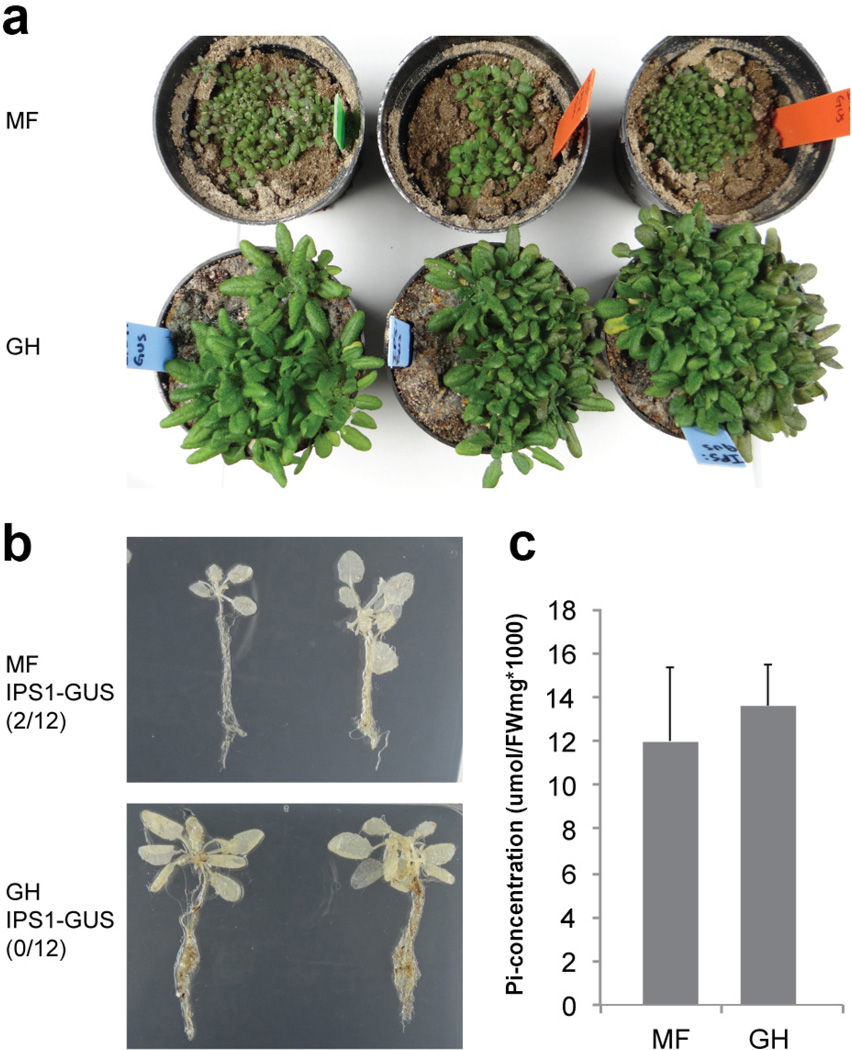
Extended Data Figure 2. Plants grown in Mason Farm wild soil or phosphate (Pi) replete potting soil do not induce PSR and accumulate the same amount of Pi.
a, Plants overexpressing the PSR reporter construct IPS1:GUS grown in Mason Farm wild soil (MF) or in phosphate (Pi) replete potting soil (GH) (250 ppm of 20-20-20 Peters Professional Fertilizer). b, Expression analysis of the reporter constructs IPS1:GUS (n= 12) shows lack of induction of PSR for both soils analyzed. In this construct, the promoter region of IPS1, highly induced by low Pi, drives the expression of GUS. Plants were grown in the conditions described in a. The number of GUS positive plants relative to the total number of plants analyzed in each condition is shown in parenthesis. c, Phosphate (Pi) concentration in shoots (n= 6) of plants grown in both soils analyzed shows no differences. Plants were grown in a growth chamber in a 15-h light/9-h dark regime (21 °C day /18 °C night). Images shown here are representative of the 12 plants analyzed in each case. Bars mean standard deviation.
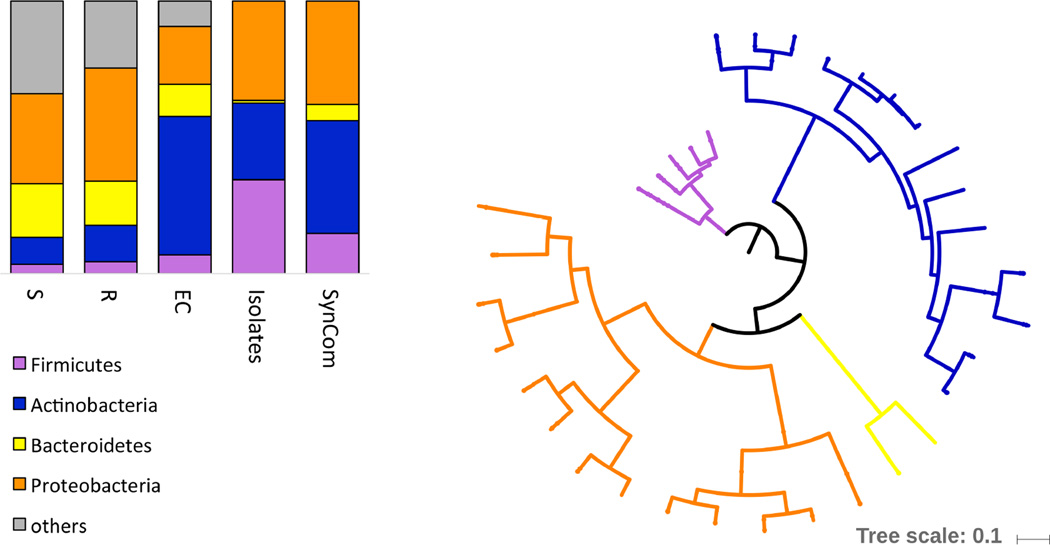
Extended Data Figure 3. Phylogenetic composition of the 35-member synthetic community (SynCom).
Left: Comparison of taxonomic composition of soil (S), rhizosphere (R) and endophyte (EC) communities from 19, with the taxonomic composition of the isolate collection obtained from the same samples and the SynCom selected from within it and used in this work. Right: Maximum likelihood phylogenetic tree of the 35-member SynCom based on a concatenated alignment of 31 single copy core proteins.
Extended Data Figure 4. Induction of the PSR triggered by the SynCom is mediated by PHR1 activity.
a, Venn diagram with the overlap among genes found up-regulated during phosphate starvation in four different gene expression experiments 13, 44–46. The intersection (193 genes) was used as a robust core set of PSR for the analysis of our transcriptional data (Supplementary Table 3). b, Expression profile of the 193 core PSR genes indicating that the SynCom triggers phosphate starvation under Low Pi conditions in a manner that depends on PHR1 activity. The RPKM expression values of these genes were z-score transformed and used to generate box and whiskers plots that show the distribution of the expression values of this gene set. Col-0, the single mutant phr1 and the double mutant phr1 phl1 were germinated at 1 mM Pi with sucrose and then transferred to low Pi (50 µM) and high Pi (625 µM Pi) alone or with the SynCom. The figure shows the average measurement of ten biological replicates for Col-0 and phr1 and six for phr1 phl1c, Percentage of genes per cluster (from figure 3) containing the PHR1 binding site (P1BS, GNATATNC) within 1000 bp of their promoters. The red line indicates the percentage of Arabidopsis genes in the whole genome that contain the analyzed feature. Asterisk denotes significant enrichment or depletion (p ≤ 0.05; hypergeometric test).
Extended Data Figure 5. The SynCom induces PSR independently of sucrose in Arabidopsis.
a, Expression analysis of a core of 193 PSR marker genes in an RNA-seq experiment using Col-0 plants. The RPKM expression values of these genes were z-score transformed and used to generate box and whiskers plots that show the distribution of the expression values of this gene set. Plants were grown in Johnson medium containing replete [1 mM Pi; (+Pi)] or stress [5 µM Pi; (−Pi)] Pi concentrations with (+Suc) or without (−Suc) 1 % sucrose. b, Expression analysis of the reporter constructs IPS1:GUS (n=20). In this construct, the promoter region of IPS1, highly induced by low Pi, drives the expression of GUS. Plants were grown in Johnson medium +Pi or -Pi at different percentages of sucrose. These results show that sucrose is required for the induction of the PSR in typical sterile conditions. Images shown are representative of the 20 plants analyzed in each case c, Top: Plants grown in sterile conditions at different Pi concentrations [left (No Bacteria)] or with a SynCom [right (+SynCom)]. Bottom: Histochemical analysis of Beta-glucoronidase (GUS) activity in overexpressing IPS1:GUS plants (n=20) from top panel. Images shown are representative of the 20 plants analyzed in each case. d, Pi concentration in plant shoots from c , in all cases n=5. Analysis of Variance indicated a significant effect of the Pi level in the media (F = 44.12, df = 1, p-value = 9.72e-8), the presence of SynCom (F = 32.61, df = 1, p-value = 1.69e-6) and a significant interaction between those two terms (F = 4.748, df = 1, p-value = 0.036) on Pi accumulation. e, Top: Plants grown in axenic conditions (No Bacteria), with a concentration gradient of heat-killed SynCom [2 h at 95 °C, (+Heat killed SynCom)] or with SynCom alive. Bottom: Histochemical analysis of GUS activity in overexpressing IPS1:GUS plants (n=15) from top panel. All plants were grown at 50 µM Pi. Images shown are representative of the 15 plants analyzed in each case f, Quantification of Pi concentration in plant shoots from e, (in call cases n=5). The SynCom effect on Pi concentration requires live bacteria. Plants were germinated on Johnson medium containing 0.5 % sucrose, with 1 mM Pi for 7 d in a vertical position, then transferred to 0, 10, 30, 50, 625 µM Pi media (without sucrose) alone or with the Synthetic Community at 105 c.f.u/mL (only for the conditions 0, 50 and 625 µM Pi), for another 12 d. For the heat-killed SynCom experiments, plants were grown as above. Heat-killed SynComs were obtained by heating different concentrations of bacteria 105 c.f.u / mL, 106 c.f.u / mL and 107 c.f.u / mL at 95 °C for 2 h in an oven. The whole content of the heat-killed SynCom solutions were add to the media. In all cases, addition of the SynCom did not change significantly the final Pi concentration or the pH in the media. Letters indicates grouping based on ANOVA and Tukey post-hoc test at 95 % confidence, conditions with the same letter are statistically indistinguishable.
Extended Data Figure 6. Bacteria induce the PSR using the canonical pathway in Arabidopsis.
a, Pi concentration in the shoot of Col-0 plants germinated in three different conditions, 5 µM Pi (−Pi) (n=14), 1 mM Pi (+Pi) (n=15) and 1 mM KH2PO3 (Phi) (n=15) for 7 days. Phi is a non-metabolizable analog of Pi; its accumulation delays the response to phosphate stress. b, Expression profile analysis of a core of PSR-marker genes in Col-0, phf1 and phr1 phl1. The RPKM expression values of these genes were z-score transformed and used to generate box and whiskers plots that show the distribution of the expression values of this gene set. Plants were germinated in three different conditions, 5 µM Pi (−Pi), 1 mM Pi (+Pi) and 1 mM KH2PO3 (Phi) and then transferred to low Pi (50 µM Pi) and high Pi (625 µM Pi) alone or with the SynCom for another 12 d. The figure shows the average measurement of four biological replicates. c, Phenotype of plants grown in axenic conditions at 625 µM Pi (Top) or at 50 µM Pi (Bottom) [left (No Bacteria)] or with a SynCom [right (+SynCom)]. Images showed here are representative of the total number of plants analyzed in each case as noted below d, Quantification of the main root elongation, e, Number of lateral roots per plant, and f, Number of lateral roots per cm of main root in plants from c. For d, e and f the number of biological replicates are: 625 uM No Bacteria (n=48), 625 uM + SynCom (n=46), 50 uM No Bacteria (n=73), and 50 uM SynCom (n=56), distributed across two independent experiments indicated with different shades of color. Measurements were analyzed with ANOVA while controlling for biological replicate. Asterisks denote a significant effect (p-value < 1e-16) of treatment with SynCom for the three phenotypes in d, e and f. In all cases, neither the interaction between Pi and Bacteria, nor Pi concentrations alone had a significant effect and were dropped from the ANOVA model. In all cases, residual quantiles from the ANOVA model were compared with residuals from a Normal distribution to confirm that the assumptions made by ANOVA hold (see code on GitHub for details, see Methods).
Extended Data Figure 7. Plant genotype and Pi concentration alter SynCom strain abundances.
a, Number of bacterial reads in samples of different types (left) and number of reads after blank normalization (right, see Methods). The number of biological replicates are: Inoculum (n=8), Agar + SynCom (n=41), Agar No Bacteria (n=2), Root + SynCom (n=36), Root No Bacteria (n=6) and Blank (n = 3), across two independent experiments b, Richness (number of isolates detected) in SynCom samples. No differences were observed between plant genotypes. The number of biological replicates per group is n=12 except for Inoculum (n=4) and phf1 (n=11) c, Exemplary SynCom strains that show quantitative abundance differences between genotypes. Genotypes with the same letter are statistically indistinguishable. d, Exemplary SynCom strains that show quantitative abundance differences depending on Pi concentration in the media. Asterisks note statistically significant differences between the two Pi concentrations. e, CAP analysis of Agar vs Root difference in SynCom communities. These differences explained 9.1 % of the variance. The number of biological replicates per fraction is: Agar (n=12) and Root (n=35), distributed across two independent experiments f, Exemplary SynCom strain that shows a statistically significant differential abundance between Root and Agar samples. Statistically significant differences are defined as FDR < 0.05. For c, d and f the number of biological replicates for every combination of genotype and Pi level is always n=6, evenly distributed across two independent experiments.
Extended Data Figure 8. PHR1 controls the balance between the SA and JA regulons during the PSR induced by a 35-member SynCom.
a, Total number of differentially expressed genes (FDR ≤ 0.01 and minimum of 1.5X fold-change) in Col-0, phr1 and phr1 phl1 with respect to Low Pi (50 µM Pi), bacteria presence and the interaction between low Pi and bacteria. In this experiment, plants were grown for 7 days in Johnson medium containing 1 mM Pi, and then transferred for 12 days to low (50 µM Pi) and high Pi (625 µM Pi) conditions alone or with the SynCom. No sucrose was added to the medium. b, Venn diagram showing the overlap between the PSR marker genes (Core Pi) and the genes that were up-regulated in Col-0 by each of the three variables analyzed. The combination of bacteria and low Pi induced the majority (85 %) of the marker genes. c, PHR1 negatively regulates the expression of a set of SA-responsive genes during co-cultivation with the SynCom. Venn diagram showing the overlap among PSR-SynCom DEGs, genes up-regulated by BTH treatment of Arabidopsis seedlings, and the direct targets of PHR1 identified by ChIP-seq. The red ellipse indicates the 468 BTH/SA-responsive genes that were differentially expressed. A total of 99 of these genes (21 %) are likely direct targets of PHR1. The yellow ellipse indicates 272 SA-responsive genes that were bound by PHR1 in a ChIP-seq experiment (see Fig. 3e). Approximately one-third of them (99/272) were differentially expressed in the SynCom experiment. d, Hierarchical clustering analysis showed that nearly half of the BTH/SA-induced genes that were differentially expressed in our experiment are more expressed in phr1 or phr1 phl1 mutants compared to Col-0 (dashed box). The columns on the right indicate those genes that belong to the core PSR marker genes (‘core’ lane) or that contain a PHR1 ChIP-seq peak (‘ChIP-seq’ lane). A subset of the SA marker genes is less expressed in the mutant lines (thin dashed box). This set of genes is also enriched in the core PSR markers and in PHR1 direct targets (p<0.001; hypergeometric test), indicating that PHR1 can function as a positive activator of a subset of SA-responsive genes. Importantly, these genes are not typical components of the plant immune system but rather encode proteins that play a role in the physiological response to low phosphate availability (e.g., phosphatases and transporters). e, Examples of typical SA-responsive genes are shown on the right along with their expression profiles in response to MeJA or BTH/SA treatment compared to Col-0. f, PHR1 activity is required for the activation of JA-responsive genes during co-cultivation with the SynCom. Venn diagram showing the overlap among DEG from this work (PSR-SynCom), genes up-regulated by MeJA treatment of Arabidopsis seedlings and the genes bound by PHR1 in a ChIP-seq analysis. Red ellipse indicates 165 JA-responsive genes that were differentially expressed. Thirty-one of these (19 %) were defined as direct targets of PHR1. The yellow ellipse indicates 96 JA-responsive genes that were bound by PHR1 in a ChIP-seq experiment. Approximately one-third of them (31/96) were differentially expressed in the SynCom experiment. g, Hierarchical clustering analysis showed that almost 75 % of the JA-induced genes that were differentially expressed in our experiment are less expressed in the phr1 mutants (dashed box). The columns on the right indicate those genes that belong to the core PSR marker genes (‘core’ lane) or that contain a PHR1 ChIP-seq peak (‘ChIP-seq’ lane). h, Examples of well-characterized JA-responsive genes are shown on the right along with their expression profiles in response to BTH and MeJA treatments obtained in an independent experiment. i, Heatmap showing the expression profile of the 18 genes that were differentially expressed in our experiment and participate in the biosynthesis of glucosinolates21. In general, these genes showed lower expression in the phr1 mutants indicating that PHR1 activity is required for the activation of a sub-set of JA-responsive genes that mediate glucosinolate biosynthesis. The transcriptional response to BTH/SA and MeJA treatments is shown on the right and was determined in an independent experiment in which Arabidopsis seedlings were sprayed with either hormone. MeJA induces the expression of these glucosinolate biosynthetic genes, whereas BTH represses many of them. The gene IDs and the enzymatic activity of the encoded proteins are shown on the right. Results presented in this figure are based on ten biological replicates for Col-0 and phr1 and six for phr1 phl1. The color key (blue to red) related to d, e, g, h, i represents gene expression as Z-scores and the color key (green to purple) related to e, h, i represents gene expression as log2 fold-changes.
Extended Data Figure 9. PHR1 activity effects on flg22 and MeJA-induced transcriptional responses.
a, Total number of differentially expressed genes (FDR ≤ 0.01 and minimum of 1.5X fold-change) in Col-0 and phr1 phl1 with respect to low Pi (50 µM Pi), flg22 treatment (1 µM) and MeJA (10 µM). In this experiment, plants were grown for 7 days in Johnson medium containing 1 mM Pi, and then transferred for 12 days to low (50 µM Pi) and high Pi (625 µM Pi) conditions alone, or in combination with each treatment. Sucrose was added to the medium at a final concentration of 1 %. b, Venn diagram showing the overlap among genes that were up-regulated by chronic exposure to flg22 in Col-0 and in phr1 phl1 and a literature-based set of genes that were up-regulated by acute exposure (between 8 to 180 min) to flg2223. The red ellipse indicates the 251 chronic flg22-responsive genes defined here. c, Venn diagram showing the overlap among genes that were up-regulated by chronic exposure to MeJA in Col-0 and in phr1 phl1 in this work and a set of genes that were up-regulated by MeJA treatment of Arabidopsis seedlings (between 1 and 8 hours). The red ellipse indicates the intersection of JA-responsive genes identified in both experiments. d, Col-0 and phr1 phl1 exhibit similar transcriptional activation of 426 common JA-marker genes (c) independent of phosphate concentration. As a control we used coi1-16, a mutant impaired in the perception of JA. The gene expression results are based on six biological replicates per condition. e, Growth inhibition of primary roots by MeJA. Root length of wild-type Col-0 (n= 125 (+ Pi - MeJA), 120 (+ Pi + MeJA), 126 (− Pi - MeJA), 125 (− Pi + MeJA)), phr1 phl1 (n=85, 103, 90, 80) and the JA perception mutant coi1-16 (n= 125, 120, 124, 119) was measured after 4 days of growth in the presence or not of MeJA with or without 1 mM Pi. Letters indicate grouping based on multiple comparisons from a Tukey post-hoc test at 95 % confidence. In agreement with the RNA-seq results, no difference in root length inhibition was observed between Col-0 and phr1 phl1.
Extended Data Figure 10. Number of mapped reads for each RNA-seq library used in this study.
The figure shows the maximum, minimum, average and median number of reads mapping per gene for all RNA-seq libraries generated. The total number of reads mapping to genes is also shown for each library. With the exception of the minimum number of mapped reads, which is zero for all libraries, all values are shown in a log scale.
Supplementary Material
Acknowledgments
Support by NSF INSPIRE grant IOS--1343020 and DOE-USDA Feedstock Award DE-SC001043 to J.L.D. S.H.P was supported by NIH Training Grant T32 GM067553--06 and is a Howard Hughes Medical Institute International Student Research Fellow. P.J.P.L.T was supported by The Pew Latin American Fellows Program in the Biomedical Sciences. J.L.D is an Investigator of the Howard Hughes Medical Institute, supported by the HHMI and the Gordon and Betty Moore Foundation (GBMF3030). M.E.F and O.M.F are supported by NIH NRSA Fellowships F32-GM112345-02 and F32-GM117758-01, respectively. N.W.B was supported by NIH NRSA Fellowship F32--GM103156. J.P-A is funded by the Spanish Ministry of Economy and Competitiveness (MINECO BIO2014-60453-R and EUI2008-03748). We thank Stratton Barth and Emily Getzen for technical assistance, the Dangl lab microbiome group for useful discussions and Drs. Sarah Grant, Derek Lundberg, Farid El Kasmi, Paul Schulze-Lefert and his colleagues for critical comments on the manuscript. Supplement contains additional data. Raw sequence data is available at the EBI Sequence Read Archive accession PRJEB15671 for microbiome 16S profiling, and at the Gene Expression Omnibus accessions GSE87339 for transcriptomic experiments. J.L.D is a co-founder of, and shareholder in, and S.H.P collaborates with, AgBiome LLC, a corporation whose goal is to use plant-associated microbes to improve plant productivity.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author contributions
G.C., P.J.P.L.T., S.H.P and J.L.D. designed the project, G.C., S.H.P, T.F.L and M.E.F. set up the experiments, harvested samples and organized construction of 16S sequencing libraries. G.C and T.F.L performed control experiments related with PSR induced by the SynCom. G.C, N.W.B, M.E.F and T.F.L set up the experiments, harvested samples and isolated RNA. P.J.P.L.T. organized, performed construction of RNA-seq libraries and analysed RNA-seq data. S.H.P. analysed 16S sequencing data. S.H.P and P.J.P.L.T oversaw data deposition. G.C, T.F.L. and P.J.P.L.T performed pathology experiments. G.C., P.J.P.L.T, S.H.P, T.F.L, O.M.F and J.L.D. analysed data and created figures. L.d.L. performed the ChIP-seq experiment. C.D.J and P.M. advised on sequencing process and statistical methods. G.C., P.J.P.L.T., S.H.P., and J.L.D. wrote the manuscript with input from O.M.F., C.D.J., and J.P.-A.
Author Information
All data generated from this project is publicly available. Raw sequences from soil census and SynCom colonization are available at the EBI Sequence Read Archive accession PRJEB15671. Count tables, metadata, taxonomic annotations and OTU representative sequences from the Mason Farm census and Syncom experiments are available as Supplementary Datasets 1 and Supplementary Datasets 2 respectively. Custom scripts for analysis and visualization are at https://github.com/surh/pbi. Raw sequences and counts from RNA-seq experiments are available at the NCBI Gene Expression Omnibus under accession number GSE87339. The corresponding metadata information is provided in Supplementary Table 15. All code is available upon request. Reprint and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interest. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 2.Lebeis SL, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 3.Hacquard S, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 2015;17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Q, Riley WJ, Tang J, Koven CD. Multiple soil nutrient competition between plants, microbes, and mineral surfaces: model development, parameterization, and example applications in several tropical forests. Biogeosciences. 2016;13:341–363. [Google Scholar]
- 5.Richardson AE, Simpson RJ. Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol. 2011;156:989–996. doi: 10.1104/pp.111.175448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghothama KG. Phosphate acquisition. Physiol. Plant Mol. Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- 7.Lambers H, Martinoia E, Renton M. Plant adaptations to severely phosphorus-impoverished soils. Current Opinion in Plant Biology. 2015;25:23–31. doi: 10.1016/j.pbi.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Hiruma K, et al. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell. 2016;165:464–474. doi: 10.1016/j.cell.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison MJ. Cellular programs for arbuscular mycorrhizal symbiosis. Current Opinion in Plant Biology. 2012;15:691–698. doi: 10.1016/j.pbi.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Hacquard S, et al. Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 2016;7:11362. doi: 10.1038/ncomms11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu YT, et al. Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defense responses. Plant Physiol. 2014;164:1456–1469. doi: 10.1104/pp.113.229740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, et al. Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant. 2013;6:301–310. doi: 10.1093/mp/sst002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bustos R, et al. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin H, Shin HS, Dewbre GR, Harrison MJ. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004;39:629–642. doi: 10.1111/j.1365-313X.2004.02161.x. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez E, Solano R, Rubio V, Leyva A, Paz-Ares J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. The Plant Cell. 2005;17:3500–3512. doi: 10.1105/tpc.105.036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang TK, et al. Identification of downstream components of ubiquitin conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell. 2013;25:4044–4060. doi: 10.1105/tpc.113.115998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin WY, Huang TK, Chiou TJ. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. The Plant Cell. 2013;25:4061–4074. doi: 10.1105/tpc.113.116012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puga MI, et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karthikeyan AS, et al. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta. 2007;225:907–918. doi: 10.1007/s00425-006-0408-8. [DOI] [PubMed] [Google Scholar]
- 21.Schweizer F, et al. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. The Plant Cell. 2013;25:3117–3132. doi: 10.1105/tpc.113.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pant BD, et al. Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ. 2014;38:172–187. doi: 10.1111/pce.12378. [DOI] [PubMed] [Google Scholar]
- 23.Rallapalli G, et al. EXPRSS: an Illumina based high-throughput expression-profiling method to reveal transcriptional dynamics. BMC genomics. 2014;15:341. doi: 10.1186/1471-2164-15-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan GA, Vogiatzaki E, Glauser G, Poirier Y. Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 2016;171:632–644. doi: 10.1104/pp.16.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 26.Barboriak DP, Padua AO, York GE, MacFall JR. Creation of DICOM—Aware Applications Using ImageJ. Journal of Digital Imaging. 2005;18:91–99. doi: 10.1007/s10278-004-1879-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arsenault JL, Pouleur S, Messier C, Guay R. WinRHIZO, a root-measuring system with a unique overlap correction method. HortScience. 1995;30:906. [Google Scholar]
- 28.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. Practical innovations for high-throughput amplicon sequencing. Nat. Methods. 2013;10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 30.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Garrity GM, Tiedje M, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yourstone SM, Lundberg DS, Dangl JL, Jones CD. MT-Toolbox: improved amplicon sequencing using molecule tags. BMC Bioinformatics. 2014;15:284. doi: 10.1186/1471-2105-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen NH, Smith D, Peay K, Kennedy P. Parsing ecological signal from noise in next generation amplicon sequencing. New Phytol. 2015;205:1389–93. doi: 10.1111/nph.12923. [DOI] [PubMed] [Google Scholar]
- 34.de Lorenzo L, et al. Arabidopsis PHOSPHATE STARVATION RESPONSE 1 acts via two cis-motifs and links plant water content with phosphate homeostasis. Genes & Development. Submitted [Google Scholar]
- 35.Hubert DA, He Y, McNulty BC, Tornero P, Dangl JL. Specific Arabidopsis HSP90.2 alleles recapitulate RAR1 co-chaperone function in plant NB-LRR disease resistance protein regulation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9556–9563. doi: 10.1073/pnas.0904877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 37.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal. 2011:17. [Google Scholar]
- 38.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 44.Morcuende R, et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007;30:85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- 45.Misson J, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castrillo G, et al. WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. The Plant Cell. 2013;25:2944–2957. doi: 10.1105/tpc.113.114009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory R, et al. gplots: Various R programming tools for plotting data. R package version. 3.0.1. 2016 [Google Scholar]
- 48.Yi X, Du Z, Su Z. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L, et al. The Pseudomonas syringae type III effector HopBB1 fine tunes pathogen virulence by degrading host transcriptional repressors. Cell Host & Microbe. doi: 10.1016/j.chom.2017.01.003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2014 URL http://www.R-project.org/
- 51.Sur Herrera Paredes. AMOR 0.0-14. Zenodo. 10.5281/zenodo. 2016:49093. [Google Scholar]
- 52.Xie Y. knitr: A feneral-purpose package for dynamic report generation in R. R package version 1.12.3. 2016 [Google Scholar]
- 53.Wickham H. ggplot2: Elegant graphics for data analysis. New York: Springer-Verlag; 2009. [Google Scholar]
- 54.Oksanen J, et al. vegan: Community ecology package. R package version 2.3–5. 2016 [Google Scholar]
- 55.Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology. 2003;84:511–525. [Google Scholar]
- 56.Lun ATL, Chen Y, Smyth GK. It’s DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol. Biol. 2016;1418:391–416. doi: 10.1007/978-1-4939-3578-9_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.