Abstract
Purpose
Cyclin E1 (CCNE1) amplification is associated with primary treatment resistance and poor outcome in high grade serous ovarian cancer (HGSC). Here, we explore approaches to target CCNE1 amplified cancers and potential strategies to overcome resistance to targeted agents.
Experiment Design
To examine dependency on CDK2 in CCNE1 amplified HGSC, we utilised siRNA and conditional shRNA gene suppression, and chemical inhibition using dinaciclib, a small molecule CDK2 inhibitor. High throughput compound screening was used to identify selective synergistic drug combinations, as well as combinations that may overcome drug resistance. An observed relationship between CCNE1 and the AKT pathway was further explored in genomic data from primary tumors, and functional studies in fallopian tube secretory cells.
Results
We validate CDK2 as a therapeutic target by demonstrating selective sensitivity to gene suppression. However, we found that dinaciclib did not trigger amplicon-dependent sensitivity in a panel of HGSC cell lines. A high throughput compound screen identified synergistic combinations in CCNE1 amplified HGSC, including dinaciclib and AKT inhibitors. Analysis of genomic data from TCGA demonstrated co-amplification of CCNE1 and AKT2. Over-expression of Cyclin E1 and AKT isoforms, in addition to mutant TP53, imparted malignant characteristics in untransformed fallopian tube secretory cells, the dominant site of origin of HGSC.
Conclusions
These findings suggest a specific dependency of CCNE1 amplified tumors for AKT activity, and point to a novel combination of dinaciclib and AKT inhibitors that may selectively target patients with CCNE1 amplified HGSC.
Keywords: Ovarian cancer, Cyclin E1, CDK2 inhibitors, AKT inhibitors
Introduction
Targeted therapies have changed the management of many cancers types, resulting in significant improvements in clinical response rates and survival (1). However, although the anti-angiogenic monoclonal antibody bevacizumab (2, 3) and the poly(ADP-ribose) polymerase (PARP) inhibitor olaparib (4, 5) have entered care in high grade serous ovarian cancer (HGSC) recently, the development of targeted therapy to this disease has been relatively slow.
HGSC are characterised by ubiquitous TP53 mutations, genomic instability and widespread copy number alterations, with relatively infrequent somatic point mutations of driver genes (6, 7). Structural aberration also contributes to loss of tumor suppressors such as RB1 and NF1 by gene breakage (8). Defects in the homologous recombination repair (HR) pathway are present in approximately 50% of HGSC, primarily associated with germline and somatic mutations in BRCA1, BRCA2 and associated proteins (7). HR deficiency imparts platinum sensitivity in HGSC, and provides the basis for the use of PARP inhibitors that target compensatory DNA repair pathways (4, 9). Of HGSC with intact HR, amplification of CCNE1, which encodes the cell-cycle regulator cyclin E1, is the best-characterised driver. CCNE1-amplification or gain occurs in 20% of all HGSC tumors and is associated with primary treatment resistance and reduced overall survival in HGSC (10, 11). Patients whose tumors have CCNE1-amplification represent a group with unmet clinical need, as they are unlikely to benefit from PARP inhibitors by virtue of the mutual exclusivity of CCNE1-amplification and BRCA1/2 mutation (7, 12), and are less likely to respond to platinum agents.
In recent pre-clinical studies, we have shown a dependency on CDK2 (13) and HR activity (12) in CCNE1-amplified cell lines. Although targeted agents have been effective in the clinical setting across many cancers, the emergence of acquired resistance is common (14). Indeed, we reported in vitro resistance to CDK2 inhibitors through selection of a polyploid population in the CCNE1-amplified cell line OVCAR3 (13). Rational drug combinations are a potential strategy to prevent resistance (15), and may also facilitate improvements in the therapeutic window by reducing the doses of drugs required to achieve efficacy, resulting in fewer side effects (16). We therefore used a high throughput drug screen to identify drug combinations that synergise with the CDK2 inhibitor dinaciclib (17) to selectively target CCNE1-amplified HGSC, and to overcome resistance in a cell line that has acquired resistance to CDK inhibitors in vitro (13). We identified several synergistic combinations, including dinaciclib and AKT inhibitors, and found that that this synergy extended more generally to CCNE1-amplified HGSC lines. Our results suggest targeting CDK2 and the AKT pathway may be an important approach to the clinical management of CCNE1-amplified HGSC.
Materials and Methods
Ethics Statement
All animal experiments were approved by the Peter MacCallum Cancer Centre Animal Experimentation Ethics Committee and conducted in accordance with the National Health and Medical Research Council Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Cell lines
Ovarian cancer cell lines were obtained from the National Cancer Institute Repository, actively passaged for less than six months and authenticated using short-tandem repeat markers to confirm their identity against the Cancer Genome Project database (Wellcome Trust Sanger Institute, Cambridgeshire, United Kingdom) prior to use in experiments. Cells were maintained at 37°C and 5% CO2 (vol/vol), and cultured in RPMI 1640 media containing 10% (vol/vol) FCS and 1% penicillin/streptomycin (P/S). Transfection and drug sensitivity assays were performed in the absence of antibiotics. Cell lines resistant to dinaciclib were generated utilising methods as described previously (13). Briefly, OVCAR3 cells were plated in 6-well plates and treated with dinaciclib at the IC50 dose for two 72-hour periods (media removed and fresh drug added). Surviving cells were allowed to repopulate for 96 hours and the process repeated once. Remaining cells were cultured in media or in the presence of drug, and regularly monitored for sensitivity to dinaciclib. Six independent cell lines were generated in this fashion, and designated OVCAR3-RD1 to –RD6.
Short hairpin mediated CDK2 knockdown
Short hairpin-mediated knockdown of CDK2 was performed by cloning CDK2-specific shRNA into a lentiviral tetracycline-inducible expression vector containing the optimized miR-E backbone (18). The modified lentiviral vector pRRL-T3G-TurboGFP-miRE-PGK-mCherry-IRES-rTA3 (also referred to as LT3GECIR) system includes a red (mCherry) fluorescent marker for transduction and a green (turboGFP) fluorescent marker for induction. Five CDK2-specific shRNA constructs were cloned into this system (see Supplementary Table S2 for sequences). For lentiviral production, HEK293T cells were transfected with plasmid DNA combined with the Lenti-X packaging system (Clontech Laboratories). Transfection, production of lentiviral particles and transduction of target cells was performed as described by the manufacturer’s protocol. Doxycycline was used to induce shRNA expression, and transfection efficiency was validated by flow cytometry (FACS), and knockdown of individual hairpins by RT-PCR and Western blot. The most efficient shRNA construct was taken forward for in vitro and in vivo experiments.
For in vivo experiments, xenograft tumors from transduced cells were generated as described below. Once tumors reached 100 mm3, mice were randomised into two groups to receive either normal food and water or doxycycline food and water (2mg/mL in 2% sucrose) as a means of reliable induction of shRNA expression. Tumors were subsequently monitored as described below.
Cyclin E1 and AKT over-expression in Fallopian tube secretory epithelial cells
The immortalised fallopian tube secretory epithelial cell (FTSEC) line FT282 was obtained from Ronny Drapkin (University of Pennsylvania) (19). Derivative cell lines were generated using pMSCV-mCherry-(empty) and pMSCV-mCherry-CCNE1, encoding full length CCNE1. Additional cell lines were generated with pMSCV-GFP-myr-AKT1, pMSCV-GFP-myr-AKT2 and pMSCV-GFP-myr-AKT3, encoding the three different isoforms of myr-AKT (20). Plasmids were validated by sequencing, and expression of CCNE1, AKT1, AKT2 and AKT3 was validated by quantitative real-time PCR and Western blotting. Primer sequences are listed in Supplementary Table S1.
High throughput compound screen
The compound library consisted of 73 targeted agents, 71 epigenetic agents, 208 kinase inhibitors and 3,707 known drugs (21). All agents were dissolved in DMSO, and diluted to concentrations from 0.01 μM to 10 μM. For targeted agents, epigenetic agents and kinase inhibitors, the primary screen was conducted using 11 concentrations; for the known drug library 3 concentrations were used. Compounds were dispensed into 384-well drug stock plates and stored at −20°C. Stock plates for dinaciclib at a fixed dose concentration (EC30) were prepared using a multichannel pipette prior to each assay.
Early passage cells were deposited into 384-well microtiter plates at 750–1,500 cells per well using a multidrop dispenser (Thermo) in 40μL of media. Cells were allowed to adhere overnight. A MiniTrak™ IX (Perkin Elmer Life Sciences) automated robotic platform was used to dispense compounds into assay plates. Compounds were added directly to assay plates using a 384, hydrophobic slotted pintool (VP Scientific) calibrated to dispense 0.1μL of DMSO compound solution. 0.1% DMSO was used as negative control. Cells were exposed to drug for 48 hours, and cell viability measured using the CellTitre-Glo luminescent assay (Promega) and the EnVision multilabel plate reader (PerkinElmer). Average viability was normalised to DMSO control wells, and EC50 dose was approximated by fitting a four-parameter dose-response curve using XLfit (IDBS).
Xenograft studies
Estrogen pellets were implanted subcutaneously into 4–6 week old female NOD/SCID mice to facilitate the growth of xenografted cells. The pellet was implanted 3 days prior to injection of cells. Cell lines were grown in vitro, washed twice with PBS and resuspended in 50% Matrigel (BD Biosciences) in PBS. Mice were injected subcutaneously with 5 × 106 cells in 100μL, and monitored at least twice weekly. Tumor volume was calculated using the equation: Volume = (width)2 × length/2. When tumors reached 100–150 mm3, mice were randomised into groups of five for treatment with vehicle alone or drug. Dinaciclib was prepared fresh prior to injection in 20% (w/v) hydroxypropyl-beta-cyclodextrin (Cyclodextrin Technologies Development, Inc) and mice dosed twice weekly as a single agent via intraperitoneal injection. MK-2206 was reconstituted in 30% (w/v) Captisol (Ligand Technology) and dosed at 60 mg/kg three times per week as a single agent via oral gavage. For combination studies, maximum tolerated doses of dinaciclib 20mg/kg and MK-2206 60mg/kg were dosed three times per week. All mice were monitored daily following drug dosing. Tumors were harvested at specific time-points for biomarker analysis or at study endpoint, with half snap frozen in liquid nitrogen and half fixed in formalin and paraffin embedded for immunohistochemistry. Percentage tumor growth inhibition (TGI) was calculated as 100 × (1−ΔT/ΔC) where ΔC and ΔT were determined by subtracting the mean tumor volume (in the vehicle control and treated groups respectively) on day 1 of treatment, from the mean tumor volume on each day of assessment. Statistical analyses were performed using GraphPad Prism Version 6.0 (GraphPad, La Jolla, CA, USA) with analysis of variance (ANOVA) followed by Dunnett’s post hoc test to compare the tumor growth between treatment groups.
CCNE1 and AKT status in primary ovarian tumor samples
Genomic alterations identified in CCNE1 and genes involved in the PI3K-AKT-mTOR pathway were obtained from The Cancer Genome Atlas (TCGA) cBioPortal (22, 23). All available data as at March 2015 was analysed, comprising 316 primary ovarian serous cystadenocarcinoma samples (7).
shRNA screen data
Data from the Project Achilles was obtained to evaluate the interaction between CCNE1- amplified ovarian cancer cell lines and genes in the AKT pathway (24). Cell line copy number data were obtained from the Cancer Cell Line Encyclopedia (25). Only cell lines known to resemble high grade serous ovarian cancer according to their genomic characteristics (26) were used in the analysis (N = 14, see Supplementary Table S3). Cell lines with a log2 copy number ratio > 0.3 over the CCNE1 locus were designated as amplified (N = 9) and cell lines with a log2 copy number ratio < 0 were designated as unamplified (N = 5). Cell lines with CCNE1 gene expression greater than the median +1 SD (N = 9) were defined as CCNE1 high expression, whereas cell lines with CCNE1 gene expression less than median (N = 5) were defined as CCNE1 low expression.
Additional methods for gene suppression studies, Western blot, immunohistochemistry, flow cytometry and drug sensitivity, clonogenic, proliferation, and anchorage independent growth assays can be found in Supplementary Methods.
Results
CCNE1-amplified HGSC cells are selectively sensitive to CDK2 knockdown
We previously demonstrated in a limited number of cell lines that CCNE1-amplified HGSC cell lines are selectively sensitive to CCNE1 and CDK2 knockdown mediated by siRNA (13). Following a recent analysis of ovarian cancer cell lines (26), we extended our analysis to a wider number of HGSC cell lines and confirmed consistent amplicon dependent sensitivity to siRNA-mediated CCNE1 and CDK2 knockdown (Fig 1A, Supplementary Fig S1A–B). The OVCAR8 cell line has a low level gain of CCNE1 and was not sensitive to CCNE1 or CDK2 knockdown (Fig 1A). However, OVCAR8 does not over-express cyclin E1 at the mRNA or protein level (Supplementary Fig S1B–C), compared to other cell lines such as OVCAR4 that have similar CCNE1 copy number. These findings suggest a threshold of CCNE1/CDK2 dependency that may be relevant to patient selection in clinical trials targeting this oncogene in HGSC.
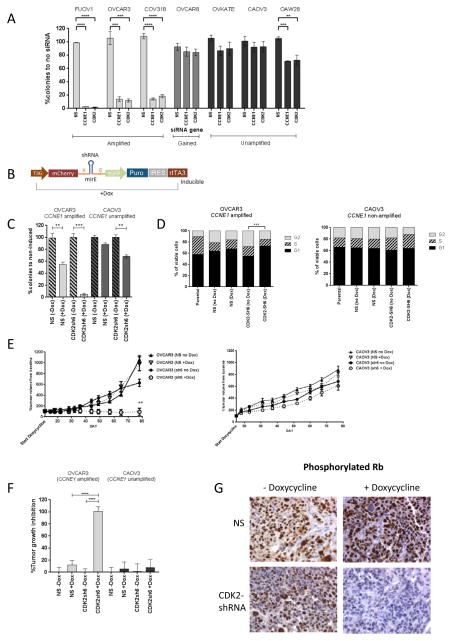
Figure 1. CDK2 knockdown via siRNA and shRNA in vitro and in vivo results in selective reduction in clonogenic survival and tumor growth arrest in CCNE1-amplified HGSC.
(A) Clonogenic survival after transfection with CCNE1 and CDK2 siRNAs in panel of HGSC cell lines. Average percentage of discrete colonies formed after 7–10 days relative to no siRNA controls shown (n=3). Error bars indicate SEM. Statistical significance (t test) calculated by comparison with non-silencing (NS) siRNA in the same cell line. **, p<0.01, ***, p<0.001, ****, p<0.0001.
(B) Schematic of conditional LT3GECIR lentiviral vector showing inducible transcripts produced by vector.
(C) Clonogenic survival after induction of a non-specific or CDK2 targeting shRNA in OVCAR3 (CCNE1-amplified) and CAOV3 (CCNE1-unamplified). Average percentage of discrete colonies formed after 7–10 days relative to no induction shown (n=3). Statistical significance (t test) calculated by comparison with non-induced (−Dox) in the same cell line. **, p<0.01, ***, p<0.001.
(D) Cell cycle analysis following CDK2 knockdown with inducible shRNA. Proportion of cells in G1, S or G2 phase for PI stained cells analysed by flow cytometry 72 hours after induction with doxycycline. Mean of 3 independently performed experiments shown. Statistical significance (t test) calculated by comparison with non-induced (−Dox) in the same cell line. ***, p<0.001.
(E) Mean percentage change in tumor volume ± SEM following induction of a non-specific (NS) or CDK2 (sh6) shRNA in subcutaneous xenograft tumors grown in immunocompromised mice, generated from OVCAR3 and CAOV3. Induced and non-induced groups as marked, n=5 per group. **, p<0.001, unpaired t-test comparison of mean percentage tumor volume change.
(F)Percentage tumor growth inhibition following induction of non-specific (NS) or CDK2 (sh6) shRNA with doxycycline. Bars represent mean ± SEM, n=5 mice per group. Statistical analysis performed with analysis of variance (ANOVA) followed by Dunnett’s post hoc test to compare the percentage tumor growth inhibition between the treatment groups. ****, p<0.0001.
(G) Immunohistochemistry assessment of phospho-Rb with or without doxycycline treatment in OVCAR3 xenograft tumor.
To validate the effect of CDK2 knockdown, we utilised a tetracycline-inducible shRNA targeting CDK2 (Fig 1B). Consistent with the siRNA data, inhibition of CDK2 by shRNA resulted in reduced clonogenic survival, more evident in the CCNE1-amplified cell line, OVCAR3 compared to the CCNE1-unamplified cell line CAOV3 (Fig 1C). Knockdown of CDK2 was validated at the protein level (Supplementary Figure S2A). Cell-cycle analysis demonstrated arrest in G1, seen only in the OVCAR3 cell line (Fig 1D). We did not observe significant levels of apoptosis following CDK2 knockdown, as assessed by percentage of Annexin V positive cells measured by FACS (Supplementary Figure S2B).
Cells transduced with CDK2-shRNA were grown as xenografts in nod/scid mice to examine the effects of CDK2 knockdown in vivo. Consistent with the in vitro data, attenuation of CDK2 expression in the OVCAR3 xenograft model resulted in significant tumor growth arrest in the group receiving doxycycline in food and water compared to controls. (Figure 1E–F). Induction of shRNA by doxycycline was monitored by CDK2 gene expression measured by RT-PCR (Supplementary Figure S2C). Reduced Rb1 phosphorylation was observed following CDK2 knockdown in OVCAR3 tumors harvested at seven days following induction (Fig 1G), providing a biomarker of targeting cyclinE1/CDK2.
Taken together, CCNE1-amplified HGSC appear selectively sensitive to siRNA- and shRNA-mediated knockdown of CDK2 both in vitro and in vivo. These findings support our previous studies and point to CDK2 as a potential therapeutic target in CCNE1-amplified HGSC.
CDK2 inhibitor dinaciclib delayed tumor growth in CCNE1-amplified HGSC xenografts
Consistent with siRNA data, we previously showed in a limited number of cell lines selective sensitivity of CCNE1-amplified cell lines to dinaciclib, a potent CDK2 inhibitor in advanced clinical development (13). However, in this study, when tested across a broader panel of HGSC cell lines, there did not appear to be a clear amplicon dependent sensitivity (Fig 2A), in contrast to the siRNA and shRNA data. Furthermore, activity in vivo was also seen in a xenograft model developed from a CCNE1-unamplified cell line, CAOV3 (Fig 2A–D). The difference in amplicon dependent sensitivity between gene suppression and pharmacological inhibition may be due to the broad activity of dinaciclib, which in addition to inhibiting CDK2, is also active against CDK1, 5, 9 and 12 (17, 27).
Figure 2. CDK inhibitor dinaciclib results in modest tumor growth inhibition in vivo but is not synergistic in combination with bortezomib in vitro.
(A) Mean IC50 values for a panel of HGSC cell lines treated with dinaciclib generated from dose-response curves following standard MTS cell proliferation assays. Error bars represent SEM, n=3 experiments.
(B) In vivo effects of dinaciclib. Immunocompromised mice bearing OVCAR3 (CCNE1-amplified) or CAOV3 (CCNE1-unamplified) tumor xenografts were treated with vehicle or drug as described in methods. Plots represent mean tumor volume change from baseline ± SEM, n=5 mice per group.
(C) Percentage tumor growth inhibition following 21 days of treatment with vehicle or dinaciclib. Bars represent mean ± SEM, n=5 mice per group. Statistical analysis performed with analysis of variance (ANOVA) followed by Dunnett’s post hoc test to compare the percentage tumor growth inhibition between the treatment groups. **, p<0.01.
(D) Immunohistochemistry analysis of Ki67 expression in OVCAR3 and CAOV3 tumor xenograft harvested 24 hours after dose of vehicle or dinaciclib.
(E) Formal assessment of synergy between dinaciclib and bortezomib using Chou-Talalay Isobologram analysis. Figures are generated with CalcuSyn 2.0. Data are normalised, with connecting line at X and Y corresponding to combination index = 1, representing line of additivity. Datapoints above the line are antagonistic, along or near the line are additive and points below the line are synergistic.
(F) Combination indexes for a panel of HGSC cell lines tested against dinaciclib in combination with bortezomib. Values represent mean ± SEM, n=3.
(G–H) Scatter plots showing EC50 values for library compounds in combination with dinaciclib from primary screen for the comparison between (G) CCNE1-amplified and unamplified; and (H) resistant versus parental. Data points in red represent compoundstaken forward for secondary screen.
In addition to CDK2 inhibitors, we previously identified use of bortezomib, a proteasome inhibitor, as a potential therapeutic strategy for CCNE1-amplified HGSC (12). Although we did not observe amplicon dependent sensitivity to dinaciclib, we investigated the interaction between dinaciclib and bortezomib to see whether the two drugs would be synergistic in combination. Using the Chou-Talalay methodology for drug combination studies (28), we did not observe a synergistic interaction with dinaciclib and bortezomib (Fig 2E–2F) in a panel of CCNE1-amplified and CCNE1-unamplified HGSC cell lines. Given this lack of synergism, we sought to identify selective synergistic drug combinations by adopting an unbiased high throughput screening approach.
A high throughput compound screen identifies synergistic drug combinations
We performed a high throughput compound screen to identify combinations that would be synergistic in CCNE1-amplified cells, as well as combinations that would be selective in a CDK-inhibitor resistant cell line OVCAR3-R1-533533 (13). In the primary screen, 4,059 compounds (including duplicates) were combined with a fixed dose of dinaciclib as described in Materials and Methods. Dose-response curves were generated and manually curated, and compounds where a curve could not be fitted were excluded from the analysis. A full list of EC50 values for each cell line and compound is found in Supplementary Table S4–S5.
EC50 values from the primary screen were used to make two pair-wise comparisons (Fig 2G–2H):
1) dinaciclib plus library compound comparing OVCAR3 (CCNE1-amplified) versus SKOV3 (CCNE1-unamplified) and 2) dinaciclib plus library compound comparing OVCAR3 (parental) and OVCAR3-R1 (CDK-inhibitor resistant). At the time of undertaking the screen SKOV3 was a commonly used ovarian cancer cell line, however recent studies have demonstrated that SKOV3 is unlikely to resemble HGSC (26). Therefore any potential hits identified in the screen were subsequently validated using only HGSC cell lines.
Library compounds where the ratio of EC50 was less than 0.5 were selected as hits for a secondary screen involving a total of 64 compounds (Supplementary Table S6–S7). Compounds that appeared to have an additive effect with dinaciclib were selected as hits from the secondary screen and carried forward for further testing.
The final part of the screen involved assessing the level of synergy between the library compound hits and dinaciclib involving an 11-point titration of each compound. Using the Chou-Talalay methodology of constant-ratio drug combinations, a series of combination indexes were generated to identify synergistic interactions.
In the OVCAR3 parental cell line, there were no synergistic combinations identified between dinaciclib and the library compounds (Supplementary Table S8). In the OVCAR3-R1 cell line, there were a number of synergistic interactions identified (Supplementary Table S8). Non-selective BH3-mimetic agents ABT-263 and ABT-737 were synergistic in combination with dinaciclib, suggestive of a class effect. This was validated further in an independently derived dinaciclib-resistant cell line, OVCAR3-RD6 (Fig 3A–B, Supplementary Fig S4A–C). There was no synergistic interaction noted in the combination between dinaciclib and ABT-199 (Fig 3C), a selective Bcl-2 antagonist. The combination of dinaciclib and ABT-737 resulted in a dose-dependent increase in apoptosis, observed only in CDK inhibitor-resistant cell lines as demonstrated by increase in PARP cleavage products on Western blot analysis (Fig 3D). Mcl-1 protein expression was not observed in the OVCAR3-RD6 cell line resistant to dinaciclib (Fig 3D). Real-time PCR demonstrated up-regulation of anti-apoptotic genes in the dinaciclib and PHA533533-resistant cell lines (Fig 3E), but down-regulation of MCL1 in the dinaciclib resistant OVCAR3-RD cell lines. Dinaciclib is reported to have a greater effect on CDK9 compared to PHA533533 (13). Given that MCL1 is regulated by CDK9 activity (29), this may explain the reduction of MCL1 levels in the presence of dinaciclib. However, it is unclear why reduced MCL1 expression is also apparent in OVCAR3-RD cell lines even when grown in the absence of dinaciclib.
Figure 3. Dinaciclib in combination with non-selective BH3 mimetics are synergistic in CDK-inhibitor resistant cell lines.
Combination indexes for parental and CDK inhibitor-resistant cell lines tested against dinaciclib in combination with (A) ABT-737, (B) ABT-263, (C) ABT-199. Values represent mean ± SEM, n=3.
(D) Western blot demonstrating protein expression of Bcl-XL, Mcl-1 and PARP cleavage products in OVCAR3 parental and CDK inhibitor-resistant cell lines after treatment with dinaciclib and ABT-737.
(E) Expression of anti-apoptotic proteins as assessed by quantitative real-time PCR. R-lines signify cell lines resistant to PHA533533. RD-lines signify cell lines resistant to dinaciclib. Bars represent mean ± SEM, n=3.
MK-2206, a pan-AKT inhibitor, was identified as a synergistic drug combination in the CDK-inhibitor resistant cell line, OVCAR3-R1. In validating this interaction between dinaciclib and MK-2206, we observed that this combination was also synergistic in CCNE1-amplified cell lines FUOV1 and parental OVCAR3 (Fig 4A). This effect was similarly observed with another AKT inhibitor, GSK-2110183 (Fig 4B), that was not included in the original high throughput screen library. Exposure to dinaciclib and MK2206 resulted in significantly higher number of apoptotic cells in CCNE1-amplified cell lines, indicated by percentage of Annexin V positive cells measured by FACS (Fig 4C). This result was similarly observed on Western blot analysis, with appearance of PARP cleavage products following treatment of OVCAR3 cells with the combination of dinaciclib and MK-2206 (Supplementary Figure S4D). As dinaciclib targets several CDKs in addition to CDK2 (17), we used siRNA knockdown of CDK2, CDK1 or CDK9 to determine the specificity of the synergistic effect of dinaciclib and MK-2206. We found that the synergy observed was predominantly mediated through CDK2 (Supplementary Fig S4E).
Figure 4. Dinaciclib in combination with two AKT inhibitors are synergistic in vitro and in vivo models of CCNE1-amplified HGSC.
Combination indexes for a panel of HGSC cell lines tested against dinaciclib in combination with (A) MK-2206 and (B) GSK2110183. Values represent mean ± SEM, n=3.
(C) HGSC cell lines were cultured in vitro with dinaciclib, MK-2206 or the combination for 24 hours and then analysed using flow cytometry for Annexin V/propidium iodide positivity. Bars represent mean ± SEM, n=3. *, p<0.05; **, p<0.01; ***, p<0.001; unpaired t-test.
(D) In vivo effects of vehicle, dinaciclib, MK-2206 or combination. Immunocompromised mice bearing OVCAR3 (CCNE1-amplified) or CAOV3 (CCNE1-unamplified) tumor xenografts were treated with vehicle or drug as described in methods. Plots represent mean tumor volume change from baseline ± SEM, n=5 mice per group.
(E) Percentage tumor growth inhibition following 21 days of treatment with vehicle, dinaciclib, MK-2206 or the combination. Bars represent mean ± SEM, n=5 mice per group. Statistical analysis performed with analysis of variance (ANOVA) followed by Dunnett’s post hoc test to compare the percentage tumor growth inhibition between the treatment groups. *,p <0.05; **, p<0.01; ****, p<0.0001.
(F) Quantitation of immunohistochemistry staining for Ki67 and cleaved caspase-3. Bars represent mean percentage of Ki67 or cleaved caspase 3 positive cells relative to background number of cells measured ± SEM, n=3 in each group. Statistical analysis performed by analysis of variance (ANOVA) with Tukey’s multiple comparison test to compare between treatment groups.
(G) Subcutaneous tumors were obtained after 24hrs of treatment and were examined by immunohistochemistry for biomarker analysis. Rb phosphorylation was inhibited by dinaciclib, but not MK-2206 treatment. AKT phosphorylation was inhibited by MK-2206, but not dinaciclib treatment. Proliferation (Ki67) was inhibited and apoptosis (cleaved caspase-3) was induced by the combination of dinaciclib and MK-2206 in CCNE1-amplified xenograft model (OVCAR3).
Dinaciclib and MK-2206 are selectively synergistic in CCNE1-amplified cell lines in vivo
The in vivo effect of dinaciclib and MK-2206 was assessed using xenograft models from CCNE1-amplified and unamplified cell lines, OVCAR3 and CAOV3 respectively. The combination was significantly more effective than each single agent alone in the CCNE1-amplified model (Fig 4D–E), whereas there was no statistically significant effect of the combination compared to single agent treatment in the CCNE1-unamplified model. Following a treatment period of three weeks with dinaciclib and MK-2206, xenograft tumors began regrowing within 10 days of treatment cessation. Re-challenge with the same drug combination resulted in significant tumor regression (Supplementary Fig S4F), indicating continued sensitivity to the combination. Consistent with this effect on tumor growth, treatment with dinaciclib and MK-2206 resulted in inhibition of cell proliferation and induction of apoptosis, as assessed by Ki67 and cleaved caspase-3 immunohistochemistry on tumors harvested at 24 hours (Fig 4F–G). Taken together, the high throughput screen identified a novel combination of dinaciclib and MK-2206 that appeared to be selectively synergistic in CCNE1-amplified HGSC cell lines both in vitro and in vivo.
CCNE1 and AKT2 are frequently co-amplified in primary HGSC samples
We sought to investigate whether there was evidence for an interaction between these CCNE1-amplification and the AKT pathway in primary tumor samples. Analysis of TCGA dataset indicated that CCNE1 and AKT2 amplification events co-occur (p<0.001, Supplementary Fig S5). This observation was not seen with other isoforms of AKT or genes in the AKT pathway. To examine the relationship between CCNE1-amplification and the AKT pathway further, we made use of data from Project Achilles, a genome-wide shRNA screen of synthetic lethality in 216 cancer cell lines (24). The abundance of shRNA sequence relative to a reference pool was measured by microarray to identify genes essential for survival. We analysed the effect of shRNA targeting genes within the AKT pathway, restricting the analysis to HGSC cell lines, classified according to CCNE1 copy number or expression. A statistically significant dependence on genes in the AKT pathway, including AKT2, was observed, indicated by a depletion of shRNAs targeting these genes in cell lines with CCNE1-amplification or overexpression (Fig 5). CDK2 was included in the analysis as a control, and consistent with our previous analysis, was shown to be required in CCNE1-amplified cells (13).
Figure 5. CCNE1 and AKT2 are co-amplified in primary HGSC samples.
Dot plots of median shRNA abundance for each gene targeted by shRNA in HGSC cell lines, stratified by CCNE1 copy number or expression. Depletion of shRNA abundance within a group suggests requirement for maintained expression of its target gene. Only genes with a statistically significant difference shown, see Supplementary Table S3 for list of genes and cell lines analysed. Statistical significance (t test) calculated by comparison between CCNE1-amplified and unamplified or CCNE1 over-expressing and low expressing cell lines. *, p<0.05, **. p<0.01.
Cyclin E1 and AKT over-expression co-operates to promote uncontrolled growth in FTSEC
Previously, Karst and colleagues demonstrated that cyclin E1 over-expression combined with TP53 mutation in FTSEC resulted in increased proliferation, colony forming ability and colony formation in soft agar (19). However, cyclin E1 over-expression alone did not result in complete transformation, suggesting that additional events are required.
We examined the interaction between cyclin E1 and AKT over-expression in FTSEC by overexpressing the myristoylated, active forms of AKT1, AKT2 and AKT3 (20). Expression of each AKT isoform and cyclin E1 was validated with Western Blot (Fig 6A) and RT-PCR (Supplementary Fig S6A). Over-expression of AKT isoforms led to increased expression of AKT downstream targets (Supplementary Fig S6B). AKT2 and cyclin E1 over-expression alone or in combination showed a trend towards increased proliferation compared to empty vector alone (Fig 6B), and AKT2 or AKT3 over-expression in combination with cyclin E1 showed a trend towards enhanced clonogenic colony formation in comparison to over-expression of cyclin E1 alone (Fig 6C). There was a significant increase in soft agar colony formation with the over-expression of AKT2 or AKT3 in combination with cyclin E1 compared to over-expression of cyclin E1 alone (Fig 6D). These findings support an interaction between cyclin E1 and AKT pathway to promote uncontrolled growth in FTSEC, and may explain synergism observed between dinaciclib and MK-2206 in CCNE1-amplified HGSC.
Figure 6. Cyclin E1 and AKT over-expression co-operates to promote uncontrolled growth in fallopian tube secretory epithelial cells.
(A) Western Blot analysis of fallopian tube secretory cells transduced with cyclin E1, empty vector and AKT1, AKT2 and AKT3 over-expression constructs. Blots are representative of three independently performed experiments.
(B) Proliferation assay of fallopian tube secretory cells (FT282) transduced with empty vector (EV), cyclin E1 (CCNE1), AKT2, and both cyclin E1 and AKT2 (CCNE1+AKT2). Plots represent mean of 3 independently performed experiments, error bars represent SEM.
(C) Clonogenic survival assay of FT282 cells transduced as labelled. Images (left) show cells fixed and stained with crystal violet. Bar chart represents mean of 3 independently performed experiments, error bars represent SEM. Statistical significance (t test) calculated by comparison with FT282 cells transduced with cyclin E1 (FT282-CCNE1).
(D) Anchorage independent assay of FT282 cells transduced as labelled. Images (left) represent cells fixed with 2% paraformaldehyde and captured using an Olympus IX81 live cell imager. Bar chart represents mean of 3 independently performed experiments, error bars represent SEM. Statistical significance (t test) calculated by comparison with FT282 cells transduced with cyclin E1 (FT282-CCNE1). *, p < 0.05, **, p < 0.01.
Discussion
HGSC patients with CCNE1-amplification have a clear unmet need in terms of effective therapies. In this study, we validate CDK2 as a selective target in CCNE1-amplified HGSC utilising shRNA-mediated gene suppression in vitro and in vivo. However, we did not observe similar amplicon dependent specificity to dinaciclib, a small molecule inhibitor targeting CDKs. This may be due to the non-specificity of inhibitors such as dinaciclib or a role for kinase independent activities of CCNE1 in amplified HGCS (30). Our findings highlight the potential differences between inhibition of kinase activity and complete suppression of CCNE1 or CDK2 gene expression.
In addition to CDK2, dinaciclib targets CDK1, 5, 9 and 12 (17, 27). CDK9 phosphorylates the carboxyl-terminal repeat domains of RNA polymerase II, and inhibition of CDK9 by dinaciclib results in rapid down-regulation of mRNA transcripts and proteins with short half-lives such as the anti-apoptotic BCL2 family member, Mcl1 (17). Pre-clinical studies have indicated dinaciclib-mediated targeting of Mcl-1 may be an effective therapeutic approach in a number of different cancers (17). Inhibition of CDK2 kinase activity may also differ significantly from complete suppression of gene expression, resulting in varying downstream and compensatory effects (31, 32). Studies with knockout experiments indicate that CDK2 functions appear redundant with CDK1, although in our studies, we did not observe up-regulation of CDK1 expression following CDK2 knockdown in vitro or in vivo (data not shown).
Although we observed a difference in the amplicon dependent sensitivity of CDK2 gene suppression compared to pharmacological inhibition, dinaciclib remains a potent CDK2 inhibitor with single agent activity in CCNE1-amplified HGSC cell lines and is one of the most clinically advanced CDK2 inhibitors (33). Therefore, in order to more effectively target CCNE1-amplified HGSC, we performed a combinatorial drug screen to identify compounds that would synergise with dinaciclib. We also sought to identify compounds that may potentially overcome resistance to dinaciclib, a common occurrence in the clinical use of targeted small molecule inhibitors, by testing a cell line that was resistant to CDK inhibitors. Dinaciclib in combination with MK-2206, an AKT inhibitor, was identified as a synergistic combination in targeting CDK inhibitor-resistant cell lines. This supported our previous work that identified increased AKT1 copy number and up-regulation of genes in the AKT pathway as a potential mechanism of resistance to CDK2 inhibitors (13). In validating this finding, we observed selective, potent synergism between dinaciclib and MK-2206 in vitro and in vivo models of CCNE1-amplified HGSC, including parental OVCAR3 cells. This interaction was not initially observed in the primary high throughput screen. However the use of SKOV3 cell line as a comparator in the screen may be a potential confounder, as the selection of compounds as hits from the primary screen was based on a difference in the EC50 values between the two cell lines tested, OVCAR3 and SKOV3. Recently, multiple studies characterising the genomic profile of commercially available ovarian cancer cell lines have shown that many of these cell lines, including SKOV3, may not accurately resemble HGSC (26, 34–36).
Synergism between dinaciclib and MK-2206, as well as another AKT-specific inhibitor GSK2110183, but an absence of a synergistic combination with other inhibitors of the PI3K/AKT/MTOR pathway suggests that the interaction with CCNE1 may be specific to AKT. Analysis of genomic data from patients demonstrated a significant co-occurrence of CCNE1 and AKT2-amplification, which may in part be explained by co-localisation on chromosome 19q. However, FUOV1, which has CCNE1-amplification without AKT2-amplification (25), was equally sensitive to the combination of dinaciclib and AKT inhibitors. Co-expression of AKT2 or AKT3 with cyclin E1 in a TP53-mutant FTSEC cell line resulted in increased proliferation and anchorage independent growth. Analysis of data from Project Achilles indicates that HGSC cell lines that have CCNE1-amplification or over-expression are dependent on multiple genes within the AKT pathway. We previously performed a pathway analysis of genes co-expressed with CCNE1-amplification and observed an enrichment of genes involved in AKT signalling (12). Collectively, these data suggests a specific dependency of CCNE1-amplified tumors for AKT activity.
Dinaciclib and MK-2206 have previously been shown to be active against pancreatic adenocarcinoma (37). In KRAS mutant pancreatic cancer patient derived xenografts, Hu and colleagues demonstrated efficacy of dinaciclib combined with MK-2206. They proposed that sensitivity was due to the effect of dinaciclib on CDK5, and in turn, inhibition of RAL pathway. Based on these results, a phase 1 clinical trial (NIH Trial NCT01783171) of dinaciclib and MK-2206 has been initiated in patients with advanced pancreatic cancer. While this trial will provide safety and recommended dosing of the combination, patients are not preselected on the basis of tumoral CCNE1-amplification, and the mechanism of interaction and biomarkers that predict response are likely to be different in pancreatic cancer compared to HGSC.
Other combinations were also identified from the high throughput screen. In particular, non-selective BH3-mimetic compounds ABT-737 and ABT-263 were synergistic in combination with dinaciclib in CDK inhibitor-resistant cell lines. There was no synergistic interaction between dinaciclib and the Bcl-2 specific antagonist, ABT-199, indicating that the targeting of multiple anti-apoptotic proteins is potentially required to overcome resistance to CDK2 inhibitors. This observation is supported by up-regulation of multiple genes in this pathway including BCL-2, BCL-XL and BCL-W in resistant cell lines. However, the use of ABT-737 or ABT-263 in combination with dinaciclib in vivo is hindered by significant toxicities, particularly haematological (Joel Leverson, personal communication), and are therefore unlikely to have clinical utility.
Biomarker driven trials in HGSC are needed to improve clinical outcomes. HGSC patients with CCNE1-amplification are a subset that requires different treatment approaches, given that they have HR proficient tumors, and as such, are likely to have poor responses to platinum-based chemotherapy and PARP inhibitors. However, targeted therapies when used alone may not be sufficient to induce selective, cytotoxic effects, and often result in the development of resistance. Combination therapies may potentially be a strategy to overcome these limitations. High throughput drug screening is an unbiased approach to identify novel therapeutic strategies, and we have identified dinaciclib and MK-2206 as a combination that may prove to selectively target patients with CCNE1-amplified HGSC. Further work incorporating additional clinically relevant models and novel combinations will inform the design of rational clinical trials targeting CCNE1-amplified HGSC.
Supplementary Material
Statement of translational relevance.
High grade serous ovarian cancer (HGSC) patients with Cyclin E1 (CCNE1) amplification represent a group with high unmet clinical need. Novel therapies are needed to improve outcomes in these patients, given that CCNE1 amplified tumors are unlikely to respond to chemotherapy or PARP inhibitors, and are associated with poor overall survival. Here, we validate CDK2 as a selective target for CCNE1 amplified cell lines. We performed a high throughput compound screen and identified a number of potential therapeutic combinations. We focused on dinaciclib and AKT inhibitors, and demonstrate selective and potent activity in CCNE1 amplified HGSC. We further show co-operation between CCNE1 and AKT, both in genomic data from TCGA and functionally in fallopian tube secretory cells. This study demonstrates approaches to targeting an important subset of solid cancers, and for the first time provides evidence to support the design of a rational clinical trial that targets CCNE1 amplified HGSC.
Acknowledgments
The authors wish to acknowledge staff from the Peter MacCallum Cancer Centre Animal facility, FACS facility and Histology core facility for their assistance. This work was supported by a National Health and Medical Research Council (NHMRC) program grant (D. Bowtell APP1092856), NHMRC project grant (APP1042358, D. Etemadmoghadam), a Pfizer Cancer Research Grant (WI80176 G. Au-Yeung), University of Melbourne Australian Postgraduate Award (G. Au-Yeung), the U.S. Army Medical Research and Materiel Command (OC140511, D. Bowtell and R. Drapkin), the National Cancer Institute at the NIH P50-CA083636 (R. Drapkin), NIH R21 CA156021 (R. Drapkin); the Honorable Tina Brozman ‘Tina’s Wish’ Foundation (R. Drapkin), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (R. Drapkin), a Canadian Institutes of Health Research Fellowship (A.M.Karst), a Kaleidoscope of Hope Foundation Young Investigator Research Grant(A.M. Karst), the Basser Center for BRCA, and Department of Obstetrics and Gynecology at the University of Pennsylvania Perelman School of Medicine (R. Drapkin). The Australian Ovarian Cancer Study is supported by the Peter MacCallum Cancer Centre Foundation, U.S. Army Medical Research and Materiel Command under DAMD17-01-1-0729, The Cancer Council Victoria, Queensland Cancer Fund, The Cancer Council New South Wales, The Cancer Council South Australia, The Cancer Foundation of Western Australia, The Cancer Council Tasmania and the National Health and Medical Research Council of Australia (NHMRC; ID#628779), Stephanie Boldeman, the Agar family, and Ovarian Cancer Australia,
Footnotes
Disclosure of Potential Conflicts of Interest
D. Rischin reports receiving research funding from Merck. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Martini M, Vecchione L, Siena S, Tejpar S, Bardelli A. Targeted therapies: how personal should we go? Nat Rev Clin Oncol. 2012;9:87–97. doi: 10.1038/nrclinonc.2011.164. [DOI] [PubMed] [Google Scholar]
- 2.Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473–83. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 3.Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365:2484–96. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 4.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 5.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–92. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–94. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 9.Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-ribose) polymerase inhibitors: recent advances and future development. J Clin Oncol. 2015;33:1397–406. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George J, Getz G, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417–27. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama N, Nakayama K, Shamima Y, Ishikawa M, Katagiri A, Iida K, et al. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer. 2010;116:2621–34. doi: 10.1002/cncr.24987. [DOI] [PubMed] [Google Scholar]
- 12.Etemadmoghadam D, Weir BA, Au-Yeung G, Alsop K, Mitchell G, George J, et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc Natl Acad Sci U S A. 2013;110:19489–94. doi: 10.1073/pnas.1314302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etemadmoghadam D, Au-Yeung G, Wall M, Mitchell C, Kansara M, Loehrer E, et al. Resistance to CDK2 inhibitors is associated with selection of polyploid cells in CCNE1-amplified ovarian cancer. Clin Cancer Res. 2013;19:5960–71. doi: 10.1158/1078-0432.CCR-13-1337. [DOI] [PubMed] [Google Scholar]
- 14.Ellis LM, Hicklin DJ. Resistance to Targeted Therapies: Refining Anticancer Therapy in the Era of Molecular Oncology. Clin Cancer Res. 2009;15:7471–8. doi: 10.1158/1078-0432.CCR-09-1070. [DOI] [PubMed] [Google Scholar]
- 15.Yap TA, Omlin A, de Bono JS. Development of therapeutic combinations targeting major cancer signaling pathways. J Clin Oncol. 2013;31:1592–605. doi: 10.1200/JCO.2011.37.6418. [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–66. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 17.Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 2010;9:2344–53. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 18.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–13. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 19.Karst AM, Jones PM, Vena N, Ligon AH, Liu JF, Hirsch MS, et al. Cyclin E1 deregulation occurs early in secretory cell transformation to promote formation of fallopian tube-derived high-grade serous ovarian cancers. Cancer Res. 2014;74:1141–52. doi: 10.1158/0008-5472.CAN-13-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Astle MV, Hannan KM, Ng PY, Lee RS, George AJ, Hsu AK, et al. AKT induces senescence in human cells via mTORC1 and p53 in the absence of DNA damage: implications for targeting mTOR during malignancy. Oncogene. 2012;31:1949–62. doi: 10.1038/onc.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lackovic K, Lessene G, Falk H, Leuchowius KJ, Baell J, Street I. A perspective on 10-years HTS experience at the Walter and Eliza Hall Institute of Medical Research - eighteen million assays and counting. Comb Chem High Throughput Screen. 2014;17:241–52. doi: 10.2174/1386207317666140109122450. [DOI] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro G. Beyond CDK4/6: Targeting additional cell cycle and transcriptional CDKs in breast cancer. [abstract]. Proceedings of the Thirty-Eigth Annual CTRC-AACR San Antonio Breast Cancer Symposium; 2015 Dec 8–12; San Antonio, TX Philadephia (PA): AACR; [Google Scholar]; Cancer Res. 2016;76(4 Suppl) Abstract nr MS1-1. [Google Scholar]
- 28.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 29.Gojo I, Zhang B, Fenton RG. The cyclin-dependent kinase inhibitor flavopiridol induces apoptosis in multiple myeloma cells through transcriptional repression and down-regulation of Mcl-1. Clin Cancer Res. 2002;8:3527–38. [PubMed] [Google Scholar]
- 30.Geng Y, Lee YM, Welcker M, Swanger J, Zagozdzon A, Winer JD, et al. Kinase-independent function of cyclin E. Mol Cell. 2007;25:127–39. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 31.Horiuchi D, Huskey NE, Kusdra L, Wohlbold L, Merrick KA, Zhang C, et al. Chemical-genetic analysis of cyclin dependent kinase 2 function reveals an important role in cellular transformation by multiple oncogenic pathways. Proc Natl Acad Sci U S A. 2012;109:E1019–27. doi: 10.1073/pnas.1111317109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Echalier A, Cot E, Camasses A, Hodimont E, Hoh F, Jay P, et al. An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity. Chem Biol. 2012;19:1028–40. doi: 10.1016/j.chembiol.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–46. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anglesio MS, Wiegand KC, Melnyk N, Chow C, Salamanca C, Prentice LM, et al. Type-specific cell line models for type-specific ovarian cancer research. PLoS One. 2013;8:e72162. doi: 10.1371/journal.pone.0072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaufort CM, Helmijr JC, Piskorz AM, Hoogstraat M, Ruigrok-Ritstier K, Besselink N, et al. Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes. PLoS One. 2014;9:e103988. doi: 10.1371/journal.pone.0103988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elias KM, Emori MM, Papp E, MacDuffie E, Konecny GE, Velculescu VE, et al. Beyond genomics: critical evaluation of cell line utility for ovarian cancer research. Gynecol Oncol. 2015;139:97–103. doi: 10.1016/j.ygyno.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu C, Dadon T, Chenna V, Yabuuchi S, Bannerji R, Booher R, et al. Combined Inhibition of Cyclin-Dependent Kinases (Dinaciclib) and AKT (MK-2206) Blocks Pancreatic Tumor Growth and Metastases in Patient-Derived Xenograft Models. Mol Cancer Ther. 2015;14:1532–9. doi: 10.1158/1535-7163.MCT-15-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.