ABSTRACT
Cholesterol-dependent cytolysins (CDCs) represent a family of homologous pore-forming proteins secreted by many Gram-positive bacterial pathogens. CDCs mediate membrane binding partly through a conserved C-terminal undecapeptide, which contains a single cysteine residue. While mutational changes to other residues in the undecapeptide typically have severe effects, mutation of the cysteine residue to alanine has minor effects on overall protein function. Thus, the role of this highly conserved reactive cysteine residue remains largely unknown. We report here that the CDC listeriolysin O (LLO), secreted by the facultative intracellular pathogen Listeria monocytogenes, was posttranslationally modified by S-glutathionylation at this conserved cysteine residue and that either endogenously synthesized or exogenously added glutathione was sufficient to form this modification. When recapitulated with purified protein in vitro, this modification completely ablated the activity of LLO, and this inhibitory effect was fully reversible by treatment with reducing agents. A cysteine-to-alanine mutation in LLO rendered the protein completely resistant to inactivation by S-glutathionylation, and a mutant expressing this mutation retained full hemolytic activity. A mutant strain of L. monocytogenes expressing the cysteine-to-alanine variant of LLO was able to infect and replicate within bone marrow-derived macrophages indistinguishably from the wild type in vitro, yet it was attenuated 4- to 6-fold in a competitive murine infection model in vivo. This study suggests that S-glutathionylation may represent a mechanism by which CDC-family proteins are posttranslationally modified and regulated and help explain an evolutionary pressure to retain the highly conserved undecapeptide cysteine.
KEYWORDS: Listeria monocytogenes, cholesterol-dependent cytolysins, thiol-dependent cytolysins, glutathione, posttranslational modification, virulence, bacterial toxins, Firmicutes, Gram-positive bacteria, pore-forming toxins
INTRODUCTION
Cholesterol-dependent cytolysins (CDCs) comprise a family of large oligomeric pore-forming toxins that are primarily secreted by pathogenic Gram-positive bacteria within the Firmicutes and Actinobacteria (1). The vast majority of CDC homologs are secreted as monomers that bind cholesterol-containing membranes, oligomerize, and then undergo significant conformational changes that allow efficient pore formation (2). Decades of study have led to a detailed understanding of the mechanism of action as well as insight into the nuanced differences between how different pathogens employ their cognate CDCs; however, a number of important questions remain unanswered.
One unresolved question relates to the original name given to this class of toxins, thiol-activated cytolysins (3). Although a few exceptions necessitated a change in nomenclature, this historical name still highlights a key feature of almost every known CDC: full in vitro activation of hemolytic activity requires pretreatment with a reducing agent (4). This requirement for reduction has been attributed to a single cysteine residue residing in a highly conserved, tryptophan-rich undecapeptide positioned within the membrane-binding domain of the toxin, yet the intermediate oxidation state, disulfide, or a modification that is sensitive to reducing agents has remained undetermined.
Recent work demonstrating the importance of glutathione (GSH) in regulating the virulence of Listeria monocytogenes (5, 6), combined with the discovery of S-bacillithiolated proteins in Bacillus subtilis (7, 8), inspired us to seek the identity of any redox-sensitive modifications to the CDC-family hemolysin listeriolysin O (LLO). LLO is an essential virulence factor secreted by the facultative intracellular pathogen L. monocytogenes that facilitates rapid bacterial escape from a phagocytic vacuole into the cytosol (9, 10). Mechanisms that affect the pore-forming ability of LLO have previously been found to have profound effects on the overall virulence of the bacteria, thus making it an attractive model with which to evaluate the role of modifications (11–13). In addition, due to the highly conserved nature of CDCs and the undecapeptide in particular, any newly identified modifications may lead to a better understanding of the reduction requirement for other CDCs. The results of this study show that LLO is rendered fully inactive by S-glutathionylation and that a mutant of LLO that is unable to be modified (a mutant in which the cysteine residue is mutated to an alanine residue at position 484 [LLOC484A]) is slightly attenuated in vivo.
RESULTS
Identification of LLO posttranslational modifications.
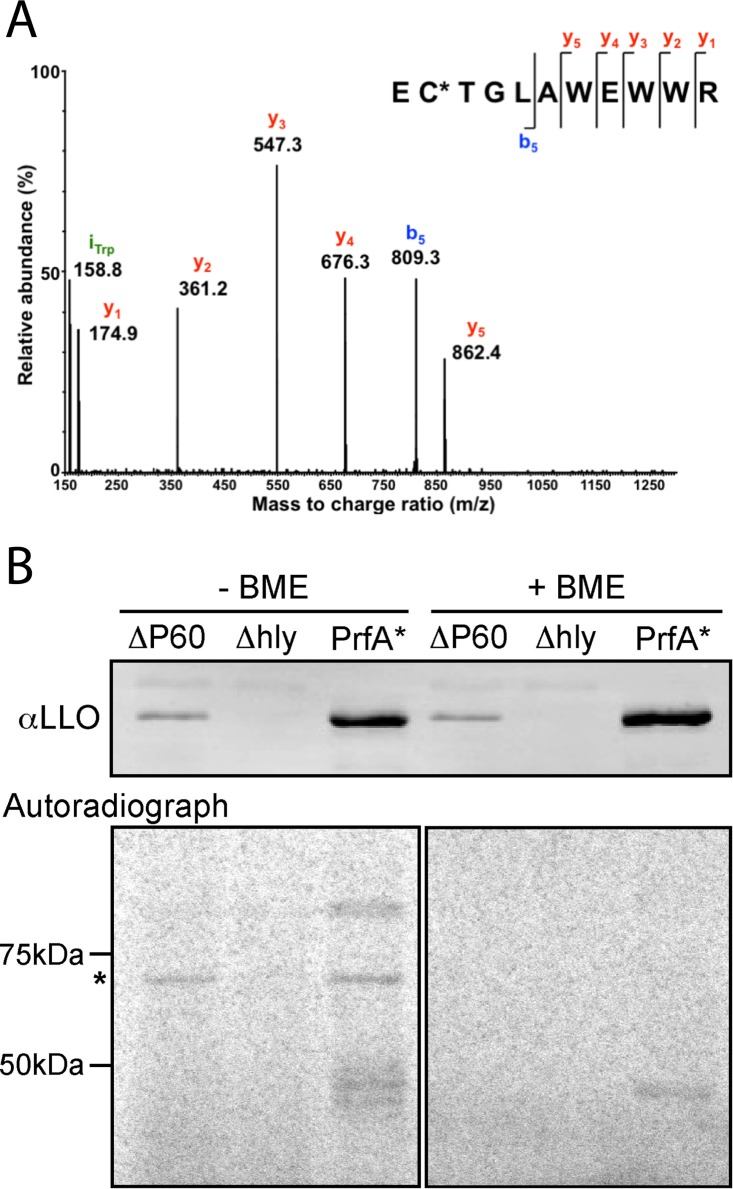
In order to identify posttranslational modifications of listeriolysin O (LLO), we performed tandem mass spectrometry (MS/MS) analysis of tryptic digests of LLO samples concentrated from broth culture. Since the tripeptide antioxidant glutathione (GSH) in L. monocytogenes pathogenesis had been recently characterized for its role in posttranslationally activating virulence gene production (5, 6), a number of these samples were prepared under native, nonreducing conditions to preserve any potential redox-sensitive modifications. Under these conditions, the single highly conserved cysteine residue (Cys484) of LLO was observed to be S-glutathionylated (Fig. 1). This modification was observed when wild-type L. monocytogenes was grown in a synthetic medium lacking GSH and when an L. monocytogenes mutant incapable of producing glutathione (the ΔgshF mutant) was grown in GSH-replete medium, demonstrating that the modification can result from either endogenous or exogenous GSH (Table 1). When the GSH-deficient mutant of L. monocytogenes was grown in synthetic medium lacking GSH, the sole LLO cysteine was S-cysteinylated (Table 1). Highlighting its propensity for oxidation, an unmodified free cysteine was never observed by mass spectrometry under nonreducing conditions.
FIG 1.
LLO is naturally S-glutathionylated at the conserved cysteine. (A) MS/MS spectrum resulting from collision-induced dissociation (CID) of the triply charged, positive precursor ion at m/z 581.2, which is due to the [M + 3H]3+ ion of the peptide EC*TGLAWEWWR (listeriolysin O residues 483 to 493), where C* denotes an S-glutathionylated cysteine residue. Glutathionylation results in an increase in molecular mass of 305.068 Da. (B) Western blot and autoradiograph of secreted proteins from cultures of L. monocytogenes grown in the presence of radiolabeled glutathione and separated by SDS-PAGE in the presence or absence of the reducing agent 2-mercaptoethanol (BME). The ΔP60 mutant is a strain of L. monocytogenes lacking the abundant autolysin P60 (or IAP [invasion-associated protein]) that comigrates with LLO and that was used here in combination with an LLO deletion strain (the Δhly mutant) to confirm that the signal observed in the autoradiogram is due to modified LLO. The PrfA* mutant is a constitutively active mutant of the master virulence transcription factor PrfA and synthesizes more LLO than the wild-type strain. The asterisk to the left of the gel denotes the band corresponding to LLO.
TABLE 1.
Comparison of LLO modifications under various conditions by MS/MS
| L. monocytogenes strain | Growth mediumb | Modification at LLO(Cys484) observed by MS/MSc |
|---|---|---|
| 10403S | BHI (GSH+) | S-Glutathionylation |
| 10403S | iLSM (GSH−) | S-Glutathionylation |
| 10403S ΔgshFa | BHI (GSH+) | S-Glutathionylation |
| 10403S ΔgshF | iLSM (GSH−) | S-Cysteinylation |
The ΔgshF mutant is a strain of 10403S lacking the bifunctional glutathione synthase gshF (lmo2770) and is unable to synthesize glutathione.
Bacteria were grown in culture medium either containing glutathione (GSH+) or lacking glutathione (GSH−).
All modifications detected from any replicates are listed. LLO(Cys484) was never detected as unmodified.
In vitro activity of modified LLO.
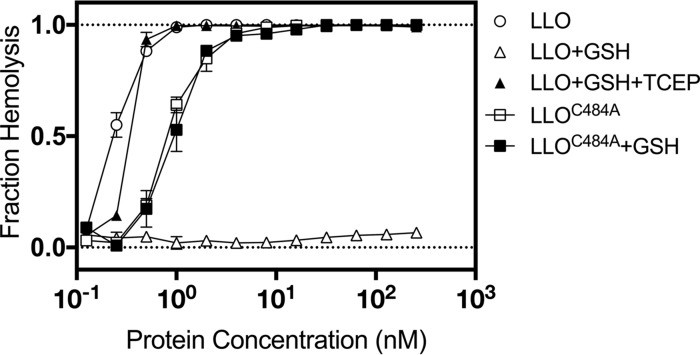
To test the effect of S-glutathionylation on activity, LLO monomers were purified under standard reducing conditions and then incubated with an optimized concentration of GSH and diamide to catalyze the synthetic S-glutathionylation of LLO monomers (LLO-SSG). Sheep red blood cells (sRBCs) incubated with LLO-SSG showed almost undetectable levels of lysis after 30 min at 37°C and were ∼1,000 times less active than sRBCs incubated with unmodified LLO (Fig. 2). The loss of activity was fully reversible by treatment with a chemical reducing agent. A modified version of LLO in which the sole cysteine residue at position 484 was mutated to an alanine residue (LLOC484A) retained nearly full activity compared to that of wild-type LLO and was completely resistant to inactivation by S-glutathionylation, confirming that S-glutathionylation of Cys484 was responsible for this loss of activity.
FIG 2.
In vitro S-glutathionylation of LLO completely blocks the hemolysis of sRBCs. Preparations of either purified LLO or LLO in which the cysteine residue was replaced by alanine (LLOC484A) were exposed to various pretreatments and then mixed with sRBCs. The fraction of lysed sRBCs was quantified after 30 min and plotted versus the concentration of LLO. Artificially S-glutathionylated LLO showed negligible lysis of sRBCs when LLO was used up to a concentration of 300 nM, and this was largely reversed upon further treatment with the reducing agent TCEP.
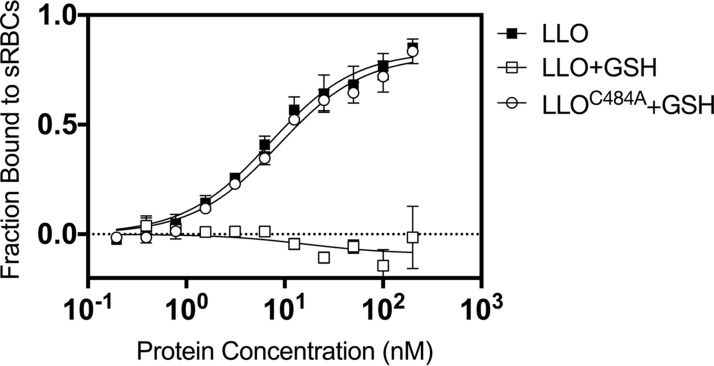
Because the modified cysteine is located at the tip of the membrane-binding domain of LLO, we hypothesized that the S-glutathionylation of cysteine might physically prevent the association of LLO with its target membrane. To test this hypothesis, LLO monomers that had been translationally fused to the fluorescent protein mCherry were compared for their ability to bind sRBCs at 4°C before and after S-glutathionylation (Fig. 3). Consistent with the hemolysis data, S-glutathionylation rendered the LLO monomers unable to bind sRBCs when they were used at concentrations up to 300 nM (i.e., there was a >35-fold loss of binding affinity compared with that of the wild type). Treatment with the chemical reducing agent tris(2-carboxyethyl)phosphine (TCEP) completely reversed the loss of binding, and the LLOC484A mutant was immune to modification.
FIG 3.
In vitro S-glutathionylated LLO is unable to bind erythrocytes. In vitro S-glutathionylation of mCherry-tagged LLO resulted in the complete loss of binding to sRBCs (open squares) compared to that for the fully reduced control (closed squares). A mutant of LLO in which the cysteine residue was replaced by alanine (LLOC484A) retained activity after identical S-glutathionylation treatment.
In vivo results.
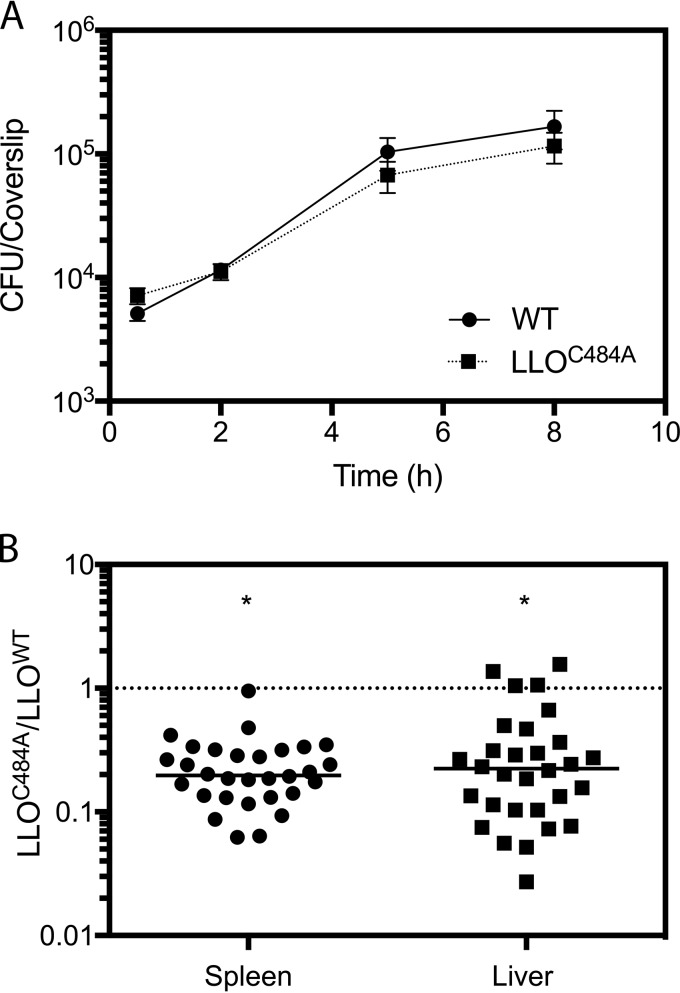
The potent yet rapidly reversible negative effect of S-glutathionylation on the function of LLO led us to hypothesize that this modification might be critical for restricting pore-forming activity to appropriate spatial and/or temporal compartments during infection. To test this hypothesis, we performed a competitive infection to compare the virulence of a wild-type strain to that of a strain expressing the LLO C484A mutation (the LLOC484A strain). At 48 h after intravenous infection of CD-1 mice, the strain expressing LLOC484A exhibited a statistically significant defect in its ability to grow in the liver and spleen compared to that of the wild type (Fig. 4B). While this defect suggests a role for Cys484 in vivo, we wanted to determine if there was another model of infection that would reveal a more specific and striking impact of losing this redox-sensitive switch of LLO. The LLOC484A mutant, however, showed no significant growth defect in bone marrow-derived macrophages (Fig. 4A) or a spreading defect in a monolayer of L2 fibroblasts (see Fig. S2A in the supplemental material). In addition, growth in resident peritoneal macrophages (Fig. S2B), in vivo oral infections (Fig. S2C), and in vivo noncompetitive infections (Fig. S1) were tested for their abilities to potentially reveal a specific role for the S-glutathionylation of LLO, yet only the in vivo competitive index was sensitive enough to yield a statistically significant defect of the LLOC484A mutant.
FIG 4.
Glutathionylation of LLO is required for maximal virulence in an animal model of infection. (A) A mutant of L. monocytogenes expressing a variant of LLO in which the cysteine residue was replaced by alanine (LLOC484A) grows similarly to the wild type (WT) during in vitro infection of bone marrow-derived macrophages. (B) Mutant bacteria expressing LLO in which the cysteine residue was replaced by alanine (LLOC484A) were attenuated for virulence compared to wild-type bacteria during in vivo competitive infection of mice. The median competitive indices for the liver and spleen were 0.1976 and 0.2240, respectively, and significance was calculated by a one-sample t test using Prism (version 7) software (GraphPad). *, P < 0.0001.
DISCUSSION
The sole cysteine residue within LLO is conserved among nearly all CDC homologs, yet the evolutionary pressure to retain such a highly reactive residue has remained a mystery. Although it is known that this cysteine confers the prototypical reduction requirement for almost all CDCs, the precise mechanism and biological relevance of this observation are still poorly understood (14, 15). Previous studies have established that artificial chemical modification of this residue, such as alkylation, prevents binding to target membranes and/or subsequent pore formation in target membranes (14, 16). The results of this study show that glutathione derived from either endogenous or exogenous sources S-glutathionylates LLO and reversibly ablates LLO activity.
The prevalence and importance of low-molecular-weight (LMW) thiols in bacterial species are becoming increasingly clear (recently reviewed in reference 17). For example, in Salmonella enterica serovar Typhimurium, more than two dozen proteins are either S-glutathionylated or S-cysteinylated, depending on the growth medium (18). In B. subtilis and Staphylococcus aureus, many proteins are modified by the analogous LMW thiol bacillithiol, while Mycobacterium tuberculosis utilizes mycothiol (19, 20). L. monocytogenes is rare among Gram-positive bacteria in that it can synthesize glutathione (5) and, as demonstrated here, utilize it as a redox-sensitive substrate for posttranslational modifications. In agreement with the findings of previous studies (21), a variant of LLO that is unable to be S-glutathionylated (LLOC484A) decreased the virulence of L. monocytogenes in vivo, despite having lytic activity similar to or, in one study (22), greater than that of the wild type.
The primary role of LLO is to mediate the escape of L. monocytogenes from a potentially hostile phagocytic vacuole (10). It is possible that S-glutathionylation protects the cysteine from irreversible oxidation in a phagosome. However, the S-glutathionylation of LLO in this context would be expected to inhibit phagosomal escape, suggesting that either the bacteria or the host possesses an oxidoreductase capable of removing the glutathione. Interestingly, macrophages possess a gamma interferon-inducible lysosomal thiol reductase (GILT; also known as Ifi30) that is capable of activating LLO in vitro (23). In addition, GILT−/− macrophages and mice are resistant to infection by wild-type L. monocytogenes but not to infection by a mutant secreting LLOC484A (23). Combined with the data presented in this study, it is reasonable to suspect that the vacuole-localized GILT is necessary to reduce S-glutathionylated LLO; however, we were unable to reproduce the in vitro growth defect of wild-type L. monocytogenes in GILT−/− macrophages (see Fig. S3 in the supplemental material). Nevertheless, the notion that S-glutathionylated LLO restricts its activity to specific host cells capable of removing this modification is appealing.
The discovery that LLO is naturally S-glutathionylated represents one of the first examples of an S-glutathionylated protein derived from a Gram-positive bacterium. In addition, these data suggest that S-thiolation of the conserved cysteine of CDCs may be a conserved posttranslational regulatory mechanism necessary for the optimal activity of different CDCs in their respective niches. Future studies with other CDC-secreting pathogens in infection models analogous to the L. monocytogenes infection model will be necessary to fully appreciate the regulatory importance of this modification.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council of the National Academy of Sciences (24). All protocols were reviewed and approved by the Animal Care and Use Committee at the University of California, Berkeley (MAUP R235-0815B).
Bacterial strains and growth.
All L. monocytogenes strains (listed in Table S2 in the supplemental material) were derivatives of strain 10403S (25) and were cultured in brain heart infusion (BHI; BD Biosciences) at 37°C with shaking and without antibiotics, unless otherwise stated. Growth was measured by determination of the optical density at a wavelength of 600 nm (OD600) using a spectrophotometer. Frozen bacterial stocks were stored at −80°C in BHI and 40% glycerol. All chemicals were purchased from Sigma-Aldrich unless otherwise stated. The following antibiotics were used at the indicated concentrations unless otherwise stated: streptomycin at 200 μg/ml, chloramphenicol at 7.5 μg/ml for L. monocytogenes and 10 μg/ml for Escherichia coli, and erythromycin at 1 μg/ml.
Identification of LLO S-glutathionylation by MS/MS.
Cultures of L. monocytogenes were grown in either incomplete Listeria synthetic medium (iLSM) (26) or BHI broth (BD Biosciences) with shaking at 37°C until entrance into stationary phase. Bacteria were separated by centrifugation, and the supernatant was filtered prior to precipitation of the proteins with 10% trichloroacetic acid (TCA). Samples were suspended in 1× lithium dodecyl sulfate (LDS) buffer (Invitrogen) without the addition of a reducing agent and separated by SDS-PAGE. After Coomassie blue staining, the LLO band (∼59 kDa) was excised, destained with 25 mM ammonium bicarbonate in 50% acetonitrile, and digested in the gel with an equal volume of trypsin gold (Promega) at 12.5 μg/ml in 80% acetonitrile with 25 mM ammonium bicarbonate for 6 to 8 h at 37°C. The digested peptides were then extracted from the gel pieces by three successive 10-min vortex treatments using a solution of 45% water, 50% acetonitrile, and 5% formic acid. The total extracted solution was reduced to 10 μl by vacuum centrifugation and analyzed by liquid chromatography (LC)-tandem mass spectrometry (MS/MS). LC-MS/MS measurements were performed using an LTQ Orbitrap XL mass spectrometer that was connected in line with an UltiMate 3000 RSLCnano liquid chromatograph (Thermo Fisher Scientific, Waltham, MA).
Natural modification of LLO by 35S-labeled glutathione.
Cultures of L. monocytogenes were incubated at 37°C in tryptic soy broth medium supplemented with [35S]glutathione (4 μCi/ml) for 5 h (PerkinElmer). Bacteria were removed by centrifugation and filtration prior to precipitation of the supernatant proteins with TCA (10%) and separation by SDS-PAGE with or without 2-mercaptoethanol. The gel was dried and exposed to a phosphor screen for 4 days before imaging on a Typhoon scanner (GE Life Sciences). Duplicate gels were transferred to a nitrocellulose membrane and analyzed for protein abundance by immunoblotting for LLO and P60 using previously described methods (27).
Intracellular growth curve.
Wild-type or GILT−/− bone marrow-derived macrophages (the latter of which were a gift from Peter Cresswell) were harvested as previously described (28), and 3 × 106 cells were plated in 60-mm-diameter non-tissue culture-treated petri dishes. Cells were infected at a multiplicity of infection (MOI) of 0.1, and growth curves were performed as described previously (9).
Determination of competitive indices in vivo.
Competitive indices were determined as previously described (29). Briefly, frozen culture stocks were thawed, grown to log phase in fresh BHI for 2 h, and mixed in a 1:1 ratio. Six- to 8-week-old female CD-1 mice (Charles River) were infected with 1 × 105 CFU. Animals were sacrificed 48 h later, and their spleens and livers were harvested. The spleens and livers were homogenized in 5 and 10 ml 0.1% NP-40, respectively, and homogenates were plated onto LB agar. At least 100 colonies per organ were replica plated or patched onto BHI agar containing erythromycin (2 μg/ml). Competitive indices were calculated by dividing the number of CFU of the test strain (erythromycin sensitive) by the number of CFU of the reference strain (erythromycin resistant).
Plasmid expressing mCherry-LLO.
The mCherry-LLO expression vector was constructed by cloning the gene encoding mCherry from pHpPL3-mCherry to pET29b-hly via Gibson assembly using the primers listed in Table S1 in the supplemental material (30). The resulting protein had mCherry (26.7 kDa) fused to the N terminus of LLO. The primers and templates are listed in Table S1.
Protein expression and purification.
The expression and purification of recombinant LLO were described previously (13). Specifically, E. coli strains harboring expression plasmids (listed in Table S2) were grown at 37°C to stationary phase and inoculated into 1 liter fresh LB medium with chloramphenicol at an initial OD600 of 0.01. When the OD600 of the culture reached 0.6 to 0.7, expression was induced by the addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and maintained for 6 h. To harvest the cells, bacteria were pelleted and resuspended in 35 ml cold lysis buffer (50 mM sodium phosphate, pH 8.0, 1 M NaCl, 20 mM imidazole, 10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride). Cells were lysed by sonication. The majority of the target protein ended up in the supernatant after centrifugation. Two milliliters of Ni-nitrilotriacetic acid resin equilibrated in washing buffer (20 mM sodium phosphate, pH 7.4, 1 M NaCl) was added to the lysate supernatant, and the mixture was mildly stirred at 4°C for 1 h. The resin was packed in a column and washed with up to 1 liter of washing buffer. LLO was eluted with elution buffer (20 mM sodium phosphate, pH 6.0, 1 M NaCl, 800 mM imidazole). The extra imidazole in the eluate was removed by dialysis against storage buffer (20 mM sodium phosphate, 1 M NaCl, pH 6.0, 5 mM dithiothreitol [DTT], 1 mM EDTA). The protocol yielded over 50 mg of LLO protein per liter of medium. mCherry-LLO proteins were purified by the same method.
Hemolysis assay.
The hemolysis assay was performed as previously described (13). Briefly, sheep red blood cells (sRBCs) were washed three times in phosphate-buffered saline (PBS) and resuspended to 2% (vol/vol) in PBS at pH 5.5 (adjusted with HCl). Protein samples were prewarmed in 0.1 mg/ml bovine serum albumin (BSA) in PBS in a 96-well plate at 37°C for 30 min; in the case of glutathionylated LLO, protein was treated with 0.2 mM glutathione (GSH), 0.5 mM diamide, and 0.1 mg/ml BSA in PBS. Where noted, LLO was subsequently reduced with 8 mM TCEP for 30 min. After the preincubation, protein samples were serially diluted to the target concentrations and mixed at a 5:1 (vol/vol) ratio with 2% sRBCs. The resulting mixtures were gently shaken and incubated at 37°C for 30 min, followed by measurement of the absorbance at 600 nm using a SpectraMax M5 microplate reader. The fractional hemolysis was calculated as (Aprotein − Abuffer)/(ATX − Abuffer), where Aprotein is the absorbance of the protein samples, Abuffer is the absorbance of the buffer, and ATX is the absorbance obtained after treatment with 0.1% Triton X-100. PBS buffer was mixed with cells (with no LLO) to yield Abuffer, and complete hemolysis (ATX) was achieved with 0.1% Triton X-100. Triplicate samples were assayed for each protein concentration.
In vitro S-glutathionylation.
Conditions for protein glutathionylation were explored with oxidants, such as hydrogen peroxide and diamide. Diamide was chosen due to its rapid conversion and little to no alteration of cell function (31). The optimized conditions were as follows: 10 μM LLO was incubated at 4°C overnight with 0.2 mM GSH and 0.5 mM diamide in PBS. Excess GSH and diamide were removed by dialysis against storage buffer without DTT. The resulting protein conjugate was verified with trypsin digestion and LC-MS/MS (32), and the percentage of each of the glutathionylated peptides was estimated to be 99.7%.
mCherry-LLO membrane-binding assay.
sRBCs were resuspended to 20% (vol/vol) in PBS (pH 7.4) after washes. mCherry-LLO was diluted in series and added to the sRBCs at a 5:1 (protein/sRBC) ratio. The mixture was incubated at 4°C for 15 min and centrifuged. The fluorescence emission of the supernatant was measured at 650 nm (excitation, 580 nm). The fraction of protein bound was calculated as (F0 − F)/F0, where F0 is the fluorescence of mCherry-LLO added to PBS buffer and F is the fluorescence of the protein added to sRBCs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants 1P01 AI063302 and 1R01 AI27655 to D.A.P. (http://www.nih.gov). The QB3/Chemistry Mass Spectrometry Facility at the University of California, Berkeley, receives support from the National Institutes of Health (grant 1S10OD020062-01).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Daniel A. Portnoy has a consulting relationship with and a financial interest in Aduro Biotech, and both he and the company stand to benefit from the commercialization of this research.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00959-16.
REFERENCES
- 1.Gilbert RJC. 2010. Cholesterol-dependent cytolysins. Adv Exp Med Biol 677:56–66. doi: 10.1007/978-1-4419-6327-7_5. [DOI] [PubMed] [Google Scholar]
- 2.Tweten RK, Hotze EM, Wade KR. 2015. The unique molecular choreography of giant pore formation by the cholesterol-dependent cytolysins of Gram-positive bacteria. Annu Rev Microbiol 69:323–340. doi: 10.1146/annurev-micro-091014-104233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smyth CJ, Duncan JL. 1978. Thiol-activated (oxygen-labile) cytolysins, p 129–183. In Jeljaszewicz J, Wadstrom T (ed), Bacterial toxins and cell membranes. Academic Press, New York, NY. [Google Scholar]
- 4.Billington SJ, Jost BH, Songer JG. 2000. Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol Lett 182:197–205. doi: 10.1016/S0378-1097(99)00536-4. [DOI] [PubMed] [Google Scholar]
- 5.Gopal S, Borovok I, Ofer A, Yanku M, Cohen G, Goebel W, Kreft J, Aharonowitz Y. 2005. A multidomain fusion protein in Listeria monocytogenes catalyzes the two primary activities for glutathione biosynthesis. J Bacteriol 187:3839–3847. doi: 10.1128/JB.187.11.3839-3847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. 2015. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517:170–173. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JW, Soonsanga S, Helmann JD. 2007. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A 104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, Fahey RC. 2009. Bacillithiol is an antioxidant thiol produced in bacilli. Nat Chem Biol 5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Portnoy DA, Jacks SP, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnupf P, Portnoy DA. 2007. Listeriolysin O: a phagosome-specific lysin. Microbes Infect 9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Decatur AL, Portnoy DA. 2000. A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science 290:992–995. doi: 10.1126/science.290.5493.992. [DOI] [PubMed] [Google Scholar]
- 12.Schnupf P, Portnoy DA, Decatur AL. 2006. Phosphorylation, ubiquitination and degradation of listeriolysin O in mammalian cells: role of the PEST-like sequence. Cell Microbiol 8:353–364. doi: 10.1111/j.1462-5822.2005.00631.x. [DOI] [PubMed] [Google Scholar]
- 13.Glomski IJ, Gedde MM, Tsang AW, Swanson JA, Portnoy DA. 2002. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J Cell Biol 156:1029–1038. doi: 10.1083/jcb.200201081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwamoto M, Ohno-Iwashita Y, Ando S. 1987. Role of the essential thiol group in the thiol-activated cytolysin from Clostridium perfringens. Eur J Biochem 167:425–430. [DOI] [PubMed] [Google Scholar]
- 15.Cowell JL, Grushoff-Kosyk PS, Bernheimer AW. 1976. Purification of cereolysin and the electrophoretic separation of the active (reduced) and inactive (oxidized) forms of the purified toxin. Infect Immun 14:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soltani CE, Hotze EM, Johnson AE, Tweten RK. 2007. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. Proc Natl Acad Sci U S A 104:20226–20231. doi: 10.1073/pnas.0708104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Loi V, Rossius M, Antelmann H. 2015. Redox regulation by reversible protein S-thiolation in bacteria. Front Microbiol 6:1748. doi: 10.3389/fmicb.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansong C, Wu S, Meng D, Liu X, Brewer HM, Deatherage Kaiser BL, Nakayasu ES, Cort JR, Pevzner P, Smith RD, Heffron F, Adkins JN, Paša-Tolić L. 2013. Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proc Natl Acad Sci U S A 110:10153–10158. doi: 10.1073/pnas.1221210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, Helmann JD. 2010. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in bacilli. Proc Natl Acad Sci U S A 107:6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spies HS, Steenkamp DJ. 1994. Thiols of intracellular pathogens. Identification of ovothiol A in Leishmania donovani and structural analysis of a novel thiol from Mycobacterium bovis. Eur J Biochem 224:203–213. [DOI] [PubMed] [Google Scholar]
- 21.Stachowiak R, Wiœniewski J, Osińska O, Bielecki J. 2009. Contribution of cysteine residue to the properties of Listeria monocytogenes listeriolysin O. Can J Microbiol 55:1153–1159. doi: 10.1139/W09-070. [DOI] [PubMed] [Google Scholar]
- 22.Walls ZF, Goodell S, Andrews CD, Mathis J, Lee K-D. 2013. Mutants of listeriolysin O for enhanced liposomal delivery of macromolecules. J Biotechnol 164:500–502. doi: 10.1016/j.jbiotec.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 23.Singh R, Jamieson A, Cresswell P. 2008. GILT is a critical host factor for Listeria monocytogenes infection. Nature 455:1244–1247. doi: 10.1038/nature07344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 25.Bécavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kühbacher A, Brisse S, Pucciarelli MG, García-del Portillo F, Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro-Cerdá J, Moszer I, Bierne H, Cossart P. 2014. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio 5:e00969-14. doi: 10.1128/mBio.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whiteley AT, Pollock AJ, Portnoy DA. 2015. The PAMP c-di-AMP is essential for Listeria growth in macrophages and rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe 17:788–798. doi: 10.1016/j.chom.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reniere ML, Whiteley AT, Portnoy DA. 2016. An in vivo selection identifies Listeria monocytogenes genes required to sense the intracellular environment and activate virulence factor expression. PLoS Pathog 12:e1005741. doi: 10.1371/journal.ppat.1005741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauer J-D, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun 79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auerbuch V, Lenz LL, Portnoy DA. 2001. Development of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect Immun 69:5953–5957. doi: 10.1128/IAI.69.9.5953-5957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosower NS, Kosower EM, Wertheim B, Correa WS. 1969. Diamide, a new reagent for the intracellular oxidation of glutathione to the disulfide. Biochem Biophys Res Commun 37:593–596. doi: 10.1016/0006-291X(69)90850-X. [DOI] [PubMed] [Google Scholar]
- 32.Rebecchi KR, Go EP, Xu L, Woodin CL, Mure M, Desaire H. 2011. A general protease digestion procedure for optimal protein sequence coverage and post-translational modifications analysis of recombinant glycoproteins: application to the characterization of human lysyl oxidase-like 2 glycosylation. Anal Chem 83:8484–8491. doi: 10.1021/ac2017037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.