ABSTRACT
Tuberculosis (TB) due to Mycobacterium tuberculosis remains a major global infectious disease problem, and a more efficacious vaccine is urgently needed for the control and prevention of disease caused by this organism. We previously reported that a genetically modified strain of Mycobacterium smegmatis called IKEPLUS is a promising TB vaccine candidate. Since protective immunity induced by IKEPLUS is dependent on antigen-specific CD4+ T cell memory, we hypothesized that the specificity of the CD4+ T cell response was a critical feature of this protection. Using in vitro assays of interferon gamma production (enzyme-linked immunosorbent spot [ELISPOT] assays) by splenocytes from IKEPLUS-immunized C57BL/6J mice, we identified an immunogenic peptide within the mycobacterial ribosomal large subunit protein RplJ, encoded by the Rv0651 gene. In a complementary approach, we generated major histocompatibility complex (MHC) class II-restricted T cell hybridomas from IKEPLUS-immunized mice. Screening of these T cell hybridomas against IKEPLUS and ribosomes enriched from IKEPLUS suggested that the CD4+ T cell response in IKEPLUS-immunized mice was dominated by the recognition of multiple components of the mycobacterial ribosome. Importantly, CD4+ T cells specific for mycobacterial ribosomes accumulate to significant levels in the lungs of IKEPLUS-immunized mice following aerosol challenge with virulent M. tuberculosis, consistent with a role for these T cells in protective host immunity in TB. The identification of CD4+ T cell responses to defined ribosomal protein epitopes expands the range of antigenic targets for adaptive immune responses to M. tuberculosis and may help to inform the design of more effective vaccines against tuberculosis.
KEYWORDS: T cells, antigen, epitope, ribosome, tuberculosis
INTRODUCTION
Tuberculosis is one of the most significant global health problems. One-third of the global population may be infected with Mycobacterium tuberculosis, with an estimated 9.6 million new cases and 1.5 million deaths annually (1). The efficacy of the only currently available vaccine, live attenuated Mycobacterium bovis bacillus Calmette-Guérin (BCG), varies greatly and is largely ineffective against adult pulmonary tuberculosis (2, 3). The poor protection afforded by BCG vaccination, low compliance with prolonged drug treatment, emergence of multidrug-resistant and extensively drug-resistant strains, and complications due to coinfection with HIV highlight the need for novel, improved vaccine strategies against M. tuberculosis (1, 4–6).
Numerous attempts to obtain an M. tuberculosis vaccine that demonstrates enhanced protection over BCG, durability, and safety have been made (7). Administration of additional doses of BCG did not boost the protection afforded by BCG (8). Candidate vaccines against M. tuberculosis have focused largely on targeting immunodominant antigens that are secreted proteins, including Ag85A (9–12), Ag85B (12–17), ESAT-6 (15, 16), TB10.4 (9, 13, 17), Rv1196, and Rv0125 (18, 19). Enhanced protection over BCG has proven to be difficult to achieve, and safety issues and adverse events have caused concern (12, 20). The development of new vaccines and diagnostics will be aided by the discovery of additional antigens relevant to both natural and vaccine-induced immune responses to M. tuberculosis infection.
The development of safe and effective vaccines against M. tuberculosis is hampered by the limited knowledge of the immune mechanisms required for protection. Previous studies, using adoptive transfer of immune CD4+ T cells (21, 22), specific in vivo antibody depletion of CD4+ T cells in mice (23–25) or macaques (26, 27), and effects of CD4+ T cell depletion due to infection with HIV (28), demonstrate the crucial role of antigen-specific CD4+ T cells in the control of M. tuberculosis infection. Furthermore, a T helper 1 (Th1)-type response and the production of interferon gamma (IFN-γ) have been linked to favorable outcomes of M. tuberculosis infection in animal models and humans (29–31).
The identification of M. tuberculosis antigens that are effective targets for protective CD4+ T cell responses remains an important focus of ongoing efforts to develop novel, effective vaccines against M. tuberculosis. Numerous mycobacterial proteins have been recognized as T cell antigens in humans (32, 33) and animals infected with M. tuberculosis (34, 35). Secreted mycobacterial proteins have been a major focus of previous studies to identify immunogenic molecules (36, 37). In addition, proteins associated with the bacterial cell wall or cell surface, such as the proline-glutamic acid and proline-proline-glutamic acid (PE/PPE) protein family in M. tuberculosis, induce cell-mediated immunity (38–40). Secreted proteins of the Esx family, including CFP-10 and ESAT-6, have been shown to be dominant T cell antigens (40–42). The antigen 85 family of mycolyl transferases comprises another group of secreted proteins, which are among the immunodominant antigens that elicit Th1-type immune responses (37, 43).
Recently, an M. tuberculosis genome-wide screen to identify targets of major histocompatibility complex (MHC) class II-restricted CD4+ T cell responses in M. tuberculosis-infected patients was conducted by using epitope prediction (44). This study highlighted the breadth of epitopes recognized during M. tuberculosis infection, emphasizing the potential importance of designing multiepitope vaccines (44). However, other recent work has shown that many known T cell epitopes of M. tuberculosis are derived from protein sequences that are hyperconserved among various M. tuberculosis isolates. This suggests that the recognition of these epitopes by the host immune system may be beneficial to the pathogen, possibly by acting as immunological decoys or driving the establishment and maturation of granulomas within the lungs to perpetuate persistence and transmission (33, 45). Alternatively, CD4+ T cell epitopes that are conserved among mycobacterial species may represent antigens not involved with the evolution of host-pathogen coexistence specific to M. tuberculosis and could represent more effective vaccine targets (33, 45).
We previously constructed a candidate vaccine strain, designated IKEPLUS, by the introduction of genes encoding the M. tuberculosis Esx-3 type VII secretion system (T7SS) into an M. smegmatis Δesx-3 mutant (IKE). Compared to standard immunization with BCG-Danish, the immunization of C57BL/6 mice with IKEPLUS gave prolonged survival after M. tuberculosis challenge and generated enhanced Th1-type responses characterized by the production of IFN-γ and interleukin-12p40 (IL-12p40) (46). Strikingly, IKEPLUS immunization of mice was associated with marked reductions in the numbers of viable bacteria in the lung and spleen and apparent sterilizing immunity in the liver. While the protection against M. tuberculosis afforded by immunization with IKEPLUS was dependent on antigen-specific CD4+ T cell memory, the specificities of CD4+ T cells in IKEPLUS-immunized mice remain to be determined. In the present study, immunization with IKEPLUS was used as a tool for uncovering possible protective epitopes that may not be effectively presented during BCG vaccination or natural M. tuberculosis infection. We used a synthetic peptide library, as well as a library of MHC class II (I-Ab)-restricted T cell hybridomas (TCHs) constructed from IKEPLUS-immunized mice, to examine the range of CD4+ T cell specificities generated upon immunization with IKEPLUS. Our findings identified the mycobacterial ribosome as a potential major source of protective antigens for immune responses against M. tuberculosis.
RESULTS
Identification of a mycobacterial ribosomal protein epitope, rplJTB146–160, for CD4+ T cells.
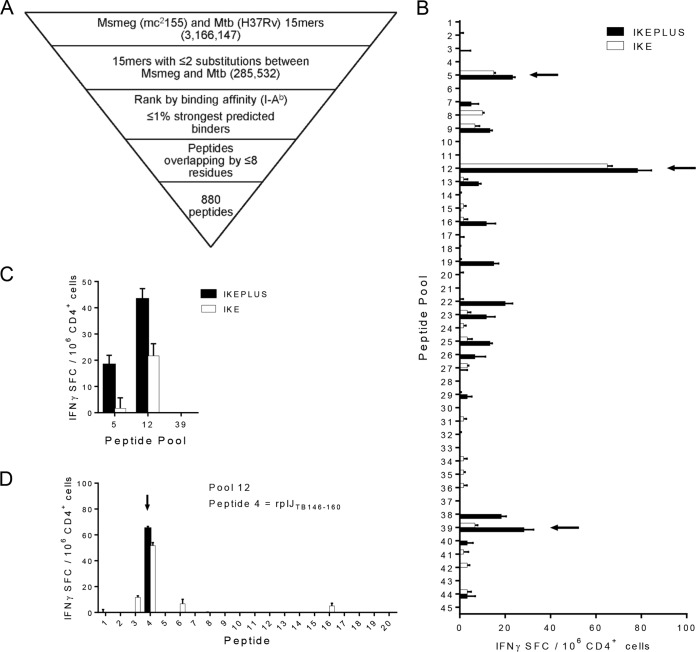
To examine the range of CD4+ T cell epitopes in IKEPLUS-immunized mice, we developed a synthetic peptide library of 880 nonoverlapping peptides designed around the homologous M. smegmatis and M. tuberculosis sequences present in IKEPLUS (Fig. 1A). Peptides were distributed into 45 pools containing 19 to 20 peptides each, and the number of IFN-γ-producing CD4+ T cells isolated from the spleens of mice immunized with either the IKE or IKEPLUS strain induced by restimulation with each of the pools was measured by using an enzyme-linked immunosorbent spot (ELISPOT) assay. Responses above background levels were observed for three pools in the primary screening (Fig. 1B), and two of these were confirmed by subsequent rescreening (Fig. 1C). Stimulation with the individual peptides comprising these pools revealed a single peptide from pool 12 that strongly and reproducibly stimulated the responses of immune CD4+ T cells (Fig. 1D). This peptide (KAAGLFNAPASQLAR) was derived from the M. tuberculosis RplJ protein, also known as the 50S large ribosomal subunit L10 protein, the product of the M. tuberculosis Rv0651 gene (see Table S1 in the supplemental material). This peptide is referred to below as rplJTB146–160.
FIG 1.
IKEPLUS peptide library construction and identification of CD4+ T cell targets. (A) Overview of the process used to generate the IKEPLUS peptide library, which resulted in 880 peptides representing predicted I-Ab-presented epitopes with homology between M. smegmatis (Msmeg) and M. tuberculosis (Mtb). See Materials and Methods for details. (B) Screening of 45 pools of peptides comprising the 880-peptide library by an IFN-γ ELISPOT assay using CD4+ T cells isolated at 2 weeks postimmunization from spleens of C57BL/6 mice immunized i.v. with 5 × 107 CFU IKE or IKEPLUS. Arrows indicate pools or peptides that were able to elicit significant responses in two independent experiments. Responses of immune CD4+ T cells to peptide pools were considered positive if the stimulation index was more than twice the mean of the values for negative-control wells (effectors, APCs, and DMSO without peptides) and at least 20 spot-forming cells (SFC) per 106 CD4+ T cells above the mean values for the negative controls. (C) Rescreening of positive pools with CD4+ T cells from additional IKE- or IKEPLUS-immunized mice confirmed responses to pools 5 and 12. (D) Deconvolution of responses to identify individual peptides in pool 12 responsible for CD4+ T cell responses. No responses to individual peptides of pool 5 were detected (not shown). Data shown in panels B to D represent mean values and standard errors for triplicate samples. All data shown are representative of results from at least two independent experiments.
The influence of the immunizing bacterial strain on the development of CD4+ T cell responses to the novel RplJ epitope was assessed by comparing responses to rplJTB146–160 by CD4+ T cells from mice immunized with M. smegmatis strains IKE, IKEPLUS, and mc2155 to those with standard BCG immunization (Fig. 2A). For comparison, we also analyzed responses to a defined CD4+ T cell epitope of the TB9.8 (Rv0287) protein, an immunodominant epitope described previously in BCG-immunized mice (47). CD4+ T cells from mice immunized with any of the three M. smegmatis strains reacted most strongly to rplJTB146–160 and only minimally to the TB9.8 peptide. In contrast, CD4+ T cells from mice immunized with BCG reacted strongly with the TB9.8 peptide but not significantly with rplJTB146–160. The moderately reduced response to rplJTB146–160 in mice immunized with mc2155 compared to those induced by IKE and IKEPLUS may reflect the lower dose of mc2155 (1 × 107 CFU for mc2155 versus 5 × 107 CFU for IKE or IKEPLUS). The lower dose of mc2155 was necessary because of lethal effects from very high doses of this strain (46). Therefore, IKE was also used for comparisons of responses in these studies as an additional representative of a modified M. smegmatis strain that could be used at the standard IKEPLUS dose. Changing the rplJTB146–160 peptide concentration gave a clear dose dependence of responses by CD4+ T cells from IKEPLUS-immunized mice, with no significant reactivity by CD4+ T cells from BCG-immunized mice at any concentration tested (Fig. 2B). Responses were dependent on MHC class II (I-Ab) expression by antigen-presenting cells (APCs), as shown by the lack of stimulation in cultures containing bone marrow-derived dendritic cells (BMDCs) from MHC class II−/− mice (Fig. 2C). Overall, these results indicated that immunization with M. smegmatis, but not BCG, induced strong MHC class II-restricted CD4+ T cell responses to the RplJ protein.
FIG 2.
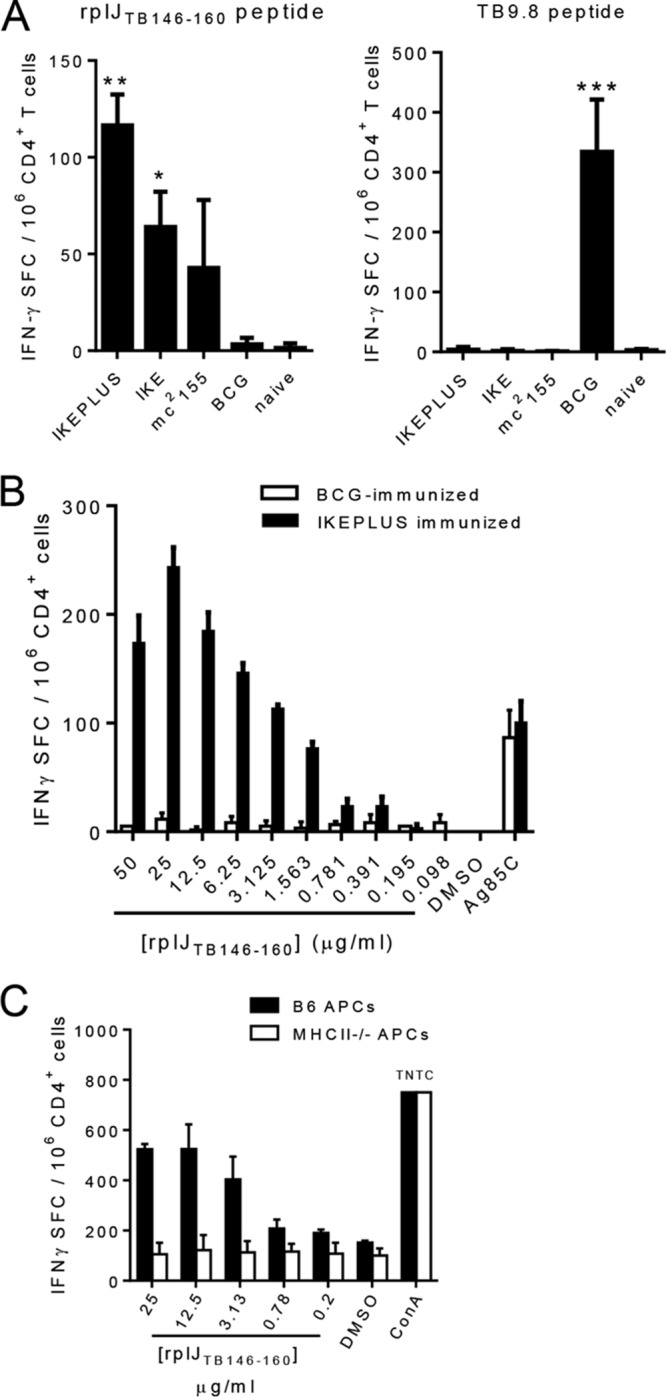
Dose dependence and MHC class II restriction of responses to the RplJ epitope following M. smegmatis immunization. (A) C57BL/6 (B6) mice were immunized with 5 × 107 CFU IKE or IKEPLUS i.v., 107 CFU mc2155 i.v., or 5 × 106 CFU BCG-Danish s.c. At 2 weeks postimmunization, CD4+ T cells were isolated from immunized splenocytes and cultured in the presence of APCs and the rplJTB146–160 (10 μg/ml) and TB9.8 (10 μg/ml) peptides. Responses of immune CD4+ T cells to peptides were quantified by an IFN-γ ELISPOT assay. *, P < 0.05 compared to BCG-immunized and naive mice; **, P < 0.0005 compared to BCG-immunized and naive mice; ***, P < 0.0001 compared to all other groups (ANOVA). (B) Dose-dependent responses of IKEPLUS-immunized splenocytes to the rplJTB146–160 peptide were assayed by an IFN-γ ELISPOT assay in comparison to BCG-immunized splenocytes. (C) The requirement for MHC class II presentation was assessed by an ELISPOT assay using APCs from wild-type mice or MHC class II−/− mice. Data shown represent mean values and standard errors for triplicate (A) or quadruplicate (B and C) samples. All data shown are representative of results from at least two independent experiments.
Mapping of the core epitope of rplJTB146–160.
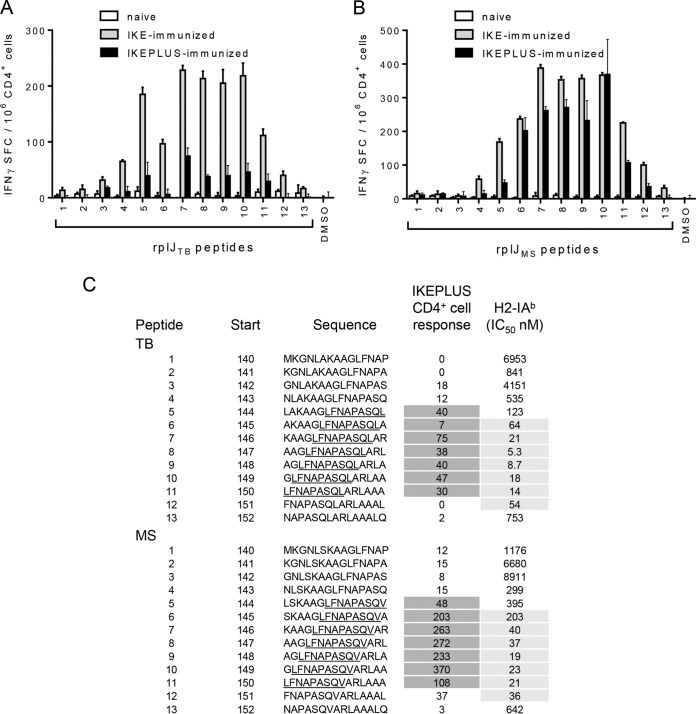
The core epitope of rplJTB146–160 was determined by defining the recognition of all possible 15-mers spanning the identified epitope and an additional six residues on either side (i.e., 15-mers overlapping by 14 amino acids covering residues 140 to 157). The peptides were derived from both the M. tuberculosis H37Rv (TB) (Fig. 3A) and M. smegmatis mc2155 (Fig. 3B) sequences of this region of RplJ, with the rplJTB146–160 peptide corresponding to peptide 7 in the TB library (Fig. 3A and C). An IFN-γ ELISPOT assay was used to quantitate responses from CD4+ T cells isolated from IKE- or IKEPLUS-immunized splenocytes 2 weeks after immunization and cultured in the presence of APCs and the peptides individually (Fig. 3A and B).
FIG 3.
Mapping of the core epitope of RplJ. C57BL/6 mice were immunized with 5 ×107 CFU IKE or IKEPLUS i.v. or 5 ×106 CFU BCG-Danish s.c. Two weeks later, CD4+ T cells were isolated from the spleen and assayed by an IFN-γ ELISPOT assay for responses to overlapping RplJ 15-mer peptides designed around the original RplJ epitope (peptide 7) and representing either M. tuberculosis H37Rv (TB) or M. smegmatis mc2155 (MS) sequences. (A) IFN-γ ELISPOT responses to M. tuberculosis peptides by CD4+ T cells from IKE-immunized, IKEPLUS-immunized, or naive mice. DMSO was used as a negative control. (B) Same as for panel A except showing responses to M. smegmatis peptides. (C) Sequences of both M. tuberculosis and M. smegmatis peptides assayed in panels A and B are shown, along with the location of their first residue (start) in the RplJ protein. The sequences are shown in single-letter amino acid code. With respect to CD4+ T cell responses, the minimal core epitopes for the M. tuberculosis and M. smegmatis sequences were defined as the common residues among the reactive peptides (underlined). Responses of immune CD4+ T cells to the peptides are summarized as the number of positive cells per 106 CD4+ T cells, quantified by an IFN-γ ELISPOT assay, as shown in panels A and B, and values for reactive peptides are indicated by dark shading. Binding affinities of the peptides for I-Ab were also determined, and the IC50s for each peptide (in nanomolar) are listed, with values indicative of significant binding being highlighted by gray shading. Data shown in panels A and B represent mean values and standard errors for triplicate samples. All data shown are representative of results from two independent experiments.
With respect to CD4+ T cell responses, the minimal core epitopes for the TB (LFNAPASQL) and M. smegmatis (LFNAPASQV) sequences were defined as the common residues among the reactive peptides (Fig. 3A and B). Binding affinities of the peptides for I-Ab were determined by using a previously described method (48), and the 50% inhibitory concentrations (IC50s) for each peptide (nanomolar) are listed in Fig. 3C. With respect to binding affinity, the minimal core regions for the TB (FNAPASQLA) and M. smegmatis (FNAPASQVA) sequences were defined as the common residues among the strongest-binding peptides. These core and extended epitope sequences were consistent with data from previously reported analyses of I-Ab-presented epitopes, which found that phenylalanine (F) is a frequent P1 anchor residue for I-Ab binding, with proline (P) and alanine (A) being highly favored residues for positions P4 and P9, respectively (49). With regard to CD4+ T cell responses, a 10-mer core epitope could be optimal, as the T cell receptor (TCR) can engage residues outside (position N-1) the 9-mer core for I-Ab binding. In the case of the RplJ core epitope peptides, leucine (L) could likely occupy the N-1 position. The lower responses to the M. tuberculosis peptides than to the M. smegmatis peptides likely reflect weaker TCR interactions, as the M. tuberculosis peptides bind more strongly to I-Ab than do the M. smegmatis peptides.
Based on protein database analyses (GenBank database using the protein Basic Local Alignment Search Tool [BLAST] algorithm), the RplJ protein and the rplJTB146–155 core epitope are highly conserved (>70% amino acid sequence identity) among mycobacterium, rhodococcus, corynebacterium, and nocardia species. The similarity between the M. tuberculosis RplJ (L10) protein and the L10 proteins in other more distantly related lung pathogens, such as Staphylococcus aureus, Streptococcus pneumoniae, and Haemophilus influenzae, is ∼40%, with no significant similarity at the RplJTB146–155 core epitope. The similarity between the M. tuberculosis RplJ (L10) protein and the L10 protein in Escherichia coli is only 31%, with no significant similarity at the rplJTB146–155 core epitope. The similarity between the M. tuberculosis RplJ protein and the homologous ribosomal protein in humans (P0) is 20%, with no significant similarity at the rplJTB146–155 core epitope. Thus, it is likely that responses to rplJTB146–155 are specific for M. tuberculosis and relatively closely related mycobacteria and are unlikely to be broadly cross-reactive with ribosomes from other bacteria or mammalian cells.
Enriched ribosomes are processed and presented to RplJ-specific CD4+ T cells.
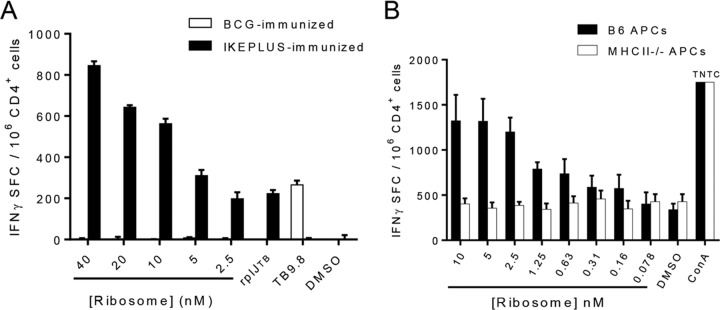
To confirm that the rplJTB146–160 epitope could be generated by the processing of intact mycobacterial ribosomes, we prepared a ribosome-enriched fraction from IKEPLUS using a standard high-performance liquid chromatography (HPLC) ion exchange and size separation method (see Fig. S1 in the supplemental material). Following immunization of mice with IKEPLUS or BCG-Danish, splenic CD4+ T cells were tested for their responses in the presence of APCs to various concentrations of a mycobacterial ribosome-enriched fraction using an IFN-γ ELISPOT assay (Fig. 4A). This showed a strong dose-dependent response in CD4+ T cells from IKEPLUS- but not BCG-immunized mice. Interestingly, the number of IFN-γ-secreting cells detected was several times larger with stimulation by a ribosome-enriched fraction than with an optimal concentration of the rplJTB146–160 peptide, suggesting the recognition of additional ribosome-associated epitopes. Responses to the ribosome-enriched fraction were also dependent on MHC class II expression by APCs (Fig. 4B).
FIG 4.
Dose dependence and MHC class II restriction of CD4+ T cell responses to mycobacterial ribosomes. C57BL/6 mice were immunized with 5 × 107 CFU IKEPLUS i.v. or 5 × 106 CFU BCG-Danish s.c., and CD4+ T cells were isolated from spleens 2 weeks later for an analysis of responses to the ribosome-enriched fraction from IKEPLUS. (A) IFN-γ ELISPOT assay showing the dose dependence of responses of CD4+ T cells from IKEPLUS-immunized mice or BCG-immunized mice to the ribosome-enriched fraction from IKEPLUS. (B) The requirement for MHC class II presentation was assessed by using APCs from wild-type mice or MHC class II−/− mice. Responses of immune CD4+ T cells from IKEPLUS-immunized mice were quantified by an IFN-γ ELISPOT assay. Data shown represent mean values and standard errors for duplicate (A) or quadruplicate (B) samples and are representative of results from three independent experiments. TNTC, too numerous to count.
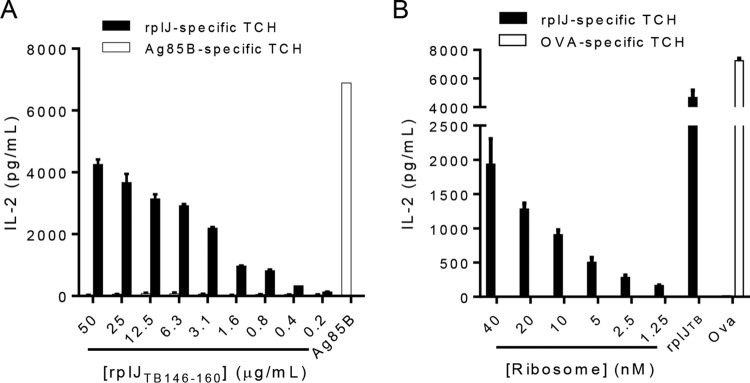
In complementary experiments, we isolated an rplJTB146–160-specific TCH clone (designated RplJ.03) from splenocytes of an IKEPLUS-immunized mouse and used this clone to assess the recognition of mycobacterial ribosomes. This TCH showed dose-dependent responses (measured as IL-2 secretion) to the rplJTB146–160 peptide (Fig. 5A) and ribosomes from IKEPLUS (Fig. 5B). These responses required the presence of MHC class II-positive APCs, and responses were not observed when MHC class II−/− BMDCs were used as APCs (data not shown). Control TCHs specific for MHC class II-restricted M. tuberculosis Ag85B or ovalbumin peptides gave no detectable responses to rplJTB146–160 or the ribosome-enriched fraction.
FIG 5.
Dose response and specificity of RplJ-specific T cell hybridomas. The rplJTB146–160-specific TCH clone “RplJ.03” was used to further establish the dose dependence and specificity of T cell responses to the rplJTB146–160 ribosomal peptide epitope (A) or the ribosome-enriched fraction from IKEPLUS (B). C57BL/6 BMDCs were used as APCs, and responses were quantified by an IL-2 ELISA. Control TCHs specific for MHC class II-restricted M. tuberculosis Ag85B or ovalbumin (OVA) peptides gave no detectable responses to either rplJTB146–160 or the ribosome-enriched fraction from IKEPLUS. Data are mean values and standard errors for duplicate samples and are representative of results from three independent experiments.
Specificity and cytokine profile of ribosome-specific CD4+ T cell responses.
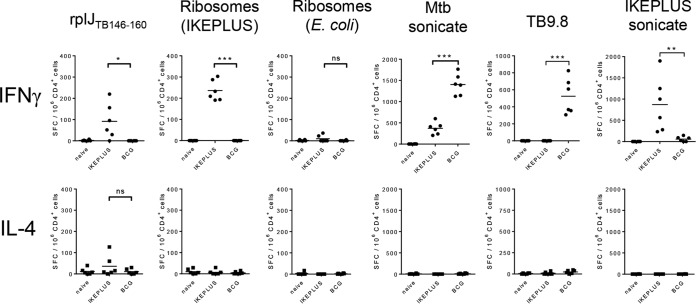
The evaluation of the CD4+ T cell responses to ribosomal antigens following immunization with BCG-Danish or IKEPLUS was extended to include additional antigens as well as other cytokines (Fig. 6). C57BL/6 mice were immunized intravenously (i.v.) with 5 × 107 CFU of either IKEPLUS or BCG-Danish. Two weeks after immunization, CD4+ T cells were purified from splenocytes and tested by an ELISPOT assay for the production of IFN-γ, IL-4, or IL-17A in response to ex vivo stimulation with the rplJTB146–160 peptide or the ribosome-enriched fraction from IKEPLUS. Stimulation with purified E. coli ribosomes was used to assess the specificity of responses, and the IKEPLUS sonicate, M. tuberculosis sonicate, or TB9.8 peptide was used an additional control. CD4+ T cells from mice immunized with IKEPLUS showed significant numbers of IFN-γ-producing cells in response to the rplJTB146–160 peptide and mycobacterial ribosomes. IL-17A-producing cells were not detected (data not shown), and an inconsistent trend in IL-4-producing cells was seen, which did not reach statistical significance (by analysis of variance [ANOVA]). We detected these cytokines in response to the positive control concanavalin A (ConA) (not shown). BCG immunization, as noted above, did not induce IFN-γ responses to the rplJTB146–160 peptide or the ribosome-enriched fraction from IKEPLUS. Ribosomes from E. coli induced only weak IFN-γ production in IKEPLUS-immunized mice, which was not statistically significant (by ANOVA). In summary, these studies showed that the responses to mycobacterial ribosomal epitopes induced by IKEPLUS immunization were predominantly Th1-like and distinguished between ribosomes from different microbial sources.
FIG 6.
Specificity and cytokine profile of ribosome-specific CD4+ T cell responses. C57BL/6 mice were immunized i.v. with 5 × 107 CFU of IKEPLUS or BCG-Danish. Two weeks later, CD4+ T cells were purified from splenocytes and tested by an ELISPOT assay for the production of IFN-γ or IL-4, as well as IL-17A (not shown), in response to ex vivo stimulation with the indicated antigens (the rplJTB146–160 peptide [10 μg/ml], the ribosome-enriched fraction from IKEPLUS [5 nM], purified E. coli ribosomes [5 nM], the M. tuberculosis sonicate [10 μg/ml], the TB9.8 peptide [10 μg/ml], or the IKEPLUS sonicate [10 μg/ml]). Responses that were significantly different between IKEPLUS- and BCG-immunized animals are indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.0001; ns, not significant (as determined by ANOVA). Positive-control wells stimulated with concanavalin A (5 μg/ml) demonstrated the ability of the assays to detect all three cytokines (not shown). Symbols represent cells harvested from individual mice (n = 6), and data shown are combined data from two independent experiments.
Prevalence of ribosome-specific CD4+ T cell responses following challenge with virulent M. tuberculosis.
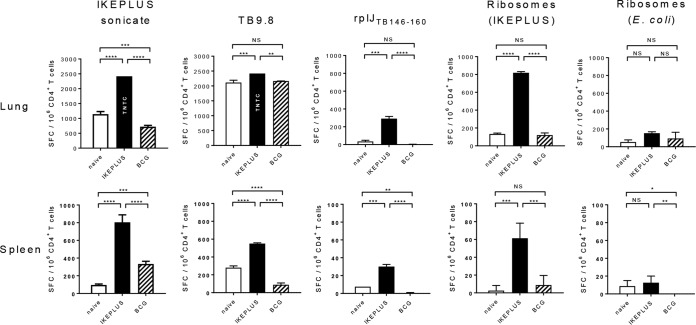
CD4+ T cell responses to ribosomal antigens in mice immunized with BCG-Danish or IKEPLUS were examined after challenge with M. tuberculosis strain H37Rv (Fig. 7). Mice were immunized with IKEPLUS or BCG-Danish and challenged 10 weeks later with a low-dose aerosol delivery of virulent M. tuberculosis (H37Rv) as described previously (50). Six weeks after challenge, CD4+ T cells were purified from spleens and lungs and tested by an ELISPOT assay for the production of IFN-γ in response to ex vivo stimulation with the TB9.8 peptide, the rplJTB146–160 peptide, or the ribosome-enriched fraction from IKEPLUS. Stimulation with purified E. coli ribosomes was used to assess the specificity of responses, and the IKEPLUS sonicate was used as an additional control. CD4+ T cells from the spleen and lung of mice immunized with IKEPLUS showed significant numbers of IFN-γ-producing cells in response to both the rplJTB146–160 peptide and mycobacterial ribosomes following challenge with M. tuberculosis. In contrast, spleen and lung CD4+ T cells from either unimmunized mice or those immunized with BCG did not respond significantly to the rplJTB146–160 peptide or mycobacterial ribosomes. Ribosomes from E. coli induced only weak responses that were not significantly increased in IKEPLUS- or BCG-immunized mice compared to those in unimmunized mice. Levels of M. tuberculosis bacilli (CFU) were not significantly diminished in the lungs of BCG- or IKEPLUS-immunized mice in comparison to unimmunized mice (see Fig. S2 in the supplemental material), likely reflecting the relatively early time postchallenge at which the animals were sacrificed in this experiment compared to previous studies showing reductions in tissue bacterial loads following IKEPLUS immunization (46). However, these results indicate that IFN-γ-producing CD4+ T cells specific for rplJTB146–160 and potentially other ribosome-associated antigens accumulate in tissues after M. tuberculosis challenge and are a prominent population in the lungs of mice that have been immunized to prime such responses.
FIG 7.
Ribosome-specific CD4+ T cell responses following M. tuberculosis challenge. Mice (C57BL/6) were immunized with 5 × 107 CFU IKEPLUS i.v. or 107 CFU BCG-Danish s.c. or sham immunized with saline s.c. (naive). Ten weeks later, immunized and naive mice were challenged with a low-dose aerosol delivery of M. tuberculosis strain H37Rv (∼100 CFU). Six weeks after challenge, CD4+ T cells were purified from spleens and lungs and tested by an ELISPOT assay for the production of IFN-γ in response to ex vivo stimulation with the indicated antigens (the IKEPLUS sonicate [10 μg/ml], the TB9.8 peptide [10 μg/ml], the rplJTB146–160 peptide [10 μg/ml], the ribosome-enriched fraction from IKEPLUS [5 nM], or purified E. coli ribosomes [5 nM]). Bars show mean numbers of IFN-γ spot-forming cells (SFC) in response to the indicated antigens for groups of 4 mice, with background values (mean number of IFN-γ spot-forming cells per 106 CD4+ T cells in response to culture medium alone) being subtracted, and error bars indicate 1 standard deviation. Background values (mean ± 1 standard deviation) were as follows: 8 ± 9 spot-forming cells for naive spleen, 11 ± 9 spot-forming cells for IKEPLUS spleen, 36 ± 9 spot-forming cells for BCG spleen, 120 ± 6 spot-forming cells for naive lung, 84 ± 19 spot-forming cells for IKEPLUS lung, and 68 ± 25 spot-forming cells for BCG lung. Responses that were significantly different between IKEPLUS-immunized, BCG-immunized, and unimmunized animals are indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (as determined by ANOVA). TNTC, too numerous to count (estimated to be 2,500 spot-forming cells).
High prevalence of ribosome-specific CD4+ T cells following immunization with IKEPLUS.
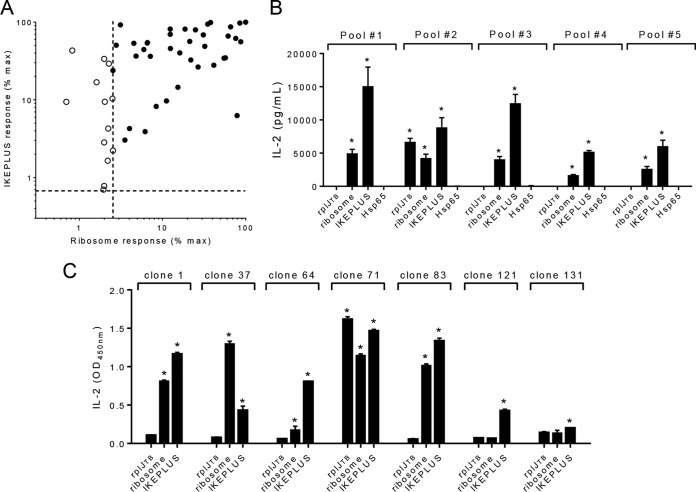
To examine the magnitude of the ribosome-specific CD4+ T cell response that develops following immunization with IKEPLUS, a TCH library was constructed by fusing splenic CD4+ T cells from IKEPLUS-immunized C57BL/6 mice with mouse thymoma cells. The TCHs that initially grew from this fusion, representing 187 separate culture wells, were tested for reactivity to the IKEPLUS sonicate in the presence of C57BL/6 BMDCs by an IL-2 enzyme-linked immunosorbent assay (ELISA) (data not shown). Positive clones were subjected to limiting dilution at a density of 0.5 cells per well to ensure the development of cell lines derived from single cells. Hybridomas that grew out of limiting dilution were again assayed as described above for reactivity to the IKEPLUS sonicate, which yielded 51 IKEPLUS-reactive TCHs, and these TCHs were further screened for reactivity against the ribosome-enriched fraction from IKEPLUS. The 51 clones were normalized based on their production of IL-2 in response to the IKEPLUS sonicate and separately by IL-2 production in response to the ribosome-enriched fraction from IKEPLUS (Fig. 8A). Of the 51 IKEPLUS TCHs, 38 (74.5%) also showed significant responses to the ribosome-enriched fraction from IKEPLUS (Fig. 8A).
FIG 8.
Predominance of ribosome-specific CD4+ T cells in IKEPLUS-immunized mice. A TCH library was constructed by fusing CD4+ T cells isolated from the spleens of IKEPLUS-immunized C57BL/6 mice with mouse thymoma cells. TCHs were screened for reactivity to the IKEPLUS sonicate, and a total of 51 IKEPLUS-specific clones were isolated for analysis (see the text for details). (A) Normalized responses of TCHs to the IKEPLUS sonicate (y axis) and the ribosome-enriched fraction from IKEPLUS (x axis). The subset of TCHs screened that responded to the IKEPLUS sonicate at a level of 3 standard deviations above the values for replicate wells with no antigen were included in this analysis. Dotted lines represent 3 standard deviations above values for replicate wells with no antigen. Each symbol represents data for a single TCH. Open symbols correspond to those TCHs that responded significantly to only the IKEPLUS sonicate, and filled symbols indicate TCHs that responded significantly to both the IKEPLUS sonicate and the ribosome-enriched fraction from IKEPLUS. Of the 51 IKEPLUS-specific TCHs, 38 were also specific for the ribosome-enriched fraction from IKEPLUS (74.5%). (B) The 38 IKEPLUS ribosome-responsive TCHs were randomly assigned to five pools for further screening. All pools responded to the IKEPLUS sonicate and the ribosome-enriched fraction from IKEPLUS, as determined by an IL-2 release assay, but only pool 2 responded to the rplJTB146–160 peptide. None of the ribosome-specific TCH pools responded to the purified hsp65 protein from M. bovis, which was included as an additional control. Asterisks indicate >3 standard deviations above values for replicate wells with no antigen. (C) Retesting individual clones from pool 2 in an IL-2 release assay revealed that one out of seven TCH clones was responsive to the rplJTB146–160 peptide. Asterisks indicate >3 standard deviations above values for replicate wells with no antigen. Data in panels B and C are mean values and standard errors for duplicate samples.
To confirm the high frequency of ribosome specificity, the 38 IKEPLUS ribosome-specific clones were randomly assigned to five pools for further screening. Since hsp65 was identified as a potential contaminant that copurified with ribosomes (data not shown), we assessed whether hsp65 contributed to CD4+ T cell responses to our ribosome preparations. Using the purified hsp65 protein from M. bovis, we found that none of the IKEPLUS ribosome-specific clones responded to the hsp65 protein (Fig. 8B). IKEPLUS ribosome-specific clones were also screened for reactivity to the rplJTB146–160 epitope (Fig. 8B). Pool 2 of the IKEPLUS ribosome-specific clones reacted to the rplJTB146–160 epitope (Fig. 8B). The seven individual clones comprising this pool were then screened for reactivity to the rplJTB146–160 epitope (Fig. 8C), revealing a single rplJTB146–160-reactive clone. Thus, the great majority of CD4+ T cells that were reactive with IKEPLUS ribosomes in our panel were directed at epitopes distinct from the rplJTB146–160 epitope or were potentially reactive with other ribosome-associated proteins.
Identification of additional mycobacterial ribosomal protein epitopes for CD4+ T cells.
To screen for additional ribosomal protein CD4+ T cell epitopes in IKEPLUS-immunized mice, we synthesized peptides with the highest predicted binding affinity for I-Ab over the entire length of the RplJ protein of M. tuberculosis and M. smegmatis. In addition, we carried out a similar in silico analysis of the sequence of a representative protein from the small ribosomal subunit RpsA (S1 protein, a product of the M. tuberculosis Rv1630 gene and the M. smegmatis MSMEG_3833 gene). Two weeks after immunization with IKEPLUS or BCG-Danish, CD4+ T cells were purified from splenocytes and tested by an ELISPOT assay for the production of IFN-γ in response to ex vivo stimulation with nine RplJ peptides and five RpsA peptides with a predicted strong affinity for I-Ab. Whereas BCG-immune CD4+ T cells did not respond to these peptides (data not shown), IKEPLUS-immune CD4+ T cells reacted strongly to the rplJMS146–160 peptide (KAAGLFNAPASQVAR), the RplJ epitope described above, and the overlapping rplJMS151–165 peptide (FNAPASQVARLAAAL) (Fig. 9). IKEPLUS-immune CD4+ T cells also reacted strongly to the rplJMS11–25 peptide (ADIAEQFKASTATVV) and the overlapping rplJMS16–30 peptide (QFKASTATVVTEYRG), representing an additional epitope within the RplJ protein (Fig. 9). In examining the peptides derived from the RpsA protein, IKEPLUS-immune CD4+ T cells were found to react strongly to the rpsAMS431–445 peptide (QMEKFAAAEAEAANA) (Fig. 9). These studies showed that the CD4+ T cell responses to the mycobacterial ribosome induced by IKEPLUS immunization included epitopes derived from both large and small ribosomal subunit proteins.
FIG 9.
Identification of CD4+ T cell targets within the RplJ and RpsA ribosomal proteins. Peptides were screened for in silico-predicted binding affinity for I-Ab. These peptides included nine peptides derived from the RplJ protein and five peptides derived from the RpsA protein. (A) IFN-γ ELISPOT responses to selected RplJ and RpsA peptides by CD4+ T cells from IKEPLUS-immunized mice. CD4+ T cells were isolated at 2 weeks postimmunization from spleens of C57BL/6 mice immunized i.v. with 5 × 107 CFU IKEPLUS and assayed by an ELISPOT assay for the production of IFN-γ in response to the 14 peptides (10 μg/ml for each peptide). Asterisks indicate >3 standard deviations above values for replicate wells with no antigen. No responses to individual peptides were detected by using CD4+ T cells from BCG-immunized mice (not shown). Data shown represent mean values and standard deviations for duplicate samples. Data shown are representative of results from two independent experiments. (B) Sequences of both M. tuberculosis (TB) and M. smegmatis (MS) peptides along with the location of their first residue (start) in the RplJ or RpsA protein. The sequences are shown in single-letter amino acid code. Shaded cells represent peptides that were synthesized and analyzed based on the I-Ab consensus percentile. Responses of immune CD4+ T cells to the peptides are summarized as the number of positive cells per 106 CD4+ T cells, quantified by an IFN-γ ELISPOT assay, as shown in panel A.
DISCUSSION
In comparison to immunization with the currently used BCG vaccine, immunization of mice with the genetically modified M. smegmatis strain IKEPLUS was previously shown to enhance survival after M. tuberculosis challenge and led to the development of a Th1-like response thought to be critical for defense against M. tuberculosis (46). While protection against M. tuberculosis afforded by immunization with IKEPLUS was shown to be dependent on antigen-specific CD4+ T cells, the specificities of CD4+ T cells in IKEPLUS-immunized mice remained to be elucidated (46). In the present study, we used immunization with the M. smegmatis IKE or IKEPLUS strain as an approach to identify novel, potentially protective, CD4+ T cell epitopes beyond those that stimulate responses following BCG vaccination or M. tuberculosis infection. Responses of CD4+ T cells generated within 2 weeks of immunization were analyzed, in order to focus on immunodominant antigens that are likely to predominate in this relatively early period after vaccination. Using a synthetic peptide library consisting of 880 nonoverlapping 15-mers representing predicted I-Ab-presented epitopes with homology between M. tuberculosis and IKEPLUS, we identified one specific epitope of IKEPLUS-immune CD4+ T cells, rplJTB146–160, derived from the RplJ protein of the mycobacterial 50S ribosomal subunit. The CD4+ T cell responses to the rplJTB146–160 epitope, as well as those to ribosomes from IKEPLUS, were found to be specific, MHC class II restricted, and dominated by the production of IFN-γ, suggesting the development of a Th1-like response thought to be critical for defense against M. tuberculosis.
We consistently observed that CD4+ T cells from mice immunized with IKEPLUS or with other M. smegmatis strains (IKE or the parent strain mc2155) responded significantly to the rplJTB146–160 epitope, while CD4+ T cells from mice immunized with a standard BCG vaccine strain did not. One hypothesis as to why this differential response to a ribosomal component occurred is that fast-growing mycobacterial strains (like M. smegmatis) have more ribosomes per bacterium than do slow-growing mycobacteria (such as BCG or M. tuberculosis), thus leading to more availability of the RplJ protein for processing and presentation by the host immune system. Alternatively, the failure of BCG, even when administered intravenously in large doses (Fig. 6), to elicit detectable responses to the RplJ epitope may reflect the presence of immune evasion mechanisms that block the induction of ribosome-specific responses in virulent mycobacterial species such as M. bovis and M. tuberculosis. This raises the possibility that directed immunization against mycobacterial ribosomal components could prime the host immune response to recognize ribosomal epitopes when challenged with virulent, ribosome-limited mycobacteria such as M. tuberculosis. Indeed, CD4+ T cells from the lungs and spleens of IKEPLUS-immunized mice responded significantly better to mycobacterial ribosomes and the rplJTB146–160 peptide than did CD4+ T cells from BCG-immunized or naive mice following M. tuberculosis challenge (Fig. 7). This suggests that ribosome-specific CD4+ T cells are recruited to and expand in the lungs of M. tuberculosis-challenged mice when animals have been primed with a suitable form of immunization, such as with the genetically modified M. smegmatis strain IKEPLUS. These data also extend previous studies in which the presence of RplJ- and ribosome-specific CD4+ T cells was assayed at 2 weeks postimmunization, here demonstrating that ribosome-specific CD4+ T cells can be recalled when challenged 10 weeks after immunization. Thus, vaccination strategies directed at priming CD4+ T cells against the rplJTB146–160 epitope or potentially other ribosome-associated determinants could be considered an approach to enhance vaccine strategies against M. tuberculosis.
Ribosome vaccines are intriguing possibilities for immunization, as they are thought to be more immunogenic, less toxic, and more defined than whole-cell vaccines (51, 52). Immunization with ribosomes could confer some cross-serotype protection, reducing the number of vaccines to be developed and delivered to patients (51, 52). Early studies, mainly from the 1960s and 1970s, demonstrated the potential of ribosomal components as protective vaccines. Studies examining protection afforded by ribosomes against M. tuberculosis in mice (53), Pasteurella multocida in mice and chickens (54), or Pseudomonas aeruginosa in mice (55) concluded that the immunogenic portion of the ribosome critical for protection was the rRNA. However, studies of ribosome vaccination against Haemophilus influenzae in mice and rabbits (56), Streptococcus pyogenes in mice (57), and Neisseria meningitidis in mice (58) identified the ribosomal protein as being critical for protection. These studies were somewhat technologically hampered, allowing only a limited determination of gross fractions (rRNA versus protein) as the immunological component of the ribosome, and no T cell epitopes were identified.
More contemporary studies have examined the potential of protective vaccines composed of specific ribosomal proteins. Immunization with the ribosomal protein L7/L12 from Brucella abortus combined with an adjuvant led to a T cell response and was protective against brucellosis in mice (59) and cattle (60). When administered to mice in the presence of a Th1 adjuvant in a prime-boost regimen, the combined recombinant ribosomal proteins L5 and L3 from Leishmania major were found to enhance Th1-like immune responses and provide cross-protection against Leishmania amazonensis and L. chagasi (61). Interestingly, a protein with homology to RplJ, the ribosomal L10-like protein from Flavobacterium psychrophilum, when administered with an adjuvant, was found to be protective in immature rainbow trout (62). While these studies indicate the potential of ribosomal proteins in protective vaccines, the mechanisms of protection were not defined, and no T cell epitopes were described.
It has been suggested that components of the ribosome itself (rRNA) or closely associated with the ribosome (recently translated protein and cell surface proteins) may act as adjuvants (51, 52). A complex particle containing rRNA, proteins, and other closely associated molecules could be highly immunogenic and thus stimulate cell-mediated as well as humoral immune responses (52). The idea of the ribosome as a self-adjuvanting, immunogenic particle for immunization is intriguing. However, the mechanism of recognition, processing, and presentation of ribosomes following infection or immunization is unclear. In addition, the kinetics of the development of ribosome-specific T cell responses following infection, and their tropism for various tissues such as the lung, have not yet been determined. Studies are under way to gain insight into the cells and pathways involved in the immune response to ribosomes.
A notable result of the present study was the observation that in the screening of a TCH library generated from IKEPLUS-immunized CD4+ T cells, a substantial majority (74.5%) of the IKEPLUS-responsive clones were also reactive to ribosomes from IKEPLUS. Only one of these ribosome-specific clones was specific for the rplJTB146–160 epitope described here, suggesting that additional components of the mycobacterial ribosome are immunogenic. To extend our examination of potential immunogenic ribosomal epitopes, we synthesized and screened the 15-mer peptides with the best predicted I-Ab-presented binding affinity from the entire RplJ protein as well as from the RpsA protein, a representative protein of the small ribosomal subunit. RpsA was chosen for further examination, as it is the largest of the ribosomal small-subunit proteins (481 amino acids [aa]), the sequence homology among mycobacteria is between 85.4% and 100% (63), and RpsA is thought to be the target of pyrazinamide, one of the commonly used first-line drugs for the treatment of tuberculosis (64). This identified one additional specific epitope of IKEPLUS-immune CD4+ T cells within the RplJ protein, rplJMS11–25, and one specific epitope derived from the RpsA protein, rpsAMS431–445.
The mycobacterial ribosome is a complex particle made up of rRNA, 57 ribosomal proteins, and other associated molecules. Our studies have identified a number of specific epitopes for CD4+ T cell responses and strongly suggest the presence of additional immunogenic ribosomal protein epitopes in mycobacteria. Although we have not yet determined that ribosome-specific CD4+ T cells contribute to protective immunity against M. tuberculosis, the identification of a broader array of immunogenic ribosome-associated CD4+ T cell epitopes has the potential to provide new targets for vaccination that are distinct from the immunodominant antigens presented during BCG vaccination or natural M. tuberculosis infection. In this regard, CD4+ T cell epitopes derived from the mycobacterial ribosome may prove successful either as completely novel immunization formulations or by enhancing current vaccine candidates to provide broader immune recognition and protection.
MATERIALS AND METHODS
Mice.
Six-week-old female C57BL/6J mice were purchased from The Jackson Laboratories (Bar Harbor, ME) and allowed to acclimate for 1 week before experiments. Mice were housed under specific-pathogen-free conditions in compliance with the procedures and regulations established by the Albert Einstein College of Medicine Institute for Animal Studies and Biosafety Committees. All experiments using animals were performed according to protocols approved by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee (IACUC).
Mycobacterial cultures and infections.
Mycobacterium smegmatis (strain mc2155) and the genetically modified variants IKE and IKEPLUS were described previously (46) and were grown in liquid cultures in Sauton medium (65). BCG-Danish was obtained from the Statens Serum Institute (Copenhagen, Denmark) and was grown in Middlebrook 7H9 medium (Difco Laboratories, BD Diagnostic Systems, Sparks, MD) with oleic acid-albumin-dextrose-catalase (OADC enrichment; Difco Laboratories, BD Diagnostic Systems, Sparks, MD) and 0.05% tyloxapol (Sigma-Aldrich, St. Louis, MO). Bacteria were grown from low-passage-number frozen stocks, cultured to mid-log phase, and then frozen in medium with 5% glycerol at −80°C. Bacteria were thawed, washed, resuspended in phosphate-buffered saline (PBS) containing 0.05% Tween 80, and sonicated to obtain a single-cell suspension prior to infection. Mice were infected with 5 × 107 CFU IKE or IKEPLUS via the tail vein, 107 CFU mc2155 via the tail vein, 5 × 106 CFU BCG-Danish subcutaneously (s.c.) in the scruff of the neck, or 5 × 107 CFU BCG-Danish via the tail vein.
Preparation of bacterial sonicates.
IKEPLUS cells grown as described above, or irradiated M. tuberculosis H37Rv cells (BEI Resources, Manassas, VA), were sonicated in lysis buffer (30 mM Tris-HCl, 0.05% tyloxapol, cOmplete mini EDTA-free protease inhibitor cocktail [Sigma] [pH 8.0]) for 20 min on ice. The culture was centrifuged at 3,000 × g for 12 min at 4°C, the supernatant was retained, and the pellet was resuspended in lysis buffer and sonicated as described above. Supernatants from three sonication/centrifugation cycles were combined and centrifuged at 30,000 × g for 30 min at 4°C. The supernatant was filtered through a 0.22-μm filter, and the protein concentration was determined by using a bicinchoninic acid (BCA) protein assay kit (ThermoFisher Scientific, Grand Island, NY).
Generation and screening of the IKEPLUS peptide library.
An overview of the process used to generate the IKEPLUS peptide library is shown in Fig. 1A. All possible 15-mer peptides were determined from the genome sequences of the Mycobacterium smegmatis (mc2155) and M. tuberculosis (H37Rv) strains used to generate IKEPLUS (a possible 3,166,147 peptides). Fifteen-mer peptides with ≤2 substitutions between the M. smegmatis and M. tuberculosis strains were included for further consideration (285,532 peptides). These peptides were then ranked for their predicted binding affinity for MHC class II (I-Ab) by using the consensus method (66). The 1% of peptides with the strongest predicted binding to I-Ab were selected, and from this set, redundant peptides overlapping by >8 residues were excluded. This yielded a library of 880 unique peptides, which were synthesized and arranged into 45 pools. Peptides were synthesized as crude material (estimated 70 to 80% purity) on a small scale (∼1 mg) by Mimotopes (Clayton, Victoria, Australia), reconstituted in dimethyl sulfoxide (DMSO), and used at a final concentration of 2.5 μg/ml per peptide within peptide pools or 10 μg/ml for peptides assayed individually.
Generation and screening of peptides from RplJ and RpsA ribosomal proteins.
The sequences from the RplJ and RpsA ribosomal proteins from the Mycobacterium smegmatis (mc2155) and M. tuberculosis (H37Rv) strains were collected from the UniProt database. The sequences were then clustered at the 30% amino acid sequence identity level to group similar sequences into separate clusters by using the epitope cluster analysis tool available at the IEDB Tools website (67). Each cluster of sequences was then aligned separately by using the ClustalW algorithm as implemented in MEGA tool (68). For these clusters, 15-mer peptides overlapping by 10 amino acids were generated, and duplicates were removed. The remaining peptides were then ranked based on their predicted binding affinity for MHC class II (I-Ab). The MHC binding affinity was predicted by using the IEDB analysis resource consensus tool (66, 69). Peptides with an IEDB consensus percentile rank of ≤10.0 were selected, and variants (peptides from the same sequence position but having amino acid mismatches) were removed by eliminating the variant with the higher consensus percentile. This resulted in nine peptides from RplJ and five from RpsA. Peptides were synthesized as crude material (estimated 70 to 80% purity) on a small scale (∼1 mg) by A&A Labs (San Diego, CA). Peptides were reconstituted in DMSO and used at a final concentration of 10 μg/ml per peptide.
Purification of CD4+ T cells.
CD4+ T cells were purified from pooled splenocytes by positive selection using magnetic antibody cell separation (MACS) CD4 (L3T4) Microbeads according to the manufacturer's instructions (Miltenyi Biotech, Auburn, CA).
MHC purification and binding assays.
The purification of H-2 I-Ab MHC class II molecules by affinity chromatography and the performance of assays based on the inhibition of binding of a high-affinity radiolabeled peptide to quantitatively measure peptide binding were performed as detailed previously (48). Briefly, the mouse B cell lymphoma line LB27.4 was used as a source of MHC molecules. A high-affinity radiolabeled peptide (0.1 to 1 nM peptide ROIV [sequence YAHAAHAAHAAHAAHAA]) was coincubated at room temperature with purified MHC in the presence of a cocktail of protease inhibitors and an inhibitor peptide. Following a 2-day incubation, MHC-bound radioactivity was determined by capturing MHC-peptide complexes on monoclonal antibody (MAb) (Y3JP)-coated Lumitrac 600 plates (Greiner Bio-One, Frickenhausen, Germany), and bound radioactivity was measured as counts per minute by using the TopCount microscintillation counter (Packard Instrument Co., Meriden, CT). The concentration of the peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated. Under the conditions used, where [radiolabeled peptide] < [MHC] and IC50 ≥ [MHC], the measured IC50s are reasonable approximations of the true Kd (equilibrium constant) values. Each competitor peptide was tested at six different concentrations covering a 100,000-fold range and in three or more independent experiments. As a positive control, the unlabeled version of the radiolabeled probe was also tested in each experiment.
Generation and screening of IKEPLUS CD4+ T cell hybridomas.
CD4+ TCHs were constructed by using a previously reported method (70). Briefly, C57BL/6 mice were immunized with 5 × 107 CFU IKEPLUS i.v. and boosted 4 weeks later with 108 CFU IKEPLUS i.v. Two days after boosting, CD4+ T cells were purified from splenocytes and fused by using polyethylene glycol 1450 (PEG 1450; ATCC) (50% [wt/vol] in unsupplemented Dulbecco's modified Eagle's medium [DMEM] [ThermoFisher Scientific]) with the BW5147.G.14 mouse lymphoma cell line (ATCC TIB 48). Following fusion, cells were plated at 105 activated CD4+ T cells per well in 96-well flat-bottom plates, followed by selection medium containing a hypoxanthine-aminopterin-thymidine (HAT) supplement (Life Technologies, ThermoFisher Scientific, Grand Island, NY). Seven days after fusion, all wells showed visible growth and were expanded in DMEM containing a hypoxanthine-thymidine (HT) supplement (Life Technologies). The resulting TCHs were screened for reactivity to a sonicate of IKEPLUS by an IL-2 ELISA. IKEPLUS-specific TCHs were then subjected to limiting dilution by plating a mean of 0.5 cells per well in 96-well flat-bottom plates to obtain single-cell-derived clonal lines. Clones that grew from these cultures were again screened for reactivity to IKEPLUS as well as to ribosomes from IKEPLUS and the recombinant M. bovis hsp65 protein (StressMarq Biosciences Inc., Victoria, BC, Canada).
Enrichment of ribosomes from IKEPLUS.
Ribosomes were enriched from IKEPLUS by using slight modifications of a previously reported protocol (71). Briefly, bacteria were cultured to mid-log phase, harvested by centrifugation at 10,000 × g for 10 min at 4°C, resuspended in lysis buffer (buffer A [70 mM KCl, 10 mM MgCl2, 10 mM Tris-HCl {pH 7.4}]), and frozen at −80°C (71). Just before ribosome enrichment, bacteria were thawed, lysed by two cycles of homogenization at 15,000 lb/in2 with an EmulsiFlex high-pressure homogenizer (Avestin Inc., Ottawa, ON, Canada), and clarified by centrifugation at 30,000 × g for 45 min at 4°C. The pellet was discarded, and the clarified lysate was passed through a 0.22-μm filter. Ribosomes were then enriched from the clarified lysate by HPLC (Agilent Technologies, Santa Clara, CA) using a quaternary amine (QA) monolithic disk ion exchange column (BIA Separations Inc., Wilmington, DE). Isolations were performed at a flow rate of 2 ml/min, using lysis buffer (buffer A) and lysis buffer containing 1 M NaCl (buffer B). Eluants from sequential elutions with 35%, 47%, and 100% buffer B were collected and further analyzed (71). Ribosome-containing fractions were concentrated by using Amicon Ultra-15 centrifugal filter units (EMD Millipore, Billerica, MA) with a 100,000-molecular-weight (100K) cutoff. The absorbance was measured at 260 nm and 280 nm by using a NanoDrop spectrophotometer (ThermoFisher Scientific). The A260/A280 ratio was found to be within 1.9 to 2.0, within the range of 1.8 to 2.0 expected for good-quality RNA. The concentration was determined by using a previously reported molar extinction coefficient at 260 nm (E260) for E. coli ribosomes of 145 ± 3 (72) and the reported predicted molecular weight of the M. smegmatis ribosome of 2.43 × 106 g/mol (73).
ELISPOT assay.
A total of 2 × 105 purified CD4+ T cells isolated from pooled immunized spleens were plated onto 96-well flat-bottom nitrocellulose ELISPOT plates (Millipore) coated with 4 μg/ml anti-IL-4 (BD Pharmingen, San Diego, CA), 4 μg/ml anti-IL-17A (BD Pharmingen), or 10 μg/ml anti-IFN-γ MAb (BD Pharmingen or Mabtech Inc., Cincinnati, OH). CD4+ T cells were incubated for 20 h in the presence of 105 lipopolysaccharide (LPS) blasts or naive T cell-depleted splenocytes as a source of APCs plus a ribosome-enriched fraction from IKEPLUS, peptide pools, or individual peptides. ConA (Sigma) was used as a positive control. After 20 h of incubation at 37°C, plates were washed with PBS containing 0.05% Tween 20. Wells were incubated with 2 μg/ml biotinylated anti-IL-4, 2 μg/ml biotinylated anti-IL-17A, or 1 μg/ml anti-IFN-γ MAb (BD Pharmingen) for 2 h at 37°C. Plates were washed as described above and then incubated with streptavidin-alkaline phosphatase (Life Technologies) for 1 h at 37°C. Plates were washed as described above and developed by the addition of 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) and nitroblue tetrazolium chloride (NBT) tablets (Sigma). Plates were counted with the aid of computer-assisted image analysis by using an AID ELISPOT reader (Autoimmun Diagnostika GmbH, Straßberg, Germany). The net number of cytokine-producing CD4+ T cells per 106 CD4+ T cells was determined as follows: ([number of spots against relevant peptide/antigen] − [number of spots against irrelevant control]) × 5.
Murine IL-2 ELISA.
Together with 105 bone marrow-derived dendritic cells, 105 ribosome-specific CD4+ TCH cells were cultured in the presence of the IKEPLUS sonicate, ribosomes from IKEPLUS, synthetic peptides, or a commercially available hsp65 protein from M. bovis (StressMarq). After 20 h of incubation at 37°C, culture supernatants were harvested, and the levels of IL-2 were quantitated by a capture ELISA as previously described (74). Absorbance values were determined at 450 nm on a Wallac 1420 Victor2 microplate reader (PerkinElmer, Waltham, MA), and values for IL-2 were determined by using a standard curve generated by using known concentrations of purified recombinant murine IL-2 (BD Biosciences).
M. tuberculosis challenge.
Mice were immunized with 5 × 107 CFU IKEPLUS i.v. or 107 CFU BCG-Danish s.c. Ten weeks later, immunized and naive mice were exposed to a low-dose aerosol of wild-type M. tuberculosis strain H37Rv. Briefly, H37Rv was cultured at 37°C in Middlebrook 7H9 medium containing OADC, 0.5% glycerol, and 0.05% tyloxapol to an A600 of 0.4 to 0.8. A total of 2 × 106 CFU/ml of bacteria in PBS-Tween plus 0.05% (vol/vol) antifoam Y-30 (Sigma) was loaded into a nebulizer attached to an airborne infection system (University of Wisconsin Mechanical Engineering Workshop). Mice were exposed to aerosolized H37Rv for 20 min, and ∼100 H37Rv bacteria were deposited into the lungs of each animal. The inoculum dose was confirmed by plating of whole-lung homogenates at 24 h post-aerosol exposure, with quantification of CFU after 4 weeks of incubation at 37°C. Lungs, livers, and spleens were harvested at 6 weeks postchallenge, and single-cell suspensions were prepared. For CFU quantification, samples were maintained separately (six mice per group), but for ELISPOT assays, samples were pooled within groups (four mice per group). CD4+ T cells were purified as described above by using MACS selection, and an IFN-γ ELISPOT assay was performed as described above. CD4+ T cells were incubated for 20 h in the presence of 105 naive T cell-depleted splenocytes as a source of APCs, plus a ribosome-enriched fraction from IKEPLUS, the rplJTB146–160 peptide, the TB9.8 peptide, and the IKEPLUS sonicate. ConA was used as a positive control.
Statistical analyses.
For ELISPOT screening of synthetic peptide pools or individual peptides, responses were considered positive if the net number of spot-forming cells per million cells was >20, the stimulation index (sample/DMSO control) was >2, and the P value was <0.05 (as determined by Student's t test for the means of triplicate values for the response against relevant pools or peptides versus the DMSO control). For ELISPOT assays comparing three or more immunization conditions, responses were considered significant if the P value was <0.05 (one-way ANOVA with Bonferroni correction for multiple-comparisons for means of triplicate or quadruplicate values against relevant antigens).
Supplementary Material
ACKNOWLEDGMENTS
Funding sources were NIH grant AI063537 to William R. Jacobs, Jr., and Steven A. Porcelli; NIH grant AI093649 to Steven A. Porcelli; contract HHSN272200900044C to Alessandro Sette; and NIH training grant T32 AI07506 to Alison J. Johnson.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.01023-16.
REFERENCES
- 1.World Health Organization. 2015. Global tuberculosis report 2015. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/en/ Accessed 9 November 2016. [Google Scholar]
- 2.Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, Mosteller F. 1994. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA 271:698–702. doi: 10.1001/jama.1994.03510330076038. [DOI] [PubMed] [Google Scholar]
- 3.Fine PE. 1995. Variation in protection by BCG: implications of and for heterologous immunity. Lancet 346:1339–1345. doi: 10.1016/S0140-6736(95)92348-9. [DOI] [PubMed] [Google Scholar]
- 4.Gandhi NR, Shah NS, Andrews JR, Vella V, Moll AP, Scott M, Weissman D, Marra C, Lalloo UG, Friedland GH, Tugela Ferry Care and Research Collaboration. 2010. HIV coinfection in multidrug- and extensively drug-resistant tuberculosis results in high early mortality. Am J Respir Crit Care Med 181:80–86. doi: 10.1164/rccm.200907-0989OC. [DOI] [PubMed] [Google Scholar]
- 5.Kalo D, Kant S, Srivastava K, Sharma AK. 2015. Pattern of drug resistance of Mycobacterium tuberculosis clinical isolates to first-line antituberculosis drugs in pulmonary cases. Lung India 32:339–341. doi: 10.4103/0970-2113.159561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, Ziazarifi AH, Hoffner SE. 2009. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest 136:420–425. doi: 10.1378/chest.08-2427. [DOI] [PubMed] [Google Scholar]
- 7.Ng TW, Saavedra-Avila NA, Kennedy SC, Carreno LJ, Porcelli SA. 2015. Current efforts and future prospects in the development of live mycobacteria as vaccines. Expert Rev Vaccines 14:1493–1507. doi: 10.1586/14760584.2015.1089175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues LC, Pereira SM, Cunha SS, Genser B, Ichihara MY, de Brito SC, Hijjar MA, Dourado I, Cruz AA, Sant'Anna C, Bierrenbach AL, Barreto ML. 2005. Effect of BCG revaccination on incidence of tuberculosis in school-aged children in Brazil: the BCG-REVAC cluster-randomised trial. Lancet 366:1290–1295. doi: 10.1016/S0140-6736(05)67145-0. [DOI] [PubMed] [Google Scholar]
- 9.Abel B, Tameris M, Mansoor N, Gelderbloem S, Hughes J, Abrahams D, Makhethe L, Erasmus M, de Kock M, van der Merwe L, Hawkridge A, Veldsman A, Hatherill M, Schirru G, Pau MG, Hendriks J, Weverling GJ, Goudsmit J, Sizemore D, McClain JB, Goetz M, Gearhart J, Mahomed H, Hussey GD, Sadoff JC, Hanekom WA. 2010. The novel tuberculosis vaccine, AERAS-402, induces robust and polyfunctional CD4+ and CD8+ T cells in adults. Am J Respir Crit Care Med 181:1407–1417. doi: 10.1164/rccm.200910-1484OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beveridge NE, Price DA, Casazza JP, Pathan AA, Sander CR, Asher TE, Ambrozak DR, Precopio ML, Scheinberg P, Alder NC, Roederer M, Koup RA, Douek DC, Hill AV, McShane H. 2007. Immunisation with BCG and recombinant MVA85A induces long-lasting, polyfunctional Mycobacterium tuberculosis-specific CD4+ memory T lymphocyte populations. Eur J Immunol 37:3089–3100. doi: 10.1002/eji.200737504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowland R, Pathan AA, Satti I, Poulton ID, Matsumiya MM, Whittaker M, Minassian AM, O'Hara GA, Hamill M, Scott JT, Harris SA, Poyntz HC, Bateman C, Meyer J, Williams N, Gilbert SC, Lawrie AM, Hill AV, McShane H. 2013. Safety and immunogenicity of an FP9-vectored candidate tuberculosis vaccine (FP85A), alone and with candidate vaccine MVA85A in BCG-vaccinated healthy adults: a phase I clinical trial. Hum Vaccin Immunother 9:50–62. doi: 10.4161/hv.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun R, Skeiky YA, Izzo A, Dheenadhayalan V, Imam Z, Penn E, Stagliano K, Haddock S, Mueller S, Fulkerson J, Scanga C, Grover A, Derrick SC, Morris S, Hone DM, Horwitz MA, Kaufmann SH, Sadoff JC. 2009. Novel recombinant BCG expressing perfringolysin O and the over-expression of key immunodominant antigens; preclinical characterization, safety and protection against challenge with Mycobacterium tuberculosis. Vaccine 27:4412–4423. doi: 10.1016/j.vaccine.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 13.Billeskov R, Elvang TT, Andersen PL, Dietrich J. 2012. The HyVac4 subunit vaccine efficiently boosts BCG-primed anti-mycobacterial protective immunity. PLoS One 7:e39909. doi: 10.1371/journal.pone.0039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horwitz MA, Harth G, Dillon BJ, Maslesa-Galic S. 2000. Recombinant bacillus Calmette-Guerin (BCG) vaccines expressing the Mycobacterium tuberculosis 30-kDa major secretory protein induce greater protective immunity against tuberculosis than conventional BCG vaccines in a highly susceptible animal model. Proc Natl Acad Sci U S A 97:13853–13858. doi: 10.1073/pnas.250480397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luabeya AK, Kagina BM, Tameris MD, Geldenhuys H, Hoff ST, Shi Z, Kromann I, Hatherill M, Mahomed H, Hanekom WA, Andersen P, Scriba TJ, H56-032 Trial Study Group, Schoeman E, Krohn C, Day CL, Africa H, Makhethe L, Smit E, Brown Y, Suliman S, Hughes EJ, Bang P, Snowden MA, McClain B, Hussey GD. 2015. First-in-human trial of the post-exposure tuberculosis vaccine H56:IC31 in Mycobacterium tuberculosis infected and non-infected healthy adults. Vaccine 33:4130–4140. doi: 10.1016/j.vaccine.2015.06.051. [DOI] [PubMed] [Google Scholar]
- 16.Ottenhoff TH, Doherty TM, van Dissel JT, Bang P, Lingnau K, Kromann I, Andersen P. 2010. First in humans: a new molecularly defined vaccine shows excellent safety and strong induction of long-lived Mycobacterium tuberculosis-specific Th1-cell like responses. Hum Vaccin 6:1007–1015. doi: 10.4161/hv.6.12.13143. [DOI] [PubMed] [Google Scholar]
- 17.Skeiky YA, Dietrich J, Lasco TM, Stagliano K, Dheenadhayalan V, Goetz MA, Cantarero L, Basaraba RJ, Bang P, Kromann I, McMclain JB, Sadoff JC, Andersen P. 2010. Non-clinical efficacy and safety of HyVac4:IC31 vaccine administered in a BCG prime-boost regimen. Vaccine 28:1084–1093. doi: 10.1016/j.vaccine.2009.10.114. [DOI] [PubMed] [Google Scholar]
- 18.Kaufmann SH. 2012. Tuberculosis vaccine development: strength lies in tenacity. Trends Immunol 33:373–379. doi: 10.1016/j.it.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Von Eschen K, Morrison R, Braun M, Ofori-Anyinam O, De Kock E, Pavithran P, Koutsoukos M, Moris P, Cain D, Dubois MC, Cohen J, Ballou WR. 2009. The candidate tuberculosis vaccine Mtb72F/AS02A: tolerability and immunogenicity in humans. Hum Vaccin 5:475–482. doi: 10.4161/hv.8570. [DOI] [PubMed] [Google Scholar]
- 20.Turner OC, Roberts AD, Frank AA, Phalen SW, McMurray DM, Content J, Denis O, D'Souza S, Tanghe A, Huygen K, Orme IM. 2000. Lack of protection in mice and necrotizing bronchointerstitial pneumonia with bronchiolitis in guinea pigs immunized with vaccines directed against the hsp60 molecule of Mycobacterium tuberculosis. Infect Immun 68:3674–3679. doi: 10.1128/IAI.68.6.3674-3679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orme IM. 1987. The kinetics of emergence and loss of mediator T lymphocytes acquired in response to infection with Mycobacterium tuberculosis. J Immunol 138:293–298. [PubMed] [Google Scholar]
- 22.Srivastava S, Ernst JD. 2013. Cutting edge: direct recognition of infected cells by CD4 T cells is required for control of intracellular Mycobacterium tuberculosis in vivo. J Immunol 191:1016–1020. doi: 10.4049/jimmunol.1301236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flory CM, Hubbard RD, Collins FM. 1992. Effects of in vivo T lymphocyte subset depletion on mycobacterial infections in mice. J Leukoc Biol 51:225–229. [DOI] [PubMed] [Google Scholar]
- 24.Leveton C, Barnass S, Champion B, Lucas S, De Souza B, Nicol M, Banerjee D, Rook G. 1989. T-cell-mediated protection of mice against virulent Mycobacterium tuberculosis. Infect Immun 57:390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller I, Cobbold SP, Waldmann H, Kaufmann SH. 1987. Impaired resistance to Mycobacterium tuberculosis infection after selective in vivo depletion of L3T4+ and Lyt-2+ T cells. Infect Immun 55:2037–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin PL, Rutledge T, Green AM, Bigbee M, Fuhrman C, Klein E, Flynn JL. 2012. CD4 T cell depletion exacerbates acute Mycobacterium tuberculosis while reactivation of latent infection is dependent on severity of tissue depletion in cynomolgus macaques. AIDS Res Hum Retroviruses 28:1693–1702. doi: 10.1089/aid.2012.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao S, Huang D, Chen CY, Halliday L, Wang RC, Chen ZW. 2014. CD4+ T cells contain early extrapulmonary tuberculosis (TB) dissemination and rapid TB progression and sustain multieffector functions of CD8+ T and CD3− lymphocytes: mechanisms of CD4+ T cell immunity. J Immunol 192:2120–2132. doi: 10.4049/jimmunol.1301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pawlowski A, Jansson M, Skold M, Rottenberg ME, Kallenius G. 2012. Tuberculosis and HIV co-infection. PLoS Pathog 8:e1002464. doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.North RJ, Jung YJ. 2004. Immunity to tuberculosis. Annu Rev Immunol 22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- 30.O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. 2013. The immune response in tuberculosis. Annu Rev Immunol 31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 31.Green AM, Difazio R, Flynn JL. 2013. IFN-gamma from CD4 T cells is essential for host survival and enhances CD8 T cell function during Mycobacterium tuberculosis infection. J Immunol 190:270–277. doi: 10.4049/jimmunol.1200061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindestam Arlehamn CS, Lewinsohn D, Sette A, Lewinsohn D. 2014. Antigens for CD4 and CD8 T cells in tuberculosis. Cold Spring Harb Perspect Med 4:a018465. doi: 10.1101/cshperspect.a018465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Commandeur S, Coppola M, Dijkman K, Friggen AH, van Meijgaarden KE, van den Eeden SJ, Wilson L, van der Ploeg-van Schip JJ, Franken KL, Geluk A, Ottenhoff TH. 2014. Clonal analysis of the T-cell response to in vivo expressed Mycobacterium tuberculosis protein Rv2034, using a CD154 expression based T-cell cloning method. PLoS One 9:e99203. doi: 10.1371/journal.pone.0099203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geluk A, van Meijgaarden KE, Joosten SA, Commandeur S, Ottenhoff TH. 2014. Innovative strategies to identify M. tuberculosis antigens and epitopes using genome-wide analyses. Front Immunol 5:256. doi: 10.3389/fimmu.2014.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blythe MJ, Zhang Q, Vaughan K, de Castro R Jr, Salimi N, Bui HH, Lewinsohn DM, Ernst JD, Peters B, Sette A. 2007. An analysis of the epitope knowledge related to mycobacteria. Immunome Res 3:10. doi: 10.1186/1745-7580-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huygen K. 2014. The immunodominant T-cell epitopes of the mycolyl-transferases of the antigen 85 complex of M. tuberculosis. Front Immunol 5:321. doi: 10.3389/fimmu.2014.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balaji KN, Goyal G, Narayana Y, Srinivas M, Chaturvedi R, Mohammad S. 2007. Apoptosis triggered by Rv1818c, a PE family gene from Mycobacterium tuberculosis is regulated by mitochondrial intermediates in T cells. Microbes Infect 9:271–281. doi: 10.1016/j.micinf.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Sampson SL. 2011. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol 2011:497203. doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sayes F, Sun L, Di Luca M, Simeone R, Degaiffier N, Fiette L, Esin S, Brosch R, Bottai D, Leclerc C, Majlessi L. 2012. Strong immunogenicity and cross-reactivity of Mycobacterium tuberculosis ESX-5 type VII secretion: encoded PE-PPE proteins predicts vaccine potential. Cell Host Microbe 11:352–363. doi: 10.1016/j.chom.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Woodworth JS, Fortune SM, Behar SM. 2008. Bacterial protein secretion is required for priming of CD8+ T cells specific for the Mycobacterium tuberculosis antigen CFP10. Infect Immun 76:4199–4205. doi: 10.1128/IAI.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, Hinds J, Neyrolles O, Butcher PD, Leclerc C, Cole ST, Brosch R. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun 74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Souza S, Rosseels V, Romano M, Tanghe A, Denis O, Jurion F, Castiglione N, Vanonckelen A, Palfliet K, Huygen K. 2003. Mapping of murine Th1 helper T-cell epitopes of mycolyl transferases Ag85A, Ag85B, and Ag85C from Mycobacterium tuberculosis. Infect Immun 71:483–493. doi: 10.1128/IAI.71.1.483-493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, Kim Y, Sidney J, James EA, Taplitz R, McKinney DM, Kwok WW, Grey H, Sallusto F, Peters B, Sette A. 2013. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog 9:e1003130. doi: 10.1371/journal.ppat.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindestam Arlehamn CS, Paul S, Mele F, Huang C, Greenbaum JA, Vita R, Sidney J, Peters B, Sallusto F, Sette A. 2015. Immunological consequences of intragenus conservation of Mycobacterium tuberculosis T-cell epitopes. Proc Natl Acad Sci U S A 112:E147–E155. doi: 10.1073/pnas.1416537112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sweeney KA, Dao DN, Goldberg MF, Hsu T, Venkataswamy MM, Henao-Tamayo M, Ordway D, Sellers RS, Jain P, Chen B, Chen M, Kim J, Lukose R, Chan J, Orme IM, Porcelli SA, Jacobs WR Jr. 2011. A recombinant Mycobacterium smegmatis induces potent bactericidal immunity against Mycobacterium tuberculosis. Nat Med 17:1261–1268. doi: 10.1038/nm.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkataswamy MM, Goldberg MF, Baena A, Chan J, Jacobs WR Jr, Porcelli SA. 2012. In vitro culture medium influences the vaccine efficacy of Mycobacterium bovis BCG. Vaccine 30:1038–1049. doi: 10.1016/j.vaccine.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, Sette A. 2013. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr Protoc Immunol Chapter 18:Unit 18.13. doi: 10.1002/0471142735.im1803s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Dai S, Crawford F, Fruge R, Marrack P, Kappler J. 2002. Alternate interactions define the binding of peptides to the MHC molecule IA (b). Proc Natl Acad Sci U S A 99:8820–8825. doi: 10.1073/pnas.132272099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hinchey J, Lee S, Jeon BY, Basaraba RJ, Venkataswamy MM, Chen B, Chan J, Braunstein M, Orme IM, Derrick SC, Morris SL, Jacobs WR Jr, Porcelli SA. 2007. Enhanced priming of adaptive immunity by a proapoptotic mutant of Mycobacterium tuberculosis. J Clin Invest 117:2279–2288. doi: 10.1172/JCI31947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregory RL. 1986. Microbial ribosomal vaccines. Rev Infect Dis 8:208–217. doi: 10.1093/clinids/8.2.208. [DOI] [PubMed] [Google Scholar]
- 52.Nikolaeva LV, Savel'ev EP. 1991. Molecular-biological and immunological properties of ribosomal vaccines. Biomed Sci 2:1–10. [PubMed] [Google Scholar]
- 53.Youmans AS, Youmans GP. 1966. Effect of trypsin and ribonuclease on the immunogenic activity of ribosomes and ribonucleic acid isolated from Mycobacterium tuberculosis. J Bacteriol 91:2146–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baba T. 1977. Immunogenic activity of a ribosomal fraction obtained from Pasteurella multocida. Infect Immun 15:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith RL, Wysocki JA, Bruun JN, De Courcy SJ Jr, Blakemore WS, Mudd S. 1974. Efficacy of ribosomal preparations from Pseudomonas aeruginosa to protect against intravenous Pseudomonas challenge in mice. J Reticuloendothel Soc 15:22–30. [PubMed] [Google Scholar]
- 56.Tewari RP, Lynn M, Birnbaum AJ, Solotorovsky M. 1978. Characterization of the immunoprotective antigen of ribosomal preparations from Haemophilus influenzae. Infect Immun 19:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schalla WO, Johnson W. 1975. Immunogenicity of ribosomal vaccines isolated from group A, type 14 Streptococcus pyogenes. Infect Immun 11:1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas DW, Weiss E. 1972. Response of mice to injection of ribosomal fraction from group B Neisseria meningitidis. Infect Immun 6:355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliveira SC, Splitter GA. 1996. Immunization of mice with recombinant L7/L12 ribosomal protein confers protection against Brucella abortus infection. Vaccine 14:959–962. doi: 10.1016/0264-410X(96)00018-7. [DOI] [PubMed] [Google Scholar]
- 60.Tabynov K, Kydyrbayev Z, Ryskeldinova S, Yespembetov B, Zinina N, Assanzhanova N, Kozhamkulov Y, Inkarbekov D, Gotskina T, Sansyzbay A. 2014. Novel influenza virus vectors expressing Brucella L7/L12 or Omp16 proteins in cattle induced a strong T-cell immune response, as well as high protectiveness against B. abortus infection. Vaccine 32:2034–2041. doi: 10.1016/j.vaccine.2014.02.058. [DOI] [PubMed] [Google Scholar]
- 61.Ramirez L, Corvo L, Duarte MC, Chavez-Fumagalli MA, Valadares DG, Santos DM, de Oliveira CI, Escutia MR, Alonso C, Bonay P, Tavares CA, Coelho EA, Soto M. 2014. Cross-protective effect of a combined L5 plus L3 Leishmania major ribosomal protein based vaccine combined with a Th1 adjuvant in murine cutaneous and visceral leishmaniasis. Parasit Vectors 7:3. doi: 10.1186/1756-3305-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crump EM, Burian J, Allen PD, Gale S, Kay WW. 2007. Identification of a ribosomal L10-like protein from Flavobacterium psychrophilum as a recombinant vaccine candidate for rainbow trout fry syndrome. J Mol Microbiol Biotechnol 13:55–64. doi: 10.1159/000103597. [DOI] [PubMed] [Google Scholar]
- 63.Wang FQ, Zhong J, Zhao Y, Xiao J, Liu J, Dai M, Zheng G, Zhang L, Yu J, Wu J, Duan B. 2014. Genome sequencing of high-penicillin producing industrial strain of Penicillium chrysogenum. BMC Genomics 15(Suppl 1):S11. doi: 10.1186/1471-2164-15-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE III, Wang H, Zhang W, Zhang Y. 2011. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333:1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatfull GF. 1996. The molecular genetics of Mycobacterium tuberculosis. Curr Top Microbiol Immunol 215:29–47. [DOI] [PubMed] [Google Scholar]
- 66.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. 2008. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, Greenbaum J, Lundegaard C, Sette A, Lund O, Bourne PE, Nielsen M, Peters B. 2012. Immune epitope database analysis resource. Nucleic Acids Res 40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. 2010. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kruisbeek AM. 2001. Production of mouse T cell hybridomas. Curr Protoc Immunol Chapter 3:Unit 3.14. doi: 10.1002/0471142735.im0314s24. [DOI] [PubMed] [Google Scholar]
- 71.Trauner A, Bennett MH, Williams HD. 2011. Isolation of bacterial ribosomes with monolith chromatography. PLoS One 6:e16273. doi: 10.1371/journal.pone.0016273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hill WE, Rossetti GP, Van Holde KE. 1969. Physical studies of ribosomes from Escherichia coli. J Mol Biol 44:263–277. doi: 10.1016/0022-2836(69)90174-0. [DOI] [PubMed] [Google Scholar]
- 73.Loge RV, Hill WE, Baker RE, Larson CL. 1974. Delayed hypersensitivity reactions provoked by ribosomes from acid-fast bacilli: physical characteristics and immunological aspects of core ribosomal proteins from Mycobacterium smegmatis. Infect Immun 9:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu KO, Im JS, Molano A, Dutronc Y, Illarionov PA, Forestier C, Fujiwara N, Arias I, Miyake S, Yamamura T, Chang YT, Besra GS, Porcelli SA. 2005. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of alpha-galactosylceramides. Proc Natl Acad Sci U S A 102:3383–3388. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.