Abstract
Methanotrophs play a key role in balancing the atmospheric methane concentration. Recently, the microbial methanotrophic diversity was extended by the discovery of thermoacidophilic methanotrophs belonging to the Verrucomicrobia phylum in geothermal areas. Here we show that a representative of this new group, Methylacidiphilum fumariolicum SolV, is able to grow as a real ‘Knallgas' bacterium on hydrogen/carbon dioxide, without addition of methane. The full genome of strain SolV revealed the presence of two hydrogen uptake hydrogenases genes, encoding an oxygen-sensitive (hup-type) and an oxygen-insensitive enzyme (hhy-type). The hhy-type hydrogenase was constitutively expressed and active and supported growth on hydrogen alone up to a growth rate of 0.03 h−1, at O2 concentrations below 1.5%. The oxygen-sensitive hup-type hydrogenase was expressed when oxygen was reduced to below 0.2%. This resulted in an increase of the growth rate to a maximum of 0.047 h−1, that is 60% of the rate on methane. The results indicate that under natural conditions where both hydrogen and methane might be limiting strain SolV may operate primarily as a methanotrophic ‘Knallgas' bacterium. These findings argue for a revision of the role of hydrogen in methanotrophic ecosystems, especially in soil and related to consumption of atmospheric methane.
Introduction
Methanotrophs form an important sink for the greenhouse gas methane and they play a key role in keeping the atmospheric methane concentration in balance (Murrell and Jetten, 2009). They limit the amount of biologically or geochemically produced methane escaping into the atmosphere. The most studied group comprises the aerobic methanotrophs that phylogenetically belong to the Gammaproteobacteria and the Alphaproteobacteria (Hanson and Hanson, 1996). However, the methanotrophic diversity was extended when three research groups independently described novel thermoacidophilic methanotrophs isolated from geothermal areas in Italy, New Zealand and Russia (Dunfield et al, 2007; Pol et al, 2007; Islam et al, 2008). These novel isolates represented a distinct phylogenetic lineage within the phylum Verrucomicrobia. They belong to a single genus for which the name Methylacidiphilum was proposed (Op den Camp et al, 2009). Recently, a second genus of mesophilic, acidophilic verrucomicrobial methanotrophs was described, Methylacidimicrobium (van Teeseling et al, 2014). In addition, data from cultivation-independent environmental studies (Sharp et al, 2012; Sharp et al, 2014) indicated that methanotrophic Verrucomicrobia may be present in many more moderate-temperature volcanic ecosystems than assumed before. The discovery of a verrucomicrobial methanotroph was exciting, since for the first time the widely distributed Verrucomicrobia phylum, from which most members remain uncultivated, was coupled to a geochemical cycle (Pol et al, 2007). Initial analyses of our isolate Methylacidiphilum fumariolicum strain SolV showed major differences with the classical methanotrophs, for example extreme acid tolerance, absence of typical membrane structures, distinct enzymes and use of rare earth elements for methane oxidation (Pol et al, 2007; Pol et al, 2013; Keltjens et al, 2014).
It has long been assumed that methanotrophs are very strict in their diet, consuming only methane or methanol and occasionally other C1 compounds (Hanson and Hanson, 1996), despite the fact that most of the time a range of multi-carbon substrates is available in their environments (Tsien et al, 1989). Stimulation of growth by multi-carbon substrates like acetate, malate and succinate has been reported already by Whittenbury et al, 1970, but claims of growth on such substrates or even sugars for several isolated strains in subsequent years have been doubted (for a review see Semrau, 2011). Only in recent years convincing evidence has shown that acetate, pyruvate, ethanol, malate and succinate can be used for growth, but this seems restricted to alpha-proteobacterial methanotrophs (Kelly et al, 2000; Kolb, 2009; Kappler and Nouwens, 2013; Knief, 2015). The ecological significance especially of acetate consumption would be the increased survival of these methanotrophs under natural conditions, where methane concentrations are very low and variable (Belova et al, 2011). Moreover, methane oxidation might be possible at much lower concentrations when additional reducing equivalents from multi-carbon compounds are available, as speculated by Theisen and Murrell, 2005.
In many ecosystems another source of reducing equivalents is available as a potential energy source for methanotrophs, that is molecular hydrogen. The presence of putative hydrogenases has been detected in many different (sub)phyla; Proteobacteria, Firmicutes, Cyanobacteria, Aquificae, Euryarchaeota, Crenarchaeota and Verrucomicrobia (Greening et al, 2016). Hydrogenase-encoding genes were also identified in the genomes of multiple proteobacterial obligate methane oxidizers and uptake of hydrogen has been reported for Methylosinus sp.de Bont (1976), Methylocystis sp. and Methylococcus capsulatus Bath (Csáki et al, 2001; Kelly et al, 2005). In methanotrophs it is believed that hydrogenases also help to ‘save' reducing equivalents by recycling the hydrogen gas that is produced during nitrogen fixation (Bothe et al, 2010). Csáki and co-workers (Csáki et al, 2001) showed increased H2 production during nitrogen fixing conditions using a hupLS mutant of M. capsulatus Bath. Furthermore, soluble and membrane-bound hydrogenases have been reported for M. capsulatus Bath and hydrogen was shown to be able to supply reducing equivalents for the methane monooxygenase (Hanczár et al, 2002).
The fact that several proteobacterial methanotrophs contain genes encoding the large and small subunit of both uptake hydrogenases and the ribulose-1,5-bisphosphate carboxylase (Rubisco) suggests the possibility of autotrophic growth. But attempts to grow M. capsulatus Bath autotrophically (on hydrogen and carbon dioxide) in liquid media were not successful (Dalton and Whittenbury, 1976; Taylor et al, 1981; Stanley and Dalton, 1982; Baxter et al, 2002). Autotrophic growth was observed on solid agar media, but no physiological studies have been reported to support this (Baxter et al, 2002).
M. fumariolicum SolV was shown to use the Calvin-Benson-Bassham (CBB) cycle for carbon fixation pathways (Khadem et al, 2011), and is capable of fixing nitrogen gas (Khadem et al, 2010). Key metabolic genes of the ribulose monophosphate and serine pathways are absent (Pol et al, 2007). The full genome of strain SolV (Anvar et al, 2014) revealed the presence of two hydrogenase types (hup- and hhy-type). Many Group 1 hydrogenases within the hup type are known to support growth. While in general hydrogenases are very sensitive towards oxygen and function only in anaerobic respiration, the Group 1d hydrogenases are known for their relative oxygen tolerance and may support aerobic growth in ‘Knallgas' bacteria. The hhy type genes encode the recently discovered Group 5 hydrogenases, which are widespread in actinobacteria from soil and were supposed to be responsible for ‘high affinity' atmospheric hydrogen uptake (Schäfer et al, 2013; Greening et al, 2014). The ‘Knallgas' bacterium Ralstonia eutropha also contains this type of hydrogenase that upon isolation appeared to be oxygen insensitive (Schäfer et al, 2013). A role for this enzyme is as yet not established. When hydrogen was added to methane-consuming cultures of M. fumariolicum SolV, the hydrogen was also oxidized (Pol et al, 2007). RNA-seq analysis of SolV cells at maximal growth rate (μmax) compared to nitrogen fixing and oxygen limiting conditions showed up-regulation of the hup-type hydrogenase at low oxygen concentrations, whereas the hhy-type enzyme was constitutively expressed (Khadem et al, 2012a). The presence of the CBB cycle, together with the uptake hydrogenase prompted us to investigate the possibility to grow strain SolV as a real ‘Knallgas' bacterium without addition of methane. We used physiological experiments in combination with phylogenetic and transcriptome analysis to show that strain SolV can grow autotrophically on hydrogen and carbon dioxide.
Materials and methods
Microorganism and medium composition
Methylacidiphilum fumariolicum strain SolV used in this study was initially isolated from the volcanic region Campi Flegrei, near Naples, Italy (Pol et al, 2007). In this study the medium was composed of 0.2 mM MgCl2.6H2O; 0.2 mM CaCl2.2H2O; 1 mM Na2SO4; 2 mM K2SO4; 2 mM (NH4)2SO4 (or 5 mM KNO3) and 1 mM NaH2PO4.H2O. A trace element solution containing 1 μM Ni, Co, Mo, Zn and Ce; 5 μM Mn and Fe; 10 μM Cu and 40–50 μM nitrilotriacetic acid. The pH of medium was adjusted to 2.7 using 1 M H2SO4. To avoid precipitation, CaCl2.2H2O and the rest of the medium were autoclaved separately and mixed after cooling. This medium composition contains all nutrients to obtain an OD600 of 1.0 and was used in batch and continuous cultures, unless stated otherwise.
Chemostat cultivation
To start a continuous culture on hydrogen, we initiated growth on methane, while hydrogen was absent. The reactor (Applikon Biotechnology, Delft, NL) was operated at 55 °C with stirring at 1000 rpm. The chemostat (liquid volume of 1.2 L) was supplied with medium at a flow rate of 30 ml h−1 (D=0.025 h−1). A gas supply of 50% CH4 (v/v) and 5% CO2 (v/v) was provided by mass flow controllers through a sterile filter and sparged into the medium at a gas flow rate of 16–17 ml min−1. The dO2 was regulated initially at 1% oxygen saturation for 18 h. Afterwards, the dO2 set-point was adjusted to 0.2% oxygen for 9 h and an OD600 value of 0.85 was obtained. At this point, methane was changed to hydrogen maintaining the total gas flow rate identical to the methane condition and keeping the dO2 value at 0.20–0.22% oxygen. The cell-containing medium was removed automatically from the chemostat by a peristaltic pump when the liquid level reached the sensor in the reactor. The pH was regulated in the steady state at 3.0 using 0.2 M NaOH.
The continuous culture with CH4 as an electron donor and nitrate (NO3−) as N-source had a liquid volume of 550 ml, and was operated at 55 °C with stirring at 900 rpm with a stirrer bar. The chemostat was supplied with medium at a flow rate of 14.5 ml h−1 (D=0.026 h−1). The cell-containing medium was removed automatically from the reactor by a peristaltic pump when the liquid level reached the sensor in the reactor. Supply of 10% CH4 (v/v), 8% O2 (v/v) and 68% CO2 (v/v) took place by mass flow controllers (total flow≈20 ml min−1) through a sterile filter and sparged into the medium just above the stirrer bar. The pH was regulated at 2.7 using a 1 M solution of bicarbonate. An O2 sensor in the liquid was coupled to a Biocontroller (Applikon) regulating the O2 mass controller.
Batch cultivation
To obtain maximum growth rate (μmax), cells were grown without any limitation in 250-ml serum bottles containing 40 ml medium (4 mM NH4+; pH 2.7), and sealed with red butyl rubber stoppers. The headspace contained air with (v/v) 10% CH4, 5% CO2 and cultures were incubated at 55 °C with shaking at 250 rpm. Incubations were performed in duplicate and exponentially growing cells (OD600=1) were collected for mRNA isolation and Ion Torrent sequencing.
Dry weight determination and elemental analysis
To determine biomass dry-weight concentration, 10 ml of the culture suspension (triplicate) were filtered through pre-weighed 0.45 μm filters and dried to stable weight in a vacuum oven at 60 °C. In order to determine the total content of carbon and nitrogen, 10 ml of the culture suspension (duplicate) was centrifuged at 4500 g for 30 min and the clear supernatant was used for the analysis. The nitrogen and carbon content in the supernatant was compared with the corresponding values in the whole cell suspension. The total carbon and nitrogen contents were measured using TOC-L and TNM-1 analysers (Shimadzu, Kyoto, Japan).
Respiration experiments
Respiration rates were measured polarographically in a respiration cell with an oxygen microsensor (RC350, Strathkelvin, Motherwell, UK) using 3 ml of whole cell suspensions of strain SolV. Methane, hydrogen or oxygen-saturated medium were injected to the respiration chamber to obtain the desired dissolved gas concentrations. The O2 signal was monitored and recorded using SensorTrace Basic software (Unisense, Aarhus, Denmark). The temperature and stirring rate in the respiration chamber was adjusted to 55 °C and 1000 rpm, respectively. Rates were expressed as nmol O2.min−I.mg DW−1 and when necessary corrected for endogenous respiration. To avoid too high oxygen concentrations at the start of an experiment, samples taken from cultures were immediately transferred into rubber septum sealed bottles under an anoxic atmosphere of nitrogen and carbon dioxide. These bottles contained medium in case dilution was necessary. A subsample was taken from the bottle by a syringe with a long needle and introduced into the respiration chamber at the bottom with the oxygen probe in place while pushing out the air via the inlet channel.
Hydrogenase activity assays
Three ml of cell suspension of strain SolV (OD600 0.3–0.4/mg DW) was incubated under an atmosphere of N2—CO2—O2 (80%: 20%: 0.4%, v/v/v) in 60-ml capped serum bottles at 55 °C and shaking at 400 rpm. Samples from cultures growing on hydrogen were preincubated under the same conditions for 30 min in order to consume any hydrogen present in the samples. The consumption of added hydrogen was measured using an HP 5890 gas chromatograph (Agilent, Santa Clara, USA) equipped with a Porapak Q column (1.8 m, ID 2 mm) and a thermal conductivity detector. For all gas analyses 250 μl gas samples were injected with a glass syringe.
Phylogenetic analysis
The gene sequences of the large and small subunit of uptake hydrogenases from different strains including Methylacidiphilum and Methylacidimicrobium species were downloaded from NCBI-GenBank. Conceptual translations into amino acids were performed and used for creating an alignment and phylogenetic analysis using MEGA 6 software (Tamura et al, 2013). The evolutionary history was interpreted using the neighbour-joining method (Saitou and Nei, 1987).
RNA isolation and sequencing
The complete genome sequence of strain SolV (Anvar et al, 2014), which is also available at the MicroScope annotation platform (https://www.genoscope.cns.fr/agc/microscope/home/), was used as the template for the transcriptome analysis (RNA-seq). A 4-ml volume of cells (OD600=1) was sampled from the continuous cultures (H2- and CH4-grown cells under O2 limitation) and from a batch culture (cells at μmax grown on methane without limitation) and harvested by centrifugation. The pellet was further used for mRNA isolation using the RiboPure™-Bacteria Kit according to the manufacturer's protocol (ThermoFisher, Waltham, USA). Briefly, cells were disrupted by cold Zirconia beads and after centrifugation 0.2 volumes of chloroform was added to the supernatant for initial RNA purification. Next, 0.5 volumes of 100% ethanol was added to the aqueous phase obtained after chloroform addition and the whole sample was transferred to a filter cartridge. After washing, the RNA was eluted from the filter cartridge. Afterwards, using MICROB express™ kit (ThermoFisher) the ribosomal RNAs were removed from the total RNA. The rRNA removal efficiency was checked using the Agilent 2100 Bioanalyzer (Agilent). Next, Ion Total RNA-Seq Kit v2 (ThermoFisher) was used to construct the cDNA libraries from rRNA-depleted total RNA. Briefly, the rRNA-depleted total RNA was fragmented using RNase III and then, reverse transcription was performed on the fragmented RNAs. The obtained cDNAs were amplified and further purified to prepare barcoded libraries. To prepare the template for the Ion Personal Genome Machine (PGM) System, a volume of 15 μl from two sample libraries with a concentration of 14 pM were mixed. This mixture of two libraries was used to prepare the template-positive Ion Sphere particles (ISPs) using the Ion OneTouch 2 instrument. Afterwards, the template-positive Ion Sphere particles were enriched using the Ion OneTouch ES instrument. Both template preparation and enrichment were performed using the Ion PGM Template OT2 200 Kit (Ion Torrent, Life technologies). Enriched templates were sequenced on an Ion 318 Chip v2 using the Ion PGM sequencing 200 Kit v2. Expression analysis was performed with the RNA-seq Analysis tool from the CLC Genomic Work bench software (version 7.0.4, CLC-Bio, Aarhus, Denmark) and values are expressed as RPKM (reads per kilo base of exon model per million mapped reads) (Mortazavi et al, 2008).
Results
Hydrogen oxidation by M. fumariolicum SolV cells grown on methane
Cells from batch cultures of strain SolV growing on methane at maximum growth rate (μ=0.07 h−1) and oxygen concentrations above 10% showed relatively high oxygen consumption rates at the expense of hydrogen, 15–20 nmol.min−1.mg DW−1, which was about 6% of the oxygen consumption with methane (280 nmol.min−1.mg DW−1; Table 1). However, growth on hydrogen and carbon dioxide without methane under such conditions was not possible. Initial batch tests in bottles showed growth only at (and below) 1% O2 concentrations. Since the oxygen consumption results in a rapid decrease of the oxygen concentration during batch cultivation, we studied the inhibiting effects of oxygen in a continuous culture for which the dO2 was regulated carefully with mass flow controllers. The hydrogen respiration rate of a continuous culture under methane limitation (D=0.03 h−1) was measured at different oxygen concentrations. The initial dO2 was regulated at 0.3% oxygen (1.5% air) and the hydrogen respiration rate was slightly higher than those in batch cultures (20–24 nmol O2.min−1.mg DW−1; Table 1), being about 7.5% of the methane respiration rates (268–317 nmol O2.min−1.mg DW−1; Table 1). When the dO2 was stepwise increased to a final value of 3.2% oxygen (16% air; each step was stabilized for at least one day), respiration rates were not significantly affected. Similarly, measurements in the respiration chamber showed that rates were more or less the same at all dO2 values and stable during measuring periods of about 2 h. This is a strong indication that the hhy-type hydrogenase responsible for this activity was constitutively expressed under these conditions and most probably oxygen insensitive.
Table 1. The oxygen respiration profile of Methylacidiphilum fumariolicum SolV cells from different growth conditions using CH4 or H2.
| Growth condition | e− donor | Limitation | Growth rate (h−1)a | dO2% | O2 (μM)b | H2 respirationc | CH4 respirationc |
|---|---|---|---|---|---|---|---|
| Batch | CH4 | — | 0.07 | >10 | — | 15–20 | 280 |
| Continuous | CH4 | CH4 | 0.03 | 0.3–16 | — | 20–24 | 244–317 |
| Continuous | CH4 | O2 | 0.015 | — | >30 5 | 12–17d 42 | 232–293 n.d. |
| Batch | H2 | — | 0.047 | 0.02–0.04 | < 5 | 66–71 | 29–39e |
| Batch | H2 | — | 0.01 | 1.5 | 1–100 | 20 | 17e |
| Continuous | H2 | O2 | 0.04 | 0.01 | 100 5 | 17 29f | >171g n.d. |
In batch and continuous cultures, growth rate refers to μ and D, respectively.
The μM units refer to O2 concentrations in the liquid phase in the respiration chamber.
The respiration rates are in nmol O2.min−1.mg DW−1.
Pre-incubated at 10% O2 for 30 min.
Batch grown after continuous growing for over a year on hydrogen gas only.
Measured after exposure to 100 μM O2 in the above respiration experiment.
After >20 generations. n.d.=not determined.
When a methane-fed continuous culture was grown under oxygen limitation (D=0.015 h−1), hydrogen respiration measurements became less straightforward with inactivation and reactivation events occurring (see Supplementary Figure S1). Hydrogen respiration rates measured were at least twice the rate obtained for the continuous culture under methane limitation at dO2 values ranging from 0.3% to 3.2% oxygen (1.5–16% air; described above). Therefore, it seems that under oxygen limitation the oxygen-sensitive hup-type hydrogenase is expressed in addition to the constitutively expressed oxygen-insensitive hhy-type.
Autotrophic growth of M. fumariolicum SolV on hydrogen
A batch culture growing at μmax (0.075 h−1) in a bioreactor on methane at low oxygen concentration (regulated at 0.2% oxygen by mass flow controllers) also showed increased hydrogen oxidizing activity. When the gas supply was changed from methane to hydrogen only, the oxygen consumption rate dropped and stabilized after 45 min at 13% of the oxygen consumption rate with methane. This hydrogen respiration rate is about double compared to batch cultures at high oxygen concentration (which was 6% of the methane respiration, see above) and also clearly higher than the rate observed in the continuous culture at dO2 above 0.3% oxygen (which was 7% of the methane respiration rate, see above). The culture immediately continued growing, since we observed a 6% biomass increase 3 h after the switch from methane to hydrogen (OD600 increased from 0.85 to 0.9). The only carbon source during growth on hydrogen was carbon dioxide.
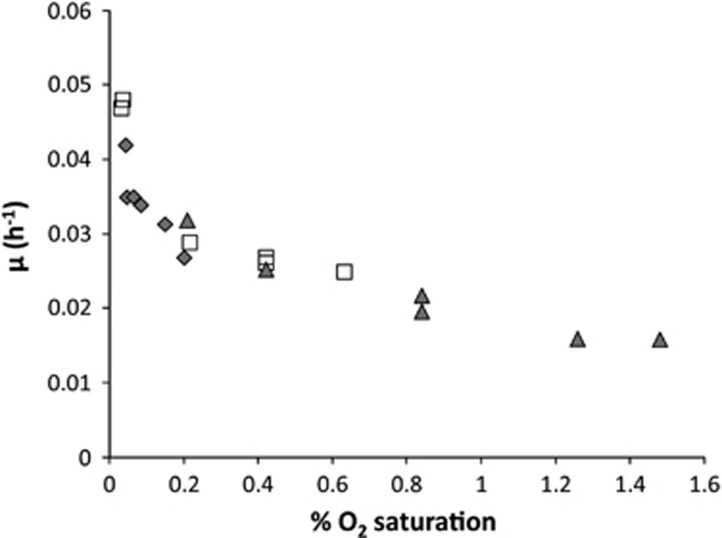
Results thus far indicated a critical oxygen concentration of about 0.2%. Below this value the oxygen-sensitive hup-type hydrogenase seems to be expressed. Therefore, the oxygen effect on growth rate was investigated in more details in a bioreactor under excess of hydrogen in which dO2 could be accurately controlled at values as low as 0.04% oxygen (equivalent to 0.3 μM O2). First growth was achieved batch-wise up to maximum OD600 values of 4 after which the culture was diluted to OD600 0.1–0.2 and restarted as a new batch. The dO2 was stepwise changed starting at 0.2% oxygen. Figure 1 shows the growth rates that were calculated when at least one generation of stable exponential growth was observed (based on OD600 measurements). The values clearly show the oxygen sensitivity of the growth on hydrogen. Highest growth rates were observed at the lowest oxygen concentrations. Decreasing the dO2 from 0.2% to 0.04% oxygen resulted in a sharp increase of the growth rate while above 0.2% oxygen, a steady decrease of the growth rate was observed. Up to a dO2 value of 1.5% oxygen we obtained stable growth rates. At a dO2 value of 2% oxygen, the growth continued for a few generations but the growth rate dropped continuously. In respiration experiments, O2 consumption was almost constant as observed in samples from the methane-limited continuous culture (only a slight reactivation was observed below 10 μM O2). Similarly, batch experiments with bottles showed growth only at (and below) 1% O2 concentrations (no growth was observed at 2.5 and 5% O2 concentrations).
Figure 1.
The effect of oxygen on the growth rate of H2-growing cells. 1% of O2 saturation is equal to 7.5 μM O2 in the liquid. Each set of symbols represents a newly started experiment in which cells were grown batch-wise up to maximum OD600=4 after which the culture was diluted to OD600 0.1–0.2 and restarted as a new batch.
Cultures at high growth rate exhibited both oxygen-sensitive and -insensitive hydrogenase activity during respiration experiments, similar to our observations for the O2-limited culture growing on methane (see above). Cells sampled from the batch cultures growing on hydrogen at the highest growth rate obtained (μ=0.047 h−1) were transferred to the respiration chamber avoiding O2 exposure. The respiration rates with hydrogen measured were as high as 66–71 nmol O2.min−1.mg DW−1 (when actual O2 concentration in the respiration chamber was below 5 μM; Table 1). When samples were incubated in bottles under an atmosphere of 0.5% hydrogen and 0.1% or 0.2% oxygen, the hydrogen consumption rate, analysed by gas chromatography, was 220 nmol H2.min−1.mg DW−1. This correlated very well with the oxygen consumption rate when assuming a H2:O2 consumption ratio of 0.32 as was determined during respiration experiments. When cells of a culture growing under similar conditions (continuous culture on hydrogen at D=0.04 h−1) were exposed to air the hydrogen respiration rate (measured at 100 μM O2) was only about 17 nmol O2.min−1.mg DW−1 and this rate was increasing again when O2 fell below 50 μM (Table 1). Methane respiration rates of cultures when changed from methane to exclusively hydrogen remained high (>171 nmol O2.min−1.mg DW−1) for many generations (>20). However, when a continuous culture was grown for over a year on hydrogen only, the methane respiration rate had dropped to 29–39 nmol O2.min−1.mg DW−1.
Yield
An oxygen-limited continuous culture on hydrogen (D=0.026 h−1) was used to assess the growth yield by accurately measuring the hydrogen consumption. Based on dry weight (DW) measurements a yield value of 3.4±0.1 g DW.mole H2−1 was obtained. Organic carbon analysis of centrifuged culture samples revealed the presence of 8–10% of the total organic matter in the supernatant, comparable to the amount excreted when growing on methane. The stoichiometry of hydrogen respiration was assessed both by the oxygen respiration measurements (H2 vs O2) and gas chromatographic analysis (for H2 vs CO2) measurements and resulted in the following stoichiometry:
Kinetics of hydrogen consumption
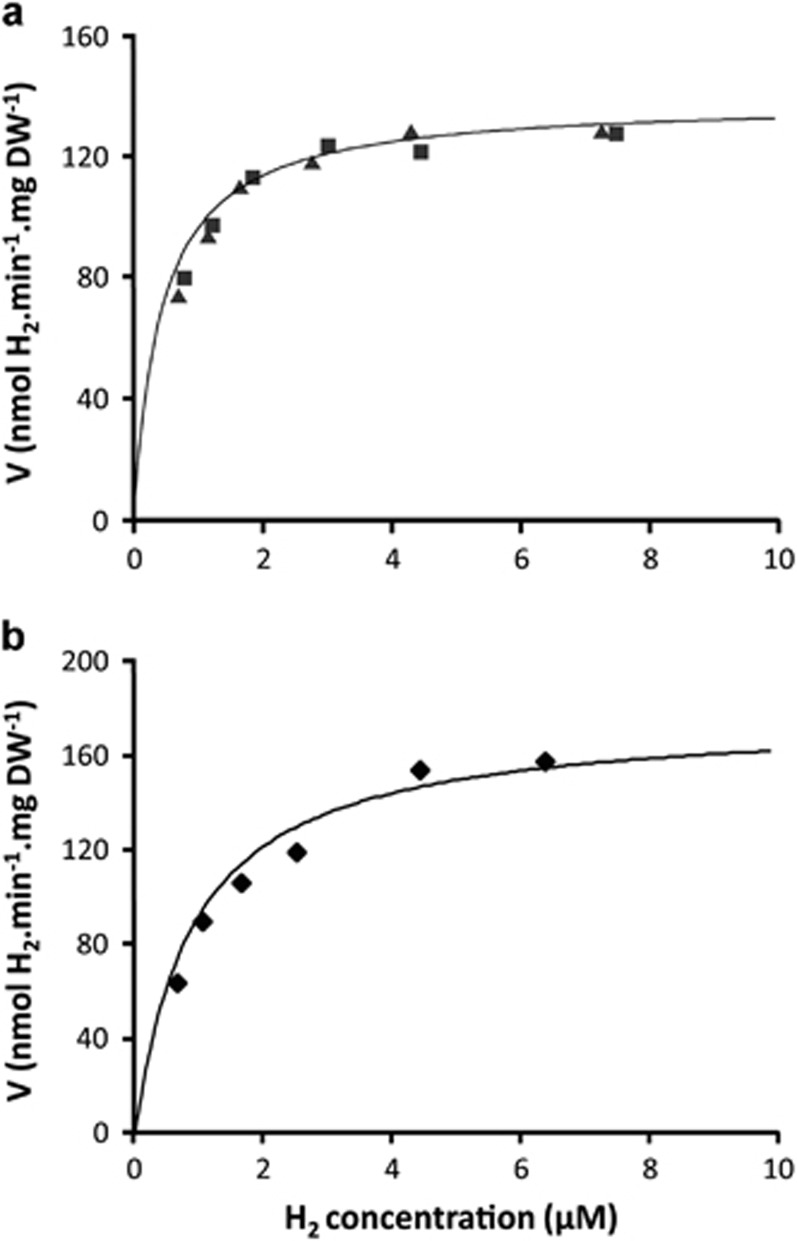
Addition of hydrogen to a methane-limited culture (D=0.013 h−1, at dO2 0.5–1%) resulted in an immediate increase of biomass (DW) and both substrates were consumed simultaneously. These conditions favour expression of the oxygen-insensitive hydrogenase. From respiration experiments it was observed that the maximum respiration rate for methane was much higher than for hydrogen. For methane the rate dropped when reaching concentrations below 15 μM, in accordance with the affinity constant for methane (Ks 6 μM) (Pol et al, 2007). In contrast hydrogen respiration rates remained constant until hydrogen was consumed completely (Supplementary Figure S2). It was estimated that this equals a hydrogen concentration well below 1 μM. The kinetics for the oxygen-insensitive hydrogenase were more accurately assessed in cells grown on hydrogen in batch at dO2 value at 0.8% (D=0.02 h−1) by measuring hydrogen consumption rates in bottles at various hydrogen concentrations. When Michaelis–Menten kinetics were fitted (Figure 2), an apparent hydrogen Ks of 0.6 μM was obtained (measured Vmax=127 nmol H2.min−1.mg DW−1). Hydrogenase kinetics of cells growing at lower dO2 of 0.2%, conditions (D=0.03 h−1) in which additional expression of the oxygen-sensitive hydrogenase is expected, showed a higher hydrogen consumption rate (Vmax=159 nmol H2.min−1.mg DW−1) but also a higher affinity constant value. Results for this mixed activity of hydrogenases were less accurate, but a Ks value of 1.1 μM H2 was estimated (Figure 2). The contribution of the oxygen-sensitive hydrogenase to the maximum measured respiration rates was estimated to be about two times higher than that of the oxygen-insensitive hydrogenase and by simulating Michaelis–Menten kinetics we estimated that the apparent affinity constant of the oxygen-sensitive hydrogenase was between 1 and 2 μM. The hydrogen consumption by SolV cells grown on methane under oxygen limitation (D=0.018 h−1) was followed in bottles with hydrogen concentrations of 0.5 μM (dO2 in bottles 0.2%). The consumption proceeded according to first order kinetics down to the GC detection level, that is 2 ppm hydrogen in the headspace (equivalent to 1.5 nM).
Figure 2.
(a) The kinetics of the oxygen-insensitive hydrogenase of the cells grown on hydrogen in a batch condition at dO2 value at 0.8% (Vmax=127 nmol H2.min−1.mg DW−1; Ks=0.6 μM). Closed squares and triangles represent two independent experiments. (b) The kinetics of both the oxygen-sensitive and -insensitive hydrogenases of cells grown on hydrogen in a batch condition at dO2 value below 0.2% (Vmax=159 nmol H2.min−1.mg DW−1; Ks=1.1 μM). Closed diamonds represent the results of three independent experiments. The solid line represents the best Michaelis–Menten kinetics fitting curve to our experimental data in both (a) and (b).
Hydrogenases in the genomes of verrucomicrobial methanotrophs
The full genome of strain SolV was analysed for hydrogenase-encoding genes (including accessory genes) and for comparison identical genes in the full and draft genomes of other verrucomicrobial methanotrophs were identified (Hou et al, 2008; Sharp et al, 2012; Sharp et al, 2014; van Teeseling et al, 2014; Erikstad and Birkeland, 2015). Both thermophilic and mesophilic strains contain a hydrogenase encoded by the genes hupS (small subunit) and hupL (large subunit; Supplementary Table S1). In addition, we identified a second hydrogenase encoded by the genes hhyS (small subunit) and hhyL (large subunit), only present in the thermophilic strain SolV and its closest relative M. kamchatkensis Kam1 (Supplementary Table S1). Survey of the genomes also revealed an electron transport unit (cytB) and several accessory genes (Supplementary Tables S2 and S3).
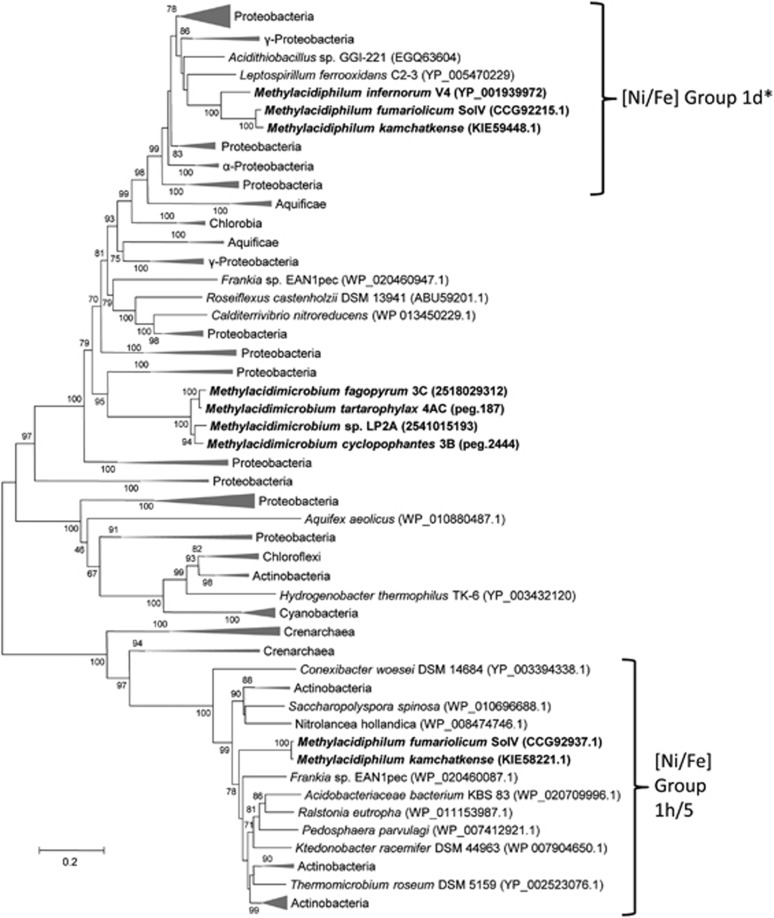
Phylogenetic analysis using the recent classification (Vignais and Billoud, 2007; Greening et al, 2016) showed that the hup-type hydrogenases of the verrucomicrobial methanotrophs belong to the membrane-bound H2-uptake [NiFe]-hydrogenases (Figure 3; Group 1). The large subunits all contain L1 and L2 motifs typical for Group 1 hydrogenases.
Figure 3.
The uptake hydrogenase gene-based phylogenetic tree of more than 130 bacterial species showing both thermophilic and mesophilic strains contain a hydrogenase encoded by the genes hupS (small subunit) and hupL (large subunit). In addition, a second hydrogenase encoded by the genes hhyS (small subunit) and hhyL (large subunit) were identified only in the thermophilic strain SolV and the close relative strain Kam1. The evolutionary history was generated using the neighbour-joining method. The percentage of replicate trees in which the associated taxa clustered in the bootstrap test is shown next to the branches. The analysis involved 136 nucleotide sequences. Evolutionary analyses were conducted in MEGA6. (*) indicates four exceptions including Thiorhodococcus drewsii AZ1, Thiocapsa marina, Salmonella enterica and Methylocystis parvus, which fit to Group 1e and 1c, in addition to Group 1d hydrogenases.
The second hhy-type hydrogenase belongs to Group 1 h/5 hydrogenases (Figure 3), based on the phylogenetic analysis of both the small and large subunits. The large subunit contains active site motifs L1 (TSRICGICGDNH) and L2 (SFDPCLPCGVH) that are highly conserved in other Group 1 h/5 hydrogenases.
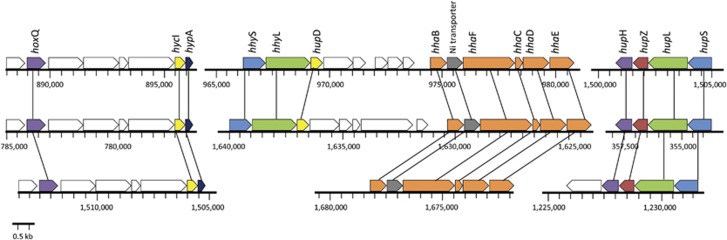
Figure 4 shows the gene arrangement of the two hydrogenase types of strain SolV. A first cluster includes the genes encoding the Group 1 h/5 hydrogenase (hhyS and hhyL), together with the gene encoding the putative maturation like protein (hupD). In close proximity (4.6 kb) the genes encoding accessory proteins (hhaBFCDE) are clustered together with a gene encoding a high affinity nickel transporter. At another location in the genome we identified genes encoding a putative expression/formation protein (hoxQ), a putative hydrogenase maturation protease (hycI) and the nickel insertion protein (hypA).
Figure 4.
Gene arrangement of the hydrogenases in the thermophilic Methylacidiphilum strains SolV (top), Kam1 (middle) and V4 (bottom). Genes are colour coded as follows: green=large subunit; blue=small subunit; yellow=putative hydrogenase maturation protease; orange=accessory proteins; red=b-type cytochrome subunit; purple=putative expression/formation protein; grey=nickel transporter; dark blue=nickel insertion protein.
The second hydrogenase cluster encoding the Group 1d type (Figure 4) includes the genes for the large and small subunit (hupSL), the expression/formation protein (hupH) and the b-type cytochrome subunit (hupZ). Comparison of the gene arrangement with the other two thermophilic strains (Figure 4) revealed an almost identical presence and clustering in strain Kam1. The genome of strain V4 does not seem to have the genes encoding the Group 1 h/5 hydrogenase but the clustering of the other genes encoding the Group 1d hydrogenase and accessory proteins is identical to strain SolV and Kam1 (Figure 4). In the mesophilic Methylacidimicrobium strains for which the most complete draft genomes are available (strains LP2A and 3C) the genes encoding the Group 1d-type hydrogenase show an identical gene clustering compared to the thermophilic Methylacidiphilum strains (Supplementary Figure S3). Unlike the thermophilic strains, the genes encoding the accessory proteins are in close proximity to the functional genes, but on the opposite strand. For the other mesophilic strains 4AC and 3B no comparison can be made since these draft genomes are too fragmented.
Transcriptome analysis
Expression levels of housekeeping genes and genes involved in hydrogen metabolism were determined for hydrogen- and methane-grown cells (both under O2-limited conditions). These values were compared to the expression values in cells growing at μmax with methane (without limitation). To determine the expression levels of the different genes under different conditions, the SolV transcriptome was characterized using IonTorrent PGM sequencing of ribosomal RNA depleted total mRNA (RNA-Seq). The sequencing reads were first mapped to the ribosomal RNA operon and all tRNA genes, and mapped reads were discarded. The remaining reads were mapped to the gene coding sequences extracted from the genome sequence of strain SolV (Anvar et al, 2014). The total number of reads obtained and mapped on the coding sequences of the genome for each sample together with the calculated expression levels (RPKM) is provided in the supplementary material (Supplementary Table S4). To compare baseline expression levels, we selected a group of 393 housekeeping genes (in total 442.8 kbp) (Khadem et al, 2012a, b) involved in energy generation, ribosome assembly, carbon fixation (CBB cycle), C1 metabolism (except for pmo), amino acid synthesis, cell wall synthesis, translation, transcription, DNA replication and tRNA synthesis. All ratios of expression levels of the housekeeping genes among these conditions were between 0.5 and 2 (Supplementary Table S5). The robustness of the transcriptome data were tested using the method of Chaudhuri et al (2011). In this method, the logarithmic value of (RPKM+1) of each condition (in duplicates) was calculated and the values were plotted against each other. This resulted in correlation coefficients of 0.80, 0.82 and 0.87 (Supplementary Figure S4), showing the high robustness of the transcriptome data.
We showed that two uptake hydrogenase encoding genes (hupL and hupS) were upregulated in both hydrogen- and methane-grown cells (both under O2 limitation) compared to cells growing at μmax. Furthermore, the expression of these genes in hydrogen-grown cells were about twofold higher compared to methane-grown cells (Table 2). The transcription of the gene encoding the [NiFe] hydrogenase b-type cytochrome subunit was significantly upregulated in the hydrogen-grown cells compared to the cells grown at μmax with methane (about 23-fold). This b-type cytochrome subunit is known to be coupled with membrane-bound hydrogenases and involved in the energy conservation. Moreover, the putative hupH hydrogenase expression gene was upregulated in the cells grown on hydrogen compared to cells grown at maximum growth rate with methane (about 22-fold). On the other hand, the genes encoding the Group 1 h/5 type hydrogenase (hhyL and hhyS) did not show different expression levels in cells grown under the three conditions tested. The same holds for the genes encoding the accessory proteins. This points to the Group 1d type [NiFe] hydrogenase being the inducible oxygen-sensitive hydrogenase.
Table 2. The transcriptome profile of the genes involved in H2 metabolism and CO2 fixation of Methylacidiphilum fumariolicum SolV.
| Protein annotation | Gene | Gene IDa |
Expression level (RPKM)b |
||
|---|---|---|---|---|---|
| H2 culturec | CH4 cultured | Cells at μmaxe | |||
| Hydrogenase expression/formation protein hoxQ | hoxQ | MfumV2_0886 | 269 | 112 | 157 |
| Putative hydrogenase maturation protease | hycL | MfumV2_0891 | 191 | 184 | 160 |
| [NiFe] hydrogenase nickel insertion protein | hypA | MfumV2_892 | 145 | 137 | 156 |
| [NiFe] hydrogenase Group 1h/5 small subunit | hhyS | MfumV2_978 | 3449 | 1853 | 2076 |
| [NiFe] hydrogenase Group 1h/5 large subunit | hhyL | MfumV2_979 | 2233 | 803 | 1746 |
| Putative maturation protein | hupD | MfumV2_980 | 328 | 388 | 580 |
| [NiFe] hydrogenase Ni-incorporation protein | hhaB | MfumV2_988 | 613 | 543 | 480 |
| High affinity Ni transporter | MfumV2_989 | 325 | 218 | 230 | |
| [NiFe] hydrogenase metallocenter assembly protein HhaF | hhaF | MfumV2_990 | 77 | 43 | 92 |
| [NiFe] hydrogenase metallocenter assembly protein HhaC | hhaC | MfumV2_991 | 240 | 353 | 524 |
| [NiFe] hydrogenase metallocenter assembly protein HhaD | hhaD | MfumV2_992 | 727 | 483 | 948 |
| [NiFe] hydrogenase metallocenter assembly protein HhaE | hhaE | MfumV2_993 | 843 | 734 | 560 |
| Hydrogenase expression protein HupH | hupH | MfumV2_1562 | 608 | 247 | 27 |
| [NiFe] hydrogenase cytochrome b subunit | hupZ | MfumV2_1563 | 818 | 266 | 36 |
| [NiFe] hydrogenase Group 1d large subunit | hupL | MfumV2_1564 | 1596 | 751 | 239 |
| [NiFe] hydrogenase Group 1d small subunit | hupS | MfumV2_1565 | 3219 | 1370 | 47 |
| RuBisCOf small subunit | cbbS | Mfumv2_1495 | 6826 | 3456 | 8520 |
| RuBisCO large subunit | cbbL | Mfumv2_1496 | 7576 | 4028 | 6419 |
Available at the MicroScope annotation platform (https://www.genoscope.cns.fr/agc/microscope/home/).
The mRNA expression is shown as RPKM according to Mortazavi et al (Saitou and Nei, 1987). Changes in expression in the continuous cultures (substrates H2 or CH4) compared to batch culture cells growing at μmax are demonstrated by shading (up-regulation>twofold, dark grey, downregulation<0.5, light grey).
Continuous culture using H2 as an electron donor.
Continuous culture using CH4 as an electron donor.
Batch culture using CH4 as an electron donor to obtain μmax=0.07 h−1.
Ribulose-1,5-bisphosphate carboxylase/oxygenase.
Furthermore, the transcriptome data showed that the hydrogen treatment had no significant influence on expression levels of the Rubisco subunits. The genes encoding the small (cbbS) and large subunit (cbbL) were highly expressed under all conditions tested (Table 2; Supplementary Table S4), supporting autotrophic growth on hydrogen and carbon dioxide.
Discussion
In their natural ecosystems the methanotroph Methylacidiphilum fumariolicum SolV and its close relatives encounter both methane and hydrogen under acidic (pH 1–2) and hot (50–70 °C) conditions with low availability of oxygen (Chiodini et al, 2001; Pol et al, 2007). Physiological experiments with both batch and continuous culture of strain SolV described in this study showed that this bacterium can grow autotrophically on hydrogen and carbon dioxide, using the ‘Knallgas' reaction to gain energy and the Rubisco pathway for carbon fixation. No supply of methane is needed. When growing on methane, hydrogen oxidation was possible under ambient oxygen concentration, but growth on hydrogen and carbon dioxide was possible only below 1.5% oxygen. Highest growth rates were obtained when oxygen was very low (dO2<0.1%). The maximum specific growth rate on hydrogen (0.047 h−1) is about 60% compared to the growth rate on methane (Pol et al, 2007). Assuming the same CO2:H2 consumption ratio as measured at D=0.0124 h−1, such a growth rate needs a specific hydrogenase activity of about 175 nmol H2.min−1.mg DW−1 (the carbon content of DW was 51%). This matches well with the in vitro measured maximum hydrogen uptake rates (220 nmol H2.min−1.mg DW−1). The measured yield on hydrogen is about half that on methane (3.4 vs 6.4 g DW/mole CH4) (Pol et al, 2007) and slightly lower than those reported for ‘Knallgas' bacteria like Ralstonia eutropha (4.6 g/mole H2) (Morinaga et al, 1978) or Hydrogenomonas eutropha (5 g/mole H2) (Bongers, 1970).
Continuous cultures fed with hydrogen under low oxygen conditions remained stable for more than a year. Although previous studies have shown that co-oxidation of hydrogen together with methane was possible (de Bont, 1976; Hanczár et al, 2002), to our knowledge this is the first time that a methanotroph is shown to grow on hydrogen/carbon dioxide only in liquid mineral medium. Baxter et al (2002) reported growth with hydrogen and carbon dioxide on agar plates but not in liquid cultures for several methanotrophs but autotrophy was not substantiated by physiological and biochemical evidence. The presence of a fully operational Calvin cycle with Rubisco as the key enzyme in verrucomicrobial methanotrophs seems to be a pre-requisite for autotrophic growth on hydrogen.
Recently, it has been shown that microorganisms from a wide diversity of ecosystems, ranging from the hypoxic hydrogen-rich habitats of animal guts and bog soils to aerated soils and waters containing small quantities of H2, possess genes encoding different types of hydrogenases (Greening et al, 2016). We showed that Methylacidiphilum sp. and Methylacidimicrobium sp. representing the acidophilic verrucomicrobial methanotrophs (Op den Camp et al, 2009; Sharp et al, 2014; van Teeseling et al, 2014) contain genes encoding hydrogen uptake hydrogenases. According to the phylogeny of both the large and small subunits, the hup-type hydrogenases in the verrucomicrobial methanotrophs belong to the membrane-bound H2-uptake [NiFe]-hydrogenases (Group 1d), and the hhy-type hydrogenases (present only in strains SolV and Kam1) belong to Group 1 h/5 hydrogenases as they are not membrane anchored (Greening et al, 2016). Based on the phylogenetic analysis, we showed that several alpha- and gammaproteobacterial methanotrophic species contain hydrogenase genes, and growth with hydrogen and carbon dioxide was reported on agar plates but not in liquid cultures for several methanotrophs (Baxter et al, 2002). In methanotrophic bacteria that cannot fix CO2, hydrogenases may help to save reducing equivalents by recycling the hydrogen gas that is produced during nitrogen fixation, or supply reducing equivalents for the methane monooxygenase (Csáki et al, 2001; Hanczár et al, 2002; Bothe et al, 2010).
The small subunits of the hup-type hydrogenases contain the typical twin-arginine signal peptide distinctive for periplasmic membrane-bound Group 1 hydrogenases (Palmer and Berks, 2012). The hup-type hydrogenases of the thermophilic strains have features typical for Group 1d. Their large subunit contains the typical L1 (xxRICGVCTxxH) and L2 (SFDPCLACxxH) motifs and their small subunit contains the conserved motif with two extra cysteines (Greening et al, 2016). In Ralstonia eutropha these extra cysteines have been shown to ligate to an ‘exclusive' proximal 4Fe3S cluster that is supposed to render hydrogenases extra oxygen tolerance (Fritsch et al, 2011). In contrast, HupS subunits of the mesophilic strains do not contain these extra cysteines. Instead cysteines seem to be arranged to coordinate a 4Fe4S proximal cluster like most Group 1 members (not being the Group 1d type), although one of the four cysteines is replaced by an asparagine. A similar change is also observed in Group 1f hydrogenases of Roseoflexis, Geobacter and Frankia spp. where the cysteine is replaced by asparagine or aspartic acid. Asparagine coordination to 4Fe4S is also present in Group 2a hydrogenases. Conserved cysteines for the medial FeS cluster (3Fe4S coordinating with three cysteines, as in Group 1b–g) and distal FeS cluster (4Fe4S, coordinating with three cysteines and one histidine, as in Group 1a–h) are present in the HupS of both mesophilic and thermophilic strains. The Group 1d hydrogenase in strain SolV was shown to be responsible for increased growth rates when dO2 level was below 0.2% (see above).
Furthermore, we showed that genes encoding an additional hydrogenase are present in the thermophilic strains SolV and Kam1. Based on the phylogenetic analysis of both its small and large subunits, this hydrogenase belongs to Group 1h/5 hydrogenases. They contain active site L1 (TSRICGICGDNH) and L2 (SFDPCLPCGVH) motifs that are highly conserved in Group 1h/5 hydrogenases and distinct from other Group 1–4 hydrogenases (Greening et al, 2016). These hydrogenases are located at the cytoplasmic side of the membrane, since they lack the twin-arginine signal peptide in the small subunit. The small subunit also lacks the C-terminal membrane span alpha helix extension to anchor in a membrane. The proximal FeS cluster is reminiscent to the mesophilic counterpart (discussed above) and others in Group 1b, f, g in that it has only three cysteines available for coordination. The fourth position for a 4Fe4S cluster would then be the conserved Asp (de Bont, 1976) as shown for the Group 1h/5 hydrogenase of Ralstonia eutropha (Schäfer, 2014) and could be a possible clue to the extreme oxygen tolerance of group 1h/5 hydrogenases (Supplementary Figure S5).
The transcriptome analysis clearly showed that the Group 1d hydrogenase gene cluster was upregulated under oxygen-limited conditions consistent with higher hydrogenase activity and growth rates found at dO2 values below 0.2%. Previously, we showed that these two genes were also upregulated under the N2-fixing condition (Khadem et al, 2012a). In contrast, the other hydrogenase encoding gene cluster (Group 1h/5) and all accessory proteins were similarly expressed under oxygen-limited conditions (continuous culture) and under oxygen excess conditions (batch culture). This clearly correlates with the constant hydrogen uptake (activity) found in respiration experiments for cells grown under higher oxygen concentrations (above 0.2% O2). Therefore we may conclude that the observed growth with hydrogen found at these higher dO2 values is supported exclusively by the Group 1h/5 uptake hydrogenase.
Growth on hydrogen only sustained by a Group 1h/5 hydrogenase has not been reported before. Other microorganisms expressing Group 1h/5 hydrogenases activity co-metabolize hydrogen but have not been shown to grow on or benefit from this hydrogen consumption. Group 1h/5 hydrogenase activity has been reported for Actinobacteria like Mycobacterium smegmatis (Greening et al, 2014; Berney et al, 2014) (2.5 nmol.min−1.g DW−1; 10 μmol.min−1.g protein−1), and Streptomyces strains (Constant et al, 2010) (0.2–2 nmol.min−1.g DW−1), whereas Meredith et al (2014) reported extremely higher Vmax values of 14–180 μmol.min−1.g protein−1 for Streptomyces species. ‘Knallgas' bacteria like Ralstonia sp. need the Group 1d hydrogenase for growth and express relatively low Group 1h/5 hydrogenase activity (Schäfer et al, 2013).
Although the Group 1h/5 hydrogenase of strain SolV was active up to at least ambient O2, growth on hydrogen was only observed below 1.5% O2. Clearly other factors also determine the oxygen tolerance during growth on hydrogen. The affinity for hydrogen of this enzyme (Ks 0.6 μM) was lower than those reported for Group 5 hydrogenases of M. smegmatis (50 nM) (Greening et al, 2014) and Streptomyces species (10 nM) (Constant et al, 2008); (40–400 nM) (Constant et al, 2010)). These Actinobacteria are held responsible for the high-affinity hydrogen uptake from the atmosphere (Constant et al, 2010; Meredith et al, 2014). The proteobacterial R. eutropha expresses only low Group 5 hydrogenase activity and showed a low affinity for hydrogen (Km 3.6 μM, after purification) (Schäfer et al, 2013).
The hydrogen kinetics of the Group 1d hydrogenase of M. fumariolicum SolV, which is active only at low oxygen concentrations, could not be determined due to the constitutive expression of Group 1 h/5 hydrogenase. However, cells with both hydrogenases active exhibited an affinity constant of about 1 μM and we estimated that the apparent affinity constant of the Group 1d hydrogenase was higher than 1 μM but less than 2 μM, similar to that of group 1 containing proteobacterial ‘Knallgas' bacteria (Ludwig et al, 2009).
It should be realized that when using whole cells (or soils) to measure hydrogenase kinetics, only a so-called apparent affinity constant is obtained, which depends on the measured Vmax. This Vmax is not necessarily the Vmax of the hydrogenase enzyme but that of the overall reaction that may be limited by the follow-up steps being respiration and this may explain the large variations found in values (nM to low μM range) of closely related strains that contain the same hydrogenase (Greening et al, 2014). As a consequence the existence of a high affinity hydrogenase remains doubtful.
The mesophilic Methylacidimicrobium strains lack a Group 1h/5 hydrogenase and an O2-tolerant Group 1d hydrogenase. They only possess a hydrogenase closely related to group 1b hydrogenases. These are found mainly in microorganisms with an anoxic H2 respiration or photosynthesis, or supposed to be involved in protection against oxygen like in Geobacter sulfurreducens (Coppi et al, 2004). Thus we assumed that these mesophilic strains only could grow on hydrogen when oxygen is limited. Preliminary tests to grow Methylacidimicrobium tartarophylax 4AC on hydrogen showed that the hydrogenase is inactivated when the sensor reading dO2 value is above detection limit (0.01% oxygen) confirming the high sensitivity of this hydrogenase to oxygen.
The presence of an oxygen-insensitive (group 1h/5) hydrogenase is of great importance for hydrogen consumption in environments like oxic upper soil layers that have been reported to take up hydrogen from the atmosphere. Although growth of methanotrophic Verrucomicrobia on ppm level ambient hydrogen may not be likely, the simultaneous uptake of hydrogen at such low concentrations may occur while these bacteria grow on methane and possibly hydrogen produced in anoxic layers beneath. Perhaps more interestingly vice versa: reducing power of hydrogen may lower the threshold for oxidizing methane as described for methanol (Benstead et al, 1998; Jensen et al, 1998). This threshold has been suggested to be determined by the availability of reductant needed for the first step in methane oxidation. This step is performed by a methane monooxygenase and hydrogen has been shown to be a suitable donor of reducing equivalents (Hanczár et al, 2002). In the volcanic environment from where M. fumariolicum SolV was isolated, hydrogen concentrations in emitted gasses are much higher than those of methane (Chiodini et al, 2001) and this hydrogen might explain the uptake of methane even at atmospheric levels that was reported by Castaldi and Tedesco (2005). Greening and co-workers (Greening et al, 2016) showed that the occurrence of [NiFe] aerobic H2-uptake hydrogenases in the phylum Verrucomicrobia is more widespread, and suggests that Verrucomicrobia play a role in the hydrogen cycle not only in areas that emit geothermal gasses, but also in ecosystems such as rice paddy soil (van Passel et al, 2011), pasture soil (Kant et al, 2011) and the human gut (Derrien et al, 2004).
In conclusion, M. fumariolicum SolV is a real ‘Knallgas' bacterium but also able to consume methane and hydrogen simultaneously using an oxygen-sensitive and oxygen-tolerant hydrogenase, respectively. Detailed physiological studies and transcriptome analysis of the Methylacidiphilum strains that can grow on hydrogen is required to understand this interesting group of hydrogenases. Oxygen sensitivity of hydrogenases is a major drawback in application (Jugder et al, 2016). In view of the high oxygen tolerance of the Group 1 h/5hydrogenase from strain SolV, biochemical and biophysical studies of the purified hydrogenase are essential since this enzyme has a high potential in biotechnological applications.
Acknowledgments
Sepehr Mohammadi was supported by the Spinoza grant of Mike SM Jetten (The Netherlands Organization for Scientific Research) and the European Research Council (ERC Advanced Grant project VOLCANO 669371), Mike S.M. Jetten by the European Research Council (ERC Advanced Grant Eco_MoM 339880) and Huub JM Op den Camp by the European Research Council (ERC Advanced Grant project VOLCANO 669371).
Author contributions
Sepehr Mohammadi and Arjan Pol and Theo A. van Alen executed experiments and analysed data. Sepehr Mohammadi and Huub JM Op den Camp contributed to transcriptome analyses. Sepehr Mohammadi, Arjan Pol, Huub JM Op den Camp and Mike SM Jetten planned research. Sepehr Mohammadi, Arjan Pol, and Huub JM Op den Camp wrote the paper. All authors discussed results and commented on the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Anvar SY, Frank J, Pol A, Schmitz A, Kraaijeveld K, den Dunnen JT et al. (2014). The genomic landscape of the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. BMC Genom 15: 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter NJ, Hirt RP, Bodrossy L, Kovacs KL, Embley TM, Prosser JI et al. (2002). The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath). Arch Microbiol 177: 279–289. [DOI] [PubMed] [Google Scholar]
- Belova SE, Baani M, Suzina NE, Bodelier PL, Liesack W, Dedysh SN. (2011). Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ Microbiol Rep 3: 36–46. [DOI] [PubMed] [Google Scholar]
- Benstead J, King GM, Williams HG. (1998). Methanol promotes atmospheric methane oxidation by methanotrophic cultures and soils. Appl Environ Microbiol 64: 1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney M, Greening C, Hards K, Collins D, Cook GM. (2014). Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ Microbiol 16: 318–330. [DOI] [PubMed] [Google Scholar]
- Bongers L. (1970). Yields of Hydrogenomonas eutropha from growth on succinate and fumarate. J Bacteriol 102: 598–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe H, Schmitz O, Yates MG, Newton WE. (2010). Nitrogen fixation and hydrogen metabolism in Cyanobacteria. Microbiol Mol Biol Rev 74: 529–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaldi S, Tedesco D. (2005). Methane production and consumption in an active volcanic environment of Southern Italy. Chemosphere 58: 131–139. [DOI] [PubMed] [Google Scholar]
- Chaudhuri RR, Yu L, Kanji A, Perkins TT, Gardner PP, Choudhary J et al. (2011). Quantitative RNA-seq analysis of the Campylobacter jejuni transcriptome. Microbiology 157: 2922–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodini G et al. (2001). CO2 degassing and energy release at Solfatara volcano, Campi Flegrei, Italy. J Geophys Res 106: 16213–16221. [Google Scholar]
- Constant P, Chowdhury SP, Pratscher J, Conrad R. (2010). Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase. Environ Microbiol 12: 821–829. [DOI] [PubMed] [Google Scholar]
- Constant P, Poissant L, Villemur R. (2008). Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. ISME J 2: 1066–1076. [DOI] [PubMed] [Google Scholar]
- Coppi MV, O'Neil RA, Lovley DR. (2004). Identification of an uptake hydrogenase required for hydrogen-dependent reduction of Fe(III) and other electron acceptors by Geobacter sulfurreducens. J Bacteriol 186: 3022–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csáki R, Hanczár T, Bodrossy L, Murrell JC, Kovács KL. (2001). Molecular characterization of structural genes coding for a membrane bound hydrogenase in Methylococcus capsulatus (Bath). FEMS Microbiol Lett 205: 203–207. [DOI] [PubMed] [Google Scholar]
- Dalton H, Whittenbury R. (1976). Acetylene reduction technique as an assay for nitrogenase activity in methane oxidizing bacterium Methylococcus capsulatus strain Bath. Arch Microbiol 109: 147–151. [Google Scholar]
- de Bont JAM. (1976). Hydrogenase activity in nitrogen-fixing methane-oxidizing bacteria. Antonie Van Leeuwenhoek 42: 255–259. [DOI] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54: 1469–1476. [DOI] [PubMed] [Google Scholar]
- Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S et al. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450: 879–883. [DOI] [PubMed] [Google Scholar]
- Erikstad HA, Birkeland NK. (2015). Draft genome sequence of “Candidatus Methylacidiphilum kamchatkense” strain Kam1, a thermoacidophilic methanotrophic Verrucomicrobium. Genome Announc 3: e00065–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch J, Scheerer P, Frielingsdorf S, Kroschinsky S, Friedrich B, Lenz O et al. (2011). The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature 479: 249–253. [DOI] [PubMed] [Google Scholar]
- Greening C, Berney M, Hards K, Cook GM, Conrad R. (2014). A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci USA 111: 4257–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB et al. (2016). Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10: 761–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanczár T, Csáki R, Bodrossy L, Murrell JC, Kovács KL. (2002). Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch Microbiol 177: 167–172. [DOI] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. (1996). Methanotrophic bacteria. Microbiol Rev 60: 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Makarova KS, Saw JH, Senin P, Ly BV, Zhou Z et al. (2008). Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol Direct 3: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam T, Jensen S, Reigstad LJ, Larsen O, Birkeland NK. (2008). Methane oxidation at 55 °C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci USA 105: 300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Priemé A, Bakken L. (1998). Methanol improves methane uptake in starved methanotrophic microorganisms. Appl Environ Microbiol 64: 1143–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugder BE, Lebhar H, Aguey-Zinsou KF, Marquis CP. (2016). Production and purification of a soluble hydrogenase from Ralstonia eutropha H16 for potential hydrogen fuel cell applications. MethodsX 3: 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant R, van Passel MW, Sangwan P, Palva A, Lucas S, Copeland A et al. (2011). Genome sequence of “Pedosphaera parvula” Ellin514, an aerobic Verrucomicrobial isolate from pasture soil. J Bacteriol 193: 2900–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler U, Nouwens AS. (2013). Metabolic adaptation and trophic strategies of soil bacteria-C1-metabolism and sulfur chemolithotrophy in Starkeya novella. Front Microbiol 17: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DP, Anthony C, Murrell JC. (2005). Insights into the obligate methanotroph Methylococcus capsulatus. Trends Microbiol 13: 195–198. [DOI] [PubMed] [Google Scholar]
- Kelly DP, McDonald IR, Wood AP. (2000). Proposal for the reclassification of Thiobacillus novellus as Starkeyanovella gen. nov., comb. nov., in the alpha- subclass of the Proteobacteria. Int J Syst Evol Microbiol 50: 1797–1802. [DOI] [PubMed] [Google Scholar]
- Keltjens JT, Pol A, Reimann J, Op den Camp HJM. (2014). PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol 98: 6163–6183. [DOI] [PubMed] [Google Scholar]
- Khadem AF, Pol A, Wieczorek A, Mohammadi SS, Francoijs KJ, Stunnenberg HG et al. (2011). Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J Bacteriol 193: 4438–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem AF, Pol A, Jetten MSM, Op den Camp HJM. (2010). Nitrogen fixation by the verrucomicrobial methanotroph “Methylacidiphilum fumariolocum” SolV. Microbiology 156: 1052–1059. [DOI] [PubMed] [Google Scholar]
- Khadem AF, Pol A, Wieczorek AS, Jetten MSM, Op den Camp HJM. (2012. a). Metabolic regulation of “Ca. Methylacidiphilum fumariolicum” SolV cells grown under different nitrogen and oxygen limitations. Front Microbiol 3: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem AF, van Teeseling MC, van Niftrik L, Jetten MS, Op den Camp HJ, Pol A. (2012. b). Genomic and physiological analysis of carbon storage in the verrucomicrobial methanotroph “Ca. Methylacidiphilum fumariolicum” SolV. Front Microbiol 3: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C. (2015). Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol 15: 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb S. (2009). Aerobic methanol- oxidizing Bacteria in soil. FEMS Microbiol Lett 300: 1–10. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Cracknell JA, Vincent KA, Armstrong FA, Lenz O. (2009). Oxygen-tolerant H2 oxidation by membrane-bound [NiFe] hydrogenases of Ralstonia species. J Biol Chem 284: 465–477. [DOI] [PubMed] [Google Scholar]
- Meredith LK, Rao D, Bosak T, Klepac-Ceraj V, Tada KR, Hansel CM et al. (2014). Consumption of atmospheric hydrogen during the life cycle of soil-dwelling actinobacteria. Environ Microbiol Rep 6: 226–238. [DOI] [PubMed] [Google Scholar]
- Morinaga Y, Yamanaka S, Ishizaki A, Hirose Y. (1978). Growth characteristics and cell composition of Alcaligenes eutrophus in chemostat culture. Agric. Biol Chem 42: 439–444. [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Murrell JC, Jetten MSM. (2009). The microbial methane cycle. Environ Microbiol Rep 1: 279–284. [DOI] [PubMed] [Google Scholar]
- Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1: 293–306. [DOI] [PubMed] [Google Scholar]
- Palmer T, Berks BC. (2012). The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 10: 483–496. [DOI] [PubMed] [Google Scholar]
- Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MS, Op den Camp HJ. (2007). Methanotrophy below pH1 by a new Verrucomicrobia species. Nature 450: 874–879. [DOI] [PubMed] [Google Scholar]
- Pol A, Barends TR, Dietl A, Khadem AF, Eygensteyn J, Jetten MS et al. (2013). Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol 16: 255–264. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schäfer C. (2014), Proteinbiochemische, spektroskopische und röntgenkristallographische Untersuchung der Actinobakteriellen [NiFe]-Hydrogenase aus Ralstonia eutropha. PhD thesis, Humboldt-University, Berlin, Germany.
- Schäfer C, Friedrich B, Lenz O. (2013). Novel, oxygen-insensitive group 5 [NiFe]-hydrogenase in Ralstonia eutropha. Appl Environ Microbiol 79: 5137–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau JD. (2011). Bioremediation via methanotrophy: overview of recent findings and suggestions for future research. Front Microbiol 2: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp CE, Smirnova AV, Graham JM, Stott MB, Khadka R, Moore TR et al. (2014). Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ Microbiol 16: 1867–1878. [DOI] [PubMed] [Google Scholar]
- Sharp CE, Stott MB, Dunfield PF. (2012). Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Evol Gen Microbiol 3: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SH, Dalton H. (1982). Role of ribulose-1,5-bisphosphate carboxylase/oxygenase in Methylococcus capsulatus (Bath). J Gen Microbiol 128: 2927–2935. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SC, Dalton H, Dow CS. (1981). Ribulose-1,5-bisphosphate carboxylase/oxygenase and carbon assimilation in Methylococcus capsulatus (Bath). J Gen Microbiol 122: 89–94. [Google Scholar]
- Theisen AR, Murrell JC. (2005). Facultative methanotrophs revisited. J Bacteriol 187: 4303–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H, Brusseau GA, Hanson RS, Wackett LP. (1989). Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol 55: 3155–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel MW, Kant R, Palva A, Copeland A, Lucas S, Lapidus A et al. (2011). Genome sequence of the verrucomicrobium Opitutus terrae PB90-1, an abundant inhabitant of rice paddy soil ecosystems. J Bacteriol 193: 2367–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling MCF, Pol A, Harhangi HR, van der Zwart S, Jetten MS, Op den Camp HJ et al. (2014). Expanding the Verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl Environ Microbiol 80: 6782–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais PM, Billoud B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev 107: 4206–4272. [DOI] [PubMed] [Google Scholar]
- Whittenbury R, Phillips KC, Wilkinson JF. (1970). Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol 61: 205–218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.