Abstract
Rare species are increasingly recognized as crucial, yet vulnerable components of Earth's ecosystems. This is also true for microbial communities, which are typically composed of a high number of relatively rare species. Recent studies have demonstrated that rare species can have an over-proportional role in biogeochemical cycles and may be a hidden driver of microbiome function. In this review, we provide an ecological overview of the rare microbial biosphere, including causes of rarity and the impacts of rare species on ecosystem functioning. We discuss how rare species can have a preponderant role for local biodiversity and species turnover with rarity potentially bound to phylogenetically conserved features. Rare microbes may therefore be overlooked keystone species regulating the functioning of host-associated, terrestrial and aquatic environments. We conclude this review with recommendations to guide scientists interested in investigating this rapidly emerging research area.
Introduction
Microbial communities typically show a skewed species abundance distribution, with relatively few dominant species co-existing alongside a high number of rare species (Nemergut et al., 2011). Although the importance of rare species is increasingly being recognized for macroorganisms (Lyons et al., 2005; Mouillot et al., 2013; Soliveres et al., 2016), many rare microbial taxa (for example, singletons) are routinely removed from data sets, thereby systemically overlooking a substantial part of the biosphere. Most of our knowledge is still based on dominant species, despite the increasing attention for the rare microbial biosphere (Pedrós-Alió, 2007; Reid and Buckley, 2011; Lynch and Neufeld, 2015). In this review, we place rare microbes in an interdisciplinary spotlight and discuss their relevance across a range of current hot topics in microbial ecology, including microbiome assembly and function, and biogeochemical cycling. We present patterns and consequences of rarity and link these to existing and emerging theoretical frameworks.
Microscopic organisms, both eukaryotic and prokaryotic, display a huge biodiversity (Locey and Lennon, 2016). Microbial communities drive Earth's biogeochemical cycles (Falkowski et al., 2008) and are essential for animal and plant survival (Russell et al., 2014). An estimated 1.5–28% of all microbes are ‘conditionally rare taxa', which are rare in most conditions but become dominant occasionally (Shade et al., 2014). These often-overlooked taxa may be key to understanding community assembly and function (Allan et al., 2011).
The increasing resolution provided by high-throughput sequencing technologies has shed new light on microbial diversity and revealed a diverse collection of microbes occurring at low densities: the ‘rare biosphere' (Sogin et al., 2006). As such, the rare biosphere is currently one of the new frontiers of microbial ecology. Recent studies have shown that low-abundance species should be considered full members of microbial communities and not treated as analytical annoyances (Nipperess and Matsen, 2013; McMurdie and Holmes, 2014; Lynch and Neufeld, 2015).
We provide an overview of theories on why species are rare (including community assembly, evolutionary processes and biogeographic patterns), and how low-abundance species can drive ecosystem processes. We also provide evidence for the importance of rare microbes across different habitats, and present approaches to examine the most pressing questions surrounding them.
Drivers of rarity
Rarity can be defined in several ways, including for instance local abundance, habitat specificity and geographical spread (Rabinowitz, 1981). These concepts are routinely applied to plants and animals, and can be easily transferred to microbial communities. Local abundance is the easiest and the most common index used to quantify species' rarity in microbial ecology. However, microbial rarity may also be expressed as a restriction to a low number of habitats, thereby reflecting habitat specificity (Barberán et al., 2014) and geographic range (Tedersoo et al., 2014).
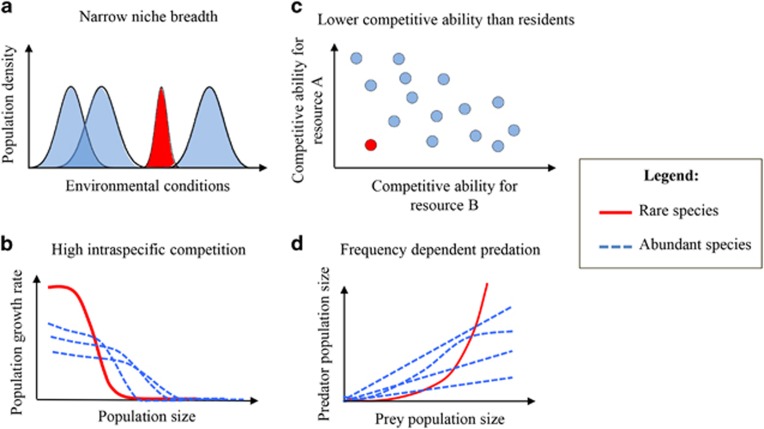
Rarity can result from stochastic processes, inherent trade-offs in life-history strategies, and biotic and abiotic interactions (Figure 1). Species rarity can for instance emerge simply by stochastic population fluctuation (Ai et al., 2013), without implying any specific physiological characteristics. Rarity is also an element of community assembly processes as a recently immigrated species is necessarily rare when it first enters a new community. External abiotic and biotic factors can have pivotal roles in species rarity. For instance, a highly specialized species showing a very narrow environmental niche may be abundant in a few habitats but remain rare in most others (Figure 1a). Local rarity may also be the result of fitness trade-offs, for instance, when stress resistance comes at the cost of a lower growth rate. Slow-growing species might not reach a high density, but may persist well under stressful conditions (Gudelj et al., 2010). An extreme case is dormancy, in which microbes completely stop growth but gain greatly in stress resistance (Gobet et al., 2012). Many microbes can remain generally inactive and at low density most of the time, only becoming dominant when more favorable conditions arise (Aanderud et al., 2015). Biotic interactions also have also an important role in explaining rarity. An uncompetitive species may often remain rare rather than going extinct. Rare persistence has been demonstrated for microbes that are sensitive to antibiotics produced by competitors (Narisawa et al., 2008), unable to build protective structures such as biofilms (Schluter et al., 2015) or incapable of using crucial resources (García-Fernández et al., 2004). The dependance of one species on another may lead to negative frequency dependency, where the fitness of the dependent phenotype decreases when it becomes more abundant. A well described example involves social cheaters depending on public goods produced by other species (Kummerli and Brown, 2010; Jousset et al., 2013). Cheats may be highly successful while rare, but their competitive advantage rapidly dwindles as they spread (Figure 1b). The strength of frequency-dependent selection may vary with environmental factors such as predators (Meyer and Kassen, 2007): the steeper the slope of the frequency-dependent fitness, the more persistent a rare species will be, but at the cost of remaining rare (Yenni et al., 2012).
Figure 1.
Potential mechanisms that can drive local rarity. Rarity can be linked to two types of mechanisms: (1) mechanisms related to species characteristics (a, b), and (2) mechanisms related to local biotic and abiotic conditions (c, d). Drivers of rarity include (a) narrow niche breadth, (b) high intraspecific competition, (c) low competitive abilities and (d) frequency-dependent predation. In all panels, rare species are indicated in red, while abundant species are indicated in blue.
Bacteriophages and protists often show frequency-dependent predation and tend to over-consume abundant prey species (Jousset et al., 2009; Winter et al., 2010), preventing them from dominating the community and creating space for rare species (Rodriguez-Valera et al., 2009; Figure 1d). Finally, rarity may be driven by changes in abiotic conditions, as rare species are more sensitive to environmental fluctuations and more prone to extinction (Gaston, 2008). Current abrupt changes linked to human activity, such as global change and intensive land use, may thus have a deep impact on the rare biosphere and associated ecosystem functions (Rodrigues et al., 2013). Monitoring changes in relative abundances or activities of rare microbes might then serve as an early warning system of environmental change (Chambers et al., 2016).
Ecological relevance of rare species
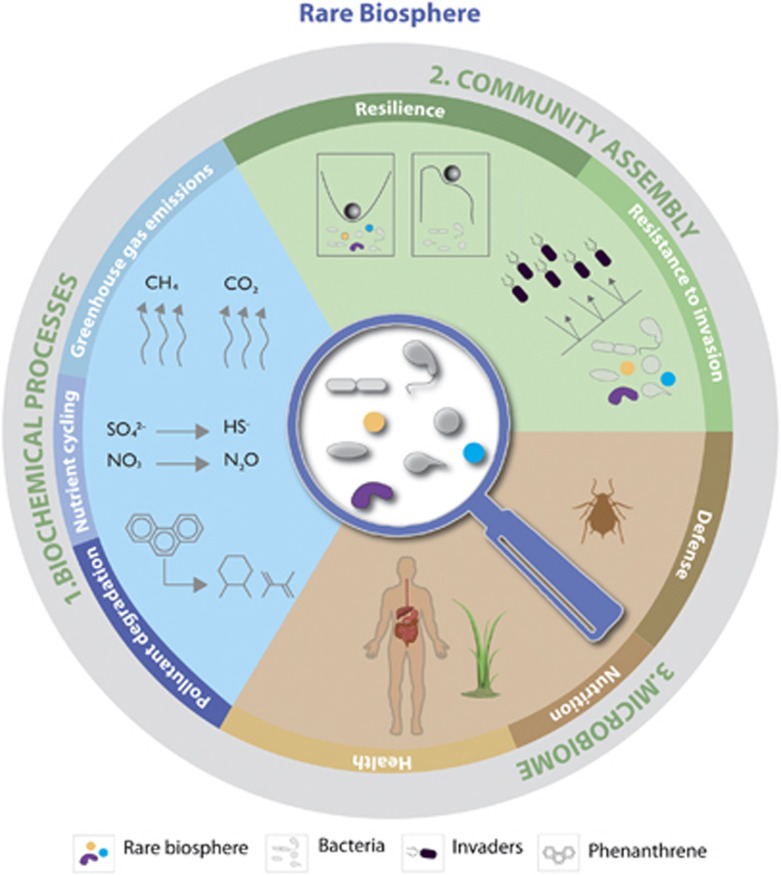
Rare species are increasingly recognized as drivers of key functions in terrestrial and aquatic ecosystems, as well as host-associated microbiomes (Figure 2; Table 1). Their functional importance may be due to effects that are disproportionately large given their abundance or via the provision of insurance effects. This may seem counter-intuitive given the expected high functional redundancy in microbial communities (Rousk et al., 2009): with many species sharing similar functions, rare species should not be necessary for maintaining function. Species that are considered functionally non-relevant under a given environmental condition may become important under changing conditions by providing necessary traits or acting as partners in new interspecific interactions (Shade et al., 2014; Fetzer et al., 2015). We propose that rare species offer a pool of genetic resources that may be activated under the appropriate conditions. This may provide insurance effects (Yachi and Loreau, 1999), as at least one species will perform a given process under a given environmental condition. Different processes vary in their sensitivity to the loss of rare species (Peter et al., 2011) with specialized functions, like pollutant degradation (Dell'Anno et al., 2012), particularly affected.
Figure 2.
Overview of ecosystem functions supported by rare species. Rare species can drive key processes in geochemical cycles and are major players in nutrient cycling greenhouse gas emissions and pollutant degradation. They can affect community assembly by preventing the invasion of new species, and stabilize community function in fluctuating environments. Further, rare microbes are essential players of host-associated microbiomes in plants and animals by preventing pathogen establishment and stimulating host immunity. References are given in Table 1.
Table 1. References for processes or functions which are supported by rare microbes.
Background information for Figure 2.
Research on the ecological importance of rare species is still in its infancy, but empirical evidence that the rare biosphere is involved in many microbial-driven processes is accumulating (Figure 2). Below, we highlight three areas in which rare microbes could be particularly influential.
Biochemical processes—nutrient cycling and pollutant degradation
Some nutrient cycling processes provide illustrative typical examples of the disproportionate effects of rare microbes. Low-abundance green and purple sulfur bacteria were found to be highly active keystone species in freshwater and crucial for nitrogen and carbon uptake (Musat et al., 2008). Similarly, Pester et al. (2010) found that the most important sulfate reducer in peatland was a rare bacterium with only 0.006% relative 16 S rRNA gene abundance. Denitrification also likely relies on rare species. A decrease of 75% of the measured species richness reduced soil denitrifying activity by a factor of 4–5 fold (Philippot et al., 2013), suggesting that dominant species cannot carry out this process alone.
Microbial communities play a key role in the degradation of organic compounds, including pollutants and the insurance provided by rare species can therefore contribute to ecosystem resilience to anthropogenic pollution. Organic pollutant degradation involves complex metabolic pathways shared across different species (Fuentes et al., 2014). Removal of rare microbes in activated sludge and freshwater greatly reduced the capacity to degrade pollutants and toxins (Dell'Anno et al., 2012; Hernandez-Raquet et al., 2013; Delgado-Baquerizo et al., 2016). Rare species probably offer the required gene pool to catalyze complex degradation processes, a hypothesis supported by observations that pollutants are often degraded by species falling below the detection limit in pristine samples (Giebler et al., 2013).
The role of rare species in the breakdown of organic matter is far less clear. Some studies found no consistent relationship between bacterial diversity and litter decomposition, concluding that a low number of species sufficed for efficient decomposition (Griffiths et al., 2001; Franklin and Mills, 2006). Other studies, however, highlighted that rare species may speed up decomposition (Salonius, 1981), especially the degradation of recalcitrant organic matter such as chitin and cellulose (Peter et al., 2011; Jimenez et al., 2014).
Community assembly
Experimental removal of rare species resulted in an increased establishment of new species (Van Elsas et al., 2012; Vivant et al., 2013), suggesting that rare species occupy a key niche and slow down invasive species establishment. A recent study demonstrated that rare species are vital in controlling invasions into soil communities by unwanted (for example, pathogenic) microbes (Mallon et al., 2015), suggesting that rare microbes may play an important role in biotechnological applications such as hygiene or crop protection.
Microbiome—host health
Rare species have been detected within the microbiomes of diverse hosts, from the rhizosphere (Nuccio et al., 2016) to human lungs (Guss et al., 2011), and evidence suggests that they can be critical to microbiome functionality and thus host health. Low-abundance plant-associated microbes are for instance involved in the production of antagonistic volatile compounds that protect the host plant against pathogens (Hol et al., 2015b). Further, removing rare species from an agricultural soil led to a higher plant biomass, albeit at the cost of a reduction of defense compounds against aboveground herbivores (Hol et al., 2010), suggesting that rare microbes can fine-tune the balance between host growth and defense.
In the human lung, a high diversity of low-abundance bacteria is associated with a reduced severity of bacterial infection in individuals with cystic fibrosis (van der Gast et al., 2011). Conversely, rare species may contribute to pathogenesis, as shown by the initiation of periodontal disease by low-abundance species that trigger changes in the oral microflora (Hajishengallis et al., 2011). Several studies have further identified low-abundance pathogens in clinically relevant samples (Guss et al., 2011; Bittinger et al., 2014).
Whilst rare species have traditionally received little attention from a clinical or biotechnological perspective, a better understanding of their ecology, and impact on microbiome function, may provide new tools to predict, and enhance, health effects of host-associated microbiomes.
Mechanisms behind disproportional effects of rare microbes
Here, we discuss three, non-mutually exclusive, mechanisms that may explain the high importance of rare microbes for ecosystem functioning: enhanced species activity, increased community functional diversity and community-wide species interactions.
Rare microbes can be more active than the abundant ones
In marine ecosystems, conditionally rare bacterial taxa tend to be more active when rare (Campbell et al., 2011). In fluctuating conditions, a rare taxon could be disproportionally active due to continuous regrowth. However, growth may not necessarily be responsible for this increased activity; Desulfosporosinus species, for instance, remain at low abundance despite increasing their ribosome content under activating conditions (Hausmann et al., 2016). A similar observation was reported for protists in lakes, where some species were always rare and yet highly active (Debroas et al., 2015). These examples utilized rRNA:rDNA ratios as a proxy for activity. Similar results have been observed using respiration as a proxy for bacterial activity. Rare species appear to sustain community activity; similar sized communities with rare microbes were shown to have higher respiration than communities with fewer rare microbes (Dimitriu et al., 2010). Thus, rare yet highly active microbes can contribute more to ecosystem functioning than expected based on their abundance, akin to some macroorganisms, such as rare but active predators or pollinators (Herrera, 1989).
Rare species represent a vast functional gene pool
Collectively, rare microbes represent a huge genetic pool and, as such, contribute to the metabolic potential of the community. Many microbial species are auxotrophs, that is, they need to obtain certain vitamins or amino acids from other organisms in their environment (Helliwell et al., 2013). Secretion of such compounds as metabolic waste by other microbes can greatly improve overall community function (Mee et al., 2014). Further, metabolism of complex products requires a large set of enzymes, which are often not present in one single organism. Potentially toxic by-products may thus accumulate if not further degraded by other organisms (Haruta et al., 2009). Dimitriu et al. (2010) measured activities of several enzymes involved in the degradation of organic material and concluded that more diverse bacterial communities harbor a greater set of functions due to the functional differences between species. They suggest that rare species are likely to be functionally dissimilar from abundant ones and therefore are likely to offer complementary, or unique, metabolic pathways to support community function. This high functional diversity of rare species can also be found in macroorganisms, for example, Amazonian fish, rainforest trees and tropical birds (Leitão et al., 2016).
Rare microbes enhance functionality of abundant microbes
Microbial physiology can be strongly influenced by the presence of other microbial species (Garbeva et al., 2011). Accordingly, the presence of rare microbes could induce metabolic responses in more abundant microbes, implying that rare microbes have indirect effects on ecosystem functioning. A comparison of communities containing and not containing rare species showed that bacterial communities including rare species strongly reduced fungal growth via production of antifungal volatiles (Hol et al., 2015b), suggesting that rare bacteria either produce or trigger dominant bacteria to synthesize these compounds. Similarly, for mammals the presence of a single predator can impact the behavior and physiology of a herd of prey. Another example of rare microbes influencing abundant microbes stems from the work of Low-Décarie et al. (2015), who showed that rare microbes rescued whole communities from lethal stress, possibly via horizontal gene transfer. Horizontal transfer of genetic material can also be found in animals and plants (Panaud, 2016), but occurs more frequently between microbes and probably has a larger role in microbial communities.
Experimental and technological approaches
Rare microbe research is a rapidly emerging field. Given the obstacles to exploring rare microbes and the rapid advances in experimental approaches, we provide a condensed summary of methods and strategies that can be employed to address the most pressing questions in this field (Table 2, see also Reid and Buckley, 2011; Lynch and Neufeld, 2015).
Table 2. Overview of the research questions and general approaches in the study of rare microbes.
| Research questions | Method | Critical issues | Options | References | |
|---|---|---|---|---|---|
| Synthetic communities | |||||
| Order of arrival | Vary order of arrival to test priority effects | Cultivation dependent | Selective/spatially structured media | Fukami and Morin, 2003 | |
| Density-dependent effects | Vary abundances to test effects of rare species | Cultivation dependent | Cell separation via microfluidic or flow cytometry | Zhang et al., 2009 | |
| Manipulation of natural communities | |||||
| Removal | Consequence of rare species loss | Dilution-to-extinction | Equal biomass in all treatments | Incubation period for recovery of biomass | Philippot et al., 2013; Mallon et al., 2015; Hol et al., 2015a,2015b; Delgado-Baquerizo et al., 2016 |
| Enrichment | Responders to changing conditions | Salinity, dry–rewet, predation, pollution, nutrient amendments | Molecular methods for composition (DNA) and activity (RNA) | DNA normalization; improve coverage rare biosphere via single-cell genomics | Giebler et al., 2013; Aanderud et al., 2015 |
| In situ | |||||
| Microbial population dynamics | Time series | Availability of data sets | Increase sampling | Shade et al., 2014 | |
| Genome recovery of rare species; predict metabolic pathways | Single-cell genomics | Selection of target | Labeling via FISH | Podar et al., 2007; Freilich et al., 2011 | |
| Function of rare species | SIP; Nano-SIMS | Low throughput | Combine with single-cell analysis | Musat et al., 2008; Pester et al., 2010 | |
Abbreviations: FISH, fluorescence in situ hybridization; SIMS, secondary ion mass spectrometry; SIP, stable isotope probing.
Synthetic communities are a powerful approach to test potential causes of rarity, such as order of arrival (Fukami and Morin, 2003), or density-dependent microbial interactions. Studies that manipulate species' abundances can directly examine the impact of rare microbial species on community functionality. The use of flow cytometry, selecting for small cells before cultivation, could increase the likelihood of obtaining rare phyla and candidate divisions (Portillo et al., 2013; Brown et al., 2015), essential to examine their role in the community.
The relevance of rare microbes can also be assessed by removal and enrichment experiments. Manipulating environmental conditions can enrich specific groups and help reveal the role of rare taxa. For example, soil spiked with alkanes harbor several previously undetectable alkane-degrading taxa (Giebler et al., 2013), suggesting that temporarily rare species have important, specific functions.
Both removal and enrichment studies can be combined with molecular methods to assess community composition and relative activity of each taxon. Although molecular methods have undergone tremendous improvement in the recent years, one should still keep in mind that estimates of relative abundances can be inaccurate due to primer bias and limited genome coverage. Overcoming primer bias (Schadt and Rosling, 2015) through DNA normalization methods (Gagic et al., 2015) and increasing coverage of genomes from candidate phyla (Brown et al., 2015) will improve our insight into the rare biosphere. To date genome reconstructions from metagenomics studies have been limited to only the most dominant microbes, but with increasing coverage and sequencing power, we are starting to make roads into recovery of genomic information from rarer members of microbial communities. Single-cell genomics (Hedlund et al., 2014) and reconstructed genomes (Brown et al., 2015; Youssef et al., 2015) can provide valuable insights regarding the metabolic capabilities, physiological preferences, genomic architecture and ecological roles of candidate phyla. This will enable predictions of metabolic pathways and inference of competitive interactions (Freilich et al., 2011) and reveal whether rare species play a role as waste product consumers or as facilitators of growth for the remaining community. Such predictions can subsequently be tested using in situ studies, for instance via stable isotope probing to identify microbes actively processing labeled compounds (Chen and Murrell, 2010). This approach may help explore the role of rare microbes for important ecosystem processes (Pester et al., 2010; Aanderud et al., 2015). Coupling of stable isotope probing with high-resolution analytic methods, for example, nano-scale secondary ion mass spectrometry facilitates analysis of the metabolic products of individual cells from cultures or complex microbial communities. Nano-scale secondary ion mass spectrometry can also be combined with in situ hybridization to link phylogeny and metabolic function in single cells (Li et al., 2008; Musat et al., 2008). This technology may offer unique insights into the ecological functions of rare microbes in both experimental set-ups and natural environments (Musat et al., 2008; Hua et al., 2015).
Conclusion
Rare microbes may represent the hidden backbone of microbial communities, and our aim was therefore to increase awareness of and stimulate research on these rare ecosystem players. Rare taxa may vary in their ecological relevance and methods are becoming increasingly available to (1) assess relatively active taxa, (2) isolate them and (3) empirically determine their influence on community functioning. We summarized how rare microbes are important for several ecosystem processes and discussed the mechanisms by which these disproportional effects may become manifest. Future studies should further investigate the life-history traits that enable such widespread yet low-abundance existence. It may be important to determine the fraction of rare microbes that are active and dormant in order to predict functional consequences of rare species loss. We propose that including rare microbes in future investigations will improve our understanding of microbial community functioning, and help explain the buffering capacities of ecosystem against environmental change.
Acknowledgments
This paper is a joint effort of the working group sRareBios and an outcome of a workshop kindly supported by sDiv, the Synthesis Centre of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig (DFG FZT 118). We thank Monique Beijaart for drawing Figure 2 and George Kowalchuk for comments and advice.
Author contributions
AJ and WHGH initiated the workshop where the ideas for this synthesis were developed. All participants of the workshop contributed ideas and text. AJ and WHGH compiled the first draft and all authors contributed substantially to revisions.
Footnotes
The authors declare no conflict of interest.
References
- Aanderud ZT, Jones S, Fierer N, Lennon JT. (2015). Resuscitation of the rare biosphere contributes to pulses of ecosystem activity. Front Microbiol 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai DXC, Chu CJ, Ellwood MDF, Hou R, Wang G. (2013). Migration and niche partitioning simultaneously increase species richness and rarity. Ecol Model 258: 33–39. [Google Scholar]
- Allan E, Weisser W, Weigelt A, Roscher C, Fischer M, Hillebrand H. (2011). More diverse plant communities have higher functioning over time due to turnover in complementary dominant species. Proc Natl Acad Sci USA 108: 17034–17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Ramirez KS, Leff JW, Bradford MA, Wall DH, Fierer N. (2014). Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol Lett 17: 794–802. [DOI] [PubMed] [Google Scholar]
- Baumann K, Dignac MF, Rumpel C, Bardoux G, Sarr A, Steffens M et al. (2013). Soil microbial diversity affects soil organic matter decomposition in a silty grassland soil. Biochemistry 114: 201–212. [Google Scholar]
- Bittinger K, Charlson ES, Loy E, Shirley DJ, Haas AR, Laughlin A et al. (2014). Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol 15: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelier PLE, Meima-Franke M, Hordijk CA, Steenbergh AK, Hefting MM, Bodrossy L et al. (2013). Microbial minorities modulate methane consumption through niche partitioning. ISME J 7: 2214–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A et al. (2015). Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523: 208–211. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers LG, Guevara R, Boyer JN, Troxler TG, Davis SE. (2016). Effects of salinity and inundation on microbial community structure and function in a mangrove peat soil. Wetlands 36: 361–371. [Google Scholar]
- Chen Y, Murrell JC. (2010). When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol 18: 157–163. [DOI] [PubMed] [Google Scholar]
- Debroas D, Hugoni M, Domaizon I. (2015). Evidence for an active rare biosphere within freshwater protists community. Mol Ecol 24: 1236–1247. [DOI] [PubMed] [Google Scholar]
- Delgado-Baquerizo M, Giaramida L, Reich PB, Khachane AN, Hamonts K, Edwards C et al. (2016). Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J Ecol 104: 936–946. [Google Scholar]
- Dell'Anno A, Beolchini F, Rocchetti L, Luna GM, Danavaro R. (2012). High bacterial biodiversity increases degradation performance of hydrocarbons during bioremediation of contaminated harbor marine sediments. Environ Pollut 167: 85–92. [DOI] [PubMed] [Google Scholar]
- Dimitriu PA, Lee D, Grayston SJ. (2010). An evaluation of the functional significance of peat microorganisms using a reciprocal transplant approach. Soil Biol Biochem 42: 65–71. [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320: 1034–1039. [DOI] [PubMed] [Google Scholar]
- Fetzer I, Johst K, Schaewe R, Banitz T, Harms H, Chatzinotas A. (2015). The extent of functional redundancy changes as species' roles shift in different environments. Proc Natl Acad Sci USA 112: 14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RB, Garland JL, Bolster CH, Mills AL. (2001). Impact of dilution on microbial community structure and functional potential: comparison of numerical simulations and batch culture experiments. Appl Environ Microbiol 67: 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RB, Mills AL. (2006). Structural and functional responses of a sewage microbial community to dilution-induced reductions in diversity. Microb Ecol 52: 280–288. [DOI] [PubMed] [Google Scholar]
- Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M et al. (2011). Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun 2: 589. [DOI] [PubMed] [Google Scholar]
- Fuentes S, Méndez V, Aguila P, Seeger M. (2014). Bioremediation of petroleum hydrocarbons: catabolic genes, microbial communities, and applications. Appl Microbiol Biotechnol 98: 4781–4794. [DOI] [PubMed] [Google Scholar]
- Fukami T, Morin PJ. (2003). Productivity-biodiversity relationships depend on the history of community assembly. Nature 424: 423–426. [DOI] [PubMed] [Google Scholar]
- Gagic D, Maclean PH, Li D, Attwood GT, Moon CD. (2015). Improving the genetic representation of rare taxa within complex microbial communities using DNA normalization methods. Mol Ecol Res 15: 464–476. [DOI] [PubMed] [Google Scholar]
- Garbeva P, Silby MW, Raaijmakers JM, Levy SB, de Boer W. (2011). Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J 5: 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Fernández JM, de Marsac NT, Diez J. (2004). Streamlined regulation and gene loss as adaptive mechanisms in Prochlorococcus for optimized nitrogen utilization in oligotrophic environments. Microbiol Mol Biol Rev 68: 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland JL, Mills AL, Morales A, Cook K. (1999). Survival of human-associated bacteria in prototype advanced life support systems. SAE Techical paper 1999-01-2061, American Society of Automotive Engineers.
- Gaston KJ. (2008). Biodiversity and extinction: the importance of being common. Prog Phys Geogr 32: 73–79. [Google Scholar]
- Giebler J, Wick LY, Chatzinotas A, Harms H. (2013). Alkane-degrading bacteria at the soil-litter interface: comparing isolates with T-RFLP-based community profiles. FEMS Microbiol Ecol 86: 45–58. [DOI] [PubMed] [Google Scholar]
- Gobet A, Boer SI, Huse SM, van Beusekom JEE, Quince C, Sogin ML et al. (2012). Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J 6: 542–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths BS, Ritz K, Wheatley R, Kuan HL, Boag B, Christensen S et al. (2001). An examination of the biodiversity-ecosystem function relationship in arable soil microbial communities. Soil Biol Biochem 33: 1713–1722. [Google Scholar]
- Griffiths BS, Kuan HL, Ritz K, Glover LA, McCaig AE, Fenwick C. (2004). The relationship between microbial community structure and functional stability, tested experimentally in an upland pasture soil. Microb Ecol 47: 104–113. [DOI] [PubMed] [Google Scholar]
- Gudelj I, Weitz JS, Ferenci T, Horner-Devine MC, Marx CJ, Meyer JR et al. (2010). An integrative approach to understanding microbial diversity: from intracellular mechanisms to community structure. Ecol Lett 13: 1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, Lory S et al. (2011). Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J 5: 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA et al. (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta S, Kato S, Yamamoto K, Igarashi Y. (2009). Intertwined interspecies relationships: approaches to untangle the microbial network. Environ Microbiol 11: 2963–2969. [DOI] [PubMed] [Google Scholar]
- Hausmann B, Knorr K-H, Schreck K, Tringe SG, Glavina del Rio T, Loy A et al. (2016). Consortia of low-abundance bacteria drive sulfate reduction-dependent degradation of fermentation products in peat soil microcosms. ISME J 10: 2365–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedlund BP, Dodsworth JA, Murugapiran SK, Rinke C, Woyke T. (2014). Impact of single-cell genomics and metagenomics on the emerging view of extremophile 'microbial dark matter'. Extremophiles 18: 865–875. [DOI] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Smith AG. (2013). Widespread decay of vitamin-related pathways: coincidence or consequence? Trends Genet 29: 469–478. [DOI] [PubMed] [Google Scholar]
- Hernandez-Raquet G, Durand E, Braun F, Cravo-Laureau C, Godon JJ. (2013). Impact of microbial diversity depletion on xenobiotic degradation by sewage-activated sludge. Environ Microbiol Rep 5: 588–594. [DOI] [PubMed] [Google Scholar]
- Herrera CM. (1989). Pollinator abundance, morphology, and flower visitation rate - analysis of the quantity component in a plant-pollinator system. Oecologia 80: 241–248. [DOI] [PubMed] [Google Scholar]
- Hol WHG, de Boer W, Termorshuizen A, Meyer KM, Schneider JHM, Van Dam NM et al. (2010). Reduction of rare soil microbes modifies plant-herbivores interactions. Ecol Lett 13: 292–301. [DOI] [PubMed] [Google Scholar]
- Hol WHG, De Boer W, de Hollander M, Kuramae EE, Meisner A, van der Putten WH. (2015. a). Context dependency and saturating effects of loss of rare soil microbes on plant productivity. Front Plant Sci 6: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol WHG, Garbeva P, Hordijk CA, Hundscheid MPJ, Klein Gunnewiek PJA, van Agtmaal M et al. (2015. b). Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology 96: 2042–2048. [DOI] [PubMed] [Google Scholar]
- Hua Z-S, Han Y-J, Chen L-X, Liu J, Hu M, Li S-J et al. (2015). Ecological roles of dominant and rare prokaryotes in acid mine drainage revealed by metagenomics and metatranscriptomics. ISME J 9: 1280–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez DJ, Korenblum E, van Elsas JD. (2014). Novel multispecies microbial consortia involved in lignocellulose and 5-hydroxymethylfurfural bioconversion. Appl Microbiol Biotechnol 98: 2789–2803. [DOI] [PubMed] [Google Scholar]
- Jousset A, Rochat L, Pechy-Tarr M, Keel C, Scheu S, Bonkowski M. (2009). Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J 3: 666–674. [DOI] [PubMed] [Google Scholar]
- Jousset A, Eisenhauer N, Materne E, Scheu S. (2013). Evolutionary history predicts the stability of cooperation in microbial communities. Nat Commun 4: 2573. [DOI] [PubMed] [Google Scholar]
- Kummerli R, Brown SP. (2010). Molecular and regulatory properties of a public good shape the evolution of cooperation. Proc Natl Acad Sci USA 107: 18921–18926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitão RP, Zuanon J, Villeger S, Williams SE, Baraloto C, Fortunel C et al. (2016). Rare species contribute disproportionately to the functional structure of species assemblages. Proc R Soc Lond Ser B Biol Sci 283: 20160084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wu T-D, Mazéas L, Toffin L, Guerquin-Kern J-L, Leblon G et al. (2008). Simultaneous analysis of microbial identity and function using NanoSIMS. Environ Microbiol 10: 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locey KJ, Lennon JT. (2016). Scaling laws predict global microbial diversity. Proc Natl Acad Sci USA 113: 5970–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Décarie E, Kolber M, Homme P, Lofano A, Dumbrell A, Gonzalez A et al. (2015). Community rescue in experimental metacommunities. Proc Natl Acad Sci USA 112: 14307–14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A, Pester M. (2010) Probing identity and physiology of uncultured microorganisms with isotopic labeling techniques. In: Barton L, Mandl M, Loy A (eds), Geomicrobiology: Molecular and Environmental Perspective. Springer: Heidelberg, pp 127–146. [Google Scholar]
- Lynch MDJ, Neufeld JD. (2015). Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13: 217–229. [DOI] [PubMed] [Google Scholar]
- Lyons KG, Brigham CA, Traut BH, Schwartz MW. (2005). Rare species and ecosystem functioning. Conserv Biol 19: 1019–1024. [Google Scholar]
- Mallon CA, Poly F, Le Roux X, Marring I, van Elsas JD, Salles JF. (2015). Resource pulses can alleviate the biodiversity-invasion relationship in soil microbial communities. Ecology 96: 915–926. [DOI] [PubMed] [Google Scholar]
- Matos A, Kerkhof L, Garland JL. (2005). Effects of microbial community diversity on the survival of Pseudomonas aeruginosa in the wheat rhizosphere. Microb Ecol 49: 257–264. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comp Biol 10: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee MT, Collins JJ, Church GM, Wang HH. (2014). Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA 111: 2149–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR, Kassen R. (2007). The effects of competition and predation on diversification in a model adaptive radiation. Nature 446: 432–435. [DOI] [PubMed] [Google Scholar]
- Mouillot D, Bellwood DR, Baraloto C, Chave J, Galzin R, Harmelin-Vivien M et al. (2013). Rare species support vulnerable functions in high diversity ecosystems. PLoS Biol 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F et al. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA 105: 17861–17866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa N, Haruta S, Arai H, Ishii M, Igarashi Y. (2008). Coexistence of antibiotic-producing and antibiotic-sensitive bacteria in biofilms is mediated by resistant bacteria. Appl Environ Microbiol 74: 3887–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut D, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nipperess DA, Matsen FA. (2013). The mean and variance of phylogenetic diversity under rarefaction. Methods Ecol Evol 4: 566–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio EE, Anderson-Furgeson J, Estera KY, Pett-Ridge J, de Valpine P, Brodie EL et al. (2016). Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology 97: 1307–1318. [DOI] [PubMed] [Google Scholar]
- Panaud O. (2016). Horizontal transfers of transposable elements in eukaryotes: the flying genes. C R Biol 339: 296–299. [DOI] [PubMed] [Google Scholar]
- Pedrós-Alió C. (2007). Dipping into the rare biosphere. Science 315: 192–193. [DOI] [PubMed] [Google Scholar]
- Pester M, Bittner N, Deevong P, Wagner M, Loy A. (2010). A 'rare biosphere' microorganism contributes to sulfate reduction in a peatland. ISME J 4: 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter H, Beier S, Bertilsson S, Lindström ES, Langenheder S, Tranvik LJ. (2011). Function-specific response to depletion of microbial diversity. ISME J 5: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Spor A, Henault C, Bru D, Bizouard F, Jones CM et al. (2013). Loss in microbial diversity affects nitrogen cycling in soil. ISME J 7: 1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA et al. (2007). Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol 73: 3205–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo MC, Leff JW, Lauber CL, Fierer N. (2013). Cell size distributions of soil bacterial and archaeal taxa. Appl Environ Microbiol 79: 7610–7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz D. (1981) Seven forms of rarity. In: Synge H (ed), The Biological Aspects of Rare Plant Conservation. John Wiley & Sons Ltd.: Chichester, pp 205–217. [Google Scholar]
- Reid A, Buckley M. (2011) The Rare Biosphere. American Academy of Microbiology: Washington, pp 1–32. [Google Scholar]
- Rodrigues JLM, Pellizari VH, Mueller R, Baek K, Jesus ED, Paula FS et al. (2013). Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc Natl Acad Sci USA 110: 988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pašić L, Thingstad TF, Rohwer F et al. (2009). Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7: 828–836. [DOI] [PubMed] [Google Scholar]
- Rousk J, Brookes PC, Bååth E. (2009). Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microbiol 75: 1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Dubilier N, Rudgers JA. (2014). Nature's microbiome: introduction. Mol Ecol 23: 1225–1237. [DOI] [PubMed] [Google Scholar]
- Salonius PO. (1981). Metabolic capabilities of forest soil microbial-populations with reduced species-diversity. Soil Biol Biochem 13: 1–10. [Google Scholar]
- Sanchez MA, Vasquez M, Gonzalez B. (2004). A previously unexposed forest soil microbial community degrades high levels of the pollutant 2,4,6-trichlorophenol. Appl Environ Microbiol 70: 7567–7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt CW, Rosling A. (2015). Comment on “Global diversity and geography of soil fungi”. Science 348: 1438–1438. [DOI] [PubMed] [Google Scholar]
- Schluter J, Nadell CD, Bassler BL, Foster KR. (2015). Adhesion as a weapon in microbial competition. ISME J 9: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N et al. (2014). Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. Mbio 5: e01371–01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored "rare biosphere". Proc Natl Acad Sci USA 103: 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliveres S, Manning P, Prati D, Gossner MM, Alt F, Arndt H et al. (2016). Locally rare species influence grassland ecosystem multifunctionality. Philos Trans R Soc Lond Ser B Biol Sci 371: 20150269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Berry D, Loy A. (2013). Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol Rev 37: 793–829. [DOI] [PubMed] [Google Scholar]
- Steger D, Wentrup C, Braunegger C, Deevong P, Hofer M, Richter A et al. (2011). Microorganisms with novel dissimilatory (bi)sulfite reductase genes are widespread and part of the core microbiota in low-sulfate peatlands. Appl Environ Microbiol 77: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardy V, Mathieu O, Leveque J, Terrat S, Chabbi A, Lemanceau P et al. (2014). Stability of soil microbial structure and activity depends on microbial diversity. Environ Microbiol Rep 6: 173–183. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R et al. (2014). Global diversity and geography of soil fungi. Science 346: 1256688. [DOI] [PubMed] [Google Scholar]
- Van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW et al. (2011). Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elsas JD, Chiurazzi M, Mallon CA, Elhottová D, Kristufek V, Salles JF. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci USA 109: 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivant AL, Garmyn D, Maron PA, Nowak V, Piveteau P. (2013). Microbial diversity and structure are drivers of the biological barrier effect against Listeria monocytogenes in soil. Plos One 8: e76991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C, Bouvier T, Weinbauer MG, Thingstad TF. (2010). Trade-offs between competition and defense specialists among unicellular planktonic organisms: the killing the winner hypothesis revisited. Microbiol Mol Biol Rev 74: 42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachi S, Loreau M. (1999). Biodiversity and ecosystem productivity in a fluctuating environment: The insurance hypothesis. Proc Natl Acad Sci USA 96: 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Kuramae EE, Klinkhamer PGL, van Veen JA. (2015). Revisiting the dilution procedure used to manipulate microbial biodiversity in terrestrial systems. Appl Environ Microbiol 81: 4246–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenni G, Adler PB, Ernest SKM. (2012). Strong self-limitation promotes the persistence of rare species. Ecology 93: 456–461. [DOI] [PubMed] [Google Scholar]
- Youssef NH, Couger MB, McCully AL, Criado AEG, Elshahed MS. (2015). Assessing the global phylum level diversity within the bacterial domain: a review. J Adv Res 6: 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q-G, Buckling A, Godfray HCJ. (2009). Quantifying the relative importance of niches and neutrality for coexistence in a model microbial system. Funct Ecol 23: 1139–1147. [Google Scholar]