Abstract
There is an increasing interest in Faecalibacterium prausnitzii, one of the most abundant bacterial species found in the gut, given its potentially important role in promoting gut health. Although some studies have phenotypically characterized strains of this species, it remains a challenge to determine which factors have a key role in maintaining the abundance of this bacterium in the gut. Besides, phylogenetic analysis has shown that at least two different F. prausnitzii phylogroups can be found within this species and their distribution is different between healthy subjects and patients with gut disorders. It also remains unknown whether or not there are other phylogroups within this species, and also if other Faecalibacterium species exist. Finally, many studies have shown that F. prausnitzii abundance is reduced in different intestinal disorders. It has been proposed that F. prausnitzii monitoring may therefore serve as biomarker to assist in gut diseases diagnostics. In this mini-review, we aim to serve as an overview of F. prausnitzii phylogeny, ecophysiology and diversity. In addition, strategies to modulate the abundance of F. prausnitzii in the gut as well as its application as a biomarker for diagnostics and prognostics of gut diseases are discussed. This species may be a useful potential biomarker to assist in ulcerative colitis and Crohn's disease discrimination.
Introduction
Faecalibacterium prausnitzii has been consistently reported as one of the main butyrate producers found in the intestine (Barcenilla et al., 2000; Duncan et al., 2002). Butyrate has a crucial role in gut physiology and host wellbeing. It is the main energy source for the colonocytes and it has protective properties against colorectal cancer (CRC) and inflammatory bowel diseases (IBD; Christl et al., 1996; Archer et al., 1998). Butyrate can reduce intestinal mucosa inflammation through inhibiting NF-κB transcription factor activation (Inan et al., 2000), upregulating PPARγ (Schwab et al., 2007) and inhibiting interferon gamma (IFN-γ Klampfer et al., 2003).
Additional anti-inflammatory properties have been attributed to this species through its capability to induce a tolerogenic cytokine profile (with very low secretion of pro-inflammatory cytokines like IL-12 and IFN-γ, and an elevated secretion of the anti-inflammatory cytokine IL-10; Sokol et al., 2008b; Qiu et al., 2013). In line with this findings, F. prausnitzii cells or their cell-free supernatant have been reported to reduce the severity of acute (Sokol et al., 2008b), chronic (Martin et al., 2014) and low grade (Martin et al., 2015) chemical-induced inflammation in murine models. These anti-inflammatory effects were partly associated with secreted metabolites capable of blocking NF-κB activation, IL-8 production (Sokol et al., 2008b) and upregulation of regulatory T cell production (Qiu et al., 2013). Recently seven peptides that derive from a single microbial anti-inflammatory molecule, a 15 kDa protein, have been identified in F. prausnitzii cultures supernatant, and their capability to block NF-κB pathway has been demonstrated (Quevrain et al., 2015).
F. prausnitzii supernatant has also been shown to attenuate the severity of inflammation through the release of metabolites that enhance the intestinal barrier function and that affect paracellular permeability (Carlsson et al., 2013; Martin et al., 2015). The mechanism by which F. prausnitzii ameliorates permeability seems to be related with expression of certain tight junction proteins, but not with an enhancement of claudin expression (Carlsson et al., 2013). Besides, a recent study performed using a gnotobiotic model has shown that F. prausnitzii could also influence gut physiology through mucus pathway and the production of the mucus O-glycans, and may help to maintain suitable proportions of different cell types of secretory linage in the intestinal epithelium (Wrzosek et al., 2013). Finally, a restoration of serotonin (a key neurotransmitter in the gastrointestinal tract that affects motility (Ohman and Simren, 2007)) to normal level has been evidenced in murine models treated with either F. prausnitzii or its supernatant (Martin et al., 2015), and this species anti-nociceptive effect in non-inflammatory irritable bowel syndrome (IBS)-like murine models has been recently evidenced (Miquel et al., 2016).
Besides, over the last few years an increasing number of studies have reported on F. prausnitzii depletion in gut diseases (Swidsinski et al., 2005; Martinez-Medina et al., 2006; Frank et al., 2007; Balamurugan et al., 2008; Sokol et al., 2008a, 2009; Swidsinski et al., 2008; Willing et al., 2009; Furet et al., 2010; Jia et al., 2010; McLaughlin et al., 2010; Qin et al., 2010; Rajilic-Stojanovic et al., 2011; Sobhani et al., 2011; Hansen et al., 2012; Vermeiren et al., 2012; de Goffau et al., 2013; Kabeerdoss et al., 2013; Karlsson et al., 2013; Machiels et al., 2013; Miquel et al., 2013), which has prompted interest in considering this bacterium as a new generation probiotic.
Taken all together these findings indicate that F. prausnitzii has a crucial role in maintaining gut physiology and host wellbeing. It still remains elusive however which gut factors modulate F. prausnitzii presence in the gut, and the extent of their influence.
Factors supporting F. prausnitzii presence in the gut
Carbon sources used by F. prausnitzii for growth
F. prausnitzii isolates can grow well using simple carbohydrates (Table 1), but some differences exist between strains in their capability to ferment more complex carbohydrates such as those that are either host or diet derived, as observed by the maximum optical density at 650 nm wavelength (OD650) that cultures can reach (Duncan et al., 2002; Lopez-Siles et al., 2012).
Table 1. Substrates of different origin metabolized by Faecalibacterium prausnitzii isolates in vitro (batch pure cultures) as reported by (Duncan, et al. 2002; Lopez-Siles, et al. 2012).
| Substrate | No. of utilizers | No. of strains tested |
|---|---|---|
| Simple carbohydratesa | ||
| Glucose | 11 | 11 |
| Fructose | 4 | 4 |
| Cellobiose | 10 | 11 |
| Maltose | 10 | 11 |
| Galactose | 9 | 10 |
| Galacturonic acid | 7 | 9 |
| Sucrose | 2 | 4 |
| Melezitose | 1 | 4 |
| Trehalose | 1 | 4 |
| Rhamnose | 1 | 11 |
| Amino acidsb | ||
| Arginine | 4 | 4 |
| Histidine arylamide | 4 | 4 |
| Glycine arylamide | 2 | 4 |
| Diet-derivedc | ||
| Fructo-oligosacharides | 4 | 4 |
| Pectin (apple) | 10 | 10 |
| Inulin (chicory) | 9 | 11 |
| Host-derivedd | ||
| Glucosamine HCl | 10 | 10 |
| N-acetylglucosamine | 9 | 10 |
| Glucuronic acid | 6 | 10 |
Other simple carbohydrates tested but non-metabolized are mannitol (0/3), melibiose (0/4), raffinose (0/4), ribose (0/4), fucose (0/10), arabinose (0/11) and xylose(0/11).
Other amino acids tested but non-metabolized are alanine (0/4), glutamic acid (0/4), glutamyl (0/4), leucine (0/4), leucine-glycine (0/4), phenylalanine (0/4), proline (0/4), pyroglutamic acid (0/4), serine (0/4), tyrosine (0/4).
Other diet-derived carbohydrates not metabolized are arabinogalactan (0/10), citrus pectin (0/10), polygalacturonic acid (0/10), xylan (0/10) and potato starch (8/11) which depends on the solubility of the starch as F. prausnitzii does not metabolize starch.
Other host-derived carbohydrates not metabolized are chondroitin sulfate (0/10), heparin (0/10), hyaluronic acid (0/10), pig gastric mucin (0/10).
Although most F. prausnitzii strains are able to ferment inulin (Table 1), the findings show that only two of them can grow well on this substrate (final OD650~0.8). This supports the observed stimulation of this species in nutritional interventions with this prebiotic (Ramirez-Farias et al., 2009), and suggests that only some members of F. prausnitzii population are selectively stimulated by inulin (Chung et al., 2016). Strains of this species have a limited ability to utilize other polysaccharides found in the gut lumen such as arabinogalactan, xylan and soluble starch (Louis et al., 2007). Most of the isolates can grow on apple pectin and are able to use some pectin derivatives (Lopez-Siles et al., 2012). In vitro studies suggested that, under physiological conditions, F. prausnitzii can have a key role in fermentation of some types of pectin and that it can compete successfully with other gut bacteria for this substrate (Lopez-Siles et al., 2012). These results are supported by the fact that pectinolytic enzymes have been found encoded in the F. prausnitzii reference genome (Heinken et al., 2014). Besides, an in vivo study has shown that Firmicutes are promoted in apple pectin-fed rats (Licht et al., 2010). Taken together this suggests that pectin or pectin derivatives could be used as a novel prebiotic approach to stimulate F. prausnitzii (Chung et al., 2016).
In addition, F. prausnitzii strains can also utilize N-acetylglucosamine (Lopez-Siles et al., 2012), a constituent of the glycoproteins found in gut mucosa (Salvatore et al., 2000). Interestingly, it has been reported that treatment with this compound may improve Crohn's disease (CD) as it will serve as a healing factor in inflamed, damaged soft tissues of the gut (Salvatore et al., 2000). Therefore, given the capability to ferment this carbohydrate by F. prausnitzii, it would be of interest to explore the effect of restoring this beneficial gut bacterium in CD patients undergoing this treatment.
Finally, F. prausnitzii isolates are unable to utilize mucin or mucopolysaccharides (Lopez-Siles et al., 2012), although some controversy exists because it has been shown that mucin may stimulate growth of this species (Sadaghian Sadabad et al., 2015). The mechanism by which F. prausnitzii would benefit from mucin metabolism remains unknown, and further studies to reveal its interaction with mucin-degraders would be of interest.
F. prausnitzii has the ability to switch between substrates derived from the diet or the host. This capability should be explored further to define novel strategies to restore F. prausnitzii populations in the diseased gut by using some of these carbohydrates alone or in combination as prebiotics. In vivo studies on healthy human volunteers revealed a clear stimulation of F. prausnitzii after various prebiotic treatments (Ramirez-Farias et al., 2009; Benus et al., 2010; Hooda et al., 2012). It remains to be established which particular subtypes of F. prausnitzii populations change under prebiotic intakes. In addition, it would be interesting to conduct metatranscriptomic studies to determine if F. prausnitzii genes participate in breakdown of these substrates. Besides, this will also provide some clues on cross-feeding relationships between F. prausnitzii and other members of the gut microbiota.
Effect of gut physicochemical conditions
Tolerance to changes in gut physiological factors can have a role in determining the ability of an organism to survive in this environment, and they contribute to the temporal/spatial organization of different gut microbes (Parfrey and Knight 2012).
The optimal pH for F. prausnitzii growth ranges between 5.7 and 6.7 (Lopez-Siles et al., 2012; Foditsch et al., 2014), the range of pH found in the colon. Although there are differences in tolerance between strains in the pH range of 5–5.7 (Lopez-Siles et al., 2012), no growth was observed at pH values between 3.5 and 4.5 (Foditsch et al., 2014). This suggests that pH influences F. prausnitzii distribution along the gut. This species has been detected also in the duodenum (pH range 5.7–6.4; Nadal et al., 2007) and in the terminal ileum (Lopez-Siles et al., 2014, 2016) in healthy subjects and patients with gut disorders. As it has been reported that ulcerative colitis (UC) and CD patients often have acidic stools (Nugent et al., 2001; Barkas et al., 2013), it remains to be demonstrated whether or not local pH in the gut is modulating F. prausnitzii abundance and composition in patients with gut disorders such as IBD.
F. prausnitzii is also highly sensitive to a slight increase in physiological concentrations of bile salts because its growth is compromised by concentrations of 0.5% (wt/vol). This provides a plausible explanation for the reduced abundance of F. prausnitzii exhibited by CD patients, as increased bilirubin concentrations have been reported in these patients, especially in those with ileal disease involvement, and who have undergone intestinal resection (Lapidus and Einarsson, 1998; Pereira et al., 2003). Besides, differences in tolerance among isolates have been reported, especially at a bile salt concentration of 0.1% (wt/vol) (Lopez-Siles et al., 2012; Foditsch et al., 2014), suggesting that alterations in bile salt concentrations may determine a variation in F. prausnitzii subtype composition. As CD patients also feature altered bile salt composition (Lapidus and Einarsson, 1998; Pereira et al., 2003), further studies need to be conducted to determine if F. prausnitzii features higher sensitivity to certain types of bile salt components, and to establish whether or not different bile salt profiles alter F. prausnitzii subtype composition.
F. prausnitzii is extremely oxygen-sensitive (Duncan et al., 2002), but it is capable of withstanding low levels of oxygen found in the intestinal mucosa by using extracellular electron transfer in the presence of flavine and cysteine or glutathione (Khan et al., 2012). Recently, it has been demonstrated that strain A2-165 can retain viability in ambient air for 24 h when formulated with these antioxidants and inulin as a cryoprotectant (Khan et al., 2014). Because oxygen gradient has an important role in defining the spatial organization of microbes in the colon (Swidsinski et al., 2005; Parfrey and Knight, 2012), it would be interesting to determine if there are differences in oxygen tolerance among F. prausnitzii subtypes, and if it correlates with inflamed state of the mucosa.
Finally, the availability of essential nutrients to support F. prausnitzii may influence the distribution of this species in the gut. A recent study based on a functional metabolic map of F. prausnitzii strain A2-165 has predicted its inability to synthesize the amino acids alanine, cysteine, methionine, serine and tryptophan (Heinken et al., 2014). Auxotrophy for vitamins and cofactors such as biotin, folate, niacin, panthothenate, pyridoxine and thiamine has been observed by further analysis of other F. prausnitzii strain genomes, and some discrepancy between strains seems to exist in relation to riboflavin production, which could be due to inter-strain differences (Heinken et al., 2014; Magnusdottir et al., 2015). In contrast, this species has been predicted as a cobalamin producer (Magnusdottir et al., 2015). Evidence that some IBD patients are predisposed to feature cobalamin deficiency has been reported (Battat et al., 2014), but the cause of this condition has not been established yet. As there is a lack of consistent clinical data that indicates predisposition of IBD patients to this deficiency (Battat et al., 2014), it would be interesting to establish if it is associated with depletion of cobalamin-producers in the gut.
Collectively, these findings provide a plausible explanation why F. prausnitzii is reduced in abundance in patients with gut disease. Besides, it points out crucial requirements in physicochemical conditions for survival of this species, which can be applied in the future to use this bacterium to treat intestinal disorders related to its depletion.
F. prausnitzii in relation to other members of gut microbiota
F. prausnitzii co-occurs with several members of the C. coccoides group and Bacteroidetes in the gut (Qin et al., 2010). It has been suggested that F. prausnitzii may rely on other species like Bacteroides for cross-feeding. In co-culture experiments it has been observed that F. prausnitzii fermentative activity continues whereas B. thetaiotaomicron is fermenting pectin (Lopez-Siles et al., 2012; Chung et al., 2016). This could partially be explained by the acetate produced by the latter, which enhances F. prausnitzii growth (Heinken et al., 2014). Besides, initial fermentation of pectin by B. thetaiotaomicron can release pectin derivatives which can then be used by F. prausnitzii.
Recent studies in rat models have revealed that F. prausnitzii needs the prior presence of B. thetaiotaomicron to colonize the gut (Wrzosek et al., 2013). The inability to maintain F. prausnitzii mono-associated animal models has been repeatedly observed (Wrzosek et al., 2013; Hoffmann et al., 2015) and a mouse model has also been described in which F. prausnitzii implantation in the gastrointestinal tract requires prior preparation with Escherichia coli (Miquel et al., 2015). Correlation between these two species has been found in IBD patients (Lopez-Siles et al., 2014). Positive or negative correlation was observed depending on the disease location. This suggests the effect of one population on the other although the influence of host factors cannot be ruled out. Depending on patients' condition, these correlations involved specifically one or both phylogroups of F. prausnitzii (Lopez-Siles et al., 2016), so future studies of co-culture experiments could further elucidate the interactions between E. coli and F. prausnitzii.
Taxonomy and phylogeny of F. prausnitzii
Duncan et al. (2002) established that the genus Faecalibacterium is related to members of Clostridium cluster IV (Clostridium leptum group), within the Firmicutes phylum, Clostridia class and Ruminococcaceae family. Currently, F. prausnitzii is the only Faecalibacterium species which has been successfully isolated.
F. prausnitzii intraspecies diversity
More recent phylogenetic characterization of isolates determined that this species includes two phylogroups, which share 97% 16S rRNA gene sequence similarity (Lopez-Siles et al., 2012). Although genomic coherence remains to be explored, in silico analyses of sequenced genomes (Table 2) reveals that the average nucleotide identity (ANI) between isolates S3L/3 (phylogroup I) and L2/6 (phylogroup II) is below 94%, thus supporting the hypothesis that these would belong to two different genomospecies (that is, species defined by genome comparisons, but without phenotypic properties defined yet (Schloter et al., 2000; Rossello-Mora and Amann, 2015)). Besides, isolates S3L/3 and M21/2 (both from phylogroup I) share ANI values over 97% confirming that they belong to the same genomospecies. The accurate sequencing and annotation of several F. prausnitzii strain genomes is required to provide conclusive information to establish whether or not the two phylogroups belong to different genomospecies or genomovars (that is, strains which are phylogenetically different but phenotypically indistinguishable (Schloter et al., 2000; Rossello-Mora and Amann, 2015)).
Table 2. ANI values for paired comparisons between F. prausnitzii strains whose genome has been fully sequenced.
|
ANIba
values |
ANImb
values |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| F. prausnitzii isolate | KLE1255 (ND) | A2-165 (II) | L2/6(II) | SL3/3(I) | F. prausnitzii isolate | KLE1255 (ND) | A2-165 (II) | L2/6(II) | SL3/3(I) |
| M21/2 (I) | 85.26 | 83.29 | 82.11 | 96.70c | M21/2(I) | 89.02 | 88.52 | 88.07 | 97.34c |
| KLE1255 (ND) | – | 82.79 | 82.46 | 84.70 | KLE1255 (ND) | – | 88.31 | 88.65 | 88.82 |
| A2-165 (II) | 82.77 | – | 82.60 | 82.74 | A2-165(II) | 88.31 | – | 88.23 | 88.28 |
| L2/6(II) | 82.33 | 82.87 | – | 81.61 | L2/6(II) | 88.65 | 88.23 | – | 87.99 |
Abbreviations: ANI, Average nucleotide identity; DDH, DNA-DNA hybridization; ND, not determined. Phylogroup for each strain is indicated in brackets.
ANIb, ANI based on BLAST searches of 1 kb genome fragments against a target genome.
ANIm, ANI based on the MUMmer algorithm that does not require the artificial generation of 1 kb fragments.
It has been shown that ANI values higher than 94% embraces organisms sharing DDH values higher than 70% which are considered to be genomospecies. ANIb has better application for distant genomes comparison, whereas both algorithms give nearly identical values in the high identity range (80–100%). Values corresponding to the same genomospecies are indicated in boldface.
With regard to phenotypic coherence, no statistically significant differences have been found concerning carbohydrate fermentation or tolerance to changes in gut environmental conditions, although there are indicators that differences do exist between the members of the two phylogroups (Table 3). For instance, F. prausnitzii S3L/3 has been shown to produce significantly higher amounts of metabolites derived from phenylalanine, tyrosine and tryptophan metabolism than strain M21/2, despite both belonging to phylogroup I (Russell et al., 2013). The link of F. prausnitzii with tyrosine metabolism has been corroborated in fecal samples of healthy subjects (Jansson et al., 2009). Because the release of different metabolites by gut bacteria can have direct effect on different host signaling pathways, it is possible that within F. prausnitzii populations there are members that interact in a different manner with the host. Supporting this hypothesis, it has been demonstrated that F. prausnitzii ATCC27768 (phylogroup I) and F. prausnitzii A2-165 (phylogroup II) are associated with the modulation of host metabolites related to different pathways (Li et al., 2008; Jansson et al., 2009; Table 3). Prevalence and/or abundance of both phylogroups varies among patients suffering gut disorders such as CD, UC and type 2 diabetes (Lopez-Siles et al., 2015, 2016; Hippe et al., 2016), and further metabolomic studies are needed to establish the effects of that in host wellbeing.
Table 3. Summary of F. prausnitzii phylogroups I and II characteristics.
| Phylogroup I | Phylogroup II | |
|---|---|---|
| Strains | ATCC27768, M21/2, S3L/3, S4L/4 | A2-165, L2-6, L2-15, L2-39, L2-61, HTF-A, HTF-B, HTF-C, HTF-E, HTF-F, HTF-I, HTF-75H, HTF-60C |
| Gut distribution | Feces and mucosa | Feces and mucosa |
| Genome size (mean Mb±s.d.)a | 3.17±0.06 | 3.21±0.16 |
| GC content (mean %±s.d.)a | 55.85±0.49 | 56.45±0.21 |
| Genes content (mean±s.d.)a | 2881.5±92.6 | 2892.5±102.5 |
| Proteins content (mean±s.d.)a | 2778.5±46.0 | 2725.5±43.1 |
| Carbohydrate utilization (mean OD650±s.d.)b | ||
| Glucose | 0.750±0.311 | 0.428±0.228 |
| Cellobiose | 0.665±0.277 | 0.383±0.312 |
| Maltose | 0.685±0.247 | 0.603±0.273 |
| Galacturonic acid | 0.373±0.208 | 0.165±0.086 |
| Galactose | 0.435±0.369 | 0.630±0.183 |
| Apple pectin | 0.408±0.108 | 0.270±0.224 |
| Inulin | 0.115±0.065 | 0.510±0.440 |
| Glucuronic acid | 0.150±0.113 | 0.360±0.410 |
| N-Acetylgucosamine | 0.615±0.224 | 0.388±0.369 |
| Glucosamine HCl | 0.345±0.177 | 0.267±0.336 |
| Tolerance to pH (mean growth rate±s.d.)b | ||
| 6.7 | 0.210±0.070 | 0.256±.0151 |
| 6.2 | 0.192±0.050 | 0.245±0.159 |
| 5.75 | 0.081±0.039 | 0.108±0.042 |
| Tolerance to bile salts (mean maximum OD650±s.d.)b | ||
| 0% | 0.717±0.427 | 0.613±0.202 |
| 0.12% | 0.174±0.223 | 0.071±0.150 |
| 0.25% | 0.032±0.037 | 0.014±0.014 |
| 0.5% | 0.026±0.033 | 0.002±0.005 |
| SCFA production (mM ±s.d.)c | ||
| Formate | 3.508±2.730 | 15.190±11.856 |
| Acetate | −8.917±11.288 | −3.192±9.256 |
| Butyrate | 18.524±11.151 | 23.882±5.386 |
| D-Lactate | 2.014±1.992 | 2.435±0.865 |
| Association with host metabolites (adapted from (Li, et al. 2008)) | Decrease in dihydrothymine and an increase in 4-hydroxyphenylacetylglycine | Decreased levels of 3-aminoisobutyrate, taurine, 3, 5-hydroxylbenzoate, dimethylamine, 2-hydroxyisobutyrate, glycolate and increased lactate and glycine |
| Abundance in gut disorders (adapted from (Hippe et al., 2016, Lopez-Siles et al., 2016)) | Depletion in IBS, CRC and IBD patients, particularly in active CD | Depletion in CD patients, especially those with intestinal resection. Associated to type 2 diabetes. |
Abbreviations: CD, Crohn's disease; CRC, colorectal cancer; GC, guanine and cytosine; IBD, inflammatory bowel disease; IBS, irritable bowel syndrome; OD, optical density; SCFA, short chain fatty acids. No statistically significant differences have been found between the members of the two phylogroups for any of the characteristics analyzed.
For these calculations phylogroup I included isolates M21/2 and S3L/3 and phylogroup II consisted of L2/6 and A2-165 isolates.
For these calculations ATCC27768, M21/2, S3L/3 and S4L/4 (phylogroup I) and A2-165, L2-15, L2-39, L2/6, HTF-F and HTF-75H (phylogroup II) were used (Lopez-Siles, et al. 2012).
Short chain fatty acids produced by strains ATCC27768, M21/2, S3L/3 and S4L/4 (phylogroup I) and A2-165 and L2-6 (phylogroup II) on yeast casitone fatty-acids medium supplemented with 0.5% (wt/vol) glucose (Lopez-Siles et al., 2012).
Approaching the real diversity of the genus Faecalibacterium
Recent studies on species diversity and abundance in healthy and diseased gut samples however suggest that other F. prausnitzii phylotypes exist (Lopez-Siles et al., 2015, 2016) and the presence of other species within the Faecalibacterium genus cannot be ruled out. These have been estimated by molecular methods analyzing the overall bacterial community in fecal samples to represent around 2% of Faecalibacterium sequences (Tap et al., 2009; Walker et al., 2011), and corroborated using species-specific primers (Lopez-Siles et al., 2015). Interestingly, rare phylotypes have been mainly recovered from subjects with gut disease (Lopez-Siles et al., 2016). Further studies based on next generation sequencing may help to corroborate the presence of these rare phylotypes, and would provide an opportunity to elucidate the taxonomy within the genus Faecalibacterium.
F. prausnitzii populations in healthy and diseased gut
F. prausnitzii population composition and richness
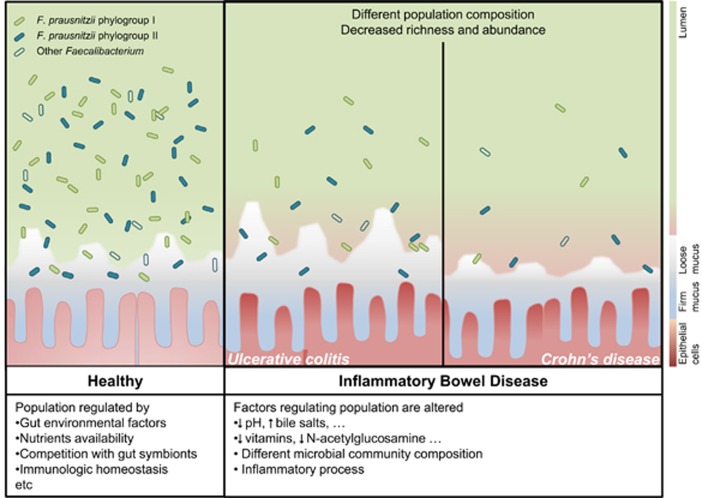
Overall a decrease in gut microbiota diversity has been reported in the mucosa of IBD patients (Tamboli et al., 2004; Seksik et al., 2006; Barnich and Darfeuille-Michaud, 2007; Ott et al., 2008; Sokol et al., 2008a; Chassaing and Darfeuille-Michaud, 2011). In particular, fewer types of Firmicutes, mostly from Ruminococcaceae, were observed in feces of CD patients (Scanlan et al., 2006). Regarding F. prausnitzii population, subtype richness is also lower in IBD patients, which frequently tends to only possess one of the two main phylogroups (Lopez-Siles et al., 2015).
IBD, CRC, IBS and healthy subjects feature a different composition of F. prausnitzii subtypes (Lopez-Siles et al., 2015). Although some phylotypes have been specifically associated to each condition, the main members of the F. prausnitzii population (four phylotypes, two phylogroups) have been detected in all the subject groups but with a different distribution between conditions (Lopez-Siles et al., 2015). As factors explaining these differences remain unknown, further studies of isolation and characterization of strains from patients suffering intestinal disorders are needed to test the effect of either host or gut physicochemical factors on different F. prausnitzii subtypes.
F. prausnitzii load
Several studies have reported F. prausnitzii depletion in adult CD (Martinez-Medina et al., 2006; Frank et al., 2007; Sokol et al., 2008b, 2009; Swidsinski et al., 2008; Willing et al., 2009; Fujimoto et al., 2013; Miquel et al., 2013), UC (Swidsinski et al., 2005; Sokol et al., 2009; McLaughlin et al., 2010; Vermeiren et al., 2012; Kabeerdoss et al., 2013; Machiels et al., 2013; Lopez-Siles et al., 2014, 2016) and CRC (Balamurugan et al., 2008; Lopez-Siles et al., 2016) subjects, and concur with the view that down-shifts in F. prausnitzii numbers occur under several pathological disorders. In contrast, other studies have reported no depletion in F. prausnitzii levels in CRC (Balamurugan et al., 2008; Sobhani et al., 2011; Wang et al., 2012), and even increased F. prausnitzii abundance in de novo pediatric CD patients (Hansen et al., 2012). Besides, a consensus on whether or not IBS patients feature a depletion of F. prausnitzii has not been reached since both studies reported normal counts (Malinen et al., 2005; Swidsinski et al., 2005, 2008; Kassinen et al., 2007; Jia et al., 2010; Duboc et al., 2012; Rigsbee et al., 2012; Lopez-Siles et al., 2014, 2016) and studies reporting lower numbers in IBS patients of alternating type (Rajilic-Stojanovic et al., 2011) have also been published. The variety of symptoms featured by IBS patients makes IBS diagnostics complex, which in turn is likely to make it difficult to establish whether or not F. prausnitzii is affected in this intestinal condition. Altogether, the exact role that F. prausnitzii has in the pathogenesis of these diseases cannot be established at this stage. On the one hand an external factor can cause a downshift in F. prausnitzii, but also this species depletion can be a contributing factor to disease aggravation. In this case, restoration of normal counts of this species should be explored as a way to achieve healing and/or attenuate disease progression.
Although the depletion of F. prausnitzii is not a specific phenomenon that occurs in a particular disease, the level of depletion as well as which components of the F. prausnitzii population are affected can be different between diseases. Depletion in phylogroup I abundance is a general feature in abnormal gut conditions, whereas phylogroup II reduction seems to be specific to CD patients, usually with ileal disease location (Lopez-Siles et al., 2016). This could be the consequence of several factors (physicochemical, host-related or microbiome-related) that may vary between disorders and can affect either some or all F. prausnitzii members. In turn, these different populations can have a direct effect in host wellbeing. For instance, a recent study has shown different F. prausnitzii profiles in obese subjects with and without developed type two diabetes (Hippe et al., 2016), suggesting that differences in phylotypes may lead to differences in inflammatory status in the host, thus having an influence on disease development. Currently, studies on anti-inflammatory properties of F. prausnitzii have been performed with strain A2-165, from phylogroup II. Similar studies conducted with strains representative of phylogroup I (for example, ATCC27768) are required to determine whether or not there are differences between phylogroups regarding anti-inflammatory activity.
Future perspectives: potential use of F. prausnitzii as a healthy gut microbiota biomarker.
F. prausnitzii load as diagnostic supporting tool
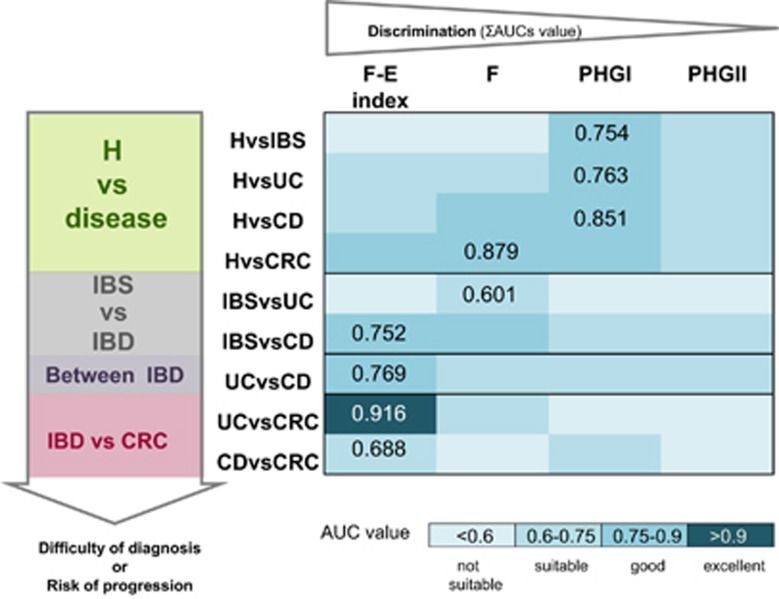
The usefulness of gut microbiota assessment to support intestinal disease diagnostics and/or prognostics has gained interest during the last few years. Some studies have pointed out that the abundance of fecal or mucosa-associated F. prausnitzii is a potential biomarker to discriminate between gut disorders (Swidsinski et al., 2008; Lopez-Siles et al., 2014, 2016). In particular, F. prausnitzii is a good biomarker to discriminate CD and CRC from healthy subjects as well as CD from IBS (Figure 1). Of interest, F. prausnitzii phylogroup I is particularly good in discriminating healthy subjects from gut disease cohorts including IBS, IBD and CRC (Lopez-Siles et al., 2016), whereas phylogroup II has a limited use as biomarker. This could be partially explained by the fact that phylogroup II load is less reduced in intestinal disease.
Figure 1.
Biomarker of choice to discriminate between conditions. Selected pair wise comparisons of conditions are represented taking into account the difficulty of diagnosis or the risk of progression. The four options of biomarkers (F. prausnitzii, the two phylogroups or the F. prausnitzii-E. coli index calculated as (Lopez-Siles et al., 2014)), have been ranked according to their discriminative power estimated as the sum of all the AUC values for all the pair wise comparisons taking into account all the conditions. For each comparison, the highest AUC value achieved is depicted. H, healthy control group; F, total F. prausnitzii load; PHG I, F. prausnitzii phylogroup I load; PHG II, F. prausnitzii phylogroup II load; F-E index, F. prausnitzii- E. coli index; AUC, area under the ROC curve; ROC, receiver operating characteristic curve.
It is difficult however to establish the use of a single bacterial species as a general biomarker for all disease types. F. prausnitzii in conjunction with E. coli abundance as a complementary indicator (F-E index) has been proven to be a better biomarker than F. prausnitzii alone (Lopez-Siles et al., 2014). This index allows good discrimination of CRC patients from other gut disorders, especially UC. The F-E index is also a good biomarker to differentiate UC and IBS patients from those with CD. However, the heterogeneity of disease subtypes is preventing discrimination between conditions.
F. prausnitzii load as IBD subtype biomarker
An accurate discrimination between UC and CD is of relevance due to differences in treatment and management between these two entities (Mowat et al., 2011). An unmet need in IBD diagnostics is to have a fast and reliable biomarker to distinguish within IBD subtypes, particularly those with shared location of inflammation, but the number of studies that have explored this issue is limited (Lopez-Siles et al., 2014, 2016).
We observed that F-E index is a suitable biomarker to discriminate ulcerative proctitis and left-sided UC from pancolitis (Lopez-Siles et al., 2014), which is of interest for clinicians to monitor risk of extension of the inflamed area in UC (Figure 2). This index was shown also to distinguish between all UC patients regardless of their disease subtypes and those with C-CD with suitable accuracy (Figure 2). In contrast, F. prausnitzii alone or phylogroup quantification showed limited ability to discriminate between IBD subtypes. Whether or not F. prausnitzii phylogroup quantification in conjunction with E. coli counts are more accurate biomarkers remains to be explored.
Figure 2.
Biomarker of choice to discriminate between IBD locations. Selected pair wise comparisons of conditions are represented taking into account the difficulty of diagnosis or the risk of progression. The four options of biomarkers (F. prausnitzii, the two phylogroups or F. prausnitzii-E. coli index calculated as (Lopez-Siles et al., 2014)), have been ranked according to their discriminative power estimated as the sum of all the AUC values for all the pair wise comparisons taking into account all the conditions. For each comparison, the highest AUC value achieved is depicted. E1, Ulcerative proctitis, E2, Distal or left-sided UC; E3, pancolitis or universal colitis; I-CD, ileal CD; IC-CD, ileocolonic CD; C-CD, colonic CD; F, total F. prausnitzii load; PHG I, F. prausnitzii phylogroup I load; PHG II, F. prausnitzii phylogroup II load; F-E index, F. prausnitzii- E. coli index; AUC, area under the ROC curve; ROC, receiver operating characteristic curve.
As the discrimination power of F-E index is limited for some disease subtypes, it could be worth to include additional biomarker characteristics of UC dysbiosis such as Roseburia hominis (Machiels et al., 2013), CD dysbiosis such as Ruminococcus gnavus, R. torques, Dialister invisus or Bifidobacterium adolescentis (Martinez-Medina et al., 2006; Png et al., 2010; Joossens et al., 2011), as well as other bacterial indicators of gut health such as Akkermansia muciniphila (Png et al., 2010). A combination of microbiological indicators with host serological data is also an approach to be further explored to improve diagnostics accuracy, since it has been reported that active CD and UC can be differentiated through monitoring fecal F. prausnitzii abundance in conjunction with leukocyte counts (Swidsinski et al., 2008).
F. prausnitzii load as a biomarker of disease progression and treatment success
Given the chronic behavior of IBD, it would be interesting to have a prognostic biomarker for flare-ups. High F. prausnitzii counts in feces have been associated with lower CD activity index and C-reactive protein levels (Fujimoto et al., 2013). F. prausnitzii level recovery has been reported in feces during remission (Swidsinski et al., 2008; Sokol et al., 2009), whereas it has been observed that in mucosa, depletion of this species occurs regardless of patients disease activity status (Willing et al., 2009; Kabeerdoss et al., 2013; Lopez-Siles et al., 2014, 2016), and particularly compromises phylogroup I (Lopez-Siles et al., 2016). Differences in the methodology or the cohort engaged as well as the type of sample analyzed may be a confounding factor that is preventing an unanimous outcome about the usefulness of F. prausnitzii to predict flare-ups. Subsequent follow-up studies are needed to conclusively establish which clinical data of the patients correlate with the quantity of F. prausnitzii colonizing the gut.
Several studies have shown that F. prausnitzii numbers are reduced in resected CD patients in comparison with those without resection (Sokol et al., 2008b; Lopez-Siles et al., 2014). We observed that this phenomenon is replicated with phylogroup counts (Lopez-Siles et al., 2016), with more evident depletion of phylogroup II. However, whether this shift is a consequence of these patients featuring a more acute disease, or if it is the outcome of the surgery is still unclear. It would be interesting to conduct follow-up studies to assess the usefulness of this biomarker to precisely predict when such interventions might be needed.
As far as therapies are concerned, treatments with infliximab and high-dose cortisol have been associated with an increase of F. prausnitzii levels (Swidsinski et al., 2008). Chemotherapy and interferon α-2b reverse the depletion of F. prausnitzii in patients with neuroendocrine tumor of the midgut, whereas somatostatin analogs have no influence on this species (Dorffel et al., 2012). These results suggest that restoration of the gut conditions due to medication can have an effect on counterbalancing F. prausnitzii depletion in the diseased intestine. In contrast, other studies have not found a medication associated with the recovery of normal levels of this species in the mucosa, suggesting that F. prausnitzii would be a poor biomarker to monitor treatment efficacy (Lopez-Siles et al., 2014, 2016; Busquets et al., 2015). However, since these studies are retrospective, further prospective studies are required to establish the usefulness of these biomarkers to monitor long-term treatment efficacy, and to relate impact of medication in this species load in the gut.
Sample of choice to implementation in diagnostics
When analyzing data by sample location, it was observed that colonic biopsies were the most suitable to distinguish disease phenotypes (Lopez-Siles et al., 2014). Although statistical significance was not reached for rectal samples, similar results were obtained. To validate these results would be of value since rectal sigmoidoscopy is a non-invasive method to collect tissue samples which will allow implementing mucosa-associated F. prausnitzii quantification in routine clinical practice. Alternatively, the validation in samples collected with rectal swabs, which have been reported to have a great similarity to biopsy specimens (Albenberg et al., 2014) would also be of interest. Nevertheless, it would be of interest to determine if fecal total abundance of F. prausnitzii and of both phylogroups can be a suitable biomarker for the detection, follow up and/or classification of IBD phenotypes. The implementation of F. prausnitzii counts in feces seems a promising strategy as a biomarker, because it has been already proven to discriminate between active UC and CD patients (Swidsinski et al., 2008) and thus would provide a straightforward method to assess IBD. However, further optimization to fine-tune this tool to achieve discrimination within IBD subtypes and also applicable in patients in remission phases is needed.
Concluding remarks
F. prausnitzii is a metabolically versatile micro-organism, and this may explain its wide distribution and high load as part of the gut microbiota in humans. Two phylogroups have been described so far within this species, although the real diversity of the genus remains unknown. F. prausnitzii is an important bacterium for human health but, members of this speceis are very sensitive to changes in gut environment which can limit its distribution, particularly in a diseased gut. Changes in this species population richness and quantity have been observed in several intestinal disorders (Figure 3). There is a lot of information still missing on which phylogroup is important under which conditions in the gut. As the depletion of this species is not homogeneous in all gut diseases however, the use of F. prausnitzii as a gold standard measure of a healthy gut microbiota is limited. Nevertheless, it is a good biomarker of certain gut conditions. It has the potential to assist in discriminating between UC and CD subtypes, particularly those with colonic disease location. Besides, discrimination between UC and CRC could be a further application of particular interest for this biomarker, to monitor disease progression since chronic colonic inflammation can lead to tumor formation. As studies in this field are somewhat limited, and a consensus has not yet been established, there is a need to conduct more studies to fully implement F. prausnitzii as a biomarker by defining in which medical condition it could be of assistance. Preferably, these studies should be conducted in larger independent cohorts of patients that include individuals from different ethnicities.
Figure 3.
F. prausnitzii populations in healthy gut and in patients with IBD. In IBD patients, alteration of gut environment may affect F. prausnitzii population composition and load. These differences can be monitored to discriminate within IBD subtypes.
Acknowledgments
We thank Dr Xavier Aldeguer and MD David Busquets from the Hospital Dr Josep Trueta (Girona, Spain) and M.D Míriam Sabat Mir from the Hospital Santa Caterina (Salt, Spain) for their help and critical discussion concerning clinical aspects. This work was partially funded by the Spanish Ministry of Education and Science through the projects SAF2010-15896 and SAF2013-43284-P, which has been co-financed with FEDER funds. Dr Sylvia H Duncan acknowledges support from the Scottish Government Food, Land and People program.
Footnotes
The authors declare no conflict of interest.
References
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A et al. (2014). Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147: 1055–1063 e1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer S, Meng S, Wu J, Johnson J, Tang R, Hodin R. (1998). Butyrate inhibits colon carcinoma cell growth through two distinct pathways. Surgery 124: 248–253. [PubMed] [Google Scholar]
- Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. (2008). Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol 23: 1298–1303. [DOI] [PubMed] [Google Scholar]
- Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C et al. (2000). Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 66: 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkas F, Liberopoulos E, Kei A, Elisaf M. (2013). Electrolyte and acid-base disorders in inflammatory bowel disease. Ann Gastroenterol 26: 23–28. [PMC free article] [PubMed] [Google Scholar]
- Barnich N, Darfeuille-Michaud A. (2007). Role of bacteria in the etiopathogenesis of inflammatory bowel disease. World J Gastroenterol 13: 5571–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battat R, Kopylov U, Szilagyi A, Saxena A, Rosenblatt DS, Warner M et al. (2014). Vitamin B12 deficiency in inflammatory bowel disease: prevalence, risk factors, evaluation, and management. Inflamm Bowel Dis 20: 1120–1128. [DOI] [PubMed] [Google Scholar]
- Benus RF, van der Werf TS, Welling GW, Judd PA, Taylor MA, Harmsen HJ et al. (2010). Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr 104: 693–700. [DOI] [PubMed] [Google Scholar]
- Busquets D, Mas-de-Xaxars T, Lopez-Siles M, Martinez-Medina M, Bahi A, Sabat M et al. (2015). Anti-tumour necrosis factor treatment with adalimumab induces changes in the microbiota of Crohn's disease. J Crohns Colitis 9: 899–906. [DOI] [PubMed] [Google Scholar]
- Carlsson AH, Yakymenko O, Olivier I, Hakansson F, Postma E, Keita AV et al. (2013). Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand J Gastroenterol 48: 1136–1144. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Darfeuille-Michaud A. (2011). The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140: 1720–1728. [DOI] [PubMed] [Google Scholar]
- Christl SU, Eisner H-D, Dusel G, Kasper H, Scheppach W. (1996). Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa. Dig Dis Sci 41: 2477–2481. [DOI] [PubMed] [Google Scholar]
- Chung WS, Walker AW, Louis P, Parkhill J, Vermeiren J, Bosscher D et al. (2016). Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol 14: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T et al. (2013). Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes 62: 1238–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorffel Y, Swidsinski A, Loening-Baucke V, Wiedenmann B, Pavel M. (2012). Common biostructure of the colonic microbiota in neuroendocrine tumors and Crohn's disease and the effect of therapy. Inflamm Bowel Dis 18: 1663–1671. [DOI] [PubMed] [Google Scholar]
- Duboc H, Rainteau D, Rajca S, Humbert L, Farabos D, Maubert M et al. (2012). Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 24: 513–520 e246-517. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. (2002). Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 52: 2141–2146. [DOI] [PubMed] [Google Scholar]
- Foditsch C, Santos TM, Teixeira AG, Pereira RV, Dias JM, Gaeta N et al. (2014). Isolation and characterization of Faecalibacterium prausnitzii from calves and piglets. PLoS One 9: e116465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DNSt, Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, Imaeda H, Takahashi K, Kasumi E, Bamba S, Fujiyama Y et al. (2013). Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn's disease. J Gastroenterol Hepatol 28: 613–619. [DOI] [PubMed] [Google Scholar]
- Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL et al. (2010). Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes 59: 3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R, Russell RK, Reiff C, Louis P, McIntosh F, Berry SH et al. (2012). Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol 107: 1913–1922. [DOI] [PubMed] [Google Scholar]
- Heinken A, Khan MT, Paglia G, Rodionov DA, Harmsen HJ, Thiele I. (2014). Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J Bacteriol 196: 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippe B, Remely M, Aumueller E, Pointner A, Magnet U, Haslberger AG. (2016). Faecalibacterium prausnitzii phylotypes in type two diabetic, obese, and lean control subjects. Benef Microbes 7: 1–8. [DOI] [PubMed] [Google Scholar]
- Hoffmann TW, Pham H-P, Bridonneau C, Aubry C, Lamas B, Martin-Gallausiaux C et al. (2015). Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. ISME J 10: 460–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooda S, Boler BM, Serao MC, Brulc JM, Staeger MA, Boileau TW et al. (2012). 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutr 142: 1259–1265. [DOI] [PubMed] [Google Scholar]
- Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, Giardina C. (2000). The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology 118: 724–734. [DOI] [PubMed] [Google Scholar]
- Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J et al. (2009). Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One 4: e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Whitehead RN, Griffiths L, Dawson C, Waring RH, Ramsden DB et al. (2010). Is the abundance of Faecalibacterium prausnitzii relevant to Crohn's disease? FEMS Microbiol Lett 310: 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P et al. (2011). Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60: 631–637. [DOI] [PubMed] [Google Scholar]
- Kabeerdoss J, Sankaran V, Pugazhendhi S, Ramakrishna BS. (2013). Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol 13: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B et al. (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498: 99–103. [DOI] [PubMed] [Google Scholar]
- Kassinen A, Krogius-Kurikka L, Makivuokko H, Rinttila T, Paulin L, Corander J et al. (2007). The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 133: 24–33. [DOI] [PubMed] [Google Scholar]
- Khan MT, Duncan SH, Stams AJ, van Dijl JM, Flint HJ, Harmsen HJ. (2012). The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J 6: 1578–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, van Dijl JM, Harmsen HJ. (2014). Antioxidants keep the potentially probiotic but highly oxygen-sensitive human gut bacterium Faecalibacterium prausnitzii alive at ambient air. PLoS One 9: e96097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfer L, Huang J, Sasazuki T, Shirasawa S, Augenlicht L. (2003). Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol Cancer Res 1: 855–862. [PubMed] [Google Scholar]
- Lapidus A, Einarsson C. (1998). Bile composition in patients with ileal resection due to Crohn's disease. Inflamm Bowel Dis 4: 89–94. [DOI] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H et al. (2008). Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA 105: 2117–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht T, Hansen M, Bergstrom A, Poulsen M, Krath B, Markowski J et al. (2010). Effects of apples and specific apple components on the cecal environment of conventional rats: role of apple pectin. BMC Microbiol 10: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. (2012). Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol 78: 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Martinez-Medina M, Busquets D, Sabat-Mir M, Duncan SH, Flint HJ et al. (2014). Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish irritable bowel syndrome and inflammatory bowel disease phenotypes. Int J Med Microbiol 304: 464–475. [DOI] [PubMed] [Google Scholar]
- Lopez-Siles M, Martinez-Medina M, Abella C, Busquets D, Sabat-Mir M, Duncan SH et al. (2015). Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl Environ Microbiol 81: 7582–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Martinez-Medina M, Suris-Valls R, Aldeguer X, Sabat-Mir M, Duncan SH et al. (2016). Changes in the abundance of Faecalibacterium prausnitzii phylogroups I and II in the intestinal mucosa of inflammatory bowel disease and patients with colorectal cancer. Inflamm Bowel Dis 22: 28–41. [DOI] [PubMed] [Google Scholar]
- Louis P, Scott KP, Duncan SH, Flint HJ. (2007). Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol 102: 1197–1208. [DOI] [PubMed] [Google Scholar]
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V et al. (2013). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63: 1275–1283. [DOI] [PubMed] [Google Scholar]
- Magnusdottir S, Ravcheev DA, de Crecy-Lagard V, Thiele I. (2015). Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Frontiers in Genetics 6: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinen E, Rinttila T, Kajander K, Matto J, Kassinen A, Krogius L et al. (2005). Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol 100: 373–382. [DOI] [PubMed] [Google Scholar]
- Martin R, Chain F, Miquel S, Lu J, Gratadoux JJ, Sokol H et al. (2014). The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm Bowel Dis. 20: 417–430. [DOI] [PubMed] [Google Scholar]
- Martin R, Miquel S, Chain F, Natividad JM, Jury J, Lu J et al. (2015). Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol 15: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. (2006). Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis 12: 1136–1145. [DOI] [PubMed] [Google Scholar]
- McLaughlin SD, Clark SK, Tekkis PP, Nicholls RJ, Ciclitira PJ. (2010). The bacterial pathogenesis and treatment of pouchitis. Therap Adv Gastroenterol 3: 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Martin R, Rossi O, Bermudez-Humaran L, Chatel J, Sokol H et al. (2013). Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol 16: 255–261. [DOI] [PubMed] [Google Scholar]
- Miquel S, Leclerc M, Martin R, Chain F, Lenoir M, Raguideau S et al. (2015). Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. MBio 6: e00300–e00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Martin R, Lashermes A, Gillet M, Meleine M, Gelot A et al. (2016). Anti-nociceptive effect of Faecalibacterium prausnitzii in non-inflammatory IBS-like models. Sci Rep 6: 19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat C, Cole A, Windsor A, Ahmad T, Arnott I, Driscoll R et al. (2011). Guidelines for the management of inflammatory bowel disease in adults. Gut 60: 571–607. [DOI] [PubMed] [Google Scholar]
- Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. (2007). Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol 56: 1669–1674. [DOI] [PubMed] [Google Scholar]
- Nugent SG, Kumar D, Rampton DS, Evans DF. (2001). Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 48: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman L, Simren M. (2007). New insights into the pathogenesis and pathophysiology of irritable bowel syndrome. Dig Liver Dis 39: 201–215. [DOI] [PubMed] [Google Scholar]
- Ott SJ, Plamondon S, Hart A, Begun A, Rehman A, Kamm MA et al. (2008). Dynamics of the mucosa-associated flora in ulcerative colitis patients during remission and clinical relapse. J Clin Microbiol 46: 3510–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Knight R. (2012). Spatial and temporal variability of the human microbiota. Clin Microbiol Infect 18(Suppl 4): 8–11. [DOI] [PubMed] [Google Scholar]
- Pereira SP, Bain IM, Kumar D, Dowling RH. (2003). Bile composition in inflammatory bowel disease: ileal disease and colectomy, but not colitis, induce lithogenic bile. Aliment Pharmacol Ther 17: 923–933. [DOI] [PubMed] [Google Scholar]
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI et al. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105: 2420–2428. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Zhang M, Yang X, Hong N, Yu C. (2013). Faecalibacterium prausnitzii upregulates regulatory T cells and anti-inflammatory cytokines in treating TNBS-induced colitis. J Crohns Colitis 7: e558–e568. [DOI] [PubMed] [Google Scholar]
- Quevrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J et al. (2015). Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut. 65: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S et al. (2011). Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141: 1792–1801. [DOI] [PubMed] [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. (2009). Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101: 541–550. [DOI] [PubMed] [Google Scholar]
- Rigsbee L, Agans R, Shankar V, Kenche H, Khamis HJ, Michail S et al. (2012). Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol 107: 1740–1751. [DOI] [PubMed] [Google Scholar]
- Rossello-Mora R, Amann R. (2015). Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol 38: 209–216. [DOI] [PubMed] [Google Scholar]
- Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG et al. (2013). Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res 57: 523–535. [DOI] [PubMed] [Google Scholar]
- Sadaghian Sadabad M, von Martels JZ, Khan MT, Blokzijl T, Paglia G, Dijkstra G et al. (2015). A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci Rep 5: 17906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore S, Heuschkel R, Tomlin S, Davies SE, Edwards S, Walker-Smith JA et al. (2000). A pilot study of N-acetyl glucosamine, a nutritional substrate for glycosaminoglycan synthesis, in paediatric chronic inflammatory bowel disease. Aliment Pharmacol Ther 14: 1567–1579. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, O'Mahony C, Marchesi JR. (2006). Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol 44: 3980–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloter M, Lebuhn M, Heulin T, Hartmann A. (2000). Ecology and evolution of bacterial microdiversity. FEMS Microbiol Rev 24: 647–660. [DOI] [PubMed] [Google Scholar]
- Schwab M, Reynders V, Loitsch S, Steinhilber D, Stein J, Schroder O. (2007). Involvement of different nuclear hormone receptors in butyrate-mediated inhibition of inducible NF kappa B signalling. Mol Immunol 44: 3625–3632. [DOI] [PubMed] [Google Scholar]
- Seksik P, Sokol H, Lepage P, Vasquez N, Manichanh C, Mangin I et al. (2006). Review article: the role of bacteria in onset and perpetuation of inflammatory bowel disease. Aliment Pharmacol Ther 24(Suppl 3): 11–18. [DOI] [PubMed] [Google Scholar]
- Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P et al. (2011). Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 6: e16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Lay C, Seksik P, Tannock GW. (2008. a). Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm Bowel Dis 14: 858–867. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ et al. (2008. b). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L et al. (2009). Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 15: 1183–1189. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. (2005). Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol 11: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. (2008). Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis 14: 147–161. [DOI] [PubMed] [Google Scholar]
- Tamboli CP, Neut C, Desreumaux P, Colombel JF. (2004). Dysbiosis in inflammatory bowel disease. Gut 53: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP et al. (2009). Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11: 2574–2584. [DOI] [PubMed] [Google Scholar]
- Vermeiren J, Van den Abbeele P, Laukens D, Vigsnaes LK, De Vos M, Boon N et al. (2012). Decreased colonization of fecal Clostridium coccoides/Eubacterium rectale species from ulcerative colitis patients in an in vitro dynamic gut model with mucin environment. FEMS Microbiol Ecol 79: 685–696. [DOI] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X et al. (2011). Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X et al. (2012). Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 6: 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L et al. (2009). Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis 15: 653–660. [DOI] [PubMed] [Google Scholar]
- Wrzosek L, Miquel S, Noordine ML, Bouet S, Chevalier-Curt MJ, Robert V et al. (2013). Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]