Summary
Liquid-liquid phase separation (LLPS) of RNA-binding proteins plays an important role in the formation of multiple membrane-less organelles involved in RNA metabolism, including stress granules. Defects in stress granule homeostasis constitute a cornerstone of ALS/FTLD pathogenesis. Polar residues (tyrosine and glutamine) have been previously demonstrated to be critical for phase separation of ALS-linked stress granule proteins. We now identify an active role for arginine-rich domains in these phase separations. Moreover, arginine-rich dipeptide repeats (DPRs) derived from C9orf72 hexanucleotide repeat expansions similarly undergo LLPS and induce phase separation of a large set of proteins involved in RNA and stress granule metabolism. Expression of arginine-rich DPRs in cells induced spontaneous stress granule assembly that required both eIF2α phosphorylation and G3BP. Together with recent reports showing that DPRs affect nucleocytoplasmic transport, our results point to an important role for arginine-rich DPRs in the pathogenesis of C9orf72 ALS/FTLD.
Keywords: amyotrophic lateral sclerosis, frontotemporal lobar degeneration, phase transition, protein aggregation, FUS, hnRNP, prion-like domain, low complexity domain, intrinsically disordered protein, LLPS
Graphical Abstract
Highlights
-
•
Arginine-rich peptides undergo LLPS dependent on counterions or polyaromates
-
•
Toxic arginine-rich DPRs perturb stress granule dynamics and protein content
-
•
PR-induced stress granule formation is dependent on eIF2α phosphorylation and G3BP
-
•
PR promotes aggregation of ALS-related proteins containing prion-like domains
Proteins high in arginine content are enriched in stress granules. Boeynaems et al. show that arginine-rich peptides can actively mediate liquid-liquid phase separation, a process underlying liquid organelle formation. Toxic C9orf72 DPRs change the arginine content and alter the properties of stress granules.
Introduction
Stress granules (SGs) are large cytoplasmic RNA-protein assemblies that form during cellular stress, and play a pivotal role in the integrated stress response (Kedersha et al., 2013). Following translational arrest, polysomes disassemble leading to a sudden increase in free mRNA levels in the cytoplasm. It is believed that this free mRNA subsequently nucleates the formation of SGs by binding of a large set of RNA-binding proteins (RBPs) (Jain et al., 2016). Interestingly, several of these SG proteins have been linked to the neurodegenerative disorders amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), in which neuronal cytoplasmic fibrillar aggregates of RBPs, e.g., TDP-43 and FUS, appear (Boeynaems et al., 2016b, Ramaswami et al., 2013). It is thought that these inclusion bodies play a key role in the disease pathogenesis (Ramaswami et al., 2013). Moreover, mutations in several of these SG proteins are also the genetic cause of the disease in a subset of familial cases (Renton et al., 2014). Hence, SGs have been suggested as a stepping stone towards pathological aggregation in ALS and FTLD (Aguzzi and Altmeyer, 2016, King et al., 2012, Ramaswami et al., 2013).
Based on previous observations from other RNA granules and nucleoli (Brangwynne et al., 2009, Brangwynne et al., 2011), recent studies have shown that the physical basis of SG formation is liquid-liquid phase separation (LLPS) (Burke et al., 2015, Conicella et al., 2016, Lin et al., 2015, Molliex et al., 2015, Murakami et al., 2015, Patel et al., 2015). Depending on in vitro conditions, recombinant TDP-43, FUS, and other hnRNPs spontaneously demix from an aqueous solution and form liquid-like protein-rich droplets. It was shown that the intrinsically disordered prion-like domains (PrLDs) of these proteins by themselves are sufficient in inducing LLPS. Interestingly, these liquid-like droplets can mature over time to more fibrillar states, and this process is accelerated by disease-causing mutations (Conicella et al., 2016, Molliex et al., 2015, Murakami et al., 2015, Patel et al., 2015). Other reports have shown that these PrLDs can also form reversible hydrogels, which have a high β sheet content (Kato et al., 2012). It was recently suggested that cellular SGs consist of a stable core surrounded by a liquid-like shell, and ATP-dependent remodeling seems necessary for maintaining SG fluidity (Jain et al., 2016, Kedersha et al., 2016). These data point at the importance of a tight control of SG dynamics in preventing pathological aggregation.
As mentioned above, PrLDs can undergo LLPS or form hydrogels in vitro (Burke et al., 2015, Kato et al., 2012, Lin et al., 2015, Molliex et al., 2015, Murakami et al., 2015, Patel et al., 2015). This type of low complexity domain (LCD) shows a strong sequence similarity to yeast prions in that they are enriched in polar uncharged amino acids (usually glutamine/asparagine) and glycine (King et al., 2012). Degenerate tyrosine-containing repeats in the PrLDs of FUS and other hnRNPs, which can mediate hydrophobic and π interactions, seem to be crucial in promoting hydrogel formation (Kato et al., 2012). The LLPS of these PrLDs indeed is inhibited by aliphatic alcohols, which interfere with hydrophobic interactions (Molliex et al., 2015). Phase separation based on hydrophobic interactions is also likely to underlie nuclear speckle and nuclear pore mesh formation (Frey et al., 2006, Hennig et al., 2015).
In this study, we investigate whether other protein domains may also contribute to RBP LLPS. We identify arginine-rich domains as regions prone to phase separation. Moreover, we find that pathogenic arginine-rich dipeptide repeats in C9orf72 ALS/FTLD also undergo LLPS in vitro and in cells, hereby uncovering a potential new toxic pathway in the disease.
Results
Arginine-Rich Peptides Phase Separate In Vitro
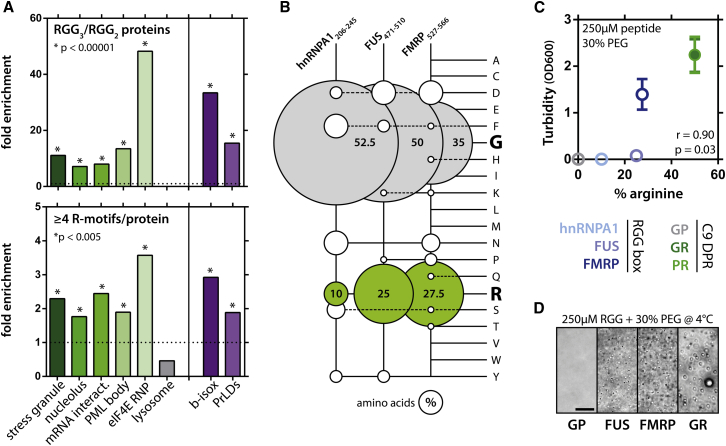
SG proteins also contain other domains that might be non-redundant in phase separation. Besides canonical RNA-binding domains, e.g., RRM, these proteins are also enriched in disordered arginine-rich sequences, e.g., RGG boxes (Aguzzi and Altmeyer, 2016) (Figure 1A), which can bind RNA or poly(ADP-ribose) and seed LLPS or aggregation of PrLDs (Altmeyer et al., 2015, Burke et al., 2015, Lin et al., 2015, Molliex et al., 2015, Patel et al., 2015, Schwartz et al., 2013). While globular RRMs have a clear RNA-binding function, these disordered arginine-rich motifs are more promiscuous and also serve as protein binding modules (Dormann et al., 2012, Thandapani et al., 2013). Given the strong conservation of both disorder and arginine content in RBPs (Varadi et al., 2015), we hypothesize that arginine-rich domains could play a role in LLPS in physiological and pathological processes. In fact, 40 amino-acid-long Arg/Gly-rich peptides from the RGG boxes of FUS, hnRNPA1, and FMRP spontaneously phase separate in the presence of a molecular crowder (Figures 1B–1D, S1A, and S1B). This emerging basic mechanism of LLPS puts particular emphasis on recent observations from C9orf72 hexanucleotide repeat expansions, which are the most common genetic cause of ALS and FTLD (DeJesus-Hernandez et al., 2011, Renton et al., 2011). A potential toxic mechanism is the generation of five species of dipeptide repeats (DPRs) through aberrant translation of the repeat RNA (Ash et al., 2013, Mori et al., 2013). Several groups have shown that two of them, namely, glycine-arginine (GR) and proline-arginine (PR) repeats, are highly toxic (Boeynaems et al., 2016a, Freibaum et al., 2015, Jovičić et al., 2015, Kwon et al., 2014, Lee et al., 2016, Mizielinska et al., 2014, Wen et al., 2014). We wondered whether these peptides exhibit a behavior similar to the RGG peptides in vitro. Synthetic GR20 and PR20, but not GP20, strongly phase separated following the addition of a molecular crowder (Figures 1C and 1D). Interestingly, the extent of phase separation for all examined peptides is significantly correlated with arginine content (Figures 1C and S2A), which indicates that arginines are likely to be the mediators of phase separation for these peptides.
Figure 1.
Arginine-Rich Motifs Are Enriched in Cellular Liquid-like Compartments and Can Phase Separate In Vitro
(A) Proteins in ribonucleoprotein complexes (Castello et al., 2012, Topisirovic et al., 2009), liquid compartments (Andersen et al., 2005, Jain et al., 2016, Liu et al., 2010), or proteins with prion-like domains (Kato et al., 2012, March et al., 2016) are enriched for both tri-RGG/di-RGG (Thandapani et al., 2013) and R motifs (Mitrea et al., 2016). Binomial test.
(B) Balloon plot indicating sequence redundancy of RGG boxes.
(C) Synthetic RGG boxes and arginine-rich C9orf72 DPRs phase separate following the addition of a molecular crowder. Measured at room temperature (RT), mean ± SD is depicted. The extent of phase separation is correlated with arginine content (Spearman).
(D) Examples of RGG and DPR phase separation. Pictures taken at 4°C. Scale bar represents 10 μm.
See also Figure S1.
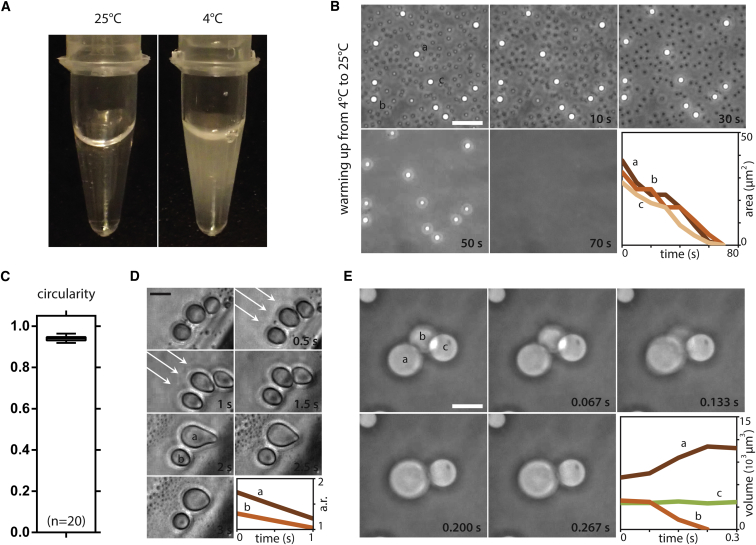
To further investigate such arginine-mediated phase separations, we used the PR DPR as a model. As shown for other proteins (Molliex et al., 2015, Nott et al., 2015, Patel et al., 2015), phase separation of PR20 is temperature dependent and reversible (Figures 2A, 2B, and S1A–S1C). PR20 droplets are highly circular (Figure 2C), suggesting that they minimize surface tension. When we applied shear stress to these droplets, we indeed observed plastic deformation with minimization of their surface area after relaxation (Figure 2D). There was also a conservation of volume after the fusion of two droplets (Figure 2E). All these observations indicate that PR20 droplets behave as a liquid.
Figure 2.
PR20 Peptide Undergoes Liquid-Liquid Phase Separation
(A) A solution of 1 mM PR20 with 20% PEG is clear at room temperature, but phase separates after cooling.
(B) Demixed PR20 droplets dissolve following a temperature increase.
(C) PR20 droplets are highly circular suggesting surface tension minimization (250 μM PR20, 30% PEG, RT).
(D) Large PR droplets deform when shear stress is applied (arrows), but take up a circular shape after stress relief as indicated by an aspect ratio approximating one (250 μM PR20, 30% PEG, RT).
(E) Volume is conserved following the fusion of two PR20 droplets (250 μM PR20, 30% PEG, RT). Scale bars represent 10 μm.
PR LLPS Is Dependent on Counterions
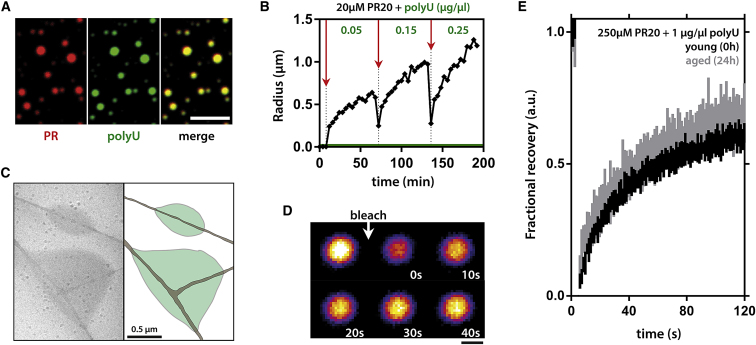
Besides temperature, LLPS of PR was also sensitive to protein concentration, molecular crowding, and the length of the peptide (Figures 3A and 3B). In agreement with the polar nature of the peptide, we found that LLPS was not perturbed by an aliphatic alcohol, but was hindered by increasing the salt concentration (Figures 3C, 3D, and S1D). Additionally, the fluorescence of the hydrophobic dye 8-anilino-1-naphthalenesulfonic acid (ANS) showed maximum intensity at 495 nm and was reduced following LLPS of PR20 (Figure S1F) (Hawe et al., 2008). These observations argue against hydrophobic interactions and indicate the importance of electrostatic forces via arginines in LLPS. Phase separation of like-charged inorganic polyelectrolytes has been reported before, and due to charge repulsion, this process is only conceivable in the presence of counteranions in solution (Brangwynne et al., 2015, Tiraferri et al., 2015). We reasoned that phosphate ions in our buffer could be a likely candidate in stabilizing PR-PR interactions. In accordance, when we incubated PR in pure water with the molecular crowder and added different salts, the extent of phase separation depended on the charge of the anions (Figure 3E). This raised the idea that biological anions could support PR LLPS as well. Indeed, addition of polyU RNA promoted LLPS in a dose-dependent manner even without molecular crowders being present (Figures 3F, 4A, and S1E); and this was also the case for other arginine-rich peptides (Figure S2A). Similar to the PEG-induced LLPS, PR20-RNA droplets were highly circular (Figure 4A). They also continuously increased in size over time, as measured by dynamic light scattering (DLS) (Figure 4B), mimicking observations for cellular SGs (Wheeler et al., 2016). Due to its polyvalent nature, polyU was much more effective than phosphate and crowding agent PEG in promoting LLPS. Besides purely electrostatic interactions, arginines can also engage in π interactions with aromatic side chains (Brangwynne et al., 2015). In support of the contribution of this mechanism, incubating PR20 with poly-tyrosine induced clustering of the insoluble poly-tyrosine polymers (Figures 3G and S1H).
Figure 3.
Molecular Determinants of PR20 Phase Separation
(A) PR20 LLPS is dependent on molecular crowding.
(B) PR LLPS is length dependent.
(C) Addition of 1,6-hexanediol does not affect PR20 LLPS.
(D) Addition of NaCl inhibits PR20 LLPS.
(E and E’) PR-PEG LLPS is dependent on inorganic counteranions and correlated to anionic charge.
(F and F’) RNA dose dependently induces PR20 LLPS in the absence of molecular crowder.
(G and G’) Addition of PR20 clusters poly-tyrosine polymers in the absence of molecular crowder.
Depict schemes of molecular interactions and phase contrast images of solutions (E’, F’, and G’). Mean ± SD, n = 3 (A–G). All measurements at RT. Scale bars represent 20 μm.
See also Figures S1 and S2.
Figure 4.
PR-RNA Droplets Are Dynamic Liquid Compartments
(A) Fluorescently tagged PR20 and polyU RNA colocalize in phase separated droplets. Scale represents 10 μm.
(B) Spontaneous fusion behavior of PR20-polyU RNA droplets over time, as expressed by changes in hydrodynamic radius over time as measured by DLS. Successive additions of polyU RNA indicated by red arrows. Final RNA concentration (green). The radius of polyU is 22nm (green line).
(C) Cryo-TEM of two PR20-polyU droplets (green) wetting carbon support (brown). No obvious structures can be observed within the droplets.
(D) Full droplet bleach of PR20 fluorescence indicates exchange between droplet and solution. Scale represents 5 μm.
(E) Within-droplet bleach on old versus young droplets indicates that PR20-polyU droplets do not mature or age. Mean ± SD, n = 12 droplets.
See also Figure S2.
PR Droplets Are Dynamic Structures
Disordered linear binding motifs often undergo induced folding following ligand binding (Tompa, 2016), but they can also remain predominantly disordered when bound in so-called “fuzzy” complexes (Tompa and Fuxreiter, 2008) and when phase separated, as already observed in the case of FUS LCD droplets (Burke et al., 2015). CD spectroscopy clearly illustrated the random coil nature of the dispersed PR20 peptide, and this was also the case for other arginine-rich peptides (Figures S2B–S2G). However, after titration with polyU RNA, the PR20 CD spectrum was lost, i.e., there is no signal for the peptide within droplets (Figure S2G). Therefore, we applied two other approaches to monitor the structural state of PR20 in droplets, both supporting its largely disordered (fuzzy) state. Fluorescence anisotropy of peptide tagged with a fluorophore (Figure S2H) is very low (0.02), and even after the addition of polyU RNA it only increases to about 0.2, far from the value measured in the highly viscous glycerol (0.38), the latter being close to the theoretical upper limit (0.4) for an immobile fluorophore. Global hydrogen-deuterium exchange (HDX) experiments (Figures S2I and S2J) show full accessibility of the free peptide, which is compatible with its disordered state. Following the addition of polyU RNA, accessibility drops only slightly (to 80% of its initial value), indicating that the peptide is still largely accessible, and suggesting that it stays disordered after LLPS. To check whether the PR-RNA droplets could have any internal fibrillar structure, we performed cryo-transmission electron microscopy (TEM), and could not discern any visible structures in the droplets (Figure 4C). Further, we performed fluorescence recovery after photobleaching (FRAP) analyses with the fluorescently tagged PR20 (Figures 4D and 4E), and found in full-droplet bleaching experiments that demixed PR readily interchanges with PR in the dispersed phase (Figure 4D). Bleaching within droplets allowed us to probe intradroplet dynamics. Aging of the PR-RNA droplets had no effect on intradroplet fluorescence recovery (Figure 4E). This result is in contrast to findings for PrLD LLPS, which undergo a liquid-to-solid transition likely through β sheet formation (Conicella et al., 2016, Molliex et al., 2015, Patel et al., 2015).
PR LLPS Recruits Specific Proteins Involved in RNA Metabolism
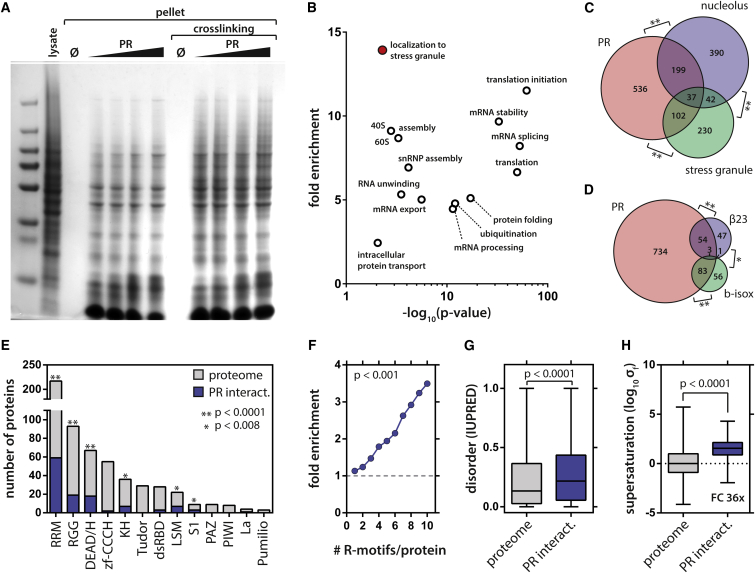
We wondered which cellular proteins could interact with PR. We took soluble HeLa cell lysate, to which we added increasing concentrations of PR30. The PR30 peptide spontaneously demixed, and could be separated from the solution by gentle centrifugation. The resultant pellet was subsequently washed and analyzed by gel electrophoresis (Figure 5A). Unexpectedly, this pellet was largely resistant to washing steps, suggesting that PR induced the precipitation of different cellular proteins to the insoluble fraction. The demixing of PR was also largely independent of the presence of RNA in the lysate, as addition of RNase only had a modest effect on the extent of protein precipitation (Figure S3). To see whether weak interactions could still be involved in the process, we performed mild crosslinking with paraformaldehyde as recently used to identify both the stable core and liquid-like shell of cellular SGs (Jain et al., 2016). The PR fraction seemed to be enriched for a specific set of proteins as was evident from the band pattern. Mass spectroscopy (MS) analysis on both crosslinked and uncrosslinked samples identified 874 proteins that were detected and quantified in both conditions.
Figure 5.
PR30 Initiates Phase Separation of RNA Granule Components and Metastable Proteins In Vitro
(A) Addition of PR30 to cleared cell lysate induces LLPS with the formation of an insoluble fraction. Background from pre-cleared lysate not treated with PR was effectively zero. Weak interactions can be stabilized by mild PFA crosslinking.
(B) The PR30 interactome (n = 874) is enriched for GO biological processes centered on RNA and protein metabolism. Benjamini-Hochberg.
(C and D) The PR30 interactome significantly overlaps with cellular liquid compartments (Andersen et al., 2005, Jain et al., 2016) (C) and metastable proteins (Kato et al., 2012, Olzscha et al., 2011) (D). Binomial test.
(E and F) The PR30 interactome is enriched for RNA binding domains (E) and arginine-rich motifs (Mitrea et al., 2016) (F). Binomial test.
(G and H) The PR30 interactome is enriched for disordered (G) and supersaturated proteins (Ciryam et al., 2013) (H). Boxplots, whiskers indicate range. Mann-Whitney.
See also Figures S3–S5 and Table S1.
We performed gene ontology (GO) enrichment analyses to identify overrepresented biological processes and cellular compartments in our hit list. Significantly overrepresented terms centered on RNA and protein metabolism (GO biological process; Figure 5B) and different cellular liquid-like compartments (GO cellular compartment; Figure S3). Indeed, when comparing the protein content of the PR30 precipitate with those of stress granules and nucleoli, we found a strong overlap with both cellular liquid compartments (Figure 5C), while this was not the case for an unrelated organelle (Figure S4). The PR30 precipitate was also enriched for metastable aggregation-prone proteins (Figure 5D). Given that ∼60% of identified proteins have been implicated in RNA metabolism (Figure S4), we evaluated overrepresentation of RNA binding domains. Several RNA binding domains were highly significantly enriched in our list. These included globular RRM or DEAD/H box domains and disordered RGG boxes (Figure 5E). Besides RGG boxes, R motifs were also highly abundant (Figure 5F), once more illustrating the importance of arginine-rich binding modules in LLPS. Lastly, we found that the PR30 precipitate is enriched for disordered (Figure 5G) and aggregation-prone supersaturated proteins (Figure 5H) (Ciryam et al., 2013).
Quantitative analysis revealed that 190 proteins were more abundant after crosslinking (Figure S5), which suggests that weak interactions between these proteins and the PR30-induced insoluble fraction could be stabilized by our mild crosslinking protocol. Interestingly, this set of cross-linking-sensitive proteins was enriched for different protein families involved in stress granule biology, such as hnRNPs (Molliex et al., 2015) and aminoacyl transferases (David et al., 2011) (Figure S5). Although both 40S and 60S ribosomal proteins were highly abundant in our protein set identified by MS, only 40S, but not 60S, constituents were overrepresented in the crosslinking-sensitive fraction. Cellular SGs also contain 40S, but exclude 60S subunits (Kedersha et al., 2002).
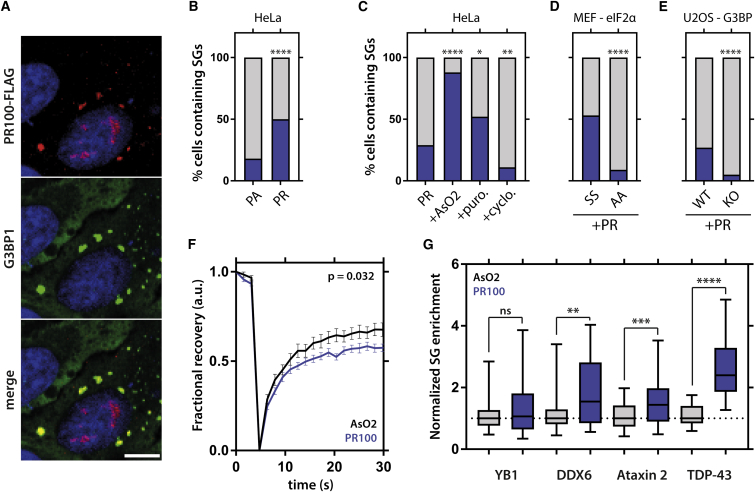
PR Induces Stress Granule Assembly in Human Cells
To investigate the biological relevance of our in vitro cell-free findings, we overexpressed a codon-optimized PR100 construct in HeLa cells (Figure 6A). As previously reported, PR100 predominantly localized to the nucleolus (Kwon et al., 2014, Tao et al., 2015, Wen et al., 2014, Yamakawa et al., 2015). Yet, the evidence whether arginine-rich DPRs affect SGs has remained ambiguous (Tao et al., 2015, Wen et al., 2014, Yamakawa et al., 2015). We found cytoplasmic PR100 granules positive for SG markers after overexpression of PR100 (Figure 6A), and this process was dose and length dependent (Figure S6). PR100 expression was more efficient at inducing SGs than PA100 expression, showing that these SGs do not just originate from the transfection protocol (Figure 6B). PR100-induced SGs were also responsive to known SG modifying compounds, i.e., arsenite and puromycin treatment enhanced SG formation, whereas cyclohexamide reduced their number (Figure 6C). By using a mutant MEF cell line carrying a non-phosphorylatable form of eIF2α, we found that PR100 requires the integrated stress response for SG induction (Figure 6D). Also the presence of G3BP1/2 seemed to be required for this, based on the use of knockout cell lines (Figure 6E) (Kedersha et al., 2016). When studying these PR100-induced SGs in more detail, we observed that they are slightly less dynamic than arsenite-induced SGs, based on FRAP analysis of a G3BP1-GFP fusion protein (Figure 6F). Moreover, the protein content of PR100 SGs was strongly altered compared to arsenite SGs. While there was no difference in the SG enrichment of a general SG marker (YB-1), both a late SG marker (DDX6) and ALS-related proteins carrying a PrLD (ataxin-2 and TDP-43) were significantly enriched in PR100 SGs compared to arsenite SGs (Figure 6G).
Figure 6.
PR100 Expression Alters Stress Granule Dynamics in Cells
(A) Expression of PR100 in HeLa cells induces cytoplasmic SGs assembly positive for PR.
(B) PR100 is more efficient at inducing SGs than PA100.
(C) Arsenite and puromycin increase number of SGs in PR100 transfected cells, whereas cyclohexamide decreases them.
(D) Non-phosphorylatable form (AA) of eIF2α prevents PR100-induced SG assembly in MEFs.
(E) G3BP1/2 knockout prevents PR100 induced SG assembly in U2OS cells.
(F) PR100-induced SGs have a mildly reduced FRAP recovery for G3BP1 compared to arsenite-induced ones. Repeated-measures ANOVA. Mean ± SEM, n = 17 SGs.
(G) SG enrichment of several RNA binding proteins is altered in PR100-induced SGs compared to arsenite-induced ones. Boxplots, whiskers indicate range. Mann-Whitney. n > 60 SGs, from three independent experiments.
Fischer’s exact test. n ≥ 100 cells, from three independent experiments (A–E).
See also Figure S6.
PR Affects Properties of PrLD Droplets
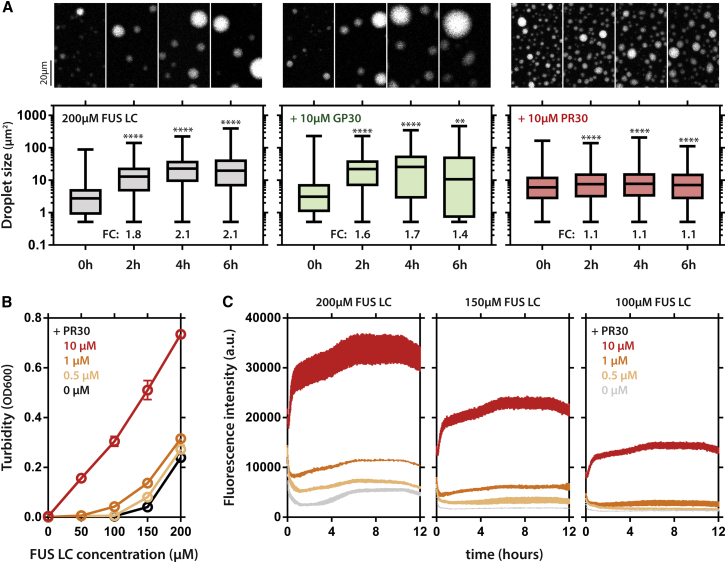
The current hypothesis for the formation of aggregates in ALS/FTLD patients is a liquid-to-solid phase transition or perturbed clearance of SGs. Although not observed in a living system so far, in vitro droplets of FUS and hnRNPs have been shown to mature to a solid state over the course of hours (Molliex et al., 2015, Patel et al., 2015). This likely corresponds to a transition of their PrLDs from a disordered fuzzy state to a rigid β sheet structure (Burke et al., 2015, Molliex et al., 2015, Patel et al., 2015). Since we found that (1) PR repeats could engage in π interactions with tyrosines, (2) PR induced the precipitation of several of these proteins in vitro, and (3) such proteins were enriched in PR100 SGs, we wondered whether PR could affect the dynamics and maturation of PrLD droplets. Addition of PR30, but not GP30, to FUS LCD droplets interfered with their spontaneous fusion dynamics (Figure 7A). Additionally, FUS LCD phase separation was also enhanced by addition of PR30 (Figures 7A and 7B). In agreement with the reduced dynamics of the droplets, kinetic thioflavin-T fluorescence analysis of FUS LCD droplets supplemented with PR30, indicated an increase in β sheet content over time (Figures 7C and S7).
Figure 7.
PR30 Affects Phase Transitions of PrLDs
(A) FUS LC droplets spontaneously fuse over time into larger droplets, as characterized by an increase in droplet size. This spontaneous fusion behavior is prevented by addition of PR30, but not GP30. Boxplots, whiskers indicate range. Kruskal-Wallis. n = (318–3,349) droplets. 50 mM MES buffer, 200 μM NaCl.
(B) The FUS LC domain (amino acids [aa] 1–163) poorly phase separates, but addition of PR30 increases phase separation dose dependently.
(C) Kinetic analysis of ThT fluorescence over a 12-hr period for different FUS LC-PR30 combinations. Addition of PR increases ThT fluorescence. Of note, PR30 by itself did not yield any increase in ThT fluorescence over time (data not shown). Mean ± SD (B and C).
See also Figure S7.
Discussion
In this study, we have investigated whether highly repetitive and disordered arginine-rich peptides generated from expanded C9orf72 repeats in ALS/FTLD could play a role in LLPS. This phenomenon is of intense interest since it could explain the biogenesis and physical underpinnings of SGs, which are the prime suspect of seeding for pathological protein aggregation in ALS and FTLD (King et al., 2012, Ramaswami et al., 2013). Until now, LLPS of PrLD containing RBPs has been mostly attributed to hydrophobic interactions. However, arginine-rich domains are highly enriched in cellular liquid-like compartments, such as stress granules and the nucleolus, and both protein disorder and arginine-content are highly conserved in RBPs (Varadi et al., 2015). This conservation suggests that these arginine-rich domains must be under selective pressure, highlighting their functionality and suggesting that a similar gain-of-function may underlie the pathological role of C9orf72 DPRs in ALS. In fact, RGG boxes have been shown to affect PrLD aggregation and LLPS by nucleating on RNA or poly(ADP-ribose) (Altmeyer et al., 2015, Lin et al., 2015, Molliex et al., 2015, Patel et al., 2015, Schwartz et al., 2013). We hence hypothesized that arginine-rich DPRs could play a more direct role in phase separation through their disorder (Zhang et al., 2015) and thus they could also contribute to developing disease.
Indeed, we found that arginine-rich DPRs derived from the C9orf72 repeat expansion undergo LLPS, a process mimicked by short synthetic peptides corresponding to the RGG boxes of three SG proteins, which also spontaneously phase separated in vitro. This phase transition was directly correlated with the arginine content of the peptides. As shown previously for inorganic polymers, such like-charged phase separation was dependent on counterions (Tiraferri et al., 2015). In accordance, we found that RNA strongly induced LLPS of PR. Recently, two other studies have also reported a similar behavior for artificial arginine-rich peptides (Aumiller and Keating, 2016) or peptides derived from nucleolar proteins (Mitrea et al., 2016).
Intriguingly, when we added cellular proteins to PR, this resulted in the formation of an insoluble pellet, rather than liquid droplets. However, crosslinking experiments showed that specific weak protein-protein interactions did occur in our test tube experiment. This is consistent with recent reports indicating that cellular SGs likely consist of a stable core surrounded by a liquid shell, rather than a homogeneous liquid droplet (Jain et al., 2016, Kedersha et al., 2016). Identifying the PR30 interactome using MS showed that PR was able to interact with a large, yet specific, set of proteins enriched for RNA-binding domains, arginine-rich motifs and protein disorder. Numerous hits were known constituents of endogenous liquid organelles, and, interestingly, several have been directly implicated in ALS/FTLD. The PR30 interactome was indeed significantly enriched for proteins associated with “neuromuscular disease” (p = 2.14 × 10−9, ingenuity pathway analysis). Hence, we suggest that DPRs may play a role in disease by upsetting the internal functional balance of membraneless organelles (Brangwynne et al., 2015). The data of our in vitro precipitation experiment are in agreement with three recent studies showing that intracellular PR and GR peptides can bind with a large set of proteins involved in RNA- and SG metabolism (Lee et al., 2016, Lin et al., 2016, Lopez-Gonzalez et al., 2016).
When overexpressed in cells, PR was indeed able to seed SG assembly, and this SG induction was dependent on phosphorylation of eIF2α and the presence of G3BP proteins. Although SG induction by PR was shown recently by others (Lee et al., 2016, Yamakawa et al., 2015), the mechanism through which this occurred remained unknown and of intense interest from a therapeutic perspective. In this light, eIF2α modulation has already been shown to ameliorate toxicity in different TDP-43 ALS models (Kim et al., 2014). G3BP proteins have been shown to be essential in eIF2α-controlled SG assembly (Kedersha et al., 2016), and the G3BP RGG domain plays a crucial role in this process, once more highlighting the importance of arginine-rich domains in SG metabolism.
PR-induced SGs displayed reduced dynamics and were enriched for ALS-related proteins. Moreover, PR was also able to accelerate the liquid-to-solid maturation of PrLD droplets, a process reminiscent of pathological aggregation (Conicella et al., 2016, Molliex et al., 2015, Patel et al., 2015). In line with our findings, two other reports also recently found that arginine-rich DPRs can affect SGs and other higher order assemblies in similar ways (Lee et al., 2016, Lin et al., 2016).
SGs are generally considered to be the stepping stone toward pathological aggregation of RBPs in disease. Identifying the mechanisms behind altered SG dynamics and a deeper appreciation of their biology, hence, will provide us with invaluable clues to better understand the etiology of disease. Disease mutations are known to target several SG proteins or proteins involved in SG dynamics (Ramaswami et al., 2013). However, the reason why wild-type proteins mislocalize to the cytoplasm and aggregate in C9orf72 cases remained elusive. Recently, we and others have shown that nucleocytoplasmic transport defects caused by arginine-rich DPRs could explain the mislocalization of these RBPs (Boeynaems et al., 2016a, Boeynaems et al., 2016b, Freibaum et al., 2015, Jovičić et al., 2015). By focusing now on the biophysical behavior of arginine-rich DPRs, we and others have found that they could also be directly implicated in the cytoplasmic aggregation of RBPs. These data suggest that DPRs could initiate a pathogenic cascade in C9orf72 ALS/FTLD, by targeting the nuclear transport-stress granule axis (Boeynaems et al., 2016b, Edbauer and Haass, 2016).
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-FLAG, mouse | Sigma-Aldrich | F3165; RRID: AB_259529 |
| Anti-FLAG, rabbit | Cell Signaling | #2368S |
| Anti-G3BP1, mouse | Abcam | Ab56574; RRID: AB_941699 |
| Anti-DDX6, rabbit | Abcam | Ab40684; RRID: AB_941268 |
| Anti-YB1, rabbit | Abcam | Ab76149; RRID: AB_2219276 |
| Anti-TDP-43, rabbit | Proteintech | 12892-1-AP; RRID: AB_2200505 |
| Anti-Ataxin-2, mouse | BD Biosciences | 611378; RRID: AB_398900 |
| Anti-TIA1, goat | Santa Cruz | sc-1751; RRID: AB_2201433 |
| Bacterial and Virus Strains | ||
| E. coli, BL21 Star (DE3) | Thermo Scientific | C601003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PR20 | This paper | N/A |
| PR30 | This paper | N/A |
| GR30 | This paper | N/A |
| GP30 | This paper | N/A |
| FUS RGG | This paper | N/A |
| hnRNPA1 RGG | This paper | N/A |
| FMRP RGG | This paper | N/A |
| FUS LC | Dr. Nicolas Fawzi | Burke et al., 2015 |
| hnRNPA2 LC | Dr. Nicolas Fawzi | Burke et al., 2015 |
| 1,6-Hexanediol | Sigma-Aldrich | 240117 |
| polyU RNA | Sigma-Aldrich | P9528 |
| poly-tyrosine | Sigma-Aldrich | P1800 |
| PEG300 | Sigma-Aldrich | 81160 |
| ANS | Sigma-Aldrich | A1028 |
| cOmplete protease inhibitor cocktail | Sigma-Aldrich | 11697498001 |
| Arsenite | Sigma-Aldrich | S7400 |
| Lipofectamine 3000 | Thermo Scientific | L3000015 |
| NucBlue | Thermo Scientific | R37605 |
| Prolong Gold Antifade | Thermo Scientific | P36934 |
| Critical Commercial Assays | ||
| Alexa Fluor 488 Protein Labeling Kit | Thermo Scientific | A10235 |
| Alexa Fluor 568 Protein Labeling Kit | Thermo Scientific | A10238 |
| Deposited Data | ||
| Mas spec data | This paper | PRIDE: PXD005509 |
| Experimental Models: Cell Lines | ||
| HeLa | ATCC | CCL-2 |
| MEF eIF2a SS | Dr. Nancy Kedersha | Emara et al., 2012 |
| MEF eIF2a SS | Dr. Nancy Kedersha | Emara et al., 2012 |
| U2OS | Dr. Nancy Kedersha | Kedersha et al., 2016 |
| U2OS G3BP1/2 KO | Dr. Nancy Kedersha | Kedersha et al., 2016 |
| U2OS G3BP1-GFP | Dr. Paul Taylor | Figley et al., 2014 |
| Oligonucleotides | ||
| polyU30-Alexa 488 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: PA100-FLAG | Dr. Daisuke Ito | Yamakawa et al., 2015 |
| Plasmid: PR100-FLAG | Dr. Daisuke Ito | Yamakawa et al., 2015 |
| Plasmid: GR100-FLAG | Dr. Daisuke Ito | Yamakawa et al., 2015 |
| Plasmid: PR100-FLAG-mCherry | This study | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Ludo Van Den Bosch (ludo.vandenbosch@kuleuven.vib.be).
Experimental Model and Subject Details
Cell culture
U2OS cells stably expressing G3BP1-GFP were a kind gift of Dr. Paul Taylor (St. Jude, USA) (Figley et al., 2014). U2OS G3BP1/2 KO cell line, and eIF2α SS and AA MEFs were a kind gift of Dr. Nancy Kedersha and Dr. Paul Anderson (Harvard, USA) (Emara et al., 2012, Kedersha et al., 2016). U2OS and HeLa cells (ATCC) were cultured in high glucose DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Greiner), 4 mM Glutamax (Invitrogen), penicillin (100 U/mL), streptomycin (100 μg/mL) and non-essencial amino acids (1%). Cells were grown at 37°C in a humidified atmosphere with 5% CO2.
Methods Details
Plasmids, peptides and recombinant proteins
FLAG-tagged DPR50 expression constructs were synthesized by Genscript (Piscataway, USA). The DPR100 constructs were a kind gift of Dr. Daisuke Ito (Department of Neurology, Keio University, Tokyo, Japan).
Peptides were chemically synthesized by Pepscan (Lelystad, Netherlands). Peptides were dissolved in milli-Q water and stored at −80°C. Peptides were fluorescently labeled using Alexa Fluor Labeling Kits (Thermo Scientific).
FUS LC domain was expressed in E. coli and purified as described previously (Burke et al., 2015). hnRNPA2 LC domain (residues 190-341) was expressed with a TEV-cleavable N-terminal hexahistidine tag, purified from the inclusion body via HisTrap (GE Healthcare) in urea containing buffers, concentrated, diluted into native buffer for TEV cleavage of hexahistidine tag, resolublized by addition of solid urea, separated from his-tagged TEV protease and cleaved his-tag by HisTrap, and concentrated to 1-2 mM in 8M urea 20mM MES pH 5.5. hnRNPA2 was aliquoted and flash frozen. Experiments were conducted by dilution into native buffer conditions.
Droplet formation
Peptides were diluted to the indicated concentrations in 100mM K2HPO4/KH2PO4 buffer at pH 7, unless otherwise indicated. PEG300, polyU RNA or poly-L-tyrosine (Sigma-Aldrich) were added at the indicated concentrations. All reactions were carried out at room temperature, unless indicated otherwise.
For fluorescence microscopy and FRAP analyses, PR-Alexa 568 and polyU30-Alexa 488 were spiked in at 200nM and 100 nM respectively.
FUS LC droplets were generated by diluting the stock solution to the desired concentration in 50mM MES buffer at pH 5, with the indicated NaCl concentrations.
hnRNPA2 LC droplets were generated by diluting the stock solution to the desired concentration in 20mM NaPi buffer with 50mM NaCl at pH 7.5.
Turbidity measurements
OD600 of 60μl samples was measured using trUView microcuvettes in a SmartSpec Plus Spectrophotometer (Bio-Rad). Turbidity of 100μl of FUS and hnRNPA2 LC droplets was measured on a Spectra Max M5 Microplate Reader (Molecular Devices).
Spontaneous fusion assays
FUS LC droplets were generated by diluting the stock solution to 250μM in 50μM MES buffer at pH 5, 200μM NaCl. 1μM of Alexa 488 tagged FUS LC was spiked into the solution.
Fluorescent droplets were incubated in plastic Cell Counter slides (Bio-Rad) at room temperature. Chambers were sealed using nail varnish to prevent evaporation during aging. Every two hours three arbitrary fields were imaged on a Zeiss LSM 510 Meta NLO confocal microscope. Droplet sizes were quantified with FIJI and analyzed with Prism.
ThT assays
ThT was added to 100μl PR30 + LC droplet mixtures at a concentration of 20μM. Fluorescence intensity over time was followed using a Spectra Max M5 Microplate Reader (Molecular Devices). Reactions were carried out in triplicate. Data was plotted using Prism software. Lag time was estimated as the time point of a local minimum in the curve.
CD measurements
CD spectra of the peptides analyzed in this study were recorded in a Jasco 715 spectropolarimeter equipped with a PTC 423S peltier element. Peptides were loaded into a 0.2mm quartz cuvette at 30μM concentration (50mM K2HPO4/KH2PO4 buffer at pH 7), and spectra were recorded with 50nm/min scan speed at 25°C with 9 acquisitions averaged on the fly. Spectra were background corrected with the buffer spectrum (50mM K2HPO4/KH2PO4 buffer at pH 7), and converted into molar ellipticity.
Cryo-TEM
PR droplet (250μM PR20 + 0.04μg/μl polyU RNA) sample (3.5 μL) was applied to a 300 mesh lacey Quantifoil grid and incubated for 30 s. Next, excess buffer was removed by blotting the grids for 3 s using a Whatman 1 filter paper and the sample was snap frozen by plunging in liquid ethane at a temperature of −180°C and stored in liquid nitrogen until visualization. Next, the samples were transferred to a Gatan 914 cryoholder and imaged at low dose conditions at −177°C, using a JEOL JEM1400 TEM equipped with an 11 Mpxl Olympus SIS Quemesa camera.
FRAP analysis
Fluorescent droplets were incubated in plastic Cell Counter slides (Bio-Rad) at room temperature. Chambers were sealed using nail varnish to prevent evaporation during aging. Fluorescence recovery after bleaching was monitored using Zen software on a Zeiss LSM 510 Meta NLO confocal microscope. For intradroplet FRAP, a circular are of 1μM radius was bleached in droplets with a radius between 5μM and 10μM. Raw data was background substracted and normalized using Excell, and plotted using Prism software. FRAP curves were fitted with a one phase exponential curve. Images were formatted FIJI and ImageJ software.
Fluorescence anisotropy measurements
Measurements were carried out in a LS55 Luminescence Spectrometer (PerkinElmer), with 200nM PR-Alexa568 N-terminally labeled protein in 50mM K2HPO4/KH2PO4 buffer at pH 7. After the addition of 10μM of bulk PR30, fluorescence anisotropy was measured at increasing RNA concentration at 578nm excitation and 603nm of emission wavelengths. Anisotropy was determined via following intensity of polarized light at 0°, 90° angles.
Dynamic light scattering (DLS)
20μM PR30 was titrated with increasing concentration of polyU RNA starting at a 0.04μg/μl concentration in a DynaPro NanoStar (Wyatt technologies) in 50mM K2HPO4/KH2PO4 buffer at pH 7. The size of particles was followed for one hour after every titration step (each titration step represented a 0.5% dilution). To control sedimentation and droplet fusion sample was mixed thoroughly with a pipet randomly between time points.
ANS titration
ANS (8-Anilinonaphthalene-1-sulfonic acid) fluorescence was measured for 40μM PR20 or 40μM GR20 peptide in the presence and absence of 0.05μg/μl polyU RNA in a BioTek MX Synergy plate reader in 50mM K2HPO4/KH2PO4 buffer at pH 7. Emission spectra were recorded between 400nm and 600nm at 380nm excitation wavelength.
Hydrogen deuterium exchange (HDX)
Global HDX experiments were performed at 4°C. PR30 and polyU RNA were dissolved in miliQ grade water and added together to make a final concentration of 400 μM PR and 4.8 mg/ml polyU RNA to induce LLPS. For experiments on the free peptide, PR was diluted to a final concentration of 400μM. The samples were then, either immediately or after an incubation time of 1h for the LLPS samples, diluted with a 20-fold excess of D2O. After the indicated time points the HDX reaction was quenched by bringing the pH of the solution to 1 using formic acid, followed by snap freezing in liquid nitrogen. The samples were stored at −80°C prior to analysis. Mass spectrometry analysis of deuterium incorporation was performed on a Synapt G2 HDMS mass spectrometer (Waters, Wilmslow, UK) by direct infusion of the undigested sample using nano-electrospray ionization with in-house prepared gold-coated borosilicate needles. All experiments were performed in triplicates.
PR30 protein precipitation
HeLa cells were trypsinized, pelleted and washed three times with 1x PBS (Thermo Scientific). Cells were resuspended in K2HPO4/KH2PO4 buffer with EDTA-free cOmplete protease inhibitor cocktail (Roche) and sonicated on ice. The lysate was cleared from the insoluble fraction by centrifugation for 15min at 10,000 rpm at 4°C. The supernatans was retrieved and protein concentration was measured using Micro BCA assay (Thermo Scientific).
1 mg of protein was added to 0.05 μmole of PR30 to a final concentration of 100μM, and left incubating for 10 min at RT. Paraformaldehyde (Sigma-Aldrich) was added to a final concentration of 0.5%, and crosslinking was left for 5 min. 500μl of 2M Glycine (Sigma-Aldrich) was added to quench paraformaldehyde, and samples were left for 5 min. Uncrosslinked samples were treated similarly, apart from paraformaldehyde addition. The volume of the samples was increased to 1.5ml with K2HPO4/KH2PO4 buffer, before gently spinning down the PR droplets at 4,000 rpm for 5 min. Pellets were subsequently washed with 1ml PBS with 0.4% Triton X-100 (Sigma-Aldrich) and vortexed before spinning down. Washing steps were repeated three times. The resulting pellets were processed for LC-MS/MS. Samples for SDS-PAGE were generated identically, but procedure was downscaled four times.
SDS-PAGE
PR LLPS pellets were resuspended in 1x reducing Laemmli containing SDS (Thermo Scientific). 1/8th of pellet or input lysate were loaded onto NuPAGE Novex 4%–12% Bis-Tris precast gels (Thermo Scientific). After running gels were stained using Coomassie Brilliant blue R-250 according to manufacturer’s instructions (Bio-Rad).
Proteomics sample preparation and LC-MS/MS analysis
Uncrosslinked (2 replicates) and crosslinked pellets (3 replicates) were redissolved in 300 mM NaCl Tris buffer, sonicated and boiled to remove crosslinks. Urea was added to a final concentration of 8M to solubilize insoluble proteins. The total sample volume was 1.5 ml.
Proteins in each sample were reduced with 5 mM DTT and incubation for 30 min at 55°C and then alkylated by addition of 10 mM iodoacetamide for 15 min at room temperature in the dark. Samples were further diluted to a final urea concentration of 2 M and proteins were digested with trypsin (Promega) (1/100, w/w) overnight at 37°C. Peptides were then purified on Omix C18 tips (Agilent), dried and re-dissolved in solvent A (25 μl 0.1% TFA in water/acetonitrile (98:2, v/v)) of which 10 μl was injected for LC-MS/MS analysis on an Ultimate 3000 RSLCnano System (Dionex, Thermo Fisher Scientific) in line connected to a Q Exactive HF mass spectrometer with a Nanospray Flex Ion source (Thermo Fisher Scientific). Trapping was performed at 10 μl/min for 4 min in solvent A (on a reverse-phase column produced in-house, 100 μm I.D. x 20 mm, 5 μm beads C18 Reprosil-Pur, Dr. Maisch) followed by loading the sample on a 40 cm column packed in the needle (produced in-house, 75 μm I.D. × 400 mm, 1.9 μm beads C18 Reprosil-HD, Dr. Maisch). Peptides were eluted by an increase in solvent B (0.1% formic acid in water/acetonitrile (2:8, v/v)) in linear gradients from 2% to 30% in 100 min, then from 30% to 56% in 40 min and finally from 56% to 99% in 5 min, all at a constant flow rate of 250 nl/min. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the 16 most abundant ion peaks per MS spectrum. Full-scan MS spectra (375-1500 m/z) were acquired at a resolution of 60,000 after accumulation to a target value of 3,000,000 with a maximum fill time of 60 ms. The 16 most intense ions above a threshold value of 22,000 were isolated (window of 1.5 Th) for fragmentation at a normalized collision energy of 32% after filling the trap at a target value of 100,000 for maximum 45 ms. The S-lens RF level was set at 55 and we excluded precursor ions with single and unassigned charge states.
Protein identification and quantification
Data analysis was performed with MaxQuant (version 1.5.3.30) (Cox and Mann, 2008) using the Andromeda search engine with default search settings including a false discovery rate set at 1% on both the peptide and protein level. Spectra were searched against the human proteins in the Uniprot/Swiss-Prot database (database release version of April 2016 containing 20,103 human protein sequences, www.uniprot.org). The mass tolerance for precursor and fragment ions were set to 4.5 and 20 ppm, respectively, during the main search. Enzyme specificity was set as C-terminal to arginine and lysine, also allowing cleavage at proline bonds with a maximum of three missed cleavages. Variable modifications were set to oxidation of methionine residues and acetylation of protein N-termini. Only proteins with at least one unique or razor peptide were retained leading to the identification of 1811 human proteins. Proteins were quantified by the MaxLFQ algorithm integrated in the MaxQuant software (Cox et al., 2014). A minimum ratio count of two unique or razor peptides was required for quantification. Further data analysis was performed with the Perseus software (version 1.5.3.0) after loading the protein groups file from MaxQuant. Proteins only identified by site and reverse database hits were removed and replicate samples of uncrosslinked and crosslinked were grouped. Proteins with less than three valid values in at least one group were removed and missing values were imputed from a normal distribution around the detection limit. Then, a t test was performed (FDR = 0.01 and S0 = 1) to compare samples uncrosslinked and crosslinked and calculate the fold change for each protein between both samples. After removal of potential contaminants, 874 proteins were quantified in total (Table S1).
Cell transfection
Cells were grown at 37°C in a humidified atmosphere with 5% CO2 for 24h. Cells were transiently transfected using Lipofectamine 3000 (Invitrogen) according to manufacturer’s instructions.
Immunohistochemistry and microscopy
Cells were fixed 24h after transfection in 4% formaldehyde in PBS and stained according standard protocols (including methanol fixation and permeabilization by PBS-T 0.04%). Following antibodies were used: anti-FLAG (F3165, Sigma), rabbit anti-FLAG (#2368S, Cell Signaling), mouse anti-G3BP1 (Ab56574, Abcam), rabbit anti-DDX6 (Ab40684, Abcam), rabbit anti-YB1 (Ab76149, Abcam), rabbit anti-TDP-43 (12892-1-AP, Proteintech), mouse anti-ataxin-2 (611378, BD Biosciences), goat anti-TIA1 (sc-1751, Santa Cruz). AlexaFluor 555 and AlexaFluor 488 secondary antibodies (Life Technologies) were used. Nuclei were visualized using NucBlue counterstaining (Thermo Scientific). Slides were mounted using ProLong Gold antifade reagent (Life Technologies).
Confocal images were obtained using a Zeiss LSM 510 Meta NLO confocal microscope. Images were analyzed, formatted and quantified with FIJI and ImageJ software. Statistics were carried out using Prism software.
Control stress granules were induced by incubating the cells for 1h with 0.5mM NaAsO2 (Sigma).
Stress granule analysis
Percentage of cells carrying stress granules was assessed by G3BP1 staining in all tested cell lines. Minimum 100 cells were counted over three independent experiments. Percentages were compared using Fisher’s Exact test in Prism.
Stress granule enrichment was quantified as the ratio of fluorescence intensity in the stress granule over the intensity of the surrounding cytoplasm. Two stress granules per cell were quantified, of at least 30 cells from 3 independent experiments. For stress granule enrichment of PR100, one stress granule per cell was counted, of 40 cells from 2 independent experiments. PR100 stress granules were visualized for quantification with FLAG staining, arsenite stress granules were visualized by TIA1 staining.
For analysis of dose- and length dependency of PR cytoplasmic structures, cells were categorized as having diffuse cytoplasmic PR, small stress granules, large stress granules or coarse/granular/heterogeneous cytoplasmic PR staining. Stress granules were assessed based on G3BP1-GFP staining in the U2OS stable cell line. Representative images of each category are shown in Figure S8. Cells were assigned to a category and plotted according to cytoplasmic PR100-FLAG intensity. Data was subsequently also plotted as the percentage of cells displaying non-diffuse cytoplasmic PR staining after manual binning according to cytoplasmic PR100-FLAG intensity. Percentages were fitted by a one-phase exponential curve, illustrating a strong dose-dependency of cytoplasmic structure formation.
Quantification and Statistical Analysis
Statistical parameters and distributions are reported in the Figures and corresponding Figure Legends. Statistical analysis was performed in Excel or GraphPad Prism.
Data and Software Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PRIDE: PXD005509.
Author Contributions
S.B., E.B., D.K., A.K., and E.T. planned and performed the experiments. A.V., M.G., M.D.D., T.J., V.H.R., A.M.J., P.B., T.V., R.-M.K., D.D., J.P.T., N.K., and P.A. provided technical support, reagents, and feedback on the project. J.P.T., N.K., F.I., F.S., J.S., F.R., N.L.F., W.R., and P.V.D. provided ideas for the project and participated in writing the paper. S.B., E.B., D.K., P.T., and L.V.D.B. wrote the paper.
Acknowledgments
Research was funded by the KU Leuven, VIB, and the European Research Council in the context of the European’s Seventh Framework Programme (FP7/2007-2013 and ERC grant agreement number 340429); the Research Foundation Flanders (FWO) (G.0983.14N); the Interuniversity Attraction Poles Programme (P7/16) initiated by the Belgian Science Policy Office; the Association Belge contre les Maladies Neuro-Musculaires (ABMM); the ALS Liga (Belgium); and the “Opening the Future” Fund. W.R. is supported through the E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders and the “Hart voor ALS” Fund, KU Leuven. P.V.D. is supported by the Alzheimer Research Foundation (SAO-FRA) (S14007), the Flemish Government initiated Flanders Impulse Program on Networks for Dementia Research (VIND), and the European Union Joint Programme-Neurodegenerative Disease Research (JPND) project RiMod-FTD. The Switch Laboratory is supported by grants from the European Research Council under the European Union's Horizon 2020 Framework Programme ERC Grant agreement (647458) (MANGO) (to J.S.), VIB, KU Leuven (“Industrieel Onderzoeksfonds”), FWO, and the Agency for Innovation by Science and Technology (IWT, SBO grant 60839). N.L.F. is supported by NIGMS of the NIH (award number R01GM118530) and a subproject as part of an Institutional Development Award (IDeA) (P20GM104937). V.H.R. was supported in part by an NIH training grant to the Neuroscience Graduate Program at Brown University (T32 MH020068). S.B. received a PhD fellowship from IWT. E.B. holds a post-doctoral fellowship, and P.V.D. a senior clinical investigatorship from FWO. F.I. was supported by an Odysseus grant (G0F8616N), and P.T. was supported by an Odysseus grant (G.0029.12) from FWO.
Published: March 16, 2017
Footnotes
Supplemental Information includes seven figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2017.02.013.
Contributor Information
Peter Tompa, Email: ptompa@vub.ac.be.
Ludo Van Den Bosch, Email: ludo.vandenbosch@vib-kuleuven.be.
Supplemental Information
References
- Aguzzi A., Altmeyer M. Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 2016;26:547–558. doi: 10.1016/j.tcb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grøfte M., Rask M.B.D., Streicher W., Jungmichel S., Nielsen M.L., Lukas J. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat. Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.S., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Ash P.E., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W., 3rd, Rademakers R. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumiller W.M., Jr., Keating C.D. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat. Chem. 2016;8:129–137. doi: 10.1038/nchem.2414. [DOI] [PubMed] [Google Scholar]
- Boeynaems S., Bogaert E., Michiels E., Gijselinck I., Sieben A., Jovičić A., De Baets G., Scheveneels W., Steyaert J., Cuijt I. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 2016;6:20877. doi: 10.1038/srep20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S., Bogaert E., Van Damme P., Van Den Bosch L. Inside out: the role of nucleocytoplasmic transport in ALS and FTLD. Acta Neuropathol. 2016;132:159–173. doi: 10.1007/s00401-016-1586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Tompa P., Pappu R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015;11:899–904. [Google Scholar]
- Burke K.A., Janke A.M., Rhine C.L., Fawzi N.L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Ciryam P., Tartaglia G.G., Morimoto R.I., Dobson C.M., Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5:781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella A.E., Zerze G.H., Mittal J., Fawzi N.L. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A., Netzer N., Strader M.B., Das S.R., Chen C.Y., Gibbs J., Pierre P., Bennink J.R., Yewdell J.W. RNA binding targets aminoacyl-tRNA synthetases to translating ribosomes. J. Biol. Chem. 2011;286:20688–20700. doi: 10.1074/jbc.M110.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Madl T., Valori C.F., Bentmann E., Tahirovic S., Abou-Ajram C., Kremmer E., Ansorge O., Mackenzie I.R.A., Neumann M., Haass C. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012;31:4258–4275. doi: 10.1038/emboj.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D., Haass C. An amyloid-like cascade hypothesis for C9orf72 ALS/FTD. Curr. Opin. Neurobiol. 2016;36:99–106. doi: 10.1016/j.conb.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Emara M.M., Fujimura K., Sciaranghella D., Ivanova V., Ivanov P., Anderson P. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 2012;423:763–769. doi: 10.1016/j.bbrc.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley M.D., Bieri G., Kolaitis R.M., Taylor J.P., Gitler A.D. Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J. Neurosci. 2014;34:8083–8097. doi: 10.1523/JNEUROSCI.0543-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum B.D., Lu Y., Lopez-Gonzalez R., Kim N.C., Almeida S., Lee K.H., Badders N., Valentine M., Miller B.L., Wong P.C. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S., Richter R.P., Görlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Hawe A., Sutter M., Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 2008;25:1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S., Kong G., Mannen T., Sadowska A., Kobelke S., Blythe A., Knott G.J., Iyer K.S., Ho D., Newcombe E.A. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovičić A., Mertens J., Boeynaems S., Bogaert E., Chai N., Yamada S.B., Paul J.W., 3rd, Sun S., Herdy J.R., Bieri G. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P., Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P., Anderson P. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Raphael A.R., LaDow E.S., McGurk L., Weber R.A., Trojanowski J.Q., Lee V.M., Finkbeiner S., Gitler A.D., Bonini N.M. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat. Genet. 2014;46:152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King O.D., Gitler A.D., Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I., Xiang S., Kato M., Wu L., Theodoropoulos P., Wang T., Kim J., Yun J., Xie Y., McKnight S.L. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Zhang P., Kim H.J., Mitrea D.M., Sarkar M., Freibaum B.D., Cika J., Coughlin M., Messing J., Molliex A. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788.e17. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Protter D.S., Rosen M.K., Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Mori E., Kato M., Xiang S., Wu L., Kwon I., McKnight S.L. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789–802.e12. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Song Y., Tian B., Qian J., Dong Y., Liu J., Liu B., Sun Z. Functional proteomic analysis of promyelocytic leukaemia nuclear bodies in irradiation-induced MCF-7 cells. J. Biochem. 2010;148:659–667. doi: 10.1093/jb/mvq105. [DOI] [PubMed] [Google Scholar]
- Lopez-Gonzalez R., Lu Y., Gendron T.F., Karydas A., Tran H., Yang D., Petrucelli L., Miller B.L., Almeida S., Gao F.B. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92:383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March Z.M., King O.D., Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 2016;1647:9–18. doi: 10.1016/j.brainres.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea D.M., Cika J.A., Guy C.S., Ban D., Banerjee P.R., Stanley C.B., Nourse A., Deniz A.A., Kriwacki R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife. 2016;5:5. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S., Grönke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O.C., Pietrzyk J. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Weng S.M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Murakami T., Qamar S., Lin J.Q., Schierle G.S., Rees E., Miyashita A., Costa A.R., Dodd R.B., Chan F.T., Michel C.H. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., Baldwin A.J. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H., Schermann S.M., Woerner A.C., Pinkert S., Hecht M.H., Tartaglia G.G., Vendruscolo M., Hayer-Hartl M., Hartl F.U., Vabulas R.M. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Ramaswami M., Taylor J.P., Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton A.E., Chiò A., Traynor B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J.C., Wang X., Podell E.R., Cech T.R. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–925. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Wang H., Xia Q., Li K., Li K., Jiang X., Xu G., Wang G., Ying Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet. 2015;24:2426–2441. doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]
- Thandapani P., O’Connor T.R., Bailey T.L., Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- Tiraferri A., Maroni P., Borkovec M. Adsorption of polyelectrolytes to like-charged substrates induced by multivalent counterions as exemplified by poly(styrene sulfonate) and silica. Phys. Chem. Chem. Phys. 2015;17:10348–10352. doi: 10.1039/c5cp00910c. [DOI] [PubMed] [Google Scholar]
- Tompa P. The principle of conformational signaling. Chem. Soc. Rev. 2016;45:4252–4284. doi: 10.1039/c6cs00011h. [DOI] [PubMed] [Google Scholar]
- Tompa P., Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Topisirovic I., Siddiqui N., Lapointe V.L., Trost M., Thibault P., Bangeranye C., Piñol-Roma S., Borden K.L.B. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009;28:1087–1098. doi: 10.1038/emboj.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M., Zsolyomi F., Guharoy M., Tompa P. Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLoS ONE. 2015;10:e0139731. doi: 10.1371/journal.pone.0139731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Tan W., Westergard T., Krishnamurthy K., Markandaiah S.S., Shi Y., Lin S., Shneider N.A., Monaghan J., Pandey U.B. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler J.R., Matheny T., Jain S., Abrisch R., Parker R. Distinct stages in stress granule assembly and disassembly. eLife. 2016;5:5. doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa M., Ito D., Honda T., Kubo K., Noda M., Nakajima K., Suzuki N. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum. Mol. Genet. 2015;24:1630–1645. doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., Gladfelter A.S. RNA controls PolyQ protein phase transitions. Mol. Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.