Abstract
The measurement of a deuterium equilibrium isotope effect (EIE) for the aryl CH···Cl– interaction of anion receptor 1H/1D is reported. Computations corroborate the results of solution measurements for a small, normal EIE in the full complex (KaH/Ka = 1.019 ± 0.010). Interestingly, isotope effects involving fragments of the anion receptor (urea, aryl ring, etc.) are predicted to produce an inverse effect. This points to an unusual and subtle structural organization effect of the anion receptor complex that changes the nature of the combined interactions to a normal isotope effect. The reversal of EIE values suggests that overall architecture of the anion receptor can dramatically impact the nature of bonding in these complexes.
The ability of isotopes to alter the strength and selectivity of binding in noncovalent interactions is well-documented for biological and nonbiological systems.1,2 Numerous methods have been developed to utilize selective labeling experiments to differentiate the importance of individual interactions on the complete symphony of binding, including relative chromatographic retention times,3 binding studies by mass spectrometry and spectroscopic titrations,4 and enzyme kinetic studies.5,6 Deuterium equilibrium isotope effect (DEIE) measurements in biological systems highlight the complex effects of deuterium labeling on noncovalent interactions.5 Recent studies combining experiments and computations provide insight into the mechanisms of DEIEs in supramolecular systems that ultimately facilitate the development of models for ab initio sensor and drug design.4,7−13
Two classic examples of such a combined experiment-and-computation approach are the acidities of deuterium-labeled pyridines and glucose binding to human brain hexokinase (HBH). Lewis and Schramm have extensively studied the EIE of tritiated glucose binding to HBH.14,15 Tritium EIEs for binding to HBH range from 0.927 to 1.027 depending on the site of CH substitution. Proximal hydrogen bonding groups in the binding site consistently explained normal EIEs, and steric interactions caused the large inverse isotope effect. In an approach that inspires our current experiments, competitive pH titrations by 13C NMR spectroscopy of 1:1 mixed H/D-pyridines allowed for the direct measurement of KaH/Ka as small as 10–4.11 DEIEs favored dx-pyridine protonation and were also dependent on the location of the deuteration (KaH/Ka range from 1.0139 to 1.0828). This example highlights the hypothesis that aggregate zero-point energy (ZPE) vibration effects are responsible for changes in DEIE rather than inductive effects. These two studies highlight the power of EIE studies in fundamentally understanding reversible interactions.
The Perrin method for competitive titration requires only a fast exchanging system with resolved resonances for the precise measurement of relative association constants by NMR spectroscopy. This method has previously been applied to guest binding in supramolecular cage complexes.7,8 We have adapted this method to the study of DEIEs on CH···Cl– hydrogen bonding by both solution measurements and computations. The synthesis of a selectively deuterated arylethynyl bis-urea anion receptor 1D enables competitive titration DEIE measurements in DMSO-d6. The observed DEIE of the singly deuterated compound 1D and quantum mechanical computations corroborate the importance of vibrational changes on the observed DEIE. We elucidate specific structural origins for DEIEs in anion receptor complexes and have discovered an emergent behavior: the DEIE of the receptor complex is uniquely different than DEIEs of various fragments of the complex. In other words, the nature of multiple hydrogen bonding in the anion receptor is uniquely different from simpler hydrogen bonding complexes.
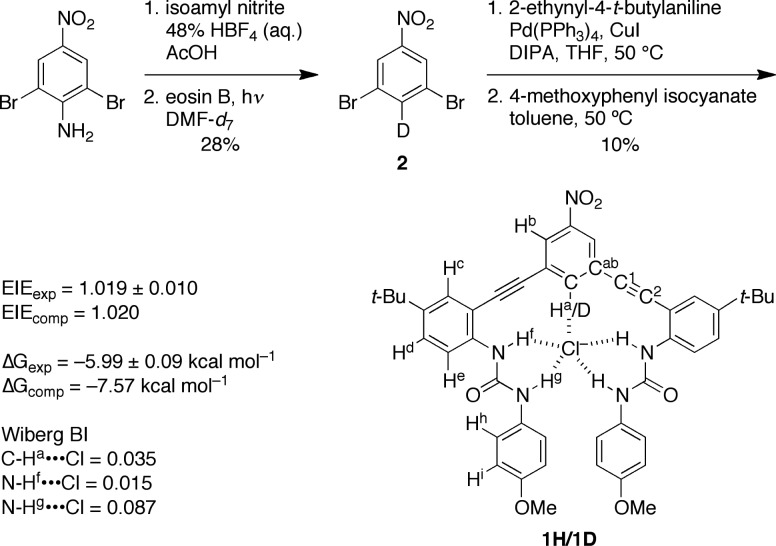
1H has been previously shown to act as a strong CH donor with log Ka for Cl– of 4.39 ± 0.62 in CHCl3.16 Isotopologue 1D is prepared by methods similar to those reported in previous papers (Scheme 1).16−18 Visible-light catalyzed deamination in DMF-d7 allows for the selective synthesis of monodeuterated intermediate 2.19 The final receptor 1D and key intermediates were characterized by 1H, 2H, and 13C NMR spectroscopy, and high-resolution mass spectrometry (see Supporting Information).
Scheme 1. Selective Deuterium Labeling of Anion Receptor 1H/1D, Summary of Equilibrium Isotope Effect, Anion Binding Energy and Wiberg Bond Index.
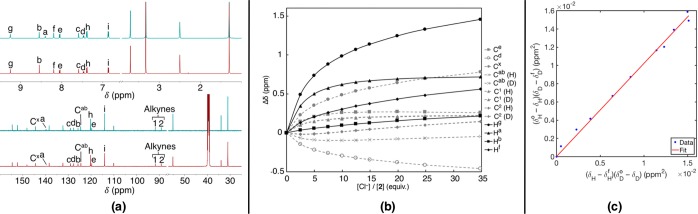
As shown by Perrin et al., 13C NMR spectroscopy is exceptionally sensitive to chemical shifts from isotopic labeling.11,20Figure 1a illustrates the similarity between 1H/1D. The two key differences are the loss of the Ha peak in 1D and small shifts in 13C peaks. Competitive titrations were performed in DMSO-d6 with ∼1:1 mixture of 1H and 1D receptors at a combined concentration of 6.1 mM, and Bu4NCl was added in 5 μL aliquots as a 1.8 M solution.21 Many 1H and 13C signals shift over the course of the titration, and four 13C peaks are resolved by isotope shifts between 1H and 1D to differentiate the two species. The labeled 13C signal (Ca) is too weak for accurate measurement of δ during the titration, which leaves three signals (alkyne C1, alkyne C2, and Cab) for possible fitting.
Figure 1.
(a) 1H (top) and 13C (bottom) NMR spectra of 1H (blue) and 1D (red) in DMSO-d6. Cab is the aryl carbon between Ha and Hb. (b) Binding isotherm with Cl– for 1:1 1H:1D in DMSO-d6 by 1H (solid lines) and 13C (dashed lines) NMR spectroscopy; lines added as a guide for the eye. Isotherm shape is consistent with weak 1:2 binding where Ka1:1 ≫ Ka. 13C NMR peaks are used for linear EIE fitting; the isotherms overlap, although the peaks are resolved. (c) Linearized plot of KaH/Ka (eq 1) for 1H:1D13C NMR peak corresponding to alkyne C1.
The binding isotherm for mixed 1H:1D is plotted in Figure 1b. Cab is the closest carbon to the labeled CH hydrogen bond donor and, along with the alkyne peaks, is well resolved between 1H and 1D with greater intensity than the labeled peak. The Cab peak and alkyne C2 are complex curves indicative of possible multistep equilibria. Alkyne C1 presents a large downfield shift and the isotherm for this carbon approximates a binding curve for a 1:1 association. A large shift in the alkyne carbons is surprising due to the large distance from the nearest hydrogen bonding site. One explanation is the through space effect of an approaching anion that shields the alkyne. An alternative effect is rotation about the alkyne upon binding of the anion, a conformational change that has also been observed in similar alkyne receptors.22,23
The remaining 1H and 13C NMR peaks produce simpler and consistent binding curves, which can be split into two categories (Figure 1b). First, several peaks reach a saturation point near 15 equiv. Cl– and remain mostly flat for the remainder of the titration (Ha, Ce, and Cab). The second category includes peaks that never appear to reach saturation in the range studied but continue to increase linearly after 15 equiv. (Hg, Hf, Cx, and Cd). The second case is consistent with a weak 1:2 complex, whereas the first case is more indicative of a 1:1 binding event. An estimated value for the overall Ka (log β) is obtained from the fitting of the 1H resonances and Cab to a 1:2 1:Cl– model using the combined [1H+1D]. The binding in DMSO-d6 is significantly weaker (log β ∼ 1.8) than in CHCl3 (log Ka = 4.39 ± 0.62), which is expected for a solvent system with high polarity and strong hydrogen bond acceptors. As well, the second Cl– association is likely very weak with Ka1:2 < 5. Based on the curve shape and position on 1 of atoms for the two binding isotherm categories, it is clear that the first Cl– binds in the expected pocket to CHa with both ureas, and then a second Cl– weakly binds to one of the ureas at high [Cl–], forcing the alkyne to rotate open to accommodate it.22,23 The complex binding isotherm, however, does not prevent the use of a linearized plot to determine the relative Ka values for 1H and 1D with greater precision than possible by direct analytical titration and nonlinear regression.
The linearized plot of KaH/Ka, eq 1, provides an accurate measure of the DEIE from a competitive titration with errors as small as 0.0004 reported.8,11 The method requires free and bound species to be in fast equilibrium with resolved peaks but is independent of receptor concentration. Typical methods of Ka determination rely on nonlinear regression, and errors are restricted to ∼5–10% by the accuracy in concentration. Least squares linear regression of the 13C NMR chemical shifts of 1H and 1D, eq 1, provides the relative KaH/Ka with greater precision.20 The chemical shifts for alkyne C1 13C peaks were fit with an R2 = 0.996 shown in Figure 1c. The weighted average KaH/Ka was 1.019 ± 0.010 for two independent titrations.
| 1 |
Quantum mechanical computations are effective at predicting kinetic and equilibrium isotope effects.10,11,24 Structures were optimized using PBE/6-31G(d) level of theory with the polarizable continuum model (PCM) for DMSO as implemented in Gaussian 09.25−27 Exhaustive conformational searches led to structures of anion receptor complexes and free receptors that corroborate known crystal structures of related compounds (see SI). The computed EIE for Cl– binding (complex [1H/1D···Cl–]) was 1.020, in good agreement with experimental values (1.019 ± 0.010). Moreover, the binding energy between the receptor and Cl– was computed to be −7.57 kcal mol–1, in good agreement with experiments (−5.99 kcal mol–1).16,28
The computed 1H/1D structure is shown in Figure 2. The chloride anion is quintuply hydrogen bonded by the central aryl CH and the four urea protons. The two substituted urea arms of the anion receptor are not planar in the presence of the anion, and the terminal urea aryl rings rest on top of each other. This structure forces an asymmetric hydrogen bond between the chloride anion and the two protons of each urea, the proton that is distal from the CH site is closer (2.19 Å) than the proton proximal to the CH (2.87 Å).
Figure 2.
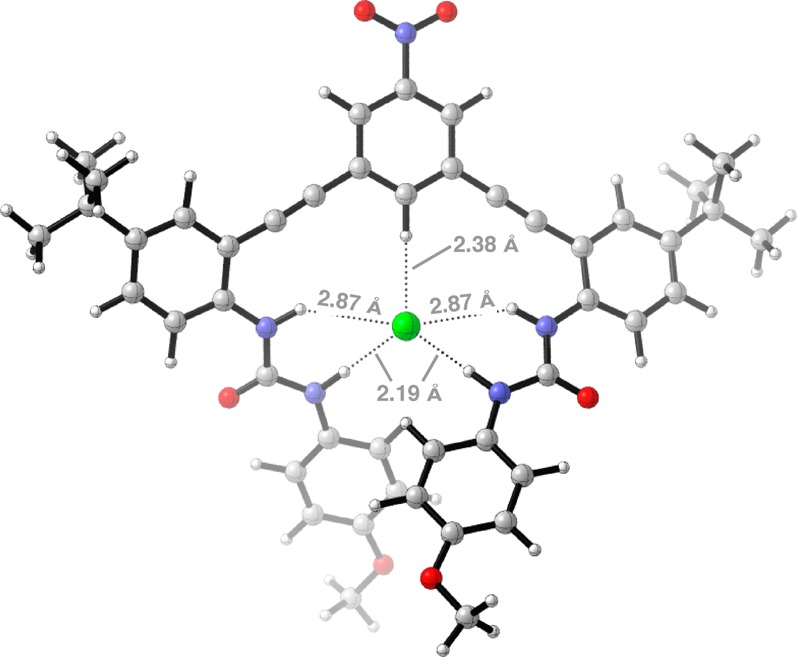
Quantum mechanically optimized structure of anion receptor 1H/1D with chloride anion (PBE/6-31G*/PCM(DMSO)).
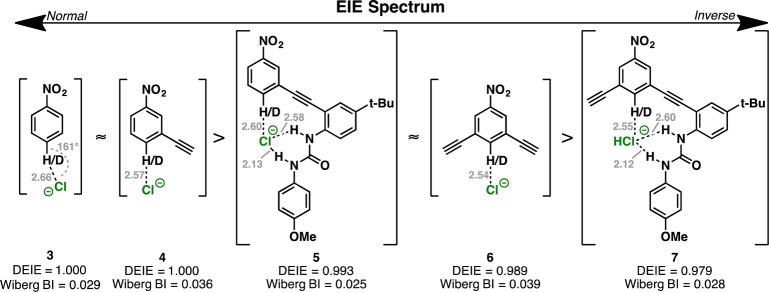
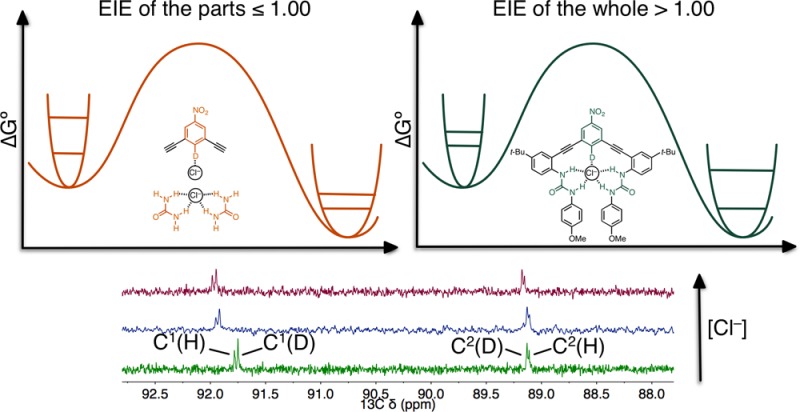
The normal EIE favors association of Cl– with the unlabeled receptor 1H. We fragmented the receptor into key components to explore the isotope effects of the basic receptor segments (Figure 3, S20). All fragments led to more inverse EIE values. For example, the addition of an alkyne to 4 (i.e., 6) lowered the DEIE from 1.000 to 0.989. Removing half of the receptor (5) also led to an inverse DEIE (0.993), and the addition of another alkyne (7) results in an even more inverse DEIE of 0.979. These results are surprising because no data from the fragments of the anion receptor would suggest that a normal EIE is possible (nonadditive), yet the experimentally measured and computed EIE is 1.020. This indicates that the isotope effect for [1H/D···Cl–] is more than the sum of its parts.
Figure 3.
Computed EIE values involving chloride complexes with fragments of the anion receptor 1H/D.
The computation of the fragment complexes revealed structural details relevant to elucidating the origin of this unexpected EIE of 1H/1D. In 3, the CH···Cl– geometry is bent (161°) with Cl– still within the plane of the aryl group. In 4 and 6, this geometry is completely linear, just as in 1H/1D. Although repulsions between the chloride anion and the alkyne groups may be responsible for this geometric change, the CH···Cl– distance in the monoalkynylated complex 4 is shorter, whereas the distance in the bis-alkynylated complex 6 is even shorter (2.66 Å vs 2.57 Å vs 2.54 Å, respectively). These data are consistent with an inductive enhancement of hydrogen bonding by the electronegative alkynes. Similar trends can be observed in complexes 5 and 7. The computed natural bond orbital (NBO) analysis revealed that the Wiberg bond index of the hydrogen bond in 3 is 0.029, whereas it is 0.036 in 4 and 0.039 in 6.29 Similarly, the bond index for complex 5 and 7 are 0.025 and 0.028, respectively. The bond index for the corresponding interaction in 1H/1D is 0.035, matching closely the alkynylated complexes 4 and 6. This would suggest the EIE does not originate from the central aryl CH hydrogen bonding.
Complexes 8 and 9 reveal that the geometry of hydrogen bonding in 1H/1D is significantly distorted (Chart 1). The ideal urea NH···Cl– distance is 2.38–2.43 Å, which deviates significantly from 1H/1D. NBO analysis also shows that the bond indices for hydrogen bonds in 8 and 9 are 0.057 and 0.044, respectively; significantly different from those of 1H/1D (0.015 and 0.087). These observations are consistent with the hypothesis that the unusual arrangement of urea NH and aryl-CH hydrogen bonds is responsible for the unexpected EIE of 1H/1D.
Chart 1.
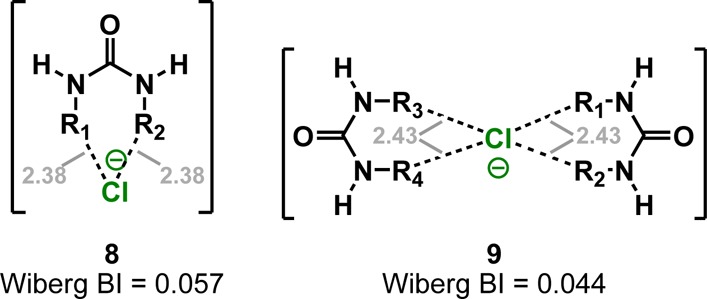
In summary, the synthesis of a selectively labeled receptor has allowed for the first accurate measurement of an EIE in an aryl CH···X– hydrogen bond. The combined evidence presented here is consistent with the categorization of certain aryl CH donors as hydrogen bond donors. The DEIE measurements from competitive titrations and computations agree on a small, normal DEIE that favors the formation of a CH hydrogen bond over a CD hydrogen bond. This study, more importantly, shows that supramolecular binding is a complex phenomenon that cannot be reduced to the sum of its parts. The reversal of EIE values between the complete anion receptor complex versus the fragments suggest that isotope effects are extremely sensitive to the overall architecture of the anion receptor. This discovery is highly relevant to recent applications of isotope labeling in pharmaceuticals, where the introduction of isotopes may improve the strength and selectivity of protein-drug interactions.
Acknowledgments
This work was supported by the NIH (R01-GM087398 to D.W.J./M.M.H.) and NSF (CHE-1352663 to P.H.-Y.C.). We thank the NSF for an NMR spectrometer grant (CHE-1427987) and for computing infrastructure in part provided by the NSF Phase-2 CCI, Center for Sustainable Materials Chemistry (CHE-1102637). P.H.-Y.C. is the Bert and Emelyn Christensen professor. A.C.B. acknowledges financial support by the Barnes Fellowship. The authors acknowledge the Biomolecular Mass Spectrometry Core of the Environmental Health Sciences Core Center at Oregon State University (NIH P30ES000210).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b00612.
Complete synthesis and characterization for all compounds, titration data, coordinates, energies, and frequencies for all computed structures (PDF)
Author Present Address
# Molecular Foundry, Lawrence Berkeley National Laboratory, Berkeley, CA 94720
The authors declare the following competing financial interest(s): NIH grant R01-GM087398 funded early-stage intellectual property that was licensed by SupraSensor Technologies, a company co-founded by M.M.H. and D.W.J.
Supplementary Material
References
- Wade D. Chem.-Biol. Interact. 1999, 117, 191–217. 10.1016/S0009-2797(98)00097-0. [DOI] [PubMed] [Google Scholar]
- Kohen A.; Limbach H.-H.. Isotope Effects In Chemistry and Biology; CRC Press: Boca Raton, FL, 2005. [Google Scholar]
- Tran C. D.; Mejac I.; Rebek J.; Hooley R. J. Anal. Chem. 2009, 81, 1244–1254. 10.1021/ac8021423. [DOI] [PubMed] [Google Scholar]
- Laughrey Z. R.; Upton T. G.; Gibb B. C. Chem. Commun. 2006, 970–972. 10.1039/b515187b. [DOI] [PubMed] [Google Scholar]
- Świderek K.; Paneth P. Chem. Rev. 2013, 113, 7851–7879. 10.1021/cr300515x. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Singh P.; Czekster C. M.; Kohen A.; Schramm V. L. J. Am. Chem. Soc. 2014, 136, 8333–8341. 10.1021/ja501936d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugridge J. S.; Bergman R. G.; Raymond K. N. Angew. Chem., Int. Ed. 2010, 49, 3635–3637. 10.1002/anie.200906569. [DOI] [PubMed] [Google Scholar]
- A similar method was applied to the supramolecular complex of benzyltrimethylphosphonium with a tetrahedral cage. The 31P NMR resonance was used to measure small normal DEIEs for association to the exterior and interior of the cage:; a Mugridge J. S.; Bergman R. G.; Raymond K. N. J. Am. Chem. Soc. 2012, 134, 2057–2066. 10.1021/ja2067324. [DOI] [PubMed] [Google Scholar]; b Mugridge J. S.; Bergman R. G.; Raymond K. N. J. Am. Chem. Soc. 2010, 132, 1182–1183. 10.1021/ja905170x. [DOI] [PubMed] [Google Scholar]
- Balevicius V.; Aidas K.; Svoboda I.; Fuess H. J. Phys. Chem. A 2012, 116, 8753–8761. 10.1021/jp305446n. [DOI] [PubMed] [Google Scholar]
- Zhao C.; Parrish R. M.; Smith M. D.; Pellechia P. J.; Sherrill C. D.; Shimizu K. D. J. Am. Chem. Soc. 2012, 134, 14306–14309. 10.1021/ja305788p. [DOI] [PubMed] [Google Scholar]
- Perrin C. L.; Karri P. J. Am. Chem. Soc. 2010, 132, 12145–12149. 10.1021/ja105331g. [DOI] [PubMed] [Google Scholar]
- Rechavi D.; Scarso A.; Rebek J. J. Am. Chem. Soc. 2004, 126, 7738–7739. 10.1021/ja048366p. [DOI] [PubMed] [Google Scholar]
- Zhao Y. L.; Houk K. N.; Rechavi D.; Scarso A.; Rebek J. J. Am. Chem. Soc. 2004, 126, 11428–11429. 10.1021/ja0471065. [DOI] [PubMed] [Google Scholar]
- Lewis B. E.; Schramm V. L. J. Am. Chem. Soc. 2001, 123, 1327–1336. 10.1021/ja003291k. [DOI] [PubMed] [Google Scholar]
- Lewis B. E.; Schramm V. L. J. Am. Chem. Soc. 2003, 125, 4785–4798. 10.1021/ja0298242. [DOI] [PubMed] [Google Scholar]
- Tresca B. W.; Hansen R. J.; Chau C. V.; Hay B. P.; Zakharov L. N.; Haley M. M.; Johnson D. W. J. Am. Chem. Soc. 2015, 137, 14959–14967. 10.1021/jacs.5b08767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C. N.; Berryman O. B.; Johnson C. A.; Zakharov L. N.; Haley M. M.; Johnson D. W. Chem. Commun. 2009, 2520–2522. 10.1039/b901643k. [DOI] [PubMed] [Google Scholar]
- Kimball D. B.; Weakley T. J. R.; Haley M. M. J. Org. Chem. 2002, 67, 6395–6405. 10.1021/jo020229s. [DOI] [PubMed] [Google Scholar]
- Majek M.; Filace F.; Jacobi von Wangelin A. Chem. - Eur. J. 2015, 21, 4518–4522. 10.1002/chem.201406461. [DOI] [PubMed] [Google Scholar]; The low yield of 2 results from a side product produced by kinetic isotope effects in DMF-d7.
- Perrin C. L.; Fabian M. A. Anal. Chem. 1996, 68, 2127–2134. 10.1021/ac960117z. [DOI] [PubMed] [Google Scholar]
- Titrations with pure 1D were also performed by UV–vis and 1H NMR; however, these are limited because differences in the raw data and Ka values are not significant enough to distinguish 1H and 1D.
- Gavette J. V.; Evoniuk C. J.; Zakharov L. N.; Carnes M. E.; Haley M. M.; Johnson D. W. Chem. Sci. 2014, 5, 2899–2905. 10.1039/c4sc00950a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavette J. V.; Mills N. S.; Zakharov L. N.; Johnson C. A.; Johnson D. W.; Haley M. M. Angew. Chem., Int. Ed. 2013, 52, 10270–10274. 10.1002/anie.201302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.; Gangopadhyay D.; Popp J.; Singh R. K. Spectrochim. Acta, Part A 2012, 99, 136–143. 10.1016/j.saa.2012.09.021. [DOI] [PubMed] [Google Scholar]
- a Adamo C.; Barone V. J. Chem. Phys. 1999, 110, 6158–6169. 10.1063/1.478522. [DOI] [Google Scholar]; b Hehre W. J.; Ditchfield R.; Pople J. A. J. Chem. Phys. 1972, 56, 2257–2261. 10.1063/1.1677527. [DOI] [Google Scholar]
- Miertǔs S.; Scrocco E.; Tomasi J. Chem. Phys. 1981, 55, 117–129. 10.1016/0301-0104(81)85090-2. [DOI] [Google Scholar]
- (For complete author list, seeSI). Frisch M. J.; Trucks G. W.; Schlegel H. B.; et al. Gaussian 09; Gaussian Inc.: Wallingford, CT, 2009.
- Computations showed that the receptor-anion-TBA complex (1H/D···Cl–···TBA+) was endergonic by 7.0 kcal mol−1, and moreover, the computed EIE of 1.005 also did not match experiments. Taken together, these suggest that the anion receptor solely binds to the Cl– anion rather than the ion pair (Cl–···TBA+).
- Foster J. P.; Weinhold F. J. Am. Chem. Soc. 1980, 102, 7211–7218. 10.1021/ja00544a007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.