Abstract
RNA drug targets are pervasive in cells, but methods to design small molecules that target them are sparse. Herein, we report a general approach to score the affinity and selectivity of RNA motif–small molecule interactions identified via selection. Named High Throughput Structure–Activity Relationships Through Sequencing (HiT-StARTS), HiT-StARTS is statistical in nature and compares input nucleic acid sequences to selected library members that bind a ligand via high throughput sequencing. The approach allowed facile definition of the fitness landscape of hundreds of thousands of RNA motif–small molecule binding partners. These results were mined against folded RNAs in the human transcriptome and identified an avid interaction between a small molecule and the Dicer nuclease-processing site in the oncogenic microRNA (miR)-18a hairpin precursor, which is a member of the miR-17-92 cluster. Application of the small molecule, Targapremir-18a, to prostate cancer cells inhibited production of miR-18a from the cluster, de-repressed serine/threonine protein kinase 4 protein (STK4), and triggered apoptosis. Profiling the cellular targets of Targapremir-18a via Chemical Cross-Linking and Isolation by Pull Down (Chem-CLIP), a covalent small molecule–RNA cellular profiling approach, and other studies showed specific binding of the compound to the miR-18a precursor, revealing broadly applicable factors that govern small molecule drugging of noncoding RNAs.
Short abstract
We developed a general approach, dubbed HiT-StARTS, to define the RNA-binding landscape of small molecules. From these studies, a small molecule was designed to inhibit oncogenic miR-18a, de-repress a downstream protein, and trigger apoptosis.
RNA has many essential functions in cells and thus is an important target for chemical probes or lead therapeutics. Developing RNA-directed chemical probes is challenging, however, due to a dearth of data describing the types of small molecules that bind RNA folds (motifs) selectively.1,2 Although a small data set, selective RNA motif–small molecule interactions have been used to inform the design of bioactive small molecules, including monomeric and modularly assembled ligands.3−13 The latter compounds bind RNAs that have multiple targetable motifs that are separated by a specific distance.8,11 In order to target the myriad of disease-causing RNAs in a cell using rational and predictable methods, much more data that describe selective interactions between small molecules and RNA motifs will be required, as well as new high throughput tools and technologies to obtain them.
A library-vs-library screen named Two-Dimensional Combinatorial Screening (2DCS) has proven to be a powerful method to identify selective RNA motif–small molecule binding partners in a high throughput fashion.14 Although screening is rapid, downstream processing of the selected interactions, such as scoring affinity and selectivity, is laborious, requiring time-consuming binding assays. Previously, a theoretical approach was developed to compute scoring functions for 2DCS selections based on the statistical confidence of subfeatures imparting binding affinity.15 Since the models are not empirical, interactions that do not obey the model could have their affinities and selectivities misassigned.
Here, we report an empirical method that rapidly defines binding landscapes, assigns RNA motif–small molecule scoring functions and generates structure–activity relationships using data derived from next-generation sequencing (RNA-seq) from a 2DCS experiment and experimentally determined affinities. The approach, named High Throughput Structure–Activity Relationships Through Sequencing (HiT-StARTS), statistically analyzes a selection by computing the enrichment that a selected RNA motif has by comparison to the starting library in an RNA-seq experiment. HiT-StARTS is likely to have broad applications. For example, SELEX is often used to screen nucleic acid libraries to identify nucleic acids that bind a ligand.14,16−18 Further, DNA can be used to encode small molecules that bind to a therapeutically important target.19−21
By using the results of 2DCS in conjunction with Inforna,22 an informatics pipeline that mines RNA motif–small molecule interactions against RNA folds in the transcriptome, a small molecule was identified that could target nuclease-processing sites in the oncogenic noncoding microRNA (miR)-17-92 cluster. Application of this compound to cells inhibited processing of miR-18a precursor, de-repressed a silenced protein, and slowed growth and triggered apoptosis in prostate cancer cells. These studies have allowed for analysis of the types of RNAs that can be targeted by small molecules, showing that targeting functional sites in an RNA and the expression of the target are important considerations.
Results and Discussion
Two-Dimensional Combinatorial Screening (2DCS) of an RNA-Focused Small Molecule Library
We designed a series of small molecules (1–8; see Figures S-1–S-46 for synthetic schemes and compound characterization) that might be privileged for selectively binding RNA. Each compound was appended with an azide tag such that they could be site-specifically immobilized onto alkyne-functionalized agarose microarray surfaces via Cu-catalyzed Huisgen dipolar cycloaddition reaction (Figure 1).23,24 The compounds have a benzimidazole core, which is privileged for RNA binding25−27 and also steric bulk such that the compounds do not bind DNA.28,29 Part of our interest in further advancing compounds such as 1–8 is that a small, related benzimidazole compound selectively inhibited biogenesis of the oncogenic miR-96 precursor in breast cancer cells at low micromolar concentrations.9 Thus, this chemotype appears privileged to target RNA in cells, the identification of which has been a significant challenge in RNA chemical biology.
Figure 1.
Structures of the small molecules and oligonucleotides used in this study. Top and middle, structures of the compounds tested for RNA binding and the Cu(I)-catalyzed “click chemistry” reaction (HDCR) to conjugate compounds onto the array surface, respectively. Bottom, secondary structures of the oligonucleotides.
The arrays were incubated with a 32P-labeled 3 × 3 nucleotide internal loop library (3 × 3 ILL), which contains 4,096 members, in the presence of excess unlabeled competitor nucleic acids (Figure 1). The competitor RNA oligonucleotides mimic regions constant to all library members and restrict binding interactions to the randomized region. DNA oligonucleotides were also used in excess to ensure selective binding to RNA. The 3 × 3 ILL was chosen because the motifs it displays have a high probability of representation in a transcriptome. Thus, 32,768 unique interactions were probed simultaneously (8 small molecules × 4,096 unique RNAs); if one considers the compounds, their densities, and the RNA library, then 163,840 interactions were probed (8 small molecules × 5 compound densities × 4,096 unique RNAs).
Initial analysis of the microarray data showed that only compounds that contained a methylphenylpiperazine moiety (1–4) bound the RNA library under these stringent conditions. A few hundred picomoles of compound delivered to the surface was sufficient to detect binding and to select bound RNA motifs (Figure 2). Furthermore, the signal on the microarray increased as the steric bulk of the functional groups on the phenyl ring increased (Figure 2).
Figure 2.

An image of the 2DCS microarray from compounds 1–8 (top) and the top three selected RNA motifs that bind to 1–4 (bottom). No binding was observed to compounds 5–8. The number before “IL” (indicates internal loop) is the small molecule to which the RNA motifs were selected to bind. Circles indicate positions from which bound RNAs were isolated and subjected to RNA-seq. The amount of compound delivered to these positions corresponds to 560, 560, 840, and 370 picomoles for 1, 2, 3, and 4, respectively.
Bound RNA motifs for each compound were harvested from the array surface, amplified, and identified by using high throughput sequencing (RNA-seq). Highly abundant RNAs in the sequencing data were then subjected to binding measurements. Typically, those RNAs that were highly enriched bound to the small molecules with affinities (Kds) that ranged from 1 to 10 μM (Figure 2 and Table 1) while binding to the starting libraries was not saturable. These affinities are promising for binding RNA, as it has been difficult to acquire low micromolar affinity with compounds of this low molecular weight. In fact, RNAs that have undergone Darwinian evolution to bind small molecules, namely, riboswitches, often bind their cognate ligand at micromolar to millimolar concentrations;30−33 bioactive compounds that mimic riboswitch ligands bind with similar affinities.30,34
Table 1. Global Analysis of Selected RNA Motif–Small Molecule Interactionsa.
| compound | range of Zobs for binders | no. of binders | average Kd of selected RNA binders | Kd to DNA hairpin |
|---|---|---|---|---|
| 1 | 8–72 | 23 | 16 ± 9 | ≫100 |
| 2 | 8–44 | 26 | 7 ± 5 | ≫100 |
| 3 | 12–29 | 64 | 8 ± 4 | ≫100 |
| 4 | 7–29 | 215 | 11 ± 2 | ≫100 |
Kds are reported in μM.
2DCS Identified RNA-Selective Small Molecule Ligands
It has been difficult to identify compounds that are heterocyclic in nature that selectively recognize RNA over DNA.35 Thus, to further gain insight into the selectivity of our RNA motif–small molecule interactions, we compared the measured affinities for selected RNA motifs to the affinities of the ligands for an AT-rich DNA hairpin, a common target of small molecules similar to those shown in Figure 1. Compounds that bound 3 × 3 ILL (1–4), however, are different from DNA binders in many key elements that suggest that selective binding to RNA is possible.36 For example, 1–4 have a benzimidazole moiety, which binds RNA,29 not a bis-benzimidazole moiety common to DNA binders. This alteration reduces DNA affinity because key stacking and hydrogen bonding interactions are not possible. Additional steric bulk was added to the compounds to ablate binding to base paired DNA as has been shown with even bis-benzimidazoles.28 Indeed, binding studies showed that 1–4 did not bind the DNA hairpin (Table 1). Thus, the interactions selected via 2DCS are RNA-selective.
Analysis of Selection Data To Define Rapidly Accurate Scoring Functions
One advantage of using next-generation sequencing to deconvolute selections is that millions of sequencing reads can be obtained. When the number of unique members of a starting library is much less than the number of reads acquired, there are high coverage and hence high confidence in the data output. Thus, we sought to leverage the large data set generated for our 2DCS selections to develop models to rapidly assign binding landscapes. Such approaches would streamline such investigations, rapidly identify privileged targets for small molecules, and determine if a small molecule has potential to discriminate effectively between desired and undesired targets. For example, a small molecule that binds tightly to an RNA that causes a disease, such as the expanded r(CUG) repeat that causes muscular dystrophy, but does not bind to the human A-site (an off-target) would be highly desirable. Such studies could preemptively eliminate nonspecific compounds from further development.
We thus sought to utilize the results obtained from high throughput sequencing to develop an effective model to quickly identify the most avid binders from the population of selected RNA motifs. Previously, we developed a framework that utilized a binding model in which highly enriched submotifs within a given RNA structure (such as a GC step) were analyzed to compute a scoring function.15 Because some interactions may not obey a theoretical model, we aimed to develop a model-free method to derive scoring functions. If successful, this approach could be generally applicable to any nucleic acid selection.
Using Frequency of Selected RNA Motifs To Score Binding
We first analyzed the frequency of each selected RNA motif in the sequencing data. RNAs were ranked from the most frequently occurring (frequency rank = 1) to the least frequently occurring (frequency rank = 4,096).37 We hypothesized that an RNA with a frequency rank of 1 would bind with higher affinity to its cognate small molecule than RNA motifs with greater frequency ranks, while RNAs at the end of the spectrum would not bind. To determine if this approach could indeed score affinities, we measured the binding of RNA motifs over a range of frequency rank values for compounds 1–4 (those that bind RNA in our 2DCS selection; Figures S-53–S-56). In total we measured the affinities of 52 RNA motif–small molecule interactions.
As shown in Figures 3 and S-52, there was poor correlation between frequency rank and the affinity of the selected RNA motif–small molecule complexes. Previously, however, this approach was used to score the relative affinities of RNA–protein interactions.37
Figure 3.
Plots of frequency rank as a function of experimentally determined Kd for compounds 1–4. RNAs were ranked from the most frequently occurring (frequency rank = 1) to the least frequently occurring (frequency rank = 4,096). Boxed data points are interactions that do not exhibit saturable binding.
Using Statistical Analysis (Zobs) of Selected RNA Motifs To Derive Scoring Functions and Structure–Activity Relationships
Since it did not appear that a simple ranking of frequency provided accurate scoring of affinities, we applied a statistical approach that we named High Throughput Structure–Activity Relationships Through Sequencing (HiT-StARTS) (Figures 4 and 5). The starting RNA motif library, 3 × 3 ILL, without selection was subjected to high throughput sequencing to define biases that occur during transcription and sequencing. Such biases are unavoidable and caused by differences in the thermodynamic stability for individual RNA motifs, among other reasons. That is, a pooled population comparison (eqs 1 and 2) was used to compare the number of reads for a given RNA and its proportion of total reads in the selection sequencing data to the same RNA’s number of reads and its proportion of total reads from the starting library’s sequencing data. This comparison affords Zobs, a metric of statistical confidence. Zobs can be converted to a two-tailed p-value, indicating the confidence that the null hypothesis (that there is no significant difference between the selected and starting RNA libraries) can be rejected. Note that Zobs can be positive or negative. A positive value indicates enrichment while a negative value indicates discrimination of the small molecule against a particular RNA. Akin to frequency rank, each RNA was also assigned a Zobs rank, ranging from 1 (greatest statistical confidence for avid binding) to 4,096 (greatest statistical confidence for not binding).
Figure 4.
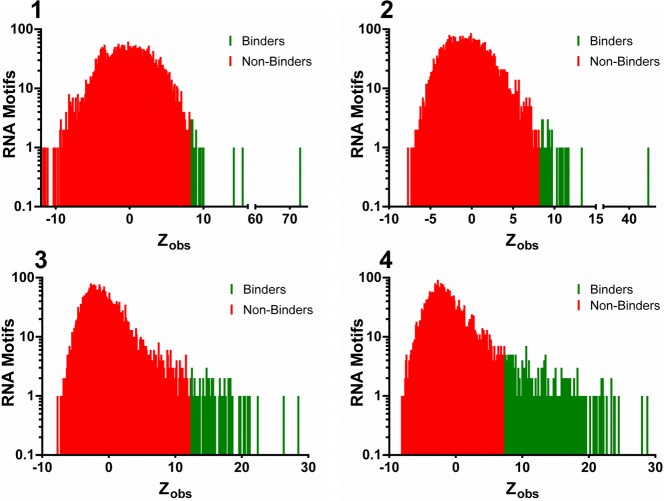
A plot of the frequency of the selected RNA motifs as a function of Zobs. Collectively, these data and the data presented in Figure 5 show that avid binders are correlated with Zobs. The signal on the microarray from 2DCS selections is directly proportional to the number of motifs with large, positive Zobs values.
Figure 5.
Statistical analysis of sequencing data correlates well with affinity. A pooled population comparison (selected RNAs vs the starting library) was used to afford Zobs, a metric of statistical confidence. Each RNA was also assigned a Zobs rank, ranging from 1 (greatest statistical confidence for avid binding; largest positive Zobs value) to 4,096 (greatest statistical confidence for not binding; largest negative Zobs value). Boxed data points are interactions that do not exhibit saturable binding.
Several features can be rapidly assessed from these data. First, there is a clear correlation between avidity of the RNA motif–small molecule interaction and Zobs rank and Zobs (Table 1 and Figures 4 and 5). Motifs with Zobs ≥ 8 (p < 0.0001) bind avidly to compounds 1 and 2; binding affinity is conferred by a Zobs ≥ 12 (p < 0.0001) and ≥7 (p < 0.0001) for compounds 3 and 4, respectively. By using these assignments, we can estimate the number of RNA motifs that bound avidly to each small molecule: 23, 26, 64, and 215 RNA motifs for 1, 2, 3, and 4, respectively (Table 1 and Figure 4). Thus, the higher signal on the array (Figure 2) is a function of the number of RNA motifs that bind each ligand rather than a function of binding affinity.
Additionally, Zobs rank (and by analogy Zobs) can also be used to estimate the affinities of binding interactions (Figure 5). Single exponential curves with an asymptote defined a correlation between Zobs and experimentally determined Kd for each compound and its cognate RNA motifs. In all cases, there is excellent correlation with R values ranging from 0.86 to 0.96. These scoring functions can provide a means to rapidly profile small molecules for binding to “on-” and “off-targets”, as mentioned above. Zobs values were normalized to the most statistically significant RNA binder (100% fitness) to afford fitness scores.
We also determined the minimal fold coverage of the starting and selected RNA libraries in RNA-seq data required to afford accurate scoring functions (Table 2 and Figure S-58). A strong correlation (R ∼ 0.8) is observed when both libraries have at least 24-fold coverage. Correlations (R > 0.5) were observed with as little as 6-fold coverage of the starting library and 12-fold coverage of the selected library. Taken together, HiT-StARTS can rapidly identify interactions that are high affinity and selective (by comparing Zobs rank for a given RNA for all four compounds), which are not predicted well by frequency rank. Additionally, the HiT-StARTS approach is able to discriminate between affinities, including those that differ by as little as 2-fold; however, the empirical relationship between Zobs and affinity can affect the sensitivity of these predictions. As seen in Figure 5, the prediction is more sensitive to differences in affinity as the slope of the curve fit line increases.
Table 2. Goodness of Fit (R Values) for Scoring Functions Relative to the Coverage of the Sequencing Data.
| coverage
of starting library |
||||
|---|---|---|---|---|
| coverage of selected library | 6-fold | 12-fold | 24-fold | 30-fold |
| 6-fold | 0.46 | 0.58 | 0.58 | 0.67 |
| 12-fold | 0.58 | 0.71 | 0.74 | 0.79 |
| 24-fold | 0.58 | 0.67 | 0.82 | 0.93 |
| 30-fold | 0.66 | 0.76 | 0.92 | 0.93 |
HiT-StARTS Applied to Other RNA Motif Libraries
To test the general applicability of these approaches to other RNA motif libraries, we used 2DCS to select RNA motifs derived from a 3 × 2 internal loop library (3 × 2 ILL; Figure 1) that bind to small molecules 1–8. This secondary structural motif displays asymmetric internal loops and bulges, the latter of which are under-represented among known RNA motif–small molecule interactions yet are highly prevalent in functional (Drosha and Dicer processing) sites in miRNA precursors.38 As was observed in selections with 3 × 3 ILL, only compounds 1–4 bind members of the RNA library. HiT-StARTS analysis of the 2DCS experiments and subsequent binding analyses of the selected interactions showed that the features identified for 3 × 3 ILL were also found when using 3 × 2 ILL. That is, selective binders have Zobs > 8 (p < 0.0001) and relative Zobs defined a scoring function for the selected RNA motif–small molecule interactions (Figure S-59).
Identification of Biologically Important RNAs That Can Be Targeted
The results of HiT-StARTS were used in conjunction with Inforna to identify RNA targets in the transcriptome that could be drugged with small molecule–RNA motif partners that we identified. Inforna mines data defining RNA motif–small molecule binding interactions to identify RNAs with targetable motifs.9,22 This approach to chemical probe discovery could be considered target agnostic because the output of 2DCS is used to infer the preferred target of the small molecule. In particular, we focused on microRNAs (miRNAs) associated with disease that have targetable motifs in Dicer or Drosha processing sites, as defined by HiT-StARTS. Previous studies have shown that small molecules can inhibit miRNA precursor processing by binding to nuclease processing sites, with affinities ranging from nM to mid to high μM.9,39−42
These studies identified that compound 4 bound with 100% fitness to the 5′G_U/3′CUA (one nucleotide U bulge) that is present in the Dicer processing sites of three microRNAs (miRNAs) in the oncogenic miR-17-92 cluster (contains six miRNAs),43 miR-17, -18a, and -20a. That is, of all RNA motifs present in the 3 × 2 ILL, 5′G_U/3′CUA is the highest affinity. Indeed, the four most fit RNAs from the 2DCS selection of 4 contain U bulges (Figure S-64). Further, other highly fit bulges for compound 4 include 5′GAU/3′C_A (91% fitness), an A bulge present in Dicer processing site of pre-miR-18a and 5′GGU/3′C_A (78% fitness), a G bulge present in the Dicer processing sites of pre-miR-17 and pre-miR-20a (Figure 6).
Figure 6.
Secondary structures of miR-17, -18a, -19a, -19b, -20a, and -92a hairpin precursors. Mature miRNAs are indicated with red letters, and binding sites for 4 in Dicer processing sites are indicated with blue circles.
The binding affinity of 4 for RNAs containing one of the three bulges was measured using a fluorescein labeled small molecule, 4-FL (Figure 7). These studies showed that 4-FL bound to the 5′G_U/3′CUA bulge common to the Dicer processing sites in miR-17, -18a, and -20a (RNA1, Figure 7) with a Kd of 30 ± 2 μM. Mutation of the U bulge to an AU pair (RNAC) eliminates binding (Kd > 100 μM). Likewise, 4 binds the A bulge present in the miR-18a precursor (RNA2) with a Kd of 32 ± 5 μM and the G bulge present in the miR-17 and miR-20a precursors (RNA3) with a Kd of 40 ± 6 μM, in agreement with predictions by HiT-StARTS. No saturable binding was observed to the motifs in the Dicer sites of miR-19a, miR-19b, or miR-92a. (Notably, the dye itself does not contribute to binding affinity, as determined by competitive binding assays with 4–FL and 4 (Table S2) in agreement with previous observations with other small molecule–fluorescein conjugates.9,29)
Figure 7.
Secondary structures of the RNAs to which the binding of small molecule 4 was assessed. RNAC is a control RNA that does not contain target sites. RNA1 contains the U bulge present in the Dicer processing site common to miR-17, -18a, and -20a. RNA2 contains the A bulge that is present in miR-18a. RNA3 contains the G bulge present in miR-17 and miR-20a. Binding affinities (Kds) are indicated in parentheses and are reported in μM.
Small Molecules Identified by Inforna Inhibit Processing of miRNAs in the Oncogenic miR-17-92 Cluster
Initial studies were completed in vitro to determine if compound 4 (Targapremir-18a) inhibits Dicer processing of each miRNA separately in the miR-17-92 cluster using radioactively labeled RNA (Figure 8A). Indeed, the compound inhibited Dicer processing of pre-miR-17, -18a, and -20 at low micromolar concentrations but had no effect on the processing of pre-miR-19a and pre-miR-19b (Figure 8A and Figures S-60–S-63), as expected from HiT-StARTS analysis. Thus, inhibition of Dicer processing by 4 is selective for miR-17, -18a, and -20a and nonselective inhibition of Dicer processing is not observed.
Figure 8.
Effect of 4 on the biogenesis of the miR-17-92 cluster in DU145 prostate cancer cells. (A) 4 inhibits in vitro Dicer processing of pre-miR-18a. Lane L indicates a hydrolysis ladder. Lane T1 indicates cleavage by nuclease T1 under denaturing conditions (cleaves G’s). (B) Effect of 4 on mature miR-17, -18a, -19a, -19b, -20a, and -92a in DU145 cells. (C) Profiling of mature miRNA levels in DU145 cells that contain potential binding sites for 4. The value indicated in parentheses is the normalized expression level, as compared to miR-18a in untreated cells. *, p < 0.05, and **, p < 0.01, as determined by a two-tailed Student t test.
Interestingly, both miR-18a and miR-20 are overexpressed in prostate cancer.44,45 Thus, the effect of 4 on processing of the miR-17-92 cluster was studied in DU145 prostate cancer cells. In particular, the mature levels of each miRNA in the cluster were studied by RT-qPCR. In agreement with our in vitro Dicer processing studies (Figure 8A and Figures S-60–S-63), 4 decreased the amount of mature miR-17, -18a, and -20a at low micromolar concentrations with an IC50 of ∼10 μM, but has no effect on miR-19a, -19b, and -92a (Figure 8B).
We further probed the selectivity of 4 by studying its effect on levels of other mature miRNAs (Figure 8C) and on pre-miR-17, -18a, and -20a. We identified miRNAs with potential binding sites for 4, as determined by HiT-StARTS, using a database of RNA folds in the human miRNA transcriptome,38 including the 5′G_U/3′CUA bulge common to the Dicer processing sites in miR-17, -18a, and -20a (n = 33). The potential binding sites for 4 can occur anywhere in the miRNA; that is, they were not confined to Drosha or Dicer processing sites. Other miRNAs that contain the 5′G_U/3′CUA motif have a much lower expression level than miR-17, miR-18a, and miR-20. Studying these RNAs could allow assessment of influence of expression level on compound potency and selectivity, an important consideration in RNA targeting that has largely gone unaddressed.
Interestingly, 4 has no statistically significant effect on the levels of any of the 27 additional mature miRNAs studied (Figure 8C). These results suggest that (i) the small molecule must bind to a functional (processing) site to inhibit miRNA biogenesis and (ii) the miRNA’s abundance in cells influences how productive an interaction with a small molecule will be. Collectively, it appears that it is easier to target and inhibit the biogenesis of highly expressed miRNAs than ones that are of lower expression. Such effects have been well-documented in the protein-targeting community but until now have not been studied in RNA targeting endeavors.
Addition of 4 Increases STK4 Expression
A previous study has shown that miR-18a represses serine/threonine protein kinase 4 (STK4) protein, a tumor suppressor, in prostate cancer cells and promotes tumorigenesis.45 Thus, we studied the ability of compound 4 to de-repress STK4 and trigger apoptosis. Treatment of DU145 cells with 20 μM 4 increases levels of STK4 by ∼2.5-fold (Figure 9A). Further, inhibition of miR-18a biogenesis and de-repression of STK4 triggers apoptosis as assessed by annexin V/PI staining (Figure 9B). Although the most common route to modulate protein expression is to inhibit them directly, here we show that it is possible to increase protein expression by targeting noncoding RNAs that repress their production.
Figure 9.
Inhibition of miR-18a biogenesis by 4 de-represses a downstream protein and triggers apoptosis. (A) Effect of 4 on STK4 protein expression, a direct target of miR-18a. (B) Ability of 4 (10 μM and 20 μM) and a miR-18a antagomir (50 nM) to trigger apoptosis. *, p < 0.05, and **, p < 0.01, as determined by a two-tailed Student t test.
Profiling the Target Selectivity of 4 by using Chemical Cross-Linking and Isolation by Pull Down (Chem-CLIP)
We previously developed a method to study the cellular targets of a small molecule called Chemical Cross-Linking and Isolation by Pull Down (Chem-CLIP).46,47 In Chem-CLIP, a small molecule is appended with a cross-linking module that reacts with nucleic acids and a purification module to allow for the facile isolation of the small molecule’s cellular targets. Thus, 4 was appended with chlorambucil (cross-linking module; CA) and biotin (purification module) to afford 4-CA-Biotin. We first studied the reaction of 4-CA-Biotin with pre-miR-18a in vitro and compared it to the reaction of the compound with tRNA. As expected, 4-CA-Biotin reacts to a greater extent with pre-miR-18a (Figure 10B). The selectivity of 4-CA-Biotin was then studied in cellular lysates via RT-qPCR by calculating the enrichment of the target of interest in the pulled-down fraction. Enrichment was studied for the 33 miRNAs with potential binding sites for 4 as described above. Incubating 10 μM 4 with DU145 cell lysate and quantification of the isolated targets showed a ∼4-fold enrichment of pre-miR-18a and ∼2-fold enrichment of pre-miR-17 and pre-miR-20a (Figure 10C). There was no enrichment of any of the other miRNAs studied (Figure 10C).
Figure 10.
Chem-CLIP to study small molecule engagement of pre-miR-18a by 4. (A) Structure of 4-CA-Biotin and Ctrl-CA-Biotin, a control compound that lacks the RNA-binding module. (B) In vitro assessment of 4-CA-Biotin and Ctrl-CA-Biotin for reacting with pre-miR-18a. (C) Top, Chem-CLIP profiling in DU145 cells with 4-CA-Biotin. Bottom, Chem-CLIP profiling in DU145 cells with Ctrl-CA-Biotin. *, p < 0.05, and **, p < 0.01, as determined by a two-tailed Student t test.
Implications and Conclusions
The HiT-StARTS approach may be general for many types of library screens that use nucleic acid sequencing for deconvolution, provided that biases in the starting library can also be obtained by sequencing with sufficient fold coverage. At present, 2DCS, with the relatively small number of nucleic acid sequences in the starting library (we have employed libraries with up to 16,384 members), and DNA-encoded small molecule libraries are ideal applications of HiT-StARTS. The approach might be applicable for SELEX or phage display, as long as the starting libraries are small enough that they can be sequenced with severalfold coverage. It is likely, however, that as sequencing technologies advance and the number of sequence reads that can be completed in a single study increases, HiT-StARTS will be more broadly applicable. Furthermore, the selected RNA motif–small molecule binding partners could be developed into lead compounds that target RNA. This has been demonstrated for a first-in-class small molecule that inhibits the oncogenic miR-17-92 cluster. One articulated goal of chemical biology is to find inhibitors and activators of all proteins in the proteome. It now appears that rational design enabled by Inforna and 2DCS can be used to develop small molecule protein activators.
Methods
Small Molecule Synthesis and Characterization
Details for the chemical synthesis and characterization of compounds 1–8 and fluorescently labeled compounds 1–FL, 2–FL, 3–FL, and 4–FL are provided in the Supporting Information.
Construction of Alkyne-Displaying Microarrays
Microarrays were constructed as described previously.18 Please see the Supporting Information for experimental details.
General Nucleic Acids
All DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. (IDT), and used without further purification. The RNA competitor oligonucleotides were purchased from Dharmacon and deprotected according to the manufacturer’s standard procedure. Competitor oligonucleotides were used to ensure that RNA–small molecule interactions were confined to the randomized region (3 × 3 or 3 × 2 nucleotide internal loop pattern; Figure 1). All aqueous solutions were made with NANOpure water. The RNA library was transcribed by in vitro transcription from the corresponding DNA template (see below).
PCR Amplification of DNA Templates Encoding 3 × 3 ILL and 3 × 2 ILL (RNA Motif Library) and Selected RNAs
The DNA templates encoding selected RNAs and 3 × 3 ILL or 3 × 2 ILL were PCR amplified using a forward primer that encodes for a T7 RNA polymerase promoter. PCR amplification was completed in 300 μL of 1× PCR Buffer (10 mM Tris-HCl, pH 9.0, 50 mM KCl, and 0.1% Triton X-100), 4.25 mM MgCl2, 0.33 mM dNTPs, 2 μM each primer (forward primer, 5′-d(GGCCGGATCCTAATACGACTCACTATAGGGAGAGGGTTTAAT), and reverse primer, 5′-d(CCTTGCGGATCCAAT)), and 1 μL of Taq DNA polymerase. The DNA was amplified by 30 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 40 s. All PCR reactions were evaluated on a 2% agarose gel stained with ethidium bromide prior to transcription.
RNA Transcription and Purification
RNAs were transcribed as previously described.48 Briefly, transcriptions were completed in a total volume of 1 mL containing 1× Transcription Buffer (40 mM Tris-HCl, pH 8.0, 1 mM spermidine, 10 mM DTT, and 0.001% Triton X-100), 2.5 mM each rNTP, 15 mM MgCl2, 300 μL of PCR-amplified DNA template, and 20 μL of 20 mg/mL T7 RNA polymerase by incubating at 37 °C overnight. After transcription, 1 unit of DNase I (Invitrogen) was added, and the sample was incubated at 37 °C for an additional 30 min. Transcribed RNAs were then purified on a denaturing 12.5% polyacrylamide gel and extracted as previously described.48 Concentrations were determined by measuring absorbance at 260 nm and the corresponding extinction coefficients, which were determined by the HyTher server49 and nearest neighbor parameters.50
RNA Screening and Selection
2DCS selections were completed as previously described.18 Please see the Supporting Information for experimental details.
Reverse Transcription and PCR Amplification To Install Barcodes for RNA-seq
The agarose containing bound RNAs excised from 2DCS arrays was placed into a thin-walled PCR tube with 18 μL of water, 2 μL of 10× RQ DNase I Buffer, and 2 units of RNase-free DNase (Promega). The solution was incubated at 37 °C for 2 h and then quenched by addition of 2 μL of 10× DNase stop solution (Promega). Samples were incubated at 65 °C for 10 min to completely inactivate the DNase and were then subjected to RT-PCR amplification to install a unique barcode. Reverse transcription reactions were completed in 1× RT Buffer, 1 mM dNTPs, 5 μM RT primer (5′-CCTCTCTATGGGCAGTCGGTGATCCTTGCGGATCCAAT; the sequence underlined is complementary to the 3′ end of the RNAs), 200 μg/mL BSA, 4 units of reverse transcriptase, and 20 μL of DNase-treated selected RNAs. Samples were incubated at 60 °C for 1 h. A 20 μL aliquot of the RT reaction was added to 6 μL of 10× PCR Buffer, 4 μL of 100 μM forward primer including barcode (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXXXXXGATGGGAGAGGGTTTAAT where X represents unique barcode, GAT is the barcode adapter, and the sequence underlined is complementary to the 5′ end of the library), 2 μL of 100 μM reverse primer, 0.6 μL of 250 mM MgCl2, and 2 μL of Taq DNA polymerase. Two-step PCR was performed at 95 °C for 1 min and 72 °C for 1 min. Aliquots of the RT-PCR product were checked every three cycles starting at cycle 10 on a denaturing 12.5% polyacrylamide gel stained with ethidium bromide to ensure that background levels of RNAs (spots excised from the array where compound was not delivered) were not amplified. RT-PCR products encoding selected RNAs were purified on a denaturing 12.5% polyacrylamide gel. Purity was assessed by a Bioanalyzer. Samples were mixed in equal amounts and sequenced using an Ion Proton deep sequencer using PI chips (60–80 million reads).
Assigning Frequency, Frequency Rank, Zobs, and Zobs Rank from the Output of Next-Generation Sequencing
A shell script was written to process the fastq sequencing files generated by Scripps’s Genomics Core. Shell functions such as grep and awk were used to find matching sequences to the 3 × 3 ILL or 3 × 2 ILL and to extract the randomized loop region in the library. Frequency of the randomized loops was calculated using awk.
Calculating Statistical Significance (Zobs) for Selected RNAs
To determine if the difference in frequency for given RNA in the starting library and in the selection was statistically significant, a pooled population comparison (Zobs) was calculated using eqs 1 and 2:
| 1 |
| 2 |
where n1 is the size of population 1 (number of reads for a selected RNA); n2 is the size of population 2 (number of reads for the same RNA from sequencing of the starting library); p1 is the observed proportion of population 1 (number of reads for a selected RNA divided by the total number of reads); and p2 is the observed proportion for population 2 (number of reads for the same RNA divided by the total number of reads in the starting library).
Binding Affinity Measurements
An in solution, fluorescence-based assay was used to determine binding affinities by monitoring the change in fluorescence intensity of 1–FL (or 2–FL/3–FL/4–FL) as a function of RNA concentration as described previously.18 Briefly, the RNA of interest was folded in 1× Folding Buffer (8 mM Na2HPO4, pH 7.0, 185 mM NaCl, and 1 mM EDTA) by heating at 60 °C for 5 min followed by slow cooling to room temperature on the benchtop. Then, the FL-conjugated compound was added into the RNA solution to a final concentration of 100 nM. Serial dilutions were completed using 1× Folding Buffer supplemented with 100 nM FL-conjugated compound. The solutions were incubated at room temperature for 30 min and then transferred to a well of a black 384-well plate. Fluorescence intensity was measured using a Bio-Tek FL×800 plate reader with an excitation wavelength of 485/20 nm and an emission wavelength of 528/20 nm. The change in fluorescence intensity as a function of the concentration of RNA was fit to eq 3:9
| 3 |
where I is the observed fluorescence intensity; I0 is the fluorescence intensity in the absence of RNA; Δε is the difference between the fluorescence intensity in the absence of RNA and in the presence of infinite RNA concentration; [FL]0 is the concentration of compound; [RNA]0 is the concentration of the selected RNA; and Kd is the dissociation constant.
Competitive binding assays were completed by incubating the RNA of interest with 100 nM 4–FL and increasing concentrations of 4. The resulting curves were fit to eq 4:
 |
where θ is the percentage of 4–FL bound, [4–FL] is the concentration of 4–FL, Kt is the dissociation constant of RNA and 4–FL, [RNA] is the concentration of RNA, Ct is the concentration of 4, Kd is the dissociation constant for 4, and A is a constant.
Dicer Inhibition Assay
The template used for pre-miR-18a (5′-GGGTGTTCTAAGGTGCATCTAGTGCAGATAGTGAAGTAGATTAGCATCTACTGCCCTAAGTGCTCCTTCTGGCA) was PCR-amplified in 1× PCR Buffer, 2 μM forward primer (5′-GGCCGAATTCTAATACGACTCACTATATCTAAGGTGCATCTAGTGCAGA), 2 μM reverse primer (5′-TGCTACAAGTGCCTTCACTGCA), 4.25 mM MgCl2, 330 μM dNTPs, and 2 μL of Taq DNA polymerase in a 50 μL reaction. Cycling conditions were 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s. Pre-miR-18a was 5′-end labeled with 32P as previously described.9 The RNA was folded in 1× Reaction Buffer (Genlantis) by heating at 60 °C for 5 min and slowly cooling to room temperature. The samples were then supplemented with 1 mM ATP and 2.5 mM MgCl2. Serially diluted concentrations of 4 were added, and the samples were incubated at room temperature for 15 min. Next, 0.005 unit/μL of recombinant human Dicer (Genlantis) was added followed by incubation at 37 °C overnight. Reactions were stopped by adding the manufacturer’s supplied stop solution.
A T1 ladder (cleaves G residues) was generated by heating the RNA in 1× RNA Sequencing Buffer (20 mM sodium citrate, pH 5.0, 1 mM EDTA, and 7 M urea) at 55 °C for 10 min followed by slowly cooling to room temperature. RNase T1 was then added to a final concentration of 3, 0.3, or 0.03 unit/μL, and the solution was incubated at room temperature for 20 min. An RNA hydrolysis ladder was generated by incubating RNA in 1× RNA Hydrolysis Buffer (50 mM NaHCO3, pH 9.4, and 1 mM EDTA) at 95 °C for 5 min. In all cases, cleavage products were separated on a denaturing 15% polyacrylamide gel and imaged using a Bio-Rad PMI phosphorimager.
Cell Culture
DU145 cells were cultured in growth medium (Roswell Park Memorial Institute medium (RPMI) supplemented with 10% fetal bovine serum (FBS)) at 37 °C and 5% CO2
RNA Isolation and Quantitative Real Time Polymerase Chain Reaction (RT-qPCR) of miRNAs
DU145 cells were transfected in either 6- or 12-well plates with a miR-17-92 cluster overexpression plasmid (Addgene plasmid #21109)51 with jetPRIME per manufacturer’s suggested protocol and treated with compound for 24 h. Total RNA was extracted from cells using a Quick-RNA Miniprep Kit (Zymo Research) per the manufacturer’s protocol. Approximately 200 ng of total RNA was used in reverse transcription (RT) reactions, which were completed using a miScript II RT kit (Qiagen) per the manufacturer’s protocol. RT-qPCR was performed on a 7900HT Fast Real Time PCR System (Applied Biosystem) using power SYBR Green Master Mix (Applied Biosystems). All primer sets were purchased from IDT or Eurofins (Table S1). The expression levels of mature miRNAs were normalized to U6 small nuclear RNA or 18s rRNA.
Reaction of 4-CA-Biotin
To determine if 4-CA-Biotin reacts with the miR-18a hairpin precursor or tRNA in vitro, 5 μL of 32P-labeled miR-18a hairpin precursor or tRNA (∼50,000 cpm) was diluted in a total volume of 300 μL of 1× PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 137 mM NaCl, and 2.7 mM KCl). The RNA was folded by heating at 60 °C for 5 min and slowly cooling to room temperature. Compound was then added at various concentrations, and the solutions were incubated overnight at room temperature. Next, 10 μL of streptavidin resin (high capacity streptavidin agarose beads; Thermo Scientific) was added to the samples, which were incubated for an additional 30 min at room temperature. After centrifugation, the supernatant was removed, and the resin was washed with 1× PBST (1× PBS + 0.1% (v/v) Tween 20). The amount of radioactivity in the supernatant plus the wash and associated with the beads was measured using a Beckman Coulter LS6500 liquid scintillation counter.
Chem-CLIP in Cell Lysates
DU145 cells were cultured as described above in 100 mm dishes and lysed with 500 μL of Cell Lysis Buffer (10 mM Tris pH 7.4, 0.25% Igepal CA-630, and 150 mM NaCl) for 5 min at room temperature. The cell lysate was then centrifuged and the supernatant collected. Next, 10 μM 4 was added to the lysate in 1× PBS and the sample was incubated for 2 h at room temperature. The reaction was then directly used for pull-down by incubating with 50 μL of streptavidin resin in 1× PBS for 30 min at room temperature. After centrifugation, the supernatant was removed, and the resin was washed twice with 1× PBS. RNA was eluted from the streptavidin beads by incubation with 100 μL of 1× Elution Buffer (10 mM EDTA and 95% formamide) at 65 °C for 20 min. The eluted RNA was cleaned up using a Quick-RNA Miniprep Kit (Zymo) per the manufacturer’s protocol. RT-qPCR was completed as described above using 50 ng of total RNA in the RT reaction. Expression levels were normalized to 18S rRNA.
Western Blotting
Cells were grown in 6-well plates to ∼80% confluency in complete growth medium and then incubated with 10 or 20 μM 4 for 48 h. Total protein was extracted using M-PER Mammalian Protein Extraction Reagent (Pierce Biotechnology) per the manufacturer’s recommended protocol. Extracted total protein was quantified using a Micro BCA Protein Assay Kit (Pierce Biotechnology). Approximately 25 μg of total protein was resolved using an 8% SDS–polyacrylamide gel and then transferred to a PVDF membrane.
The membrane was briefly washed with 1× Tris-buffered saline (TBS; 50 mM Tris-Cl, pH 7.5. 150 mM NaCl) and blocked with 5% milk in 1× TBST (1× TBS containing 0.05% Tween-20) for 1 h at room temperature. The membrane was then incubated with 1:1000 STK4 primary antibody in 1× TBST containing 5% milk overnight at 4 °C. The membrane was washed with 1× TBST and incubated with 1:2000 anti-rabbit IgG horseradish-peroxidase secondary antibody conjugate in 1× TBST for 1 h at room temperature. After washing with 1× TBST, protein expression was quantified using SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology) per the manufacturer’s protocol.
The membrane was then stripped using 1× Stripping Buffer (200 mM glycine, 1% Tween-20, and 0.1% SDS, pH 2.2) followed by washing in 1× TBST. The membrane was blocked and probed for β-actin following the same procedure described above using 1:5000 β-actin primary antibody in 1× TBST containing 5% milk overnight at 4 °C. The membrane was washed with 1× TBST and incubated with 1:10,000 anti-rabbit IgG horseradish-peroxidase secondary antibody conjugate in 1× TBST for 1 h at room temperature. ImageJ software from the National Institutes of Health was used to quantify band intensities.
Acknowledgments
This work was funded by the US National Institutes of Health Grant R01-GM097455 to M.D.D.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00009.
Synthesis and characterization details and experimental details for small molecule microarray construction and 2DCS selections (PDF)
Author Present Address
⊥ G.J.M.: Department of Microbiology and Immunology, Chicago Medical School, Rosalind Franklin University of Medicine and Science, 3333 Green Bay Rd, North Chicago, Illinois 60064, USA.
Author Contributions
‡ S.P.V. and Y.L. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Thomas J. R.; Hergenrother P. J. Targeting RNA with small molecules. Chem. Rev. 2008, 108 (4), 1171–1224. 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- Guan L.; Disney M. D. Recent advances in developing small molecules targeting RNA. ACS Chem. Biol. 2012, 7 (1), 73–86. 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- Gallego J.; Varani G. Targeting RNA with small-molecule drugs: therapeutic promise and chemical challenges. Acc. Chem. Res. 2001, 34 (10), 836–843. 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- Moehle K.; Athanassiou Z.; Patora K.; Davidson A.; Varani G.; Robinson J. A. Design of beta-hairpin peptidomimetics that inhibit binding of alpha-helical HIV-1 Rev protein to the Rev response element RNA. Angew. Chem., Int. Ed. 2007, 46 (47), 9101–9114. 10.1002/anie.200702801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanassiou Z.; Patora K.; Dias R. L.; Moehle K.; Robinson J. A.; Varani G. Structure-guided peptidomimetic design leads to nanomolar beta-hairpin inhibitors of the Tat-TAR interaction of bovine immunodeficiency virus. Biochemistry 2007, 46 (3), 741–751. 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
- Davidson A.; Leeper T. C.; Athanassiou Z.; Patora-Komisarska K.; Karn J.; Robinson J. A.; Varani G. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (29), 11931–11936. 10.1073/pnas.0900629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werstuck G.; Green M. R. Controlling gene expression in living cells through small molecule-RNA interactions. Science 1998, 282 (5387), 296–298. 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- Pushechnikov A.; Lee M. M.; Childs-Disney J. L.; Sobczak K.; French J. M.; Thornton C. A.; Disney M. D. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 2009, 131 (28), 9767–9779. 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Gallo S. M.; Disney M. D. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol. 2014, 10 (4), 291–297. 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Disney M. D. Two-dimensional combinatorial screening enables the bottom-up design of a microRNA-10b inhibitor. Chem. Commun. (Cambridge, U. K.) 2014, 50 (23), 3027–3029. 10.1039/c3cc00173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. M.; Pushechnikov A.; Disney M. D. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem. Biol. 2009, 4, 345–355. 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arambula J. F.; Ramisetty S. R.; Baranger A. M.; Zimmerman S. C. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (38), 16068–16073. 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahromi A. H.; Nguyen L.; Fu Y.; Miller K. A.; Baranger A. M.; Zimmerman S. C. A novel CUGexp·MBNL1 inhibitor with therapeutic potential for myotonic dystrophy type 1. ACS Chem. Biol. 2013, 8 (5), 1037–1043. 10.1021/cb400046u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney J. L.; Wu M.; Pushechnikov A.; Aminova O.; Disney M. D. A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem. Biol. 2007, 2 (11), 745–754. 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- Velagapudi S. P.; Seedhouse S. J.; Disney M. D. Structure-activity relationships through sequencing (StARTS) defines optimal and suboptimal RNA motif targets for small molecules. Angew. Chem., Int. Ed. 2010, 49 (22), 3816–3818. 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249 (4968), 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Robertson D. L.; Joyce G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344 (6265), 467–468. 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- Disney M. D.; Labuda L. P.; Paul D. J.; Poplawski S. G.; Pushechnikov A.; Tran T.; Velagapudi S. P.; Wu M.; Childs-Disney J. L. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J. Am. Chem. Soc. 2008, 130 (33), 11185–11194. 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- Franzini R. M.; Ekblad T.; Zhong N.; Wichert M.; Decurtins W.; Nauer A.; Zimmermann M.; Samain F.; Scheuermann J.; Brown P. J.; Hall J.; Graslund S.; Schuler H.; Neri D. Identification of structure-activity relationships from screening a structurally compact DNA-encoded chemical library. Angew. Chem., Int. Ed. 2015, 54 (13), 3927–3931. 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- Zambaldo C.; Barluenga S.; Winssinger N. PNA-encoded chemical libraries. Curr. Opin. Chem. Biol. 2015, 26, 8–15. 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Mannocci L.; Leimbacher M.; Wichert M.; Scheuermann J.; Neri D. 20 years of DNA-encoded chemical libraries. Chem. Commun. (Cambridge, U. K.) 2011, 47 (48), 12747–12753. 10.1039/c1cc15634a. [DOI] [PubMed] [Google Scholar]
- Disney M. D.; Winkelsas A. M.; Velagapudi S. P.; Southern M.; Fallahi M.; Childs-Disney J. L. Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem. Biol. 2016, 11 (6), 1720–1728. 10.1021/acschembio.6b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T. R.; Hilgraf R.; Sharpless K. B.; Fokin V. V. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org. Lett. 2004, 6 (17), 2853–2855. 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- Kolb H. C.; Finn M. G.; Sharpless K. B. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem., Int. Ed. 2001, 40 (11), 2004–2021. . [DOI] [PubMed] [Google Scholar]
- Seth P. P.; Miyaji A.; Jefferson E. A.; Sannes-Lowery K. A.; Osgood S. A.; Propp S. S.; Ranken R.; Massire C.; Sampath R.; Ecker D. J.; et al. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J. Med. Chem. 2005, 48 (23), 7099–7102. 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- Zhou S.; Rynearson K. D.; Ding K.; Brunn N. D.; Hermann T. Screening for inhibitors of the hepatitis C virus internal ribosome entry site RNA. Bioorg. Med. Chem. 2013, 21, 6139–6144. 10.1016/j.bmc.2013.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.; Castaldi M. P.; Dutta S.; Dibrov S. M.; Wyles D. L.; Hermann T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat. Chem. Biol. 2009, 5 (11), 823–825. 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando S.; Narita A.; Aoyama Y. Light-up Hoechst-DNA aptamer pair: generation of an aptamer-selective fluorophore from a conventional DNA-staining dye. ChemBioChem 2007, 8 (15), 1795–1803. 10.1002/cbic.200700325. [DOI] [PubMed] [Google Scholar]
- Velagapudi S. P.; Seedhouse S. J.; French J.; Disney M. D. Defining the RNA internal loops preferred by benzimidazole derivatives via 2D combinatorial screening and computational analysis. J. Am. Chem. Soc. 2011, 133 (26), 10111–10118. 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount K. F.; Wang J. X.; Lim J.; Sudarsan N.; Breaker R. R. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007, 3 (1), 44–9. 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- Winkler W. C.; Cohen-Chalamish S.; Breaker R. R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. U. S. A. 2002, 99 (25), 15908–15913. 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W. C.; Breaker R. R. Genetic control by metabolite-binding riboswitches. ChemBioChem 2003, 4 (10), 1024–1032. 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- Nahvi A.; Sudarsan N.; Ebert M. S.; Zou X.; Brown K. L.; Breaker R. R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002, 9 (9), 1043–1049. 10.1016/S1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Daldrop P.; Reyes F. E.; Robinson D. A.; Hammond C. M.; Lilley D. M.; Batey R. T.; Brenk R. Novel ligands for a purine riboswitch discovered by RNA-ligand docking. Chem. Biol. 2011, 18 (3), 324–335. 10.1016/j.chembiol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W.; He H. W.; Wang Z. M.; Zhao H.; Lian X. Q.; Wang Y. S.; Zhu J.; Yan J. J.; Zhang D. G.; Yang Z. J.; et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012, 11, 55. 10.1186/1476-511X-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fede A.; Billeter M.; Leupin W.; Wüthrich K. Determination of the NMR solution structure of the Hoechst 33258-d(GTGGAATTCCAC)2 complex and comparison with the X-ray crystal structure. Structure 1993, 1 (3), 177–186. 10.1016/0969-2126(93)90019-D. [DOI] [PubMed] [Google Scholar]
- Lambert N.; Robertson A.; Jangi M.; McGeary S.; Sharp P. A.; Burge C. B. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell 2014, 54 (5), 887–900. 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Childs-Disney J. L.; Znosko B. M.; Wang D.; Fallahi M.; Gallo S. M.; Disney M. D. Analysis of secondary structural elements in human microRNA hairpin precursors. BMC Bioinformatics 2016, 17, 112. 10.1186/s12859-016-0960-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga C. L.; Velagapudi S. P.; Strivelli J. R.; Yang W. Y.; Disney M. D.; Phinney D. G. Small molecule inhibition of mir-544 biogenesis disrupts adaptive responses to hypoxia by modulating atm-mtor signaling. ACS Chem. Biol. 2015, 10 (10), 2267–2276. 10.1021/acschembio.5b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T. P.; Vo D. D.; Di Giorgio A.; Duca M. Ribosome-targeting antibiotics as inhibitors of oncogenic microRNAs biogenesis: Old scaffolds for new perspectives in RNA targeting. Bioorg. Med. Chem. 2015, 23 (17), 5334–5344. 10.1016/j.bmc.2015.07.062. [DOI] [PubMed] [Google Scholar]
- Vo D. D.; Staedel C.; Zehnacker L.; Benhida R.; Darfeuille F.; Duca M. Targeting the production of oncogenic microRNAs with multimodal synthetic small molecules. ACS Chem. Biol. 2014, 9 (3), 711–721. 10.1021/cb400668h. [DOI] [PubMed] [Google Scholar]
- Bose D.; Jayaraj G.; Suryawanshi H.; Agarwala P.; Pore S. K.; Banerjee R.; Maiti S. The tuberculosis drug streptomycin as a potential cancer therapeutic: inhibition of miR-21 function by directly targeting its precursor. Angew. Chem., Int. Ed. 2012, 51 (4), 1019–1023. 10.1002/anie.201106455. [DOI] [PubMed] [Google Scholar]
- He L.; Thomson J. M.; Hemann M. T.; Hernando-Monge E.; Mu D.; Goodson S.; Powers S.; Cordon-Cardo C.; Lowe S. W.; Hannon G. J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435 (7043), 828–833. 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang X. F.; Zhang Z. W.; Liu Q.; Sun N.; Pan L. L.; Shen J.; Li T.; Yun C.; Li H.; Shi L. H. miR-20a promotes prostate cancer invasion and migration through targeting ABL2. J. Cell. Biochem. 2014, 115 (7), 1269–1276. 10.1002/jcb.24778. [DOI] [PubMed] [Google Scholar]
- Hsu T. I.; Hsu C. H.; Lee K. H.; Lin J. T.; Chen C. S.; Chang K. C.; Su C. Y.; Hsiao M.; Lu P. J. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis 2014, 3, e99. 10.1038/oncsis.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L.; Disney M. D. Covalent small molecule-RNA complex formation enables cellular profiling of small molecule-RNA interactions. Angew. Chem. Int. Ed. Engl. 2013, 52 (38), 10010–10013. 10.1002/anie.201301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z.; Zhang Y.; Gendron T. F.; Bauer P. O.; Chew J.; Yang W. Y.; Fostvedt E.; Jansen-West K.; Belzil V. V.; Desaro P.; et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 2014, 83 (5), 1043–1050. 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Pushechnikov A.; Labuda L. P.; French J. M.; Disney M. D. Probing a 2-aminobenzimidazole library for binding to RNA internal loops via two-dimensional combinatorial screening. ACS Chem. Biol. 2012, 7 (11), 1902–1909. 10.1021/cb300213g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyret N.; Seneviratne P. A.; Allawi H. T.; SantaLucia J. Jr. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry 1999, 38 (12), 3468–3477. 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D.; Tinoco I. Jr. Absorbance melting curves of RNA. Methods Enzymol. 1989, 180, 304–325. 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- O’Donnell K. A.; Wentzel E. A.; Zeller K. I.; Dang C. V.; Mendell J. T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435 (7043), 839–843. 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.