Abstract
To study molecular epidemiology of CTX-M-55-carrying Escherichia coli isolates from urinary tract infections (UTIs) in China. 111 blaCTX-M-55-positive E.coli isolates from UTIs patients in China were studied. Pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) were used to analyze the homologies among the strains. Conjugation experiments, S1nuclease PFGE and PCR analysis were performed to characterize plasmids harboring blaCTX-M-55 and their genetic environment. 111 isolates were clustered into 86 individual pulsotypes and three clusters by PFGE. Fifty-five (49.5%) of the isolates belonged to 8 STs. Most of the ST1193 isolates belonged to one PFGE cluster. Transconjugants (n = 45) derived from randomly selected blaCTX-M-55 donors (n = 58), were found to contain a single 90-kb conjugative plasmid, which mainly belonged to the IncI1 groups (34, 76%). Among the IncI1 plasmids, the blaCTX-M-55/IncI1/ST16 predominated (23/34, 68%). The blaTEM-1 and aac (3′)-II genes were frequently detected on the IncI1 plasmids, and the insertion of ISEcp1 or IS26 was observed at the 48 bp or 45 bp upstream of the start codon of blaCTX-M-55 gene. The dissemination of blaCTX-M-55 gene among E. coli UTI isolates, appeared to be due to both the major clonal lineage of ST1193 and the horizontal transfer of epidemic plasmid IncI1/ST16.
Urinary tract infections (UTIs) are the most common bacterial infection and have a significant economic impact on the health care system. About 70–95% of community-acquired, and 50% of nosocomial UTIs are caused by E. coli1. There is increasing evidence that epidemic clonal groups of E. coli are responsible for a rise in antimicrobial-resistance in UTIs. For example, the prevalence of extended spectrum beta lactamase (ESBL)-producing E. coli (ESBL-EC) sequence type (ST131) is increasing worldwide, accounting for 25% and 54% of all ESBLs in Mexico and the United States, respectively2,3. Moreover, the emergence of drug-resistant epidemic plasmids, that confer antimicrobial resistance among bacteria, also contributes to increased resistance, through the horizontal transmission of ESBL-encoding genes. IncFII, IncA/C, IncL/M, IncN and IncI1 plasmids, carrying blaCTX-M genes and acquired ampC genes, are frequently detected in Enterobacteriaceae of different origin and source4. Specifically, the IncFII-like type plasmids have been reported to be closely associated with E. coli ST131 carrying blaCTX-M-15, while the IncI1 plasmids carrying blaCTX-M-15 or blaCMY-2 are frequently isolated from non-ST131 E. coli strains, some of which are shared between humans and animals5,6.
In China, ST131 was the most abundant type (12.7%) in 256 ESBL-producer isolates based on a study on ESBL-EC isolates from patients with community-onset infections in Chinese country hospitals from 2010 to 20117, and its prevalence increased to 29.5% in another study on bloodstream infections patients in three hospitals in China from 2013 to 20148. However, E. coli isolates from UTIs show a significantly different drug resistant profile than those from Europe, with more than 50% of isolates from China estimated to be resistant to both ciprofloxacin and cefotaxime by the year 2000. Since 1998, the predominant ESBL type in E. coli in China has been CTX-M-14, which was detected in 71% of the ESBL-ECs in 20089,10. However, a shift in ESBL genotypes has been reported recently, with the prevalence of CTX-M-15 and CTX-M-55, which are members of the CTX-M-1 group, increasing significantly in China since 201111. According to the national surveillance program in China, the blaCTX-M-15 positive strains accounted for 24% of all ESBL-EC UTI isolates that harbored blaCTX-M genes in 2011. But as of 2013,the situation had changed, with blaCTX-M-15 and blaCTX-M-55 positive strains accounting for 15% and 21% of all ESBL-EC UTI isolates, respectively10,11. Interestingly, the emerging blaCTX-M-55 ESBL genotype was also detected in E. coli strains from food animals or pets, according to different survey programs conducted from 2006–201212,13. These findings suggest that the high prevalence of blaCTX-M-55 in animals might contribute to the spread of blaCTX-M-55 genes between animals and humans. Unfortunately, there is limited data to clarify the clonal relationship between these blaCTX-M-55-carrying E. coli isolates from humans and animals. Furthermore, to our knowledge, compared to several molecular studies on blaCTX-M-55 in ESBL-EC strains of animal origin, there are limited studies on blaCTX-M-55 from clinical ESBL-EC strains from humans both in China and worldwide.
Consequently, in the present study, we examined the occurrence and characteristics of blaCTX-M-55 carrying ESBL-EC from UTIs following our previous study11. The plasmids harbored by these isolates, as well as the association between blaCTX-M-55 and other resistance genes, were also investigated.
Results
Distribution of isolates in demographics and source
In this study, a total of 111 CTX-M-55-producing E. coli isolates were obtained from urine specimens from 21 general hospitals in 19 provinces in China. These included 29 isolates from the Eastern (26%), 6 from the Southern (5%), 26 from the Northern (23%), 11 from the Central (10%), 12 from the Northeast (11%), 12 from the Southwest (11%), and 15 from the Northwest (13%) regions (data not shown). The majority of the isolates, 62(56%) were from hospital acquired UTIs, and the rest from community onset infection. Most of the isolates, 87(78%) were from females, and the median age was 60 years (range, 10 months to 91years).
Antimicrobial susceptibility patterns
All the 111 CTX-M-55- producing E. coli isolates were highly susceptible to imipenem, meropenem,ertapenem, piperacillin/tazobactam, tigecycline, and colistin, at approximately 93–100%. However, 76–98% of the isolates were resistant to ciprofloxacin, levofloxacin, cefotaxime, cefepime, ceftazidime, aztreonam, and ceftriaxone (Table S1).
PFGE profiles
Using PFGE, the CTX-M-55-producing E. coli isolates were discriminated into 89 pulsotypes (figure not shown), composed of 86 individual pulsotypes, and three clusters (I, II, III) for the remaining 25 isolates. Cluster I, which was defined at the 80% similarity level, encompassed 20(18%) isolates, and covered 14 hospitals in all the seven geographical regions across China (Figure S1). Cluster II and cluster III encompassed two and three isolates each, respectively.
MLST results
The CTX-M-55-producing E. coli isolates were grouped into 48 distinct STs, with the predominant ST being ST1193 (n = 20, 18%), followed by ST405 (n = 9, 8.1%), ST156 and ST2003 (5, 4.5% each), ST12, ST95, ST167, and ST354 (4, 3.6% each). These eight major STs accounted for 49.5% of the studied isolates (Table S2). A further 4 STs (ST10, ST 38, ST101, ST131) comprised of 3 isolates each, whilst the rest (36 STs) occurred in one or 2 isolates each. Three STs, including ST4207, ST4208 and ST4210, were novel to this study. An overwhelming majority of ST1193 isolates (18 of 20) were classified into the cluster I pulsotype by PFGE, whilst the isolates from other STs were scattered in diverse pulsotypes. Greater diversity was observed in isolates from hospital acquired UTIs, belonging to 35 different STs, than among isolates from community acquired UTIs, which belonged to 23 STs.
eBURST analysis revealed one clonal complex CC(CC10) encompassing 8 STs including ST10, ST167, ST44, ST744, ST1634, ST1488, ST617, and ST4204, which represented 13 isolates, whereas the remaining 40 profiles were observed in a single isolate each (Figure S2). Interestingly, ST1193(and other 38) were not assigned to a clonal complex, even when less stringent conditions were employed (i.e. STs sharing four or more alleles; data not shown). We further examined the clonality of our STs in an eBURST comparison with all E. coli STs in the international MLST web database (as at April 2014). With this wider comparison, ST1193 appears to belong to a prevalent clonal complex (CC14) (Figure S3). In addition, a novel single-locus variant (SLV) of ST1193, designated ST4207 was identified in one isolate. Furthermore, with regards to the drug susceptibility test results, all ST1193 isolates were susceptible to imipenem, meropenem, ertapenem, tigecycline, colistin and amikacin, and resistant to ciprofloxacin, ceftriaxone, ceftazidime, cefepime, and aztreonam, respectively (data not shown).
Plasmid analysis
The 20 ST1193 isolates were classified into 12 pulsotypes by PFGE. Among them, 5 isolates originating from one hospital belonged to cluster I, and the remaining 15 isolates belonged to other 11 clusters. So, one out of the five isolates from one hospital, and 15 isolates belonging to other 11 clusters were selected for conjugation studies. Additionally, another 42 randomly selected isolates belonging to other ST types were studied. Successful transfer of plasmids occurred in 77.6% (45 of 58) isolates. The 45 blaCTX-M-55 donors were the 16 ST1193 strains and the remaining 29 donors were distributed as follows: ST95 (n = 3), ST156 (n = 3), ST167 (n = 3), ST405 (n = 3), ST10 (n = 2), ST131 (n = 2), and 13 other individual ST donors. Plasmid DNA was isolated from 45 transconjugants (Table 1), and subsequent characterization of plasmids revealed that they belonged to the IncI1 plasmid class (n = 34; 76%), the F plasmid family (n = 10; 22%) and IncP (n = 1; 2%). Notably, 88% (14/16) of ST1193 isolates carried IncI1 plasmids (Table 1).
Table 1. Characteristics of plasmids in transconjugants analysed in this study.
| Plasmid | pMLST(Kp)/RSTa | Isolate | Region | ST | Other antibiotic resistance genes | Insertion fragments | Intergenic spacer region(bp)b |
|---|---|---|---|---|---|---|---|
| IncI1 | ST16(97) | AH1 | east | 1193 | TEM-1 | ISEcp1 | 48 |
| ST16(97) | AH14 | east | 12 | DHA | ISEcp1 | 48 | |
| ST16(97) | AH30 | east | 1193 | TEM-1 | ISEcp1 | 48 | |
| ST16(97) | SD28 | east | 998 | — | ISEcp1 | 48 | |
| ST15 | SD33 | east | 167 | TEM-1, aac(3′)-II | IS26 | 45 | |
| ST16(97) | Z1-8 | east | 10 | TEM-1,qnrS | ISEcp1 | 48 | |
| ST23 | Z1-40 | east | 4208 | CMY-2 | IS26 | 45 | |
| ST16(97) | ZS45 | east | 1193 | TEM-1, aac(3′)-II, qnrA | ISEcp1 | 48 | |
| ST16(97) | GP20 | south | 44 | aac-(6′)-Ib | IS26 | 48 | |
| ST136 | HN40 | south | 410 | TEM-1, aac(3′)-II, aac-(6′)-Ib,VEB, | ISEcp1 | 48 | |
| ST16(97) | HN47 | south | 1193 | TEM-1 | ISEcp1 | 48 | |
| ST16(97) | HB41 | north | 1193 | TEM-1, aac(3′)-II | NDd | ND | |
| ST16(97) | HB65 | north | 95 | TEM-1, aac(3′)-II,VEB | IS26 | 45 | |
| NTc | SX3 | north | 2003 | aac(3′)-II | ISEcp1 | 48 | |
| NT | SX4 | north | 1193 | — | IS26 | 48 | |
| NT | TZ17 | north | 4204 | — | ISEcp1 | 48 | |
| ST16(97) | TZ48 | north | 1193 | — | ND | ND | |
| ST16(97) | XA41 | north | 10 | TEM-1, aac(3′)-II | ISEcp1 | 48 | |
| ST16(97) | TJ4 | central | 1193 | TEM-1, aac(3′)-II | IS26 | 45 | |
| NT | TJ24 | central | 648 | TEM-1,aac(3′)-II,DHA | ISEcp1 | 48 | |
| ST136 | ZZ7 | central | 156 | TEM-1, aac(3′)-II,qnrA, | ISEcp1 | 45 | |
| ST16(97) | ZZ25 | central | 131 | TEM-1,aac(3′)-II,aac-(6′)-Ib | ISEcp1 | 48 | |
| NT | ZZ39 | central | 405 | aac(3′)-II | ISEcp1 | 48 | |
| ST16(97) | 1502 | southwest | 1193 | TEM-1, aac(3′)-II | ISEcp1 | 48 | |
| ST16(97) | 1642 | southwest | 1193 | TEM-1 | IS26 | 45 | |
| ST16(97) | NX18 | northwest | 95 | — | ISEcp1 | 48 | |
| ST16(97) | NX32 | northwest | 167 | TEM-1, aac(3′)-II, qnrA, qnrB | IS26 | 48 | |
| ST16(97) | HZ2 | northwest | 1193 | TEM-1 | ISEcp1 | 48 | |
| ST16(97) | QH11 | northwest | 1193 | TEM-1 | IS26 | 48 | |
| ST16(97) | QR29 | northwest | 1193 | TEM-1, ARM | IS26 | 45 | |
| ST16(97) | JL13 | northeast | 648 | TEM-1, aac(3′)(3′)-II | ISEcp1 | 48 | |
| ST16(97) | JL38 | northeast | 1193 | — | ISEcp1 | 48 | |
| NT | SJ13 | northeast | 167 | aac(3′)-II | IS26 | 48 | |
| NT | SJ22 | northeast | 38 | TEM-1,aac(3′)-II | ISEcp1 | 48 | |
| IncF | F18:A-:B1 (145) | Z1-26 | east | 156 | aac(3′)-II | ISEcp1 | 48 |
| F18:A-:B1 (145) | ZS13 | east | 617 | aac(3′)-II | ISEcp1 | 48 | |
| FII,FIA,FIB (NT) | HN34 | south | 405 | aac(3′)-II, qnrA,OXA-1,ARM | ISEcp1 | 48 | |
| F-:A1:B1 | HB24 | north | 1193 | qnrA | ISEcp1 | 45 | |
| FII,FIA(NT) | ZR36 | north | 46 | qnrA | ISEcp1 | 48 | |
| F-:A-:B20 | 1660 | southwest | 131 | aac(3′)-II | ISEcp1 | 48 | |
| F18:A-:B1 (145) | SC19 | southwest | 354 | TEM-1 | ISEcp1 | 48 | |
| F18:A-:B1 (145) | SC34 | southwest | 156 | aac(3′)-II, | ISEcp1 | 48 | |
| FII,FIA(NT) | JL25 | northeast | 405 | TEM-1, aac(3′)-II, | IS26 | 45 | |
| F51:A-:B10 | SJ38 | northeast | 1193 | — | ISEcp1 | 48 | |
| IncP | NX45 | northwest | 95 | TEM-1, aac(3′)-II, IMP | ISEcp1 | 48 |
aPlasmids were identified in transconjugants. The allele number of the IncI1 pMLST profile (repI, ardA, trbA, sogS and pill) is indicated. NT, not typed. aPlasmids were identified in transconjugants. Plasmid subtypes were classified according to their plasmidic MLST (pMLST) for IncI1 plasmids or to their FAB (FII, FIA, and FIB) formulas for IncF plasmids.
bIntergenic spacer region between insertion fragments and blaCTX-M-55.
cNT, not typed.
dND, not detected.
The replicon types of IncF plasmids were FIB (n = 1), FII (n = 1), FIA + FIB (n = 1), FIA + FII (n = 2), FIB + FII (n = 2), FIA + FIB + FI (n = 1) and FIB + FIC + FI (n = 2). IncI1 plasmids were classified into five STs according to the database of plasmid Multi-Locus Sequence Typing (http://pubmlst.org/plasmid/), with IncI1/ST16 the predominant subtype (23/34, 68%), with size of 97 kb. The remainder were ST15 and ST23 (one isolate each), ST136 (n = 2), and 7 were untypable. Of the seven untypable plasmids, a novel combination of alleles (repI1, 1; ardA, 5; trbA, 10; sogS, 8; pilL, 1) was detected in five plasmids, and another novel combination (repI1, 1; ardA, 2; trbA, 8; sogS,4; pilL, 10) was detected in one plasmid, while three alleles of the remaining one plasmid could not be typed. Further typing of the lncF plasmids (10/45, 22%) revealed a variety of subgroups: F18:A-:B1(n = 4,145 kb); F-:A1:B1(n = 1); F-:A-:B20(n = 1); F51:A-:B10(n = 1); and three were untypable.
Characterization of the antibiotic resistance genes and genetic environment of blaCTX-M-55 revealed a variable genetic context of the IncI1 plasmid. Except for six IncI1 plasmids which contained only blaCTX-M-55 gene, the remaining plasmids (n = 17) contained at least two other antibiotic resistance genes. The blaTEM-1 gene and aac (3′)-II gene were detected in 65% and 44% of IncI1 plasmids, respectively(Table 1). Other antibiotic resistance genes were variably found (data not shown) while the blaSHV gene was not detected.
PCR identified the insertion sequence ISEcp1 or IS26 in its entirety or partially truncated upstream of the blaCTX-M-55 gene in 32 of 45 (71%) transconjugants. Nucleotide sequence analysis of the upstream region of blaCTX-M-55 genes revealed that the spacer region between the right inverted repeat (IRR) and the start codon of blaCTX-M-55 genes were 45 bp or 48 bp.
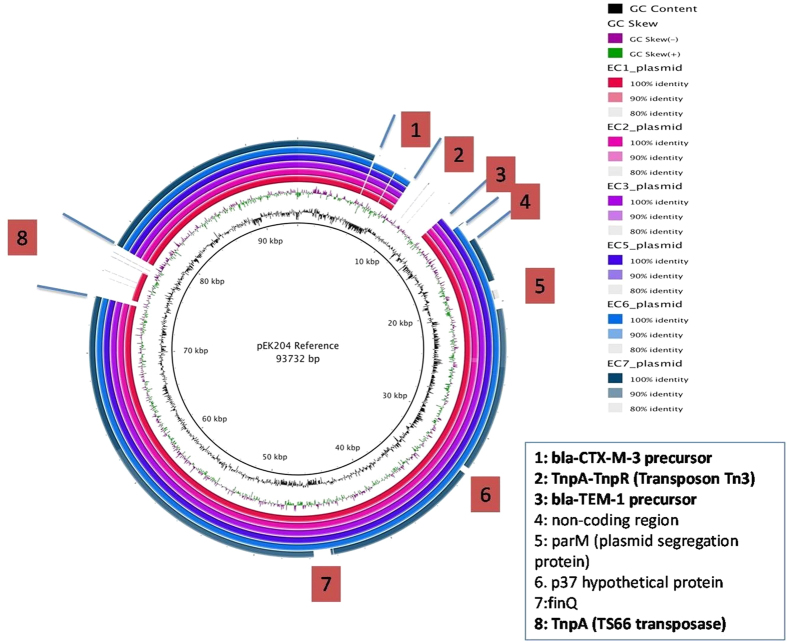
To understand if the IncI1/ST16 type plasmid was a possible epidemic plasmid, six of the IncI1/ST16 plasmids were selected randomly to determine their complete nucleotide sequences. The six plasmids consisted of a large backbone with considerable homology to the pEK204 plasmid isolated from E. coli in the UK, except for 8 regions missing in some of the plasmids compared to the reference. As shown in Fig. 1, BRIG output image of pseudo-plasmids compared to E. coli bla-CTX-M-15 plasmid pEK204, the innermost rings show GC content (black) and GC skew (purple/green). The remaining rings show BLAST comparisons of the 6 plasmids against the reference plasmid pEK204. The numbers 1–8 highlight the regions missing in some of the plasmids compared to the reference.
Figure 1. BRIG output image of pseudo-plasmids compared to E.coli blaCTX-15 plasmid pEK204.
Discussion
The emergence of ESBL-E.coli strains carrying blaCTX-M-55 gene across China poses a great threat to the health care systems, as these strains have been shown to cause UTIs more frequently than those carrying blaCTX-M-15 gene11. Our study provides some evidence that dissemination of the blaCTX-M-55 in China is partly attributed to the expansion of the E. coli ST1193 clone carrying the IncI1/ST16 plasmids. Twenty (18%) of 111 ESBL-EC strains possessing the blaCTX-M-55 gene and isolated from 24 hospitals among seven regions of China, belonged to a single sequence type (ST), ST1193 and a single PFGE cluster (cluster I). Furthermore, 88% (14 of 16) ST1193 isolates harbored the IncI1 plasmids of the same sequence type, ST16.
Incidentally, ST1193 was the most common E. coli ST isolated from both humans and dogs in Australia, and the isolates from both sources had similar serotypes, drug resistance and virulence genes, suggesting transmission between owners and dogs14. ST1193 was also prevalent in UTI isolates in Korea15. Furthermore, these 2 previous studies showed that ciprofloxacin resistance and lactose non-fermenting are two common phenotypic characteristics of the ST1193 E.coli.
Our present work reveals novel features of the ST1193 E.coli strains, including carriage of plasmids encoding drug-resistance genes such as tem-1, aac(3′) - II, qnrA and arm, in addition to the CTX-M-55 gene. We have also previously reported on ST1193 strains possessing the blaCTX-M-15, blaCTX-M-14 and blaCMY-2 genes11. Thus the multiple drug resistance due to presence of several antibiotic resistance determinants, might contribute significantly to the successful dissemination of the ST1193 E.coli in both the community and hospital environment. On the other hand, pandemic E. coli clones such as ST131 often encode multiple virulence gene products associated with mobile genetic elements (MGEs)16. It is worth investigating the whole genomic structure of ST1193 in future studies to explain its nationwide dissemination in China.
Besides ST1193, E. coli strains from several other STs were also found in about 4% of the isolates in the present study, including ST405(8%), ST156 and ST2003(4.5% each), ST12, ST95, ST167 and ST354(3.6% each). These findings suggest possible dissemination of these clones in China, though on a lower scale than ST1193. Unfortunately, due to cost limitation, we were not able to fully characterize and compare isolates in each of these STs, which would have given further insights. It is highly possible that in future, we will see an increase in the prevalence of these STs as they disseminate further in the community.
Interestingly, there were only three blaCTX-M-55 carrying ESBL-EC strains in our study which belonged to the international pandemic ST131. This is in sharp contrast to our previous study in 2014, which showed a high percentage of blaCTX-M-14 carrying UTI E. coli ST13111. Thus, it is likely that the STs of ESBL-ECs are greatly diversified, perhaps driven by multiple factors, including clinical antibiotic pressure, public health conditions in different regions, and food-borne animal origin.
Plasmids play a key role in horizontal transfer of ESBL genes among E. coli strains and thus can significantly contribute to the rise in antibiotic resistance in UTIs. Through typing of ESBL-carrying plasmids, we observed that 76% (34/45) were IncI1 type plasmids, 22% (10/45) Inc F plasmids and 2% (1/45) IncP plasmids, suggesting that these plasmids were contributors to antibiotic resistance and hence dissemination of these strains. IncI1 plasmids belong to narrow-host-range (NHR) type, and carry genes for synthesis of type IV pili, which mediate bacterial adhesion and invasion17. The IncI1 plasmids often carry a variety of β-lactamase antibiotic resistant genes such as blaCMY-2, blaCTX-M-1 and blaSHV-126,18. Our present findings also show that IncI1 plasmids can also carry resistance determining genes of other antibiotic classes, such as blaTEM-1, aac(3′)-II, aac-(6′)-Ib, qnrA and qnrB, which may contribute to their long-term existence in the intestinal flora of humans and animals, and thus become a reservoir for continual transmission of bacterial strains carrying these genes.
In order to understand the epidemiology of the IncI1 plasmids, Garcia and colleagues established an IncI1 plasmid MLST database (http://pubmlst.org/plasmid/), in which 158 subtypes have been registered. Among these subtypes, the blaCTX-M-1/IncI1/ST3, blaCTX-M-1/IncI1/ST7 and blaCMY-2/IncI1/ST2, are the frequently reported subtypes for ESBL-EC isolates of animal origin (such as poultry and pets) in Europe and America6,18,19. IncI1 plasmids carrying the blaCTX-M-55 were reported in ESBL-EC isolates of animal origin (dogs, chickens, ducks, etc.) from surveillance studies in Southern China, but unfortunately the MLST subtypes of these plasmids remain unknown13.
Contrary to these previous studies, the IncI1/ST16 subtype was the predominant subtype in the present study, constituting 68% of IncI1 carrying the blaCTX-M-55 gene.
We also identified another five IncI1/ST16 closely related plasmids that might be IncI1/ST16 variants due to one single allele gene mutation (pilL6 → 1), compared to the sequence of IncI1/ST16. The IncI1/ST16 subtype was detected first in the British bovine E. coli isolate (pEK204), and was subsequently reported in a French dog E. coli isolate18,20. Complete sequencing of the six IncI1/ST16 plasmids revealed that they consisted of a similar large backbone with considerable homology to the pEK204 plasmid, except for 8 regions missing in some of the plasmids compared to the reference, suggesting a possible similar origin.
Furthermore, five pairs of the IncI1/ST16 alleles in the present study(repI1, ardA, trbA, sogS and pill), are different with those of the IncI1/ST3 subtype, and four pairs of the IncI1/ST16 alleles (ardA, trbA, sogS and pill) are different to those of the IncI1/ST2 subtype, suggesting a lower genetic relationship between IncI1/ST16 and these two commonly described subtypes. Notably, the IncI2 (pHN1122–1) subtype plasmids that are commonly found in strains of animal origin in China and harbor blaCTX-M-55 were not detected in this study13. The IncI2 and IncI1 plasmids have an identical backbone gene encoding fimbriae that play an important role in engaging the plasmid and metastasis21. So far, the cause of high incidence of IncI1/ST16 carrying blaCTX-M-55 from the ESBL isolates remains unclear, and the relationship between IncI1/ST16 and other ESBL genotypes in clinical and animal-derived strains remains to be further studied.
Another significant plasmid subtype carrying the blaCTX-M-55 in our study was the IncF that accounted for 22%(10/45) of all blaCTX-M-55 carrying plasmids. IncF plasmids harboring blaCTX-M-15 are frequently detected among Enterobacteriaceae worldwide, and the acquisition of these plasmids is known to contribute to the dissemination of antimicrobial resistance and virulence genes22. There are currently 35 subtypes of IncF reported in plasmid MLST database. In the present study, we detected the IncF subtype F18: A-: B1, which is characterized by iron uptake (missing eitABCD), haemagglutinin and serum survival gene, and was firstly reported in the Avian Pathogenic E. coli (APEC)23. Other studies from Tunisia and China also reported high prevalence of the F18: A-: B1 subtype in E. coli strains of animal origin, and harbored the oqxAB or blaCTX-M-1 resistance genes24,25. These findings again suggest the possibility of horizontal transfer of the F18: A-: B1 plasmids between humans and animals.
An important feature of the ESBL-carrying plasmids is the existence of insertion fragments such as ISEcp1, IS26 and ISCR in these plasmids. For example, the ISEcp1 gene, often located at the 48 bp upstream of the blaCTX-M-55 gene, can be used to determine the homology of different CTX-M-ESBL genes26. Furthermore, the ISEcp1 gene was reported to be responsible for the mobility and expression of the blaCTX-M gene. Qu and colleagues reported that the insertion of ISEcp1 gene at 45 bp, 48 bp and 127 bp upstream of the blaCTX-M-55 gene, induces the expression of this gene in plasmids27. In this study, 94% of IncI1 plasmids contained the ISEcp1 or the IS26 insertion at 45 bp or 48 bp upstream of the blaCTX-M-55 gene, which suggests that acquisition of the blaCTX-M-55 gene by the Inc I1 plasmids might be mediated by the insertion fragments.
There are several limitations in our study. Firstly, only UTI ESBL-EC strains were analyzed; it is unclear whether the blaCTX-M-55 gene carrying ESBL-EC strains from other specimens such as blood belong to ST1193 and have IncI1 plasmids. Secondly, due to limited time and financial resources, a limited number (58 of 111, 52%) of isolates were used in conjugation studies and hence in plasmid analysis, which introduces some bias to our study. It’s arguable that the untested isolates would have yielded different results. Thirdly, to the best of our knowledge, this is the first report on the molecular epidemiology of the blaCTX-M-55-carrying major clone ST1193 and epidemic plasmid IncI1F/ST16 and F18: A-: B1. A whole genome analysis would greatly facilitate our understanding of the prevalence of the ST1193 and epidemic plasmid IncI1F/ST16 and F18: A-: B1 among the ESBL-EC strains. And finally, although blaCTX-M-55-carrying strains have been isolated from animal samples, there is insufficient evidence to conclude that there is dissemination of the blaCTX-M-55 gene between humans and animals.
In conclusion, the high prevalence of the blaCTX-M-55 gene among E. coli isolates from UTI patients was mainly attributed to the clonal dissemination of ST1193 strains, carrying the Inc I1/ST16 plasmids and mobile genetic element in China. This information can facilitate monitoring and controlling dissemination of the blaCTX-M-55 gene carrying strains so as to limit the rise in antibiotic resistance, which is a public health concern.
Materials and Methods
Ethics statement
This study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Guangzhou Medical University. All subjects signed written informed consent prior to the study. Patient information was anonymized and de-identified prior to analysis. All methods were carried out in accordance with relevant guidelines and regulations that are stated in the methods below.
Bacterial isolate collection
In our previous study, 137 out of 660 clinical ESBL-EC isolates from cases of UTI were shown to harbor blaCTX-M-55 gene. Of these137 isolates, only those that harbored a single blaCTX-M-55 gene (n = 111), were included in the current study. The isolates were collected from 21 general hospitals from 19 cities in China. These hospitals are distributed in seven geographic regions across China: four in Eastern China, two in Southern China, five in Northern China, two in Central China, two in Northeast China, two in Southwest, and four in Northwest. Clinical information of patients from whom the isolates were obtained, was reviewed retrospectively, including age, gender, underlying diseases, site and type of infection. All the bacterial isolates were re-identified using a VITEK2 Compact automated microbial identification system (bioMérieux, France). E. coli ATCC 25922 was used as the control strain.
Antimicrobial susceptibility
The minimum inhibitory concentrations (MICs) of common antibiotics (National Institutes for Food and Drug Control, Beijing, China) against ESBL-producing E. coli were determined using the broth microdilution method with E. coli ATCC 25922 as the control strain. Interpretation of results followed the criteria of CLSI 201328. The interpretation breakpoint of cefoperazone/Sulbactam (Pfizer Inc., USA) was extrapolated from that of cefoperazone, and those of tigecycline and colistin (Pfizer Inc.) were based on the criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST-2012).
Pulsed-field gel electrophoresis (PFGE)
PFGE analysis was conducted on the 111 isolates following a standardized protocol29, with electrophoresis performed using a CHEF DRIII System (Bio-Rad). The results were analysed by BioNumerics software (Applied Maths, St-Martens-Latern, Belgium) using the Dice similarity coefficient.
Multilocus Sequence Typing (MLST)
MLST was carried out as previously described30. Different sequences of a given locus were assigned an allele number based on E. coli MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). Sequence types (STs) were obtained via the electronic database at the above website. The allelic profiles and defined clonal complexes (CCs) of all STs were further analyzed with the Based upon Related Sequence Types (BURST) clustering algorithm (eburst.mlst.net).
Conjugation experiments and plasmid analysis
Conjugation experiments were performed on 16 of 20 ST1193 isolates, and a random selection (n = 42), of other isolates, making sure that at least one to three isolates from each hospital was included. Transferability of blaCTX-M-55 genes was determined by conjugation experiments using rifampin -resistant E. coli C600 as the recipient strain as described previously31. Transconjugants were selected on Mueller-Hinton agar plates containing rifampin (300 mg/L) and ceftazidime (2 mg/L). Incompatibility (Inc) groups, antibiotic resistance genes, and genetic environment were detected as described previously32,33. The sizes of plasmids of transconjugants were determined using S1-treated genomic DNA followed by PFGE (S1-PFGE). The epidemiological relatedness of IncI and IncF plasmids was determined by MLST and referred to the database (http://pubmlst.org/plasmid/)23,34.
Complete nucleotide sequencing of the epidemic plasmids
A random selection of the same ST Inc I1 plasmids (n = 6) was sequenced by next generation sequencing. Total DNA (includes genome and plasmid) from the transconjugants was sequenced using Illumina MiSeq 300 PE platform.The sequence reads were de novo assembled using CLC genomics workbench version 8.0 (Qiagen).
Additional Information
How to cite this article: Xia, L. et al. Prevalence of ST1193 clone and IncI1/ST16 plasmid in E-coli isolates carrying blaCTX-M-55 gene from urinary tract infections patients in China. Sci. Rep. 7, 44866; doi: 10.1038/srep44866 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Natural Science Foundation of China (No. 81271881) and The State Major Infectious Disease Research Program (China Central Government, 2012ZX10004–213). We thank Yang Liang (Nanyang Technological University, Singapore.) for his suggestion and revision for this study and all the members of National surveillance of antimicrobial resistance program for supply of the isolates.
Footnotes
The authors declare no competing financial interests.
Author Contributions Chao Zhuo, Nan-shan Zhong, Guo-sheng Ren, designed the study; Liang Xia, Yang Liu, Shu Xia, Shu-nian Xiao steered literature search, data collection and analysis; Liang Xia drafted the manuscript; Chao Zhuo, Nan-shan Zhong, Guo-sheng Ren reviewed and approved the submission of the manuscript. Timothy Kudinha reviewed and revised the manuscript.
References
- Kucheria R., Dasgupta P., Sacks S. H., Khan M. S. & Sheerin N. S. Urinary tract infections: new insights into a common problem. Postgrad Med J. 81, 83–86 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna-Flores F. et al. Molecular epidemiology of Escherichia coli O25b-ST131 isolates causing community-acquired UTIs in Mexico. Diagn Microbiol Infect Dis. 76, 396–398 (2013). [DOI] [PubMed] [Google Scholar]
- Doi Y. et al. Community-associated extended-spectrum β-lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis. 56, 641–648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol. 301, 654–658 (2011). [DOI] [PubMed] [Google Scholar]
- Naseer U. et al. Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS. 117, 526–536 (2009). [DOI] [PubMed] [Google Scholar]
- Accogli M. et al. IncI1 plasmids associated with the spread of CMY-2, CTX-M-1 and SHV-12 in Escherichia coli of animal and human origin. Clin Microbiol Infect. 19, E238–240 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis. 14, 659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. Antimicrobial Resistance and Molecular Epidemiology of Escherichia coli Causing Bloodstream Infections in Three Hospitals in Shanghai, China. PLoS one. 11(1), e0147740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanawong A., M’Zali F. H., Heritage J., Xiong J. H. & Hawkey P. M. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People’s Republic of China. Antimicrob Agents Chemother. 46, 630–637 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. et al. Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J Clin Microbiol. 49, 2496–2501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S. et al. Dominance of CTX-M-Type Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli Isolated from Patients with Community-Onset and Hospital-Onset Infection in China. PLoS One 9, e100707 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L. et al. Genetic characterization of IncI2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China. Antimicrob Agents Chemother. 57, 2824–2827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo C. et al. Study on CTX-M type ESBLs-producing Escherichia coli and Klebsiella pneumoiae in Guangzhou. Chin J Lab Med. 32, 1114–1119 (2009). [Google Scholar]
- Platell J. L. et al. Prominence of an O75 clonal group (clonal complex 14) among non-ST131 fluoroquinolone-resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrob Agents Chemother. 56, 3898–3904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. et al. Prevalence and characteristics of lactose non-fermenting Escherichia coli in urinary isolates. J Infect Chemother. 20, 738–740 (2014). [DOI] [PubMed] [Google Scholar]
- Petty N. K. et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci USA 111, 5694–5699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M. S. & Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 47, 565–596 (1993). [DOI] [PubMed] [Google Scholar]
- Haenni M., Saras E., Métayer V., Médaille C. & Madec J. Y. High prevalence of blaCTX-M-1/IncI1/ST3 and blaCMY-2/IncI1/ST2 plasmids in healthy urban dogs in France. Antimicrob Agents Chemother. 58, 5358–5362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverstein-van Hall M. A. et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 17, 873–880 (2011). [DOI] [PubMed] [Google Scholar]
- Woodford N. et al. Complete nucheotide sequences of Plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the internationgal O25:H4-ST131 clone. Antimicrob Agents Chemother. 53, 4472–4482 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K and Z. J Gen Microbiol. 130, 1489–1502 (1984). [DOI] [PubMed] [Google Scholar]
- Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L., Garcia-Fernandez A., Fortini D. & Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 65, 2518–2529 (2010). [DOI] [PubMed] [Google Scholar]
- Ben Sallem R. et al. IncI1 plasmids carrying bla(CTX-M-1) or bla(CMY-2) genes in Escherichia coli from healthy humans and animals in Tunisia. Microb Drug Resist. 20, 495–500 (2014). [DOI] [PubMed] [Google Scholar]
- Liu B. T. et al. Dissemination and characterization of plasmids carrying oqxAB-blaCTXM genes in Escherichia coli isolates from food-producing animals. PLoS One 8, e73947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madec J. Y. et al. Non-ST131 Escherichia coli from cattle harbouring human-like bla(CTX-M-15)-carrying plasmids. J Antimicrob Chemother. 67, 578–581 (2012). [DOI] [PubMed] [Google Scholar]
- Qu F. et al. Plasmid-encoding extended-spectrum β-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future Microbiol. 9, 1143–1150 (2014). [DOI] [PubMed] [Google Scholar]
- Wayne P. A. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: Twenty-Third Informational Supplement M100-S23 [J]. CLSI USA (2013).
- Gautom R. K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coliO157:H7 and other Gram-negative organisms in 1 day. J Clin Microbiol. 35, 2977–2980 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T. et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 60, 1136–1151 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. et al. High prevalence of blaCTX-M extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin Microbiol Infect. 16, 1475–1481 (2010). [DOI] [PubMed] [Google Scholar]
- Carattoli A. et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 63, 219–228 (2005). [DOI] [PubMed] [Google Scholar]
- Zong Z., Partridge S. R., Thomas L. & Iredell J. R. Dominance of blaCTX-M within an Australian extended-spectrum beta-lactamase gene pool. Antimicrob Agents Chemother. 52, 4198–4202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Fernández A. et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 61, 1229–1233 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.