Abstract
Shotgun (gel-free) proteomics is a useful approach to perform identification and relative quantification of protein in complex mixtures such as tissue homogenates, biological fluids, cell lysates, and extracellular proteins. Incorporation of separative and analytical techniques such as two-dimensional liquid chromatography at nanoscale (2D-nanoLC) coupled to tandem mass spectrometry (MS/MS analysis) into the shotgun protocol provides an excellent strategy. This chapter describes the application of the shotgun proteomics protocol to evaluate the identity and expression analysis of proteins from rat uterus after estrogen (ethinylestradiol) treatment. The steps of the protocol involve sample preparation (digestion), 2D-nanoLC-MS/MS analysis, and shotgun proteomics analysis including bioinformatics tools for data conversion, organization, and interpretation.
Keywords: Proteomics, “Shotgun” proteomics, Liquid chromatography, Mass spectrometry, Proteins, Peptides, Rat, Uterus, Estrogen
1 Introduction
The effects of ovarian hormones on the uterus of rats have been well studied. Actions mediated by estrogens contribute to the adult reproductive life through the cycles of proliferation and degeneration of the uterine tissue [1–3, 16]. The large-scale identification of the proteins that are involved directly and/or indirectly with estrogen actions is a useful tool that allows understanding the complexity of such hormonal responses [4]. One of the best approaches to resolve the proteins in complex mixtures present in tissues such as the uterus is the use of “shotgun” proteomics protocols, also known as “gel-free” proteomics [4, 5]. This strategy permits the investigator to perform protein identification, characterization, and relative quantification from tissue homogenates, cell lysates, biological fluids, and similar sample types [4–7]. The technique uses a bottom-up strategy in which the proteins are digested using a protease, such as trypsin, that recognizes specific cleavage sites. The tryptic digest peptides that are obtained are separated and analyzed using tandem mass spectrometry, and used to identify the most probable protein/s present in the sample through a combination of specific bioinformatics tools and protein databases [8].
The combination of protein digestion with trypsin in solution, separation of the peptides obtained through two-dimensional liquid chromatography at nanoscale (2D-nanoLC) [9], and analysis of mass by tandem mass spectrometry (MS and MS/MS analysis) allows improvement of the shotgun proteomics protocol by increasing the number of proteins identified from complex mixtures, extends the molecular weight (MW) range, covers the entire range of isoelectric points (pI) from pH 0 to 14, and resolves the protein dynamic range to 105. Moreover, it can provide an approximation of the protein relative abundance when it is combined with protocols such as “label free” technology through spectral counting strategy [10, 11]. Also, the gel-free method can be used as a complementary approach to two-dimensional gel electrophoresis (2DGE). An example of the application of shotgun proteomics for the analysis of proteins obtained from uterine tissue of rats ovariectomized with and without treatment with estrogen (ethinylestradiol) is provided in this chapter. The proteins extracted from the uterus were digested in solution using trypsin and the peptides analyzed by a 2D-nanoLC-MS/MS protocol through a 2D-nanoAcquity UPLC with dilution coupled to a quadrupole time-of-flight (Q-Tof) Synapt Gl MS (Waters) [6, 7]. The peptide and fragment masses were submitted against the Swiss Protein database with taxonomy filter for Rattus norvcgicus database using a combination of bioinformatics tools that included MassLynx v4.1, ProteinLynx Global Server v3.0 (Waters, Milford, MA) and Mascot +Mascot Daemon toolbox v2.5 local license (www.matrix-science.com). The post-mass spectrometry analysis to organize the proteins into different sets and the relative abundance data were performed using a ProteoIQ software v2.7 (www.premierbiosoft.com). A brief summary of the entire protocol is shown in the schematic flow chart of Fig. 1. From a total number of 800 proteins identified, 498 were expressed in common between the control and estrogen-treated groups, 182 proteins were exclusively present in the control, and 162 proteins in the estrogen-treated group (Fig. 2). The 20 most differentially expressed proteins observed, either up- or downregulated by estrogen, have been previously reported in the literature [12–15].
Fig. 1.
Schematic representation of the “shotgun” proteomics protocol applied to the sample analysis through the 2D-nanoLC-MS/MS analysis (1st D.=first dimension, 2nd D. = second dimension)
Fig. 2.
Venn diagram representing the number of unique proteins identified in the uterus from vehicle control-treated and ethinylestradiol-treated rats, and those proteins common to both groups. The numbers in each Venn diagram and/or intersections correspond to the proteins analyzed by 2D-nanoLC-MS/MS analysis
2 Materials
Prepare all solutions using ultrapure water (purified deionized water with a resistivity of 18 MΩ cm at 25 °C), and LC-MS-grade reagents.
Extract protein samples from animals or cells that had been treated with vehicle or with estrogen (see Note 1).
Reaction buffer stock solution: 250 mM ammonium bicarbonate. Mix 98.0 mL water and 1.977 g ammonium bicarbonate. Adjust the pH with 0.1 M ammonium hydroxide to pH 8.0. Raise the volume to 100 mL.
Reaction buffer working solution (AB): 100 mM ammonium bicarbonate, 5 % acetonitrile.
Reducing agent: 50 mM dithiothreitol (DTT). Add 7.7 mg DTT to 1 mL water. Store in aliquots at −20 °C until use.
Alkylating agent: 100 mM iodoacetamide. Add 18.5 mg of iodoacetamide to 1 mL water (see Note 2).
Trypsin resuspension buffer: 50 mM acetic acid.
Sequencing-grade modified trypsin: 1 µg/µL trypsin. Dissolve 20 µg trypsin in 20 µL trypsin resuspension buffer. Keep on ice until use (see Note 3).
Sample injection and mobile-phase buffer stock solution: 200 mM ammonium formate, pH 10. Place 450 mL of purified water in a 500 mL beaker. Using a pipette, add 6.95 mL 28 % ammonium hydroxide and mix well. Carefully add 0.81 mL formic acid and mix well. Adjust the solution to pH 10 using ammonium hydroxide or formic acid in water. Add water to bring the volume to 500 mL (see Note 4).
Sample injection buffer working solution: 100 mM ammonium formate, pH 10. Mix one volume of 200 mM ammonium formate, pH 10, with one volume of purified water.
First-dimension mobile phases for liquid chromatography, solution Al: 20 mM ammonium formate, pH 10. Prepare 400 mL solution A by mixing 360 mL of purified water with 40 mL 200 mM ammonium formate, pH 10. Degas the solvent with an ultrasonic liquid processor (sonicator) for 5 min or with Argon gas.
First-dimension mobile phases for liquid chromatography, solution B1: 100 % acetonitrile. Degas the solvent with a sonicator for 5 min or with Argon gas.
Second-dimension mobile phases for liquid chromatography, solution A2: 0.1 % formic acid in water. Add 400 µL formic acid to 400 mL water. Degas the solution for 5 min in a sonicator.
Second-dimension mobile phases for liquid chromatography, solution B2: Acetonitrile with 0.1 % formic acid. Add 400 µL formic acid to 400 mL acetonitrile. Degas the solution for 5 min in a sonicator.
First-dimension column 1: XBridge BEH130 C18, 5 µm, 300 µm × 50 mm NanoEase reversed phase column (Waters).
First-dimension column 2 for trapping and desalting online: 180 µm × 20 mm, 5 µm Symmetry C18 nanoAcquity UPLC reversed phase trap column (Waters).
Second-dimension column: BEH130C18 1.7 µm, 100 µm × 100 mm nanoAcquity UPLC reversed phase column (Waters).
Liquid chromatography instrumentation: 2D-nanoAcquity Ultra Performance Liquid Chromatography (UPLC) with dilution (Waters).
Mass spectrometry and liquid chromatography standards for MS calibration and LC analysis: Human [Glu1]-fibrinopeptide B human; phosphorylase B (rabbit), alcohol dehydrogenase (ADH, Saccharomyces cerevisiae), enolase 1 (Saccharomyces cerevisiae), and bovine serum albumin (BSA, Bos taurus) (see Note 5).
Mass spectrometry (MS) instrumentation: Q-Tof Synapt Gl MS with nano electrospray ionization and lock mass (Waters) (see Note 6).
Bioinformatics tools: MassLynx v4.1 (LC-MS control and operation, MS, and MS/MS data acquisition) (Waters) (see Note 7).
Bioinformatics tools: ProteinLynx Global Server v3.0 (PLGS 3.0) (Waters) (raw file processing, centering, smoothing, and de-isotoping, and generation of list of masses: peak list files = PKL files (see Note 7)).
Bioinformatics tools: Mascot server v2.5 and Mascot Daemon toolbox v2.4 (data searching against databases) in MS/MS Ion Search Mode local license (www.matrixscience.com).
Bioinformatics tools: ProteoIQ v2.7 software (www.premier-biosoft.com) (post-mass spectrometry data analysis).
Digital dry bath incubator.
Refrigerated microcentrifuge.
Centrifugal vacuum concentrator such as the SPIOIO SpeedVac (Thermo) with variable range of vacuum.
Sonicator.
3 Methods
Perform all procedures at room temperature unless otherwise specified.
3.1 Sample Preparation and Protein Digestion
For lyophilized proteins, dissolve the samples in reaction buffer (100 mM ammonium bicarbonate/5 % acetonitrile) to obtain a protein solution of 1 µg/µL. If the proteins are already in solution in another buffer (such as 20 mM Tris—HCl, pH 7.4, 2 mM EDTA, 5 mM EGTA, 0.25 M sucrose, 50 mM β-mercaptoethanol and 10 µg/µL phenylmethylsulfonyl fluoride), dilute the sample by addition of reaction buffer to reach a final concentration of 100 mM ammonium bicarbonate/5 % acetonitrile as well as 1 µg/µL protein. Titrate the pH of the solution to 8.0 and place on ice until the next step (see Notes 8 and 9).
Perform the reduction of the sample in solution by adding 1/10 (v/v) of 50 mM DTT, and incubate at 65 °C for 5 min (see Note 10).
Cool, centrifuge (10,000 × g for 10 s), and return the sample to ice.
Place the sample in a rack at room temperature and add 1/10 (v/v) 100 mM iodoacetamide. Incubate at room temperature for 30 min and protect the samples from direct exposure to light with aluminum foil (see Note 11).
Place the sample on ice for at least 20 min.
Add 1 µg/µL trypsin solution 1:40 ratio of µg trypsin:µg of sample protein (see Note 12).
Incubate at 37 °C overnight or at least 15 h.
Centrifuge the sample (10,000 × g for 10 s).
Add 0.5 % acetic acid and titrate to pH 2.0 to halt the digestion (a pH tape can be used to follow the change in pH).
Place the tube on dry ice and concentrate in SpeedVac until almost dry. SpeedVac conditions: temperature: Off, vacuum: 24 Torr/min, vacuum ramp: level 4, time: 35 min, auto run mode (see Note 13) (Fig. 3).
Fig. 3.
Schematic representation of the in-solution digestion protocol for preparation of digested peptide sample for “shotgun” proteomics analysis
3.2 Peptide Analysis by 2D-nanoLC and Tandem Mass Spectrometry
Dissolve the tryptic-digested peptides obtained from the digestion in Subheading 3.1 above in sample injection buffer (100 mM ammonium formate pH = 10) to a final concentration of 1 µg/µL of digested peptide.
Place the peptides in a total recovery vial and inject 1 µg of digested peptide into the 2D-nanoLC system using the autosampler (see Notes 14–16).
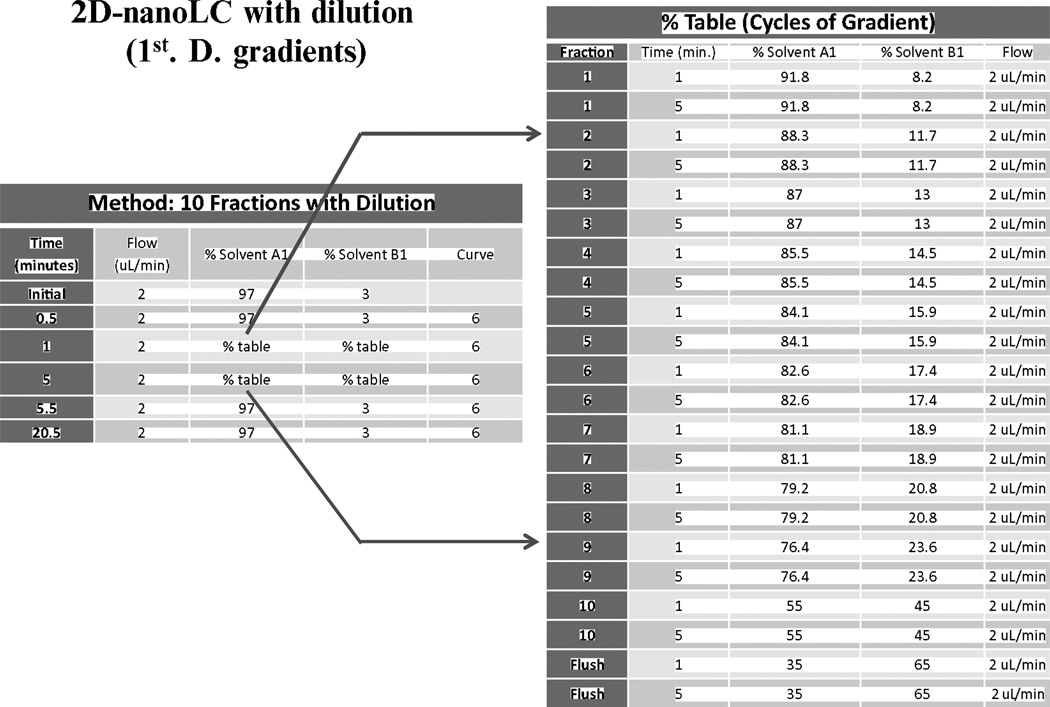
Set up the conditions for the 2D-nanoLC-MS/MS using a 10 fraction + flushing protocol. Perform the first dimension on an XBridge BEH130 C18, 5 µm, 300 µm × 50 mm NanoEase Column using solvents A1 and B1. Set the flow in the first dimension at 2 µL/min and program 11 step gradients (dilution method) for 20 min each.
Include a second column for trapping and desalting online. Set the flow at 20 µL/min through the 180 µm × 20 mm, 5 µm Symmetry C18 nanoAcquity UPLC trap column. Use column buffers 99 % A2 to 1 % B2 for 20 min.
After the peptides are desalted and concentrated, separate them online in the second dimension through a BEH130C18 1.7 µm, 100 µm × 100 mm nanoAcquity UPLC column using a flow rate of 400 nL/min for 60 min through the following gradient program: begin with the standard solvent gradient of 0–2 min, 3 % B2 isocratic, 2–40 min, 3–85 % B2 linear, and return to the original gradient of 3 % B2 until the 60-min separation is complete. For specific information on the plumbing and gradients corresponding to each dimension, see Fig. 4, Tables 1 and 2, respectively.
Acquire the data coming from each fraction with MassLynx v4.1 program through the data depend acquisition (DDA) mode and uses the following parameters: MS and MS/MS range = 400–2000 and 50–2000 m/z, respectively, with a time scanning of 1 s and 0.02 s for inter-scan delay, total acquisition time = 59 min (see Notes 7 and 17).
Fig. 4.
Schematic representation of the plumbing for 2D with dilution system configuration. An Xbridge BEH130, 5 µm, 300 µm × 50 mm NanoEase RP (pH = 10) column was used as the first dimension. A180 µm × 20 mm, 5 µm Symmetry nanoAcquity UPLC and BEH130, 5 µm, 100 µm × 100 mm nanoAcquity UPLC (pH = 2) columns were used in the second dimension for the trapping and desalting and analytical, respectively (©Waters Corporation. Used with permission)
Table 1.
First-dimension separation gradient setup using a protocol for 10 fractions+flushing
Solvent A1 and B1 are 20 nM ammonium formate and acetonitrile 0.1 %, respectively. The gradient step in each fraction was run for 5 min (©Waters Corporation. Used with permission)
Table 2.
Second-dimension separation gradient setup using a protocol for 10 fractions+flushing
| Time (min) | Flow (nl/min) | %A | %B | Curve |
|---|---|---|---|---|
| 0 | 400 | 99 | 1 | 6 |
| 1 | 400 | 99 | 1 | 6 |
| 30 | 400 | 50 | 50 | 6 |
| 31 | 400 | 15 | 85 | 6 |
| 35 | 400 | 15 | 85 | 6 |
| 36 | 400 | 99 | 1 | 6 |
Solvent A2 and B2 are Water/0.1 % formic acid and acetonitrile 0.1 % formic acid, respectively. The gradient step in each fraction is similar in the second dimension, and the standard solvent gradient used is 0–2 min, 3 % B2 isocratic; 2–40 min, 3–85 % B2 linear, at a flow rate of 400 nL/min for 60 min run for 5 min (©Waters Corporation. Used with permission)
3.3 Bioinformatics Analysis of Data
Transfer the raw files generated from each fraction onto ProteinLynx Global Server v3.0 (PLGS 3.0) and convert the spectrum into a list of masses (peak list or PKL files). Use the same processing parameters as the mass spectrometer conditions for the data acquisition (de-isotoping, background correction, centering and smoothing) (see Notes 7, 18 and 19).
Move the PKL files from PLGS 3.0 to a Mascot Daemon toolbox (v2.5) to perform a search against NCBInr and Swiss Protein databases using a taxonomy filter for Rattus norvegicus.
Browse all of the PKLs corresponding to each fraction from the sample into Mascot Daemon toolbox v2.5 using the following parameter set up for the search: (a) MS/MS ion search mode, (b) enzyme = trypsin, (c) missed cleavage = 1, (d) fixed modifications = carbamidomethyl (C), (e) variable modifications = deamidation (N and Q), and oxidation (M), (f) peptide and MS/MS tolerance = 50 ppm and 0.3 Da, respectively, (g) monoisotopic, (h) data format = micromass (.PKL), (i) instrument =ESI-QUAD-ToF, and error-tolerant mode. Before submitting the search, select “merge” to combine the partial data into one unique report of the most probable proteins identified in the sample (see Note 20).
Export the data obtained from Mascot (Fig. 5) as Mascot.dat file format to ProteoIQ software to perform a post-mass spectrometry analysis of the proteins identified as well as their relative abundance (see Notes 21 and 22) (Figs. 2 and 6).
Fig. 5.
MS/MS fragmentation map of GLGTDEDSILNLLTAR found in Annexin A5 [Rattus norvegicus]. Peptide mass 1686.8818, and charge state = +2
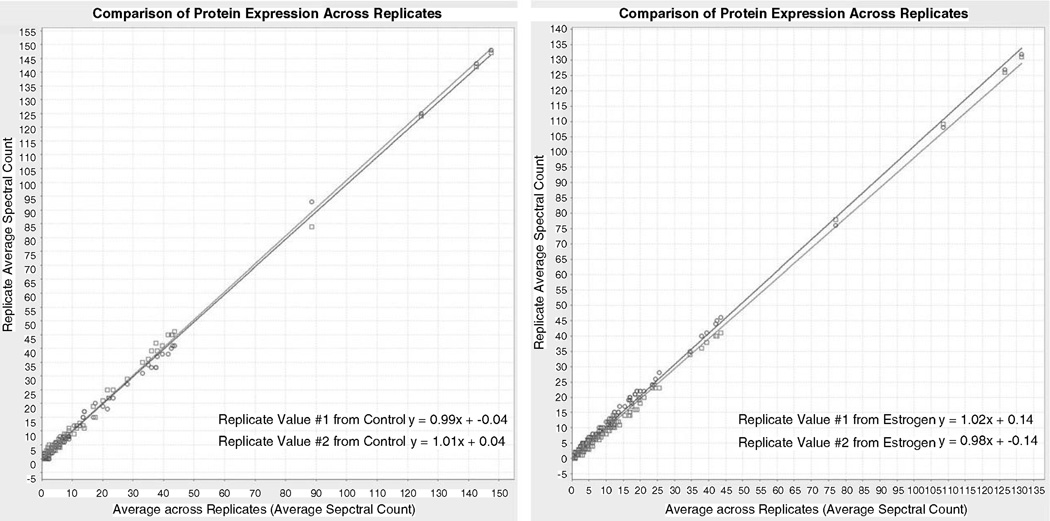
Fig. 6.
Replicate scatterplot analysis showing the comparison of protein expression across sample control and estrogen replicates through the average of spectral count. “y”- and “x”-axes represent the replicate average spectral count, and the average across replicates (average spectral count), respectively
Acknowledgments
This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103443. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health. The author would like to thank Dr. Kathleen Eyster for her generously providing the samples, and Dr. Barbara Goodman for her critical reading of the chapter. All sample analysis was performed in the SD BRIN Proteomics Core Facility. Mr. Douglas Jennewein, through the ITS Research Computing staff, contributed valuable technical expertise to this project.
References
- 1.Groothuis PG, Dassen HH, Romano A, Punyadee RAC. Estrogen and the endometrium: lessons learned from gene expression profiling in rodents and human. Hum Reprod Update. 2007;13(4):405–417. doi: 10.1093/humupd/dmm009. [DOI] [PubMed] [Google Scholar]
- 2.Blake CA, Brown LM, Duncan MW, Hunsucker SW, Helmike SM. Estrogen regulation of the rat anterior pitutitary gland proteome. Exp Biol Med (Maywood) 2005;230(11):800–807. doi: 10.1177/153537020523001104. [DOI] [PubMed] [Google Scholar]
- 3.Rhen T, Grissom S, Afshari C, Cidlowski JA. Dexamethasone blocks the rapid biological effects of 17beta-estradiol in the rat uterus without antagonizing its global genomic actions. FASEB J. 2003;17(13):1849–1870. doi: 10.1096/fj.02-1099com. [DOI] [PubMed] [Google Scholar]
- 4.Mayne J, Starr AE, Ning Z, Chen R, Chiang CR, Figeys D. Fine tuning of proteomics technologies to improve biological findings: advancements in 2011–2013. Anal Chem. 2014;86:176–195. doi: 10.1021/ac403551f. [DOI] [PubMed] [Google Scholar]
- 5.Prokai L, Stevens SM, Rauniyar N, Nguyen V. Rapid label-free identification of estrogen-induced differential protein expression in vivo from mouse brain and uterine tissue. J Proteome Res. 2009;8(8):3862–3871. doi: 10.1021/pr900083v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong G, Callegari EA, Gloeckner CJ, Ueffing M, Wang H. Prothymosin-alpha interacts with mutant huntingtin and suppresses its cytotoxicity in cell culture. J Biol Chem. 2012;287(2):1279–1289. doi: 10.1074/jbc.M111.294280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong G, Callegari EA, Gloeckner CJ, Ueffing M, Wang H. Mass spectrometric identification of novel posttranslational modification sites in Huntingtin. Proteomics. 2012;12(12):2060–2064. doi: 10.1002/pmic.201100380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen GY, Chen SH, Huang SY, Tsai ML. Trypsin digest coupled with two-dimensional shotgun proteomics reveals the involvement of multiple signaling pathways in functional remodeling of late-gestation uteri in rats. Proteomics. 2008;8:3173–3184. doi: 10.1002/pmic.200701040. [DOI] [PubMed] [Google Scholar]
- 9.Gilar M, Olivora P, Daly AE, Gebler JC. Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J Sep Sci. 2005;28:1964–2703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- 10.Resaul K, Thumar JK, Lundgren DH, et al. Differential protein expression profiles in estrogen receptor-positive and –negative breast cancer tissues using label-free quantitative proteomics. Genes Cancer. 2010;1(3):251–271. doi: 10.1177/1947601910365896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nahnsen S, Bielow C, Reinert K, Kohlbacher O. Tools for label-free peptide quantification. Mol Cell Proteomics. 2013;12(3):549–556. doi: 10.1074/mcp.R112.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilf R, McDonald E, Sartini J, Rector WD, Richards AH. Response of uterine glucose-6-phosphate dehydrogenase isoenzymes to estrogen. Endocrinology. 1972;91(1):280–286. doi: 10.1210/endo-91-1-280. [DOI] [PubMed] [Google Scholar]
- 13.Thomas T, Macpherson A, Rogers P. Ceruloplasmin gene expression in the rat uterus. Biochim Biophys Acta. 1995;1261(1):77–82. doi: 10.1016/0167-4781(94)00224-q. [DOI] [PubMed] [Google Scholar]
- 14.Katayama ML, Federico MH, Brentani RR, Brentani MM. Eosinophil accumulation in rat uterus following estradiol administration is modulated by laminin and its integrin receptors. Cell Adhes Commun. 1998;5(5):409–424. doi: 10.3109/15419069809010785. [DOI] [PubMed] [Google Scholar]
- 15.Boverhof DR, Burgoon LD, Williams KJ, Zacharewski TR. Inhibition of estrogen-mediated uterine gene expression responses by dioxin. Mol Pharmacol. 2008;73:82–93. doi: 10.1124/mol.107.040451. [DOI] [PubMed] [Google Scholar]
- 16.Montoya TI, Maldonado PA, Acevedo JF, Word RA. Effect of vaginal or systemic estrogen on dynamics of collagen assembly in the rat vaginal wall. Biol Reprod. 2015;92(2):43. doi: 10.1095/biolreprod.114.118638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turini P, Kurooka S, Steer M, Corbascio N, Singer TP. The action of penylmethylsulfonyl fluoride on human acetylcholinesterase, chymotrypsin and trypsin. J Pharmacol Exp Ther. 1969;167(1):98–104. [PubMed] [Google Scholar]
- 18.Neff PA, Serr A, Wunderlich BK, Bausch AR. Label-free electrical determination of trypsin activity by a silicon-on-insulator based thin film resistor. Chemphyschem. 2007;8(14):2133–2137. doi: 10.1002/cphc.200700279. [DOI] [PubMed] [Google Scholar]
- 19.Willard BB, Kinter M. Effects of the position of the internal histidine residues on the collision-induced fragmentation of triply protonated tryptic peptides. J Am Mass Spectrom. 2001;12(12):1262–1271. doi: 10.1016/S1044-0305(01)00312-9. [DOI] [PubMed] [Google Scholar]
End Notes
- 1.The samples that were used in this example are came from protein homogenates extracted from the uterus of vehicle-treated control and ethinylestradiol-treated female rats that had been ovariectomized 2 weeks prior to treatment. The proteins were originally in a buffer containing 20 mM Tris-HCl, pH 7.4, 2 mM EDTA, 5 mM EGTA, 0.25 M sucrose, 50 mM β-mercaptoethanol, and 10 µg/µL phenylmethylsulfonyl fluoride (PMSF). However, to avoid the interference that the β-mercaptoethanol and PMSF could cause to the trypsin activity, reaction buffer was added to the sample to adjust the concentrations of PMSF and β-mercaptoethanol prior to the digestion (see Notes 8 and 9).
- 2.The iodoacetamide solution should be covered with aluminum foil to protect it from light as it is light sensitive. The iodoacetamide must be prepared fresh.
- 3.The trypsin powder must be stored at −20 °C. In solution it should be stored at −80 °C. It can be frozen and thawed for no more than two cycles.
- 4.Work in a fume hood and add carefully to avoid inhaling potential formic acid released from the reaction between ammonium hydroxide and formic acid.
- 5.Human [Glu1]-fibrinopeptide B human is used for mass calibration in MS and MS/MS modes at the mass spectrometer. Phosphorylase B (rabbit), alcohol dehydrogenase (ADH, Saccharomyces cerevisiae), enolase 1 (Saccharomyces cerevisiae), and BSA (Bos taurus) digestion standards are used for quality control of the chromatography step (retention time, peak shapes and areas, and separation reproducibility prior the sample analysis), as well as to evaluate the sensitivity and mass accuracy of the mass spectrometer prior the analysis.
- 6.The MS used in this protocol is purchased from a specific vendor, but the procedures could be carried out on any Q-Tof mass spectrometer.
- 7.The tools MassLynx 4.1 and ProteinLynx 3.0 (PLGS 3.0), as well as the processing parameters used for the data acquisition and mass list extraction mentioned above are related to the vendor of the mass spectrometer; however, these tools can be used with other instruments.
- 8.The sample preparation step involves the resuspension or dilution of the proteins present in the samples, depending on whether they are lyophilized or in solution. The reaction buffer used for the protein digestion is ammonium bicarbonate + acetonitrile, which provides the proper environment for the protease action. If the proteins are lyophilized, the reaction buffer should be used directly to dissolve it. However, if the proteins are in solution with an original extraction buffer, a volume of reaction buffer stock solution should be added to reach a final concentration of 100 mM ammonium bicarbonate/5 % acetonitrile.
- 9.If the protein sample contains β-mercaptoethanol or protease inhibitors such as PMSF, they need to be diluted so they do not inhibit the ability of the protease (e.g., trypsin) to digest proteins. The current sample contained 10 µg/µL PMSF and 50 mM β-mercaptoethanol in order to inhibit endogenous serine protease activities. PMSF will not affect trypsin activity at a concentration of 0.17 µg/µL, so dilution of the current sample to this concentration with reaction buffer was necessary to avoid potential inhibition of the trypsin activity during protein digestion [17, 18].
- 10.The proteins in solution are reduced with dithiothreitol (DTT) to reduce the disulfide bonds between cysteines located in the loop of the molecule. This action allows the molecule to expand and exposes the cleavage sites to be recognized by the protease. In general, the DTT is prepared fresh; however, in the case of having many samples from different batches, DTT could be stored in aliquots at −20 °C, avoiding more than one cycle of thaw.
- 11.Immediately after the reduction, an alkylating agent such as iodoacetamide is added to block the sulfhydryl groups to prevent the bonds from reforming. This reagent must be protected from the action of light before and during the process of alkylation of the samples by covering the tubes with aluminum foil.
- 12.Trypsin is a universal protease utilized for protein digestion that recognizes cleavage sites at lysine (K) and arginine (R) when they are present in the protein, except when those amino acids are surrounded by an amino acid such as proline that can produce an impediment to trypsin recognition and cleavage. Hydrolysis mediated by trypsin converts the amine bonds of the C-terminus side of the R or K residues to a strong basic site, and moderates the N-terminus. The combination of those kinds of peptides and the acidic conditions used with the electrospray ionization (ESI) produces protonation at both C-termini, either at the side chain of K or R [19], thus becoming more effective positive ions.
- 13.Concentrate the samples to almost, but not completely dry, to make it easier to resuspend the peptides in the sample injection buffer.
- 14.The volume of tryptic-digested peptides to be injected is 1 µg/µL) in sample injection buffer as the initial conditions. If the initial analysis indicates that the peptide concentration is too low, a second run can be performed with a higher concentration of peptides.
- 15.The volume of the sample loop used at the injection valve is 5 µL and the partial loop method allows injection of volumes between 0.2 and 4.9 µL.
- 16.The 2D-nanoLC with dilution is a new technology that permits the performance of orthogonal separation using two reversed-phase columns working at high and low pH respectively. The tryptic peptide sample is solubilized in 100 mM ammonium formate buffer (AF) (sample injection buffer, pH 10, to improve the peptide bound to the stationary phase of the first column, and loaded into the first dimension column = pH 10) in which the unwanted solutes are washed and thus removed. Setting a gradient for the first dimension with 20 mM AF buffer and 100 % acetonitrile as mobile phases (solutions A1 and B1, respectively), the pH from the first-dimension column can be decreased, releasing peptides into the second-dimension column according to the pH of each peptide. The second-dimension column works as an analytical column, resolving the peptide mixtures coming from the first dimension column, and infusing them into the ESI of the mass spectrometer. A total number of 10 fractions + 3 flushing washes are set up for running each sample. Throughout the ten fractions, the gradient changes by increasing the percentage of the mobile phase corresponding to the organic solvent (acetonitrile) and reducing the percentage of buffer. The flushing permits cleaning the column for the next sample to be injected and avoids potential carryover of sample.
- 17.The data acquired for each fraction are saved in an individual raw data file. PKL is a format from Waters and consists of a base text file with specific syntax and order of the data (precursor mass, intensity, charge state and fragment masses) corresponding to each precursor or parent fragmented.
- 18.MassLynx v4.1, Mascot Distiller or similar programs can be used to extract the list of masses to be exported in different formats (PKL was used specifically in this case because it is the Waters format).
-
19.After finishing the data acquisition, the list of masses is extracted using the tool ProteinLynx Global Server v3.0 Expression Analysis and exported as a PKL format. The processing parameters applied during the extraction of the list of masses must match the conditions present in the mass spectrometer during the data acquisition. For the programs described herein, the following list describes the parameters that should be matched. Set the processing parameters as in the following example (also see
Note 7):
Mass accuracy Electrospray survey Attribute Value Perform lock spray calibration Yes Lock spray lock mass 785.8427 Da/e Lock spray scan 10 Lock mass tolerance 0.1 Da MSMS Attribute Value Perform lock spray calibration Yes Lock spray lock mass 785.8427 Da/e Lock spray scan 10 Lock mass tolerance 0.1 Da Noise reduction Electrospray survey Attribute Value Background subtraction type Normal Background threshold 35% Background polynomial 5 Perform smoothing Yes Smoothing type Savitzky-Golay Smoothing iterations 2 Smoothing window 3 channels MSMS Attribute Value Background subtraction type Normal Background threshold 35% Background polynomial 5 Perform smoothing Yes Smoothing type Savitzky-Golay Smoothing iterations 2 Smoothing window 3 channels Peptide filter De-isotoping and centroiding Electrospray survey Attribute Value Perform de-isotoping Yes De-isotoping type Fast Iterations 30 Automatic thresholds No Threshold 3% Minimum peak width 4 channels Centroid top 80% TOF resolution 10,000 NP multiplier 0.7 MSMS Attribute Value Perform de-isotoping Yes De-isotoping type Fast Iterations 30 Automatic thresholds No Threshold 3% Minimum peak width 4 channels Centroid top 80% TOF resolution 10,000 NP multiplier 0.7 - 20.All of the PKL files corresponding to the 10 fractions and flushes are imported into the Mascot Daemon toolbox v2.5 to perform searches against the databases. Each submission is searched as an individual event for each fraction coming from the sample, and all of the data are merged into one unique protein list. The explanation for each parameter set at Mascot server is the following: (a) MS/MS ion search mode = the data is acquired using tandem mass spectrometry (the mass of the precursor or parent peptides is determining followed by fragmentation of the peptide and the determination of the fragment masses (MS/MS analysis); (b) enzyme: trypsin: the proteins in the sample are digested with this particular protease; (c) missed cleavage = 1, means the probability that the enzyme fails in recognizing one site for cleavage of the proteins during the digestion; (d) fixed modifications: carbamidomethyl (C): the proteins are reduced and alkylated prior to digestion with the purpose of breaing the disulfide bridge between cysteines, making the recognition sites more available to the enzyme to perform cleavage (DTT and iodoacetamide are used during the reduction and alkylation step, respectively, generating a fixed modification transduced in the addition of a carbamidomethyl group modification at the cysteine group, which produced a shift of 57 Da at the total peptide mass); (e) variable modifications: deamidation (N and Q), and oxidation (M), are variable because they may formed after the reduction and alkylation of the proteins and their presence could produce the additions of mass (oxidation in M = 15.9949 Da, and deamidation in N or Q=0.9840 Da); (f) peptide and MS/MS tolerance = 50 ppm and 0.3 Da, respectively, represent the potential error permitted in the differences of masses between the experimental (at the mass spec) and theoretical from the database; (g) monoisotopic: this parameter indicate that the mass measured is related to 12C); (h) data format: micromass (.PKL) corresponds to the type and brand of instrument used; (i) Instrument = ESI-QUAD-ToF, means that the mass spectrometer used is hybrid Q-ToF; (j) error-tolerant mode = allows the search to be more flexible in terms of the potential modifications detection in the peptides that were not selected prior at the fixed and/or variable mode.
- 21.This tool allows accurate and organized data management for statistical analysis between replicates or samples and the normalization, as well as the relative quantification through label free using spectral counts according to the criteria from the Paris Proteomics Guidelines. Moreover, data can be exported in different formats such as Venn diagrams, heat maps (absolute normalization or log 2 values), scatter, bar, and pie charts, also introducing gene ontology (GO) and pathway (KEGG) analysis.
- 22.The data organization as well as relative quantification through a “label-free” protocol was performed using ProteoIQ software v2.7. The protein list coming from Mascot can be exported as a Mascot.Dat File (format compatible with ProteoIQ) into the ProteoIQ. Also, the database to be used in the program mentioned earlier must be the same as it was for the Mascot search.