Abstract
Proteins with Tetratricopeptide-repeat (TPR) domains are encoded by large gene families and distributed in all plant lineages. In this study, the Soluble NSF-Attachment Protein (SNAP) subfamily of TPR containing proteins is characterized. In soybean, five members constitute the SNAP gene family: GmSNAP18, GmSNAP11, GmSNAP14, GmSNAP02, and GmSNAP09. Recently, GmSNAP18 has been reported to mediate resistance to soybean cyst nematode (SCN). Using a population of recombinant inbred lines from resistant and susceptible parents, the divergence of the SNAP gene family is analysed over time. Phylogenetic analysis of SNAP genes from 22 diverse plant species showed that SNAPs were distributed in six monophyletic clades corresponding to the major plant lineages. Conservation of the four TPR motifs in all species, including ancestral lineages, supports the hypothesis that SNAPs were duplicated and derived from a common ancestor and unique gene still present in chlorophytic algae. Syntenic analysis of regions harbouring GmSNAP genes in soybean reveals that this family expanded from segmental and tandem duplications following a tetraploidization event. qRT-PCR analysis of GmSNAPs indicates a co-regulation following SCN infection. Finally, genetic analysis demonstrates that GmSNAP11 contributes to an additive resistance to SCN. Thus, GmSNAP11 is identified as a novel minor gene conferring resistance to SCN.
The majority of crop species appears to be polyploids as a result of duplication or hybridization events. It is generally accepted that polyploidy has conferred distinct advantages to the development of agronomically important traits1,2,3. Polyploidization, for example, has been associated with an increased size of harvested organs, novel gene interactions leading to new traits, and the formation of new crop species1. In the plant model Arabidopsis thaliana, at least four different large-scale duplication events occurred 100 to 200 million years ago, favouring the diversification of this species4. Recently, numerous studies have reported large segmental duplication events and subsequent divergent selection across many gene families in soybean5,6,7. Soybean has a paleopolyploid genome and nearly 75% of predicted soybean genes are present in multiple copies due to two duplication events that occurred 13 and 59 mya8.
In recent years, proteins containing TPRs have been shown to be essential for responses to hormones such as ethylene, cytokinin, gibberellin, salicylate, and auxin in Arabidopsis. Thus, proteins containing TPRs are emerging as essential determinants for signal transduction pathways9. Several studies have reported that proteins containing TPRs are involved in a plethora of cellular functions including cell cycle regulation, neurogenesis, and mitochondrial/peroxisomal protein transport10,11. Interestingly, mutations in TPR proteins have been found to produce several human diseases, indicating essential roles in cell function. Importantly, the TPR domain facilitates specific interactions with a partner protein(s)11. Moreover, TPR domains also play important roles in aspects of plant development; being essential for gametophytic viability as well as root growth and integrity under osmotic stress12. In addition, a different member of TPR containing proteins, TTL3, was found to interact with the constitutively active VH1/BRL2, a protein homologous to the brassinosteroid receptor BRI1, and play a role in vasculature development13. There are a large number of proteins containing the TPR motif, and they are found in many organisms, including humans, yeast, bacteria, and plants10. The TPR gene family is divided into several subfamilies including tetratricopeptide thioredoxin like (TTL), Cyclophilin (CYP), and Soluble NSF attachement proteins (SNAP).
SNAPs have been widely studied in both plants and animals. SNAP protein, a member of soluble NSF attachment protein receptor (SNARE) complex, has been reported to be involved in vesicular trafficking, plasma membrane stability, cytokinesis (involving KNOLLE), calcium binding (involving Synaptotagmin), membrane repair, and human genetic diseases including certain cancers14,15,16,17,18. Additionally, an α-SNAP has also been linked to disease resistance in plants19,20. SNAPs are characterized by the presence of a tetratricopeptide repeat (TPR) domain. The TPR domain was identified and named in 1990, with a name denoting the 34 amino acids comprising the basic repeat and was reported to be involved in the cell cycle in yeast21,22. Proteins do not normally contain an individual TPR motif, but consist of three to 16 tandem repeats that can be grouped or dispersed throughout the protein10,23. In soybean, the SNAP gene family is composed of five members; of which GmSNAP18 is required for SCN resistance20. However, the other GmSNAP members were not investigated for their role in SCN resistance.
Soybean (Glycine max (L) Merr.) is considered one of the most economically important crops worldwide. It is a valuable source of protein, edible oil, and biodiesel, and represents more than 56% of the world’s oilseed production (http://SoyStats.com, 2016). Soybean production is severely endangered by diseases such as soybean cyst nematode (Heterodera glycines Ichinohe), a microscopic worm which causes over $1.2 billion yield losses annually in the U.S alone24. Planting resistant cultivars is the preferred disease management strategy against SCN. Two types of SCN resistant soybean lines have been used by soybean breeders, the PI88788 type of resistance which requires three genes together at the rhg1 locus for its function: a Soluble NSF Attachment Protein (α-SNAP), an Amino Acid Transporter (AAT) and a Wound-Inducible domain (WI12)20; and the Peking type of resistance requires two genes: the GmSNAP18 at the rhg1 locus and the GmSHMT08 at the Rhg4 locus25,26. However, the molecular mechanisms of how SNAP proteins mediate SCN resistance remain unclear. Recently, it has been described that elevated expression of resistance-type Rhg1 α-SNAP negatively affected the abundance of SNARE-recycling 20 S complexes, disrupting vesicle trafficking, and induced elevated abundance of NSF causing cytotoxicity27. However, expression of other loci encoding a canonical wild-type α-SNAPs counteracted the cytotoxicity of resistance-type Rhg1 α-SNAP27. Furthermore, a SCN gene encoding a bacterial-like protein containing a putative SNARE domain (HgSLP-1), an esophageal-gland protein that is secreted by the nematode during plant parasitism, has been suggested to physically interact with the Rhg1 α-SNAP in SCN resistance28. The authors suggested that HgSLP-1 protein may function as an avirulence protein and it helps SCN evade host defenses when absent.
In this study, we conducted a detailed phylogenetic and structural characterization of the GmSNAP subfamily from various species. Furthermore, we tested the contribution of soybean segmental duplications of SNAP and investigated if they originated from an ancestral gene in plants with subsequent duplication events. Results obtained suggest that this family evolved from an early land plant ancestor, and was subject to duplications followed by subfunctionalization and neofunctionalization events. Expression profiling and functional analysis of GmSNAP genes were also performed. Finally, we demonstrate that in addition to GmSNAP18, the GmSNAP11 is a novel minor gene in SCN resistance, contributing to an additive effect.
Results
Duplication of GmSNAP genes in soybean genome
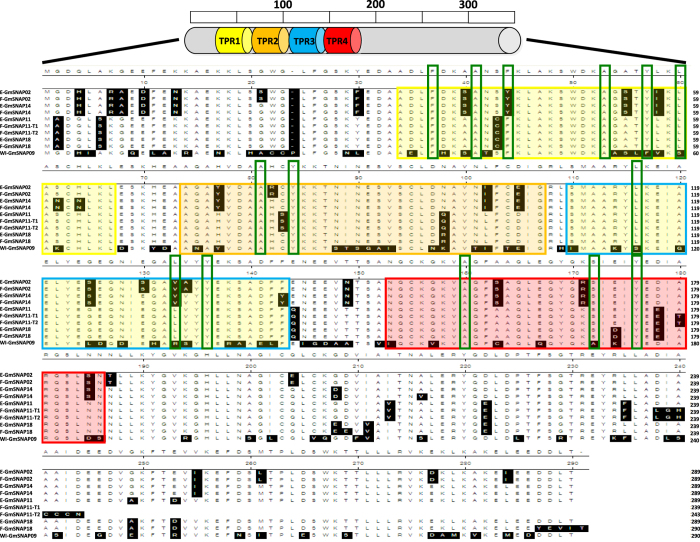
In silico analysis reveals that SNAPs constitute a family of proteins with a common modular architecture containing four tetratricopeptide repeat (TPR) motifs conserved and distributed in specific positions throughout the sequence (Fig. 1). The TPR motifs are common modules in molecular chaperones and are required for the establishment of protein–protein interactions during the formation of multi-protein complexes11. Extensive searches employing a variety of sequenced genomes using the typical distribution of the four TPR motifs of the soybean SNAPs failed to identify any members of this protein family in red algae (Hemiselmis andersenii). However, SNAPs were found to be present in all plant genomes analyzed, including chlorophytic algae (Supplementary Table S2).
Figure 1. Comparative analysis of GmSNAP’s TPR proteins.
Amino acid alignment for the four TPR domains of the five predicted GmSNAP protein members in soybean. In silico analysis showing a high similarity between TPR domains. Yellow, orange, blue, and red boxes present the details of the alignment of the TPR1, TPR2, TPR3, and TPR4 domains, respectively. Green boxes indicate carboxylate clamp residues highly conserved in TPR domains. The alignment shows a high similarity between GmSNAP18 and GmSNAP11, followed by GmSNAP14 and GmSNAP02. However, GmSNAP09 presented the highest polymorphisms.
Investigation of the Williams 82 soybean genome indicates that the GmSNAP gene family is composed of five members located on chromosomes 02 (Glyma.02g260400), 14 (Glyma.14g054900), 11 (Glyma.11g234500), and 18 (Glyma.18g022500), all encoding 289 amino acid (aa) proteins, except one in chromosome 09 (Glyma.09g279400) encoding a 290aa protein. GmSNAP18 was previously reported to be involved in SCN resistance, along with an amino acid transporter (AAT) and a wound inducible protein (WI12)20. However, no study has reported any function of the other four GmSNAP members (GmSNAP11, GmSNAP14, GmSNAP02, GmSNAP09) in SCN resistance.
In order to test the contribution of the soybean duplication events in the number of SNAP genes, the soybean genome was analysed for duplicated chromosomal segments containing GmSNAPs using the Plant Genome Duplication Database29,30,31. Using the locus surrounding GmSNAP18 as bait, three independent duplicate blocks (±100 kb) were discovered to harbour SNAP11, SNAP14, and SNAP02 genes (Table 1, Supplementary Fig. S1). GmSNAPs intragenome syntenic relationship calculations for all the conserved genes surrounding GmSNAPs reveal that the GmSNAP duplication between chr18 and chr11 belongs to a very large inverted duplicated segments containing 386 additional conserved duplicated genes or anchors (Supplementary Table S2). However, GmSNAP duplication between chr18/chr14, and chr18/chr02 was not as conserved, with only 72 and 23 duplicated genes or anchors retained, respectively (Supplementary Table S2). Syntenic analysis showed that GmSNAP18 and GmSNAP09 were not located on duplicated blocks together. Similarity analyses showed that the GmSNAP09 protein shared the lowest identity with GmSNAP18 (only 68%), as compared to the other members GmSNAP02, GmSNAP14, and GmSNAP11, which shared 84.8%, 86.9%, and 92.4% identity with GmSNAP18, respectively (Fig. 1).
Table 1. Gene divergence of duplicated regions around GmSNAP gene members in soybean between chr18 (1.54–1.74 Kb), chr11 (32.87–33.07 Kb), chr14 (4.28–4.48 Kb), and chr02 (44.6–44.8 Kb).
| Locus 1 | Annotation 1 | Block order | Locus 2 | Ka | Ks | Ka/Ks | Block order | Locus 3 | Ka | Ks | Ka/Ks | Block order | Locus 4 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glyma.18G021200 | EXPRESSED PROTEIN | 200 | Glyma.11G235700 | 0.03 | 0.2 | 0.15 | ||||||||||
| Glyma.18G021300 | Weak chloroplast movement under blue light (WEMBL) | 201 | Glyma.11G235600 | 0.04 | 0.16 | 0.25 | 1 | Glyma.02G259100 | 0.22 | 0.71 | 0.31 | |||||
| Glyma.18G021400 | HALOACID DEHALOGENASE-LIKE HYDROLASE | 202 | Glyma.11G235500 | 0.08 | 0.21 | 0.38 | ||||||||||
| Glyma.18G021500 | Uroporphyrinogen decarboxylase | 203 | Glyma.11G235400 | 0.06 | 0.1 | 0.60 | ||||||||||
| Glyma.18G021600 | CBL-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | 204 | Glyma.11G235300 | 0.02 | 0.1 | 0.20 | ||||||||||
| Glyma.18G021700 | PROTEIN NRT1/PTR FAMILY 5.1 | 205 | Glyma.11G235200 | 0.01 | 0.12 | 0.08 | 2 | Glyma.02G259400 | 0.11 | 0.6 | 0.18 | |||||
| Glyma.18G022000 | Hypoxia induced protein conserved region (HIG_1_N) | 206 | Glyma.11G235000 | 0.09 | 0.15 | 0.60 | 3 | Glyma.02G259700 | 0.11 | 0.79 | 0.14 | |||||
| Glyma.18G022100 | BTB/POZ domain | 207 | Glyma.11G234900 | 0.05 | 0.06 | 0.83 | ||||||||||
| Glyma.18G022200 | Unknown Function | 208 | Glyma.11G234800 | 0.04 | 0.13 | 0.31 | 4 | Glyma.02G259800 | 0.12 | 0.37 | 0.32 | 4 | Glyma.14G054100 | 0.15 | 0.67 | 0.22 |
| Glyma.18G022300 | BETA CATENIN-RELATED ARMADILLO REPEAT-CONTAINING | 209 | Glyma.11G234700 | 0.03 | 0.14 | 0.21 | ||||||||||
| Glyma.18G022400 | AMINO ACID TRANSPORTER | 210 | Glyma.11G234600 | 0.06 | 0.14 | 0.43 | 5 | Glyma.02G260100 | 0.16 | 0.63 | 0.25 | |||||
| Glyma.18G022500 | SOLUBLE NSF ATTACHMENT PROTEIN (GmSNAP18) | 211 | Glyma.11G234500 | 0.01 | 0.13 | 0.08 | 6 | Glyma.02G260400 | 0.07 | 0.56 | 0.13 | 5 | Glyma.14G054900 | 0.06 | 0.53 | 0.11 |
| Glyma.18G022600 | (Z)-GAMMA-BISABOLENE SYNTHASE 1-RELATED | 212 | Glyma.11G234400 | 0.02 | 0.13 | 0.15 | 7 | Glyma.02G260500 | 0.14 | 0.47 | 0.30 | |||||
| Glyma.18G022700 | SnoaL-like domain | 213 | Glyma.11G234300 | 0.02 | 0.14 | 0.14 | ||||||||||
| Glyma.18G022800 | Unknown Function | 214 | Glyma.11G234200 | 0.04 | 0.09 | 0.44 | ||||||||||
| Glyma.18G022900 | HEAVY METAL TRANSPORT/DETOXIFICATION SUPERFAMILY | 215 | Glyma.11G234100 | 0.02 | 0.15 | 0.13 | 8 | Glyma.02G260700 | 0.13 | 0.65 | 0.20 | 6 | Glyma.14G055100 | 0.1 | 0.91 | 0.11 |
| Glyma.18G023000 | Sodium/hydrogen exchanger family (Na_H_Exchanger) | 216 | Glyma.11G234000 | 0.03 | 0.03 | 1.00 | ||||||||||
| Glyma.18G023100 | Arogenate dehydrogenase (NADP(+))/TyrAAT2 | 217 | Glyma.11G233900 | 0.01 | 0.12 | 0.08 | 9 | Glyma.02G260900 | 0.2 | 0.78 | 0.26 | 7 | Glyma.14G055300 | 0.2 | 1.14 | 0.18 |
| Glyma.18G023200 | Haem-binding uptake, Tiki superfamily, ChaN (Cofac_haem_bdg) | 218 | Glyma.11G233800 | 0.02 | 0.09 | 0.22 | ||||||||||
| Glyma.18G023300 | CARBOXYLASE:PYRUVATE/ACETYL-COA/PROPIONYL-COA CARBOXYLASE | 219 | Glyma.11G233700 | 0.01 | 0.07 | 0.14 | ||||||||||
| Glyma.18G023400 | Unknown Function | 220 | Glyma.11G233600 | 0.01 | 0.07 | 0.14 | 10 | Glyma.02G261000 | 0.24 | 0.8 | 0.30 | 8 | Glyma.14G055400 | 0.23 | 0.83 | 0.28 |
| Glyma.18G023500 | LEUCINE-RICH REPEAT RECEPTOR-LIKE PROTEIN KINASE IMK3-RELATED | 221 | Glyma.11G233500 | 0.04 | 0.13 | 0.31 | 11 | Glyma.02G261400 | 0.12 | 0.57 | 0.21 | 9 | Glyma.14G055900 | 0.12 | 0.56 | 0.21 |
| Glyma.18G023600 | LACCASE-12-RELATED | 222 | Glyma.11G233400 | 0.07 | 0.18 | 0.39 | 12 | Glyma.02G261600 | 0.07 | 0.74 | 0.09 | 10 | Glyma.14G056100 | 0.07 | 0.73 | 0.10 |
Analysis represent +/− 100 kb duplicated region centered in the GmSNAP genes.
Because GmSNAP18/GmSNAP11 and GmSNAP14/GmSNAP02 blocks shared the highest amount of conserved genes (with 386 and 37 anchors) (Supplementary Table S2), this finding suggests that GmSNAP18/GmSNAP11 and GmSNAP14/GmSNAP02 were the result of a recent duplication event (13 mya)8. In addition, Glycine max intraspecific synteny data in Soybase suggests that GmSNAP on chromosome 18 was present in the ancient duplication event; however, GmSNAP on chromosome 11 has appeared only in the recent duplication.
In silico and structural analysis of GmSNAP proteins reveals that GmSNAP18 and GmSNAP11 have conserved the carboxylate clamp residues
Within a TPR motif, eight amino acids at positions 4 (W/L/F), 7 (L/I/M), 8 (G/A/S), 11 (Y/L/F), 20 (A/S/E), 24 (F/Y/L), 27 (A/S/L), and 32 (P/K/E) are conserved and important in maintaining the α-helical structures32. In silico analysis showed that TPR1 of both GmSNAP18 and GmSNAP11 presents six conserved amino acids, 4 (F), 8 (A), 11 (F), 20 (A), 24 (Y), and 27 (L); TPR2 presents two amino acids conserved at positions 8 (A) and 11 (Y); TPR3 presents three conserved amino acids at positions 7 (L), 24 (L), and 27 (Y); and TPR4 presents three conserved amino acids at positions 8 (A), 20 (S), and 24 (Y) (Fig. 1). This structural analysis supports the functionality of the TPR domain in recognizing its target proteins32. However, GmSNAP02 and GmSNAP14 did not conserve the F11Y and L24V in the TPR1 and TPR3, respectively (Fig. 1). Interestingly, GmSNAP09 did not conserve these carboxylate clamp residues in most TPR domains. Positions 24 (F24Y) and 27 (L27S) in TPR1, positions 7 (L7S) and 24 (L24R) in TPR3, and 20 (S20A) in TPR4 were not conserved. In addition, GmSNAP09 presented the highest number of polymorphisms compared to GmSNAP18 and GmSNAP11 in TPR1 (35%), TPR2 (44%), TPR3 (41%), and TPR4 (26%) (Supplementary Table S3), followed by GmSNAP14 (8 to 20%) and GmSNAP02 (11 to 17%). No polymorphisms were observed between GmSNAP18 and GmSNAP11 at TPR1 and TPR3, only one polymorphism at TPR2 (2%) and two polymorphisms at TPR4 (5%) were present. However, no polymorphisms affected the conserved carboxylate clamp residues. Thus, GmSNAP11 share the most identity to GmSNAP18.
Evolution of GmSNAP gene family members
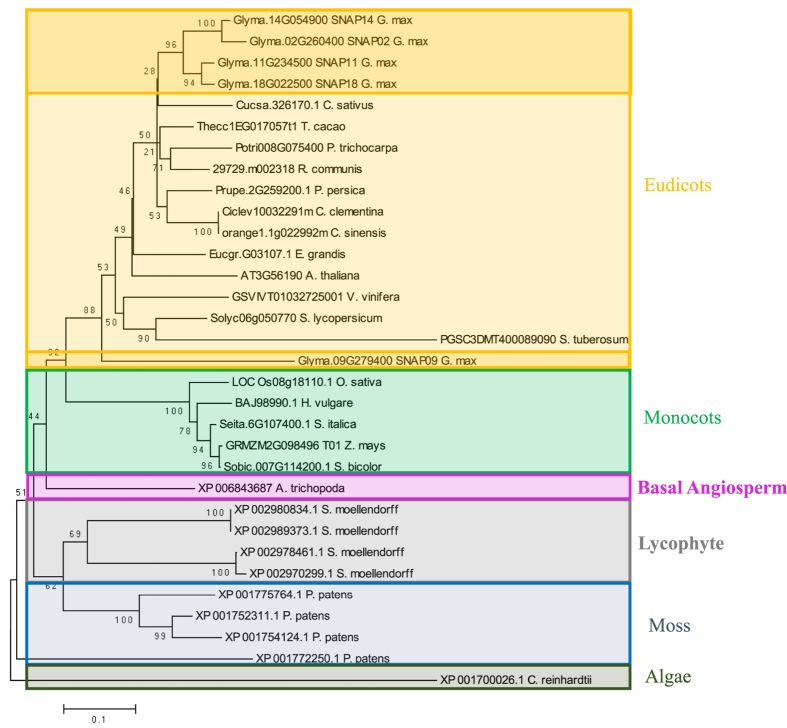
Phylogenetic analysis was conducted to elucidate the evolution of the GmSNAP gene family in soybean. The analysis showed that SNAPs were distributed in six subclades corresponding to eudicots, monocots, basal angiosperms, lycophytes, moss, and chlorophytic algae. This suggests that GmSNAPs derived from a common ancestor as SNAP genes are distributed in all plant lineages (Fig. 2). Monocot, eudicot, and basal angiosperm SNAPs were separately grouped from ancestral SNAPs from S. moellendorfii, P. patens, and C. reinhardtii. We will consider the C. reinhardtii SNAP as the root of this phylogenetic tree.
Figure 2. Phylogenetic tree of SNAP from 22 plants species.
All SNAP proteins identified in six model plants; C. reinhardtii (algae), P. patens (moss), S. moellendorfii (lycophyte), Amborella (basal angiosperm), O. sativa (monocot), and A. thaliana (eudicot), in addition to G. max (soybean), were included in the phylogenetic analysis. However, only the SNAP proteins most similar to soybean GmSNAP18, were included from the rest of the other species. The phylogenetic tree was generated using MEGA4 software package and the ClustalW algorithm, and calculated using the neighbor-joining method. The tree bootstrap values are indicated at the nodes (n = 1000).
From the phylogenetic analysis, four GmSNAPs clustered together with GmSNAP18/GmSNAP11 and GmSNAP14/GmSNAP02 localized in subclades. Interestingly, GmSNAP09 did not cluster with the other GmSNAPs. This suggests that this gene diverged from the others or has become pseudogenized. Branch length distances suggest that GmSNAP18 is the most closely related to the ancestral SNAPs, and GmSNAP14 is more closely related to the ancestral SNAPs than GmSNAP02. Furthermore, the large conserved anchor number of GmSNAP18 from the syntenic analysis suggests that GmSNAP18 may be most similar to the ancestral SNAP, which also gave rise to GmSNAP14 in the oldest duplication event (59 mya), followed by the divergence of GmSNAP11 from GmSNAP18 and GmSNAP02 from GmSNAP14 in the most recent duplication event (13 mya) (Supplementary Table S2).
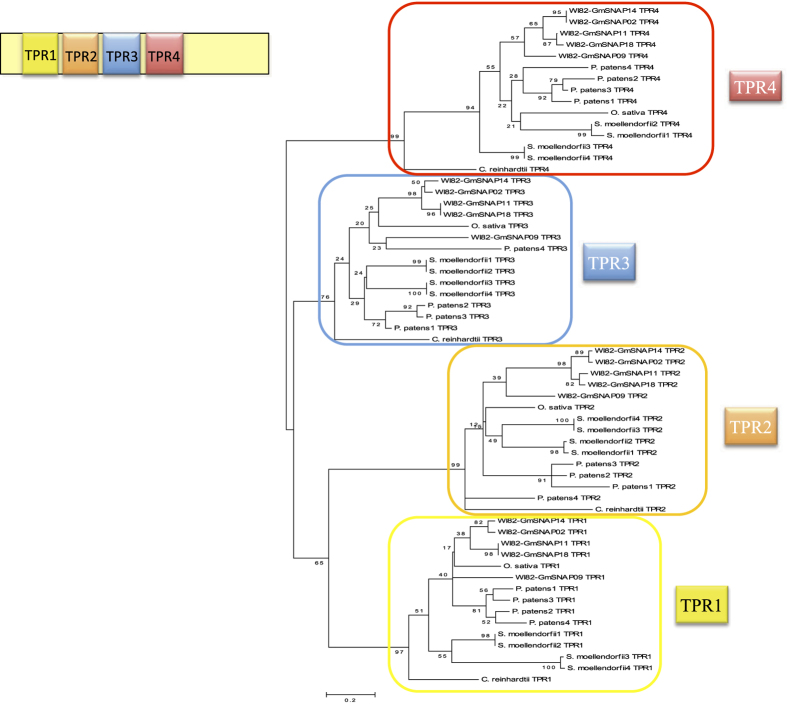
Next, we investigated whether SNAP genes originated from an ancestral gene in plants with subsequent duplications or resulted from convergent evolution. Because TPRs are located in similar positions in SNAP proteins from phylogenetically distant species, we aligned the four TPR motifs (TPR1, TPR2, TPR3, and TPR4) of SNAPs from various species and obtained the corresponding phylogenetic tree. The result indicates that TPRs in equivalent positions from different species have higher similarity than TPRs within a SNAP protein (Fig. 3). For example, TPR1 from soybean GmSNAP18 is more closely related to TPR1 from the oldest plant lineage (algae C. reinhardtii) than to any other TPRs (TPR2 and/or TPR3 and/or TPR4) within GmSNAP18. This indicates that an original SNAP protein from the most ancestral plant lineage expanded among different species by duplication, and the same TPR motifs have been relatively well conserved throughout evolution, most likely due to functional constraints. These data suggests that SNAPs derived from a common ancestor and unique gene still present in chlorophytic algae.
Figure 3. Phylogenetic relationships among individual TPR motifs in GmSNAP.
All GmSNAPs in soybean contain 4 TPR motifs (TPR1, TPR2, TPR3, and TPR4). The unrooted tree was generated using the four TPR motifs sequences from the 5 soybean SNAP proteins, the SNAP orthologs from monocots and eudicots, and from phylogenetically distant species such algae, moss, and lycophyte. The phylogenetic tree was generated using MEGA4 software package and the ClustalW algorithm, and calculated using the neighbor-joining method. The tree bootstrap values are indicated at the nodes (n = 1000).
Soybean GmSNAP genes display overlapping and divergent functions in resistance to SCN
To investigate the specific evolutionary path(s) of different GmSNAP family members in soybean, we studied their expression patterns and evaluated their specific roles in response to SCN. We first compiled expression data for the GmSNAP genes using the public RNAseq data available at Soybase. As shown in Supplementary Figure S2, GmSNAP gene members presented three gene expression patterns. GmSNAP18 and GmSNAP11 were both ubiquitously highly expressed throughout most tissues, GmSNAP14 had lower expression in all tissues with no expression in the leaves, and GmSNAP02 expression only appeared in the flowers and seeds. The RNAseq analyses obtained from soybase point to a possible neofunctionalization or subfunctionalization event in the GmSNAP gene family in soybeans.
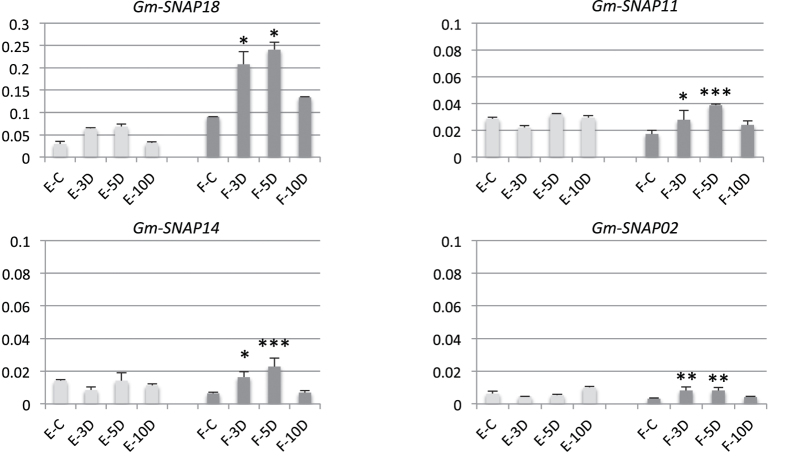
Next, we analysed the expression using qRT-PCR of the GmSNAP gene members in the SCN susceptible line Essex and the SCN resistant line Forrest (Peking-type) following SCN infection. Surprisingly, the expression analysis shows that the transcripts of the four identified GmSNAP members in Forrest were significantly upregulated under SCN infection. Specifically, GmSNAP18 transcripts were the most upregulated, followed by GmSNAP11. Furthermore, GmSNAP14 and GmSNAP02 presented much lower expression levels. All GmSNAP gene expression levels in Forrest reached the highest at five days post infection (dpi). Transcript levels were upregulated by 1.6, 1.23, 2.43, and 1.5 times for GmSNAP18, GmSNAP11, GmSNAP14, and GmSNAP02, respectively. On the contrary, a slight but not significant induction of all GmSNAP members was observed in the susceptible line Essex throughout the time series. Furthermore, expression levels were about 3 times more upregulated in Forrest than in Essex at 3, 5, and 10 dpi. This expression profile was maintained regardless of nematode presence (Fig. 4).
Figure 4. qRT-PCR of GmSNAP gene family in soybean in Forrest and Essex wild types.
Quantitative RT-PCR analysis of four GmSNAP gene family members in chromosomes 02, 11, 14 and 18. Expressions were normalized using Ubiquitin as reference. (E) Essex, (F) Forrest, (C) without SCN infection, and (D) SCN infection at 3, 5 and 10 days post inoculation. The gene-specific primers designed to amplify cDNA fragments are detailed in Supplementary Table S5. *Asterisks indicate significant differences between samples as determined by t-test (***P < 0.0001, **P < 0.001, *P < 0.01).
Both GmSNAP18 and GmSNAP11 contribute to SCN resistance but not GmSNAP14 and GmSNAP02
α-SNAP, corresponding to Peking-type GmSNAP18, has been reported to play a major role in resistance to SCN in PI88788-type soybeans (Cook et al.20). Expression analysis showed that the rest of the GmSNAP gene family in Forrest also responds to SCN infection. In order to test whether GmSNAP11, GmSNAP14, and GmSNAP02 have the same function in SCN resistance as the identified GmSNAP18, and determine whether the contribution of the GmSNAP gene family members to SCN resistance is redundant, partially redundant, or additive, we quantified their corresponding female index (FI) using a recombinant inbred line (RIL) ExF RIL population under SCN infection33. In this study, the F5 derived RILs from the ExF population were genotyped for the following four genes: GmSNAP18, GmSNAP11, GmSNAP14, and GmSNAP02. Because GmSNAP02 did not present any polymorphism between Forrest and Essex, the lines were classified into four different genotypes according to their allelic combinations (Supplementary Table S4). Forward screening showed a range of FI among the different ExF genotypes tested. The genotypes containing SNAP18+/SNAP11+ alleles were deemed as resistant to SCN regardless of the GmSNAP14 allele, and presented the lowest female index of 4.01% (SNAP18+/SNAP11+/SNAP14−) and 4.48% (SNAP18+/SNAP11+/SNAP14+) among the four ExF lines. A similar result was obtained in the Forrest wild type (FI = 3.65%) (Fig. 5). Surprisingly, GmSNAP18+ alone was not able to confer a complete resistance to SCN, presenting a moderate susceptibility. Thus, the female index FI = 11.05% of SNAP18+/SNAP11−/SNAP14− and FI = 12.59% of SNAP18+/SNAP11−/SNAP14+ were significantly higher than the female index in the genotypes SNAP18+/SNAP11+/SNAP14− and SNAP18+/SNAP11+/SNAP14+ (Fig. 5). These data point to an additive effect presented by GmSNAP11, but not by GmSNAP14. Therefore, the GmSNAP11 at linkage group B1 is considered a novel minor contributor to SCN resistance in Peking-type alleles. However, no significant differences in FI were observed when GmSNAP14 was present as the Forrest allele compared to the Essex allele.
Figure 5. Female index presented by the four different ExF genotypes, Essex and Forrest wild types.
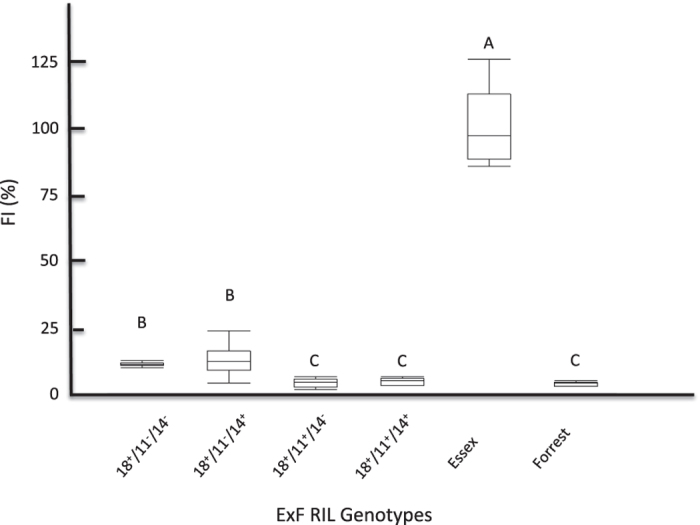
Two lines from each genotype were analyzed. Five replicates were included for each line: n = 10 per genotype.
Discussion
The genome distribution of GmSNAP genes from syntenic analysis indicates the existence of segmental duplications and tandem rearrangement events, which occurred following an allotetraploidy event5,6,8,34. This is further supported by the evolutionary conservation of the internal modular domains among soybean GmSNAPs and phylogenetically separated SNAP proteins, and enhanced by the identification of a single SNAP gene in an early land plant species, i.e. C. reinhardtii (CrSNAP). The presence of a single SNAP gene in C. reinhardtii suggests that an ancestral aquatic SNAP protein in chlorophytic algae may have given rise to all the divergent SNAP proteins found in land plants.
Consequently, we suggest that successive duplications of a unique SNAP gene derived from an ancestral chlorophytic algae led to the subfunctionalization and neofunctionalization of the SNAP gene family in the soybean genome, contributing to the fine-tuning of soybean responses against biotic stresses, e.g. SCN resistance. Thus, the substantial changes in SNAP gene expression, as well as functional changes of the SNAP gene duplicates over time are likely due to both gene duplication and selection pressure imposed by stressful conditions. This may contribute to the physiological complexity of the soybean SNAP response against multiple stresses. These results support the hypothesis that gene duplication is an important evolutionary mechanism in the generation of novel functions and phenotypes, contributing to the adaptation of land plants to stressful environments35,36.
Furthermore, large conserved anchor numbers from the syntenic analysis revealed that GmSNAP18 may be the most similar to the ancestral SNAP (which also gave rise to GmSNAP14 through a duplication event that occurred (59 mya), followed by the divergence of GmSNAP11 from GmSNAP18, and GmSNAP02 from GmSNAP14 in the most recent duplication event (13 mya). Due to the four GmSNAP family members: GmSNAP02, GmSNAP11, GmSNAP14, and GmSNAP18 localized on syntenic genomic regions, our data suggests that the GmSNAP gene family evolved from a common ancestor. However, syntenic analysis did not show any duplicated block or segment between GmSNAP18 and GmSNAP09. GmSNAP09 shared the lowest identity with GmSNAP18 (68%) and the highest polymorphisms on the four TPR domains compared to the rest of the four GmSNAP members. GmSNAP09 also did not conserve most of the essential carboxylate clamp residues that maintain the activity and functionality of the TPR domain. In addition, phylogenetic analysis showed that GmSNAP09 did not cluster with the other four GmSNAPs, or with the rest of the eudicot SNAPs. This suggests that GmSNAP09 may have become pseudogenized.
TPR proteins consist of three to sixteen tandem repeats of TPRs that can be grouped or dispersed throughout the protein23. Because most TPR proteins contain three repeats, it is likely that three is the minimum number required to form a functional domain. Our structural analysis has shown that GmSNAP proteins in soybeans are characterized by the presence of four tetratricopeptide repeats (TPR) in conserved positions along the protein (Fig. 1). This structure containing four TPR motifs supports the functionality of the TPR domain in the GmSNAPs.
Sequence analysis of many TPR proteins indicates that TPRs are defined by a pattern of small and large hydrophobic amino acids rather than a pattern of conserved amino acid residues. Although no invariant positions are found in TPRs, some amino acids tend to be conserved9. Three-dimensional structures have shown that a TPR motif contains two antiparallel α-helices (helix A and B), such that tandem arrays of TPR motifs generate a right-handed helical structure with an amphipathic channel that might accommodate the complementary region of a target protein11. Within a TPR motif, eight amino acids at positions 4 (W/L/F), 7 (L/I/M), 8 (G/A/S), 11 (Y/L/F), 20 (A/S/E), 24 (F/Y/L), 27 (A/S/L), and 32 (P/K/E) have a higher frequency of conservation and are important in maintaining the α-helical structures within a TPR motif 32. Clustering of several α-helices within a tandem array of TPR motifs generates an amphipathic channel with a large surface area, allowing the TPR domain to recognize its target protein11. Within these highly conserved amino acids, our in silico analysis shows that GmSNAP18 and GmSNAP11 conserved most of the carboxylate clamp residues (Fig. 1, Supplementary Table S3). However, GmSNAP14 and GmSNAP02 conserved less carboxylate clamp residues. This structural analysis supports the functionality of the TPR domain at GmSNAP18 and GmSNAP11 in order to recognize its target proteins and trigger the SCN resistance32. Interestingly, GmSNAP09 did not conserve these residues in most TPR domains. In addition, GmSNAP09 presented the highest number of polymorphisms compared to GmSNAP18 and GmSNAP11 in TPR1 (35%), TPR2 (44%), TPR3 (41%), and TPR4 (26%) (Supplementary Table S3). These data further support the evolution of GmSNAP09 obtained from the syntenic and phylogenetic analysis, and indicate that this member may have become pseudogenized.
Forward screening showed that lines with Forrest alleles (+) at both GmSNAP18 and GmSNAP11 were highly resistant to SCN. However, the presence of GmSNAP18+ alone was not able to confer complete resistance to SCN when the Essex allele (−) GmSNAP11− was present. In fact, the female index of 11.05% and 12.59% obtained was significantly higher in the genotypes with SNAP18+/SNAP11−/SNAP14+ and SNAP18+/SNAP11−/SNAP14−, respectively, presenting a moderate susceptibility. Interestingly, lines containing the GmSNAP11+ allele were significantly more resistant than the ones that possessed the GmSNAP11− allele (Fig. 5). The presence of the GmSNAP11+ Forrest allele decreases the FI in the genotypes (SNAP18+/SNAP14+), and (SNAP18+/SNAP14−) from 11.05% to 4.03%; and from 12.59% to 4.48%%, respectively.
The correlation between GmSNAP11+ and SCN resistance, indicate a direct link between the presence of the Forrest allele at GmSNAP11+ and increased resistance to SCN in the tested allelic combinations (Supplementary Table S4). However, this contribution is marginal and minor because of the large effect of GmSNAP18+. This finding suggests that GmSNAP11+ contributes a minor resistance to SCN, but cannot substitute the major contribution of GmSNAP18+. These data are in accordance with the results reported recently that the over-expression of an α-SNAP gene suppresses plant parasitic nematode infection in soybeans19. Using the same primers these authors used to overexpress this α-SNAP, we cloned this gene from Forrest and found that the predicted protein does not correspond to the previously reported GmSNAP18 (289 aa), but that it corresponds to a truncated GmSNAP11, which is present in Forrest Peking-type under two different types: GmSNAP11-T1 and GmSNAP11-T2, encoding a 239 aa and a 244 aa protein, respectively (Fig. 1). This difference in structure and accumulation of mutations that occurred during evolution between the two SNAP members, GmSNAP18 and GmSNAP11, explain their specific contributions to SCN resistance reported previously. Because GmSNAP11 conserves the carboxylate clamp residues and the nonsense mutations occur after the four TPR motifs, but before the two reported polymorphisms between resistant and susceptible lines at the C-terminus, both GmSNAP11 truncated proteins may conserve protein-protein interaction capabilities, but may lose the downstream activity and affect its function (Fig. 5). Thus, the GmSNAP11 expression profiles, structural, and SCN phenotypic analysis point to the discovery of a minor contribution of GmSNAP11 for resistance to SCN.
After duplication, the predominant fate of duplicated genes is pseudogenization37. Still, a significant fraction of duplicated genes in plants are preserved and follow different evolutionary paths including retention, neofunctionalization, and subfunctionalization. The phylogenetic analysis within the soybean SNAP family indicates that GmSNAP18 and GmSNAP11 proteins cluster together and form a separate clade from GmSNAP14 and GmSNAP02 (Fig. 2). Because GmSNAP11 and GmSNAP18 began diverging recently, they are closely related with a 92.4% amino acid sequence similarity. However, nonsense mutations in GmSNAP11 at positions E244* and A240* result in a truncated protein which may affect and reduce the function of GmSNAP11-T1 and GmSNAP11-T2 in Peking-type Forrest, without entirely supressing its function. Previous studies in the plant model Arabidopsis thaliana have shown that a TPR protein family named Tetratricopeptide repeat thioredoxin-like (TTL), formed by four members. TTL1, TTL3, and TTL4, but not TTL2, presented an additive effect in response to osmotic stress tolerance and are essential for root growth and integrity, while all present the same amino acid sequence length with high identity12. This is an example of a duplicated gene resulting in a truncated protein (50 aa less) that presents an additive effect in resistance to a pathogen.
Furthermore, the reported low differences in GmSNAP14 and GmSNAP02 expression patterns and function suggest that its subfunctionalization is ongoing. It has been reported that in addition to GmSNAP18, GmSNAP11 and GmSNAP14 were associated with QTL for resistance to SCN38,39. GmSNAP02 was reported to be associated with QTL for resistance to Phytophtora40. Moreover, GmSNAP02 RNAseq data (soybase RNAseq expression) show that its expression was confined only to flowers and seeds, and qRT-PCR data showed that GmSNAP02 had a very low expression in the roots, which suggests that it may have been subfunctionalized. Similar results were reported in the TPR protein TTL2, which was demonstrated to have neofunctionalized to be involved in male sporogenesis12. TTL2 is essential for gametophytic viability but not in root growth and integrity under osmotic stress, as is the rest of the TTL gene family12. Furthermore, the expression data shows that the four closely related GmSNAPs may have acquired an accumulation of mutations that lead to a difference in their regulatory or protein sequences, ultimately leading to a neofunctionalization or subfunctionalization. However, these genes are generally responding to the same controlling elements, which could directly or indirectly trigger their expression. The presence of the cumulated mutations within the TPR domains of each reported GmSNAP member and the differences within their C-termini and the rest of the protein sequence are most likely to dictate potential interaction and localization preferences (Fig. 1).
Although the roles of GmSNAP14 and GmSNAP02 members needs to be elucidated, this study shows evidence of the neofunctionalization and subfunctionalization of the GmSNAP gene family in Peking-type soybeans. Considering the function of soybean GmSNAP18 in SCN resistance20, and the newly discovered function of GmSNAP11 as a minor contributor to SCN resistance, the duplication and retention of SNAP genes in plants suggests that SNAP genes may be a source of diversity which is important for proper responses to stressful environments.
Material and Methods
SCN-infection phenotyping
SCN-infection phenotyping was performed on the M3 lines as described by ref. 41. Seedlings were inoculated with infective eggs from the PA3 population (HG type 0). Briefly, cysts were extracted from Essex infested roots and soil by flotation in water and collected on a 250-μm sieve. Harvested cysts were gently crushed using a drill press and the eggs were collected on a 25-μm sieve42. The eggs were further diluted to 1,000 eggs/ml of water. Individual seedlings were inoculated with 1 ml of the egg suspension. Plants were maintained in the growth chamber at 27 °C. Cyst counts were performed at 30 days post inoculation.
Phylogenetic Analysis and Genomic Structure
Multiple sequence alignments were performed using the MEGA4 software package and the Clustal W algorithm. An unrooted phylogenetic tree was calculated with the neighbor-joining method43, and tree topology robustness was tested by bootstrap analysis of 1,000 replicates. Alignment analysis of SNAPs in soybean (Glycine max (L.) Merr.), Arabidopsis Arabidopsis thaliana (dicotyledons model), rice Oryza sativa (monocotyledons model), Physcomitrella patens (moss model), Selaginella moellendorffii (lycophyte model), Chlamydomonas reinhardtii (algae model), in addition to other monocots (H. vulgare, S. italica, Z. mays, S. bicolor), and eudicots (C. sativus, T. cacao, P. trichocarpa, R. communis, P. persica, C. clementine, C. sinensis, E. grandis, A. thaliana, V. vinifera, S. lycopersicum), were obtained using MegAlign 4 software. SNAP and TPR domain in silico analysis was performed using the MegAlign (DNASTAR Lasergene 8) software package and the Clustal W algorithm. All parameter values correspond to default definitions.
Genotyping of ExF RILs population
The ExF population containing 100 RILs used in this study was developed at Southern Illinois University Carbondale44. The Eco TILLING marker GmSNAP18, GmSNAP11, GmSNAP14, and GmSNAP02 whose primers were listed in Supplementary Table S5, were developed and used to identify the genotype of each ExF RIL at the genes GmSNAP18 (Glyma.18G022500), GmSNAP11 (Glyma.11G234500), GmSNAP02 (Glyma.02G260400), and GmSNAP14 (Glyma.14G054900) by EcoTILLING. The EcoTILLING was conducted as described by ref. 41.
SNAP Analysis
The mining of SNAP genes was performed by searching sequences homologous to the GmSNAP18 proteins using the Phytozome database (www.phytozome.net). SNAP sequences obtained from different species were also analyzed using the NCBI Conserved Domain Database for TPR domains (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Generation of the unrooted phylogenetic tree was performed by the alignment of the full-length amino acid sequences using ClustalW and the DNASTAR Lasergene 8 software.
qRT-PCR of GmSNAP gene family
Soybean seedlings from the susceptible line Essex and the resistant lines Forrest (wild types) were grown in autoclaved sandy soil in the growth chamber for one week and then infected with eggs from the PA3 population. Total RNA was isolated from root samples after three, five, and ten days following SCN infection using a Qiagen RNeasy Plant Mini Kit (cat# 74904). Total RNA was DNase treated and purified using Turbo DNA-free Kit (QAmbion/life technologies AM1907). RNA was quantified using Nanodrop 1000 (V3.7). Then, a total of 400 nanograms of treated RNA was used to generate cDNA, using the cDNA synthesis kit (Thermoscript, life technologies, 11146-025) with random hexamers and 1/10th of a 20 microliter RT reaction was used in gene-specific quantitative PCR with the Power SYBR® Green PCR Master Mix kit (Applied Biosystems™ #4368706). A list of primers used in this work is found in Supplementary Table S5. For each genotype/primer pair, RNA from three individual biological replicates was used for quantitation and then normalized using the delta Cq method with Ubiquitin used as a reference gene (ΔCq = Cq(TAR) − Cq(REF)). Each gene’s expression was exponentially transformed to the expression level using the formula (ΔCq Expression = 2−ΔCq). A –RT reaction was also performed in all the samples.
Bioinformatics Analysis
Bioinformatics analysis was performed using the following databases: The Soybean database (www.soybase.org), NCBI (www.ncbi.nlm.nih.gov), Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/), phytosome (www.phytozome.net) and the European Bioinformatics Institute (www.ebi.ac.uk/Tools/clustalw2/index.html).
Statistical analysis
All presented results were performed with the analysis of variance by a T-student test means comparison, using JMP Pro V12 software.
Ethics and consent to participate
This study did not involve humans, human data or animals; no ethics approval or consent is required to publish the results.
Availability of data and materials
The developed cross between the resistant Forrest (+) and the susceptible Essex (−) using both recombinant inbred lines (RILs) and near-isogenic lines (NILs) seeds, are property of Southern Illinois University (SIU). Access to the germplasm is subjected to Transfer Material Agreement Form.
Additional Information
How to cite this article: Lakhssassi, N. et al. Characterization of the Soluble NSF Attachment Protein gene family identifies two members involved in additive resistance to a plant pathogen. Sci. Rep. 7, 45226; doi: 10.1038/srep45226 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This research was supported by the United Soybean Board. The authors would like to thank Dr. Richard Thomas for his helpful comments and Dr. Judy Davie for providing the qRT-PCR machine.
Footnotes
The authors declare no competing financial interests.
Author Contributions N.L. conceived and drafted the manuscript, performed qRT-PCR analysis, evolution analysis, statistical analysis, haplotyping studies, in silico analysis, phylogenetic analysis, syntenic and structural analysis, SCN phenotyping, and ExF genotyping. S.L. carried out the EcoTILLING of the RIL and gene sequencing. S.B. performed qRT-PCR analysis and the SCN infection. Z.Z. did the RNA extractions and measured SCN female index. V.C. performed data analysis, evolution analysis, and drafted the manuscript. K.L. provided the nematode assay. A.B. did the phylogenetic analysis, participated in the data analyses and edited the manuscript. K.M. designed the experiments, planned and supervised the work and edited the manuscript. All authors reviewed and commented on the manuscript.
References
- Eckardt N. A. Two genomes are better than one: widespread paleopolyploidy in plants and evolutionary effects. Plant Cell 16, 1647–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P. W. J. Polyploid Incidence And Evolution. Annu. Rev. Genet. 34, 401–37 (2000). [DOI] [PubMed] [Google Scholar]
- Wendel J. F. Genome evolution in polyploids. Plant Mol Biol 42, 225–49 (2000). [PubMed] [Google Scholar]
- Vision T. J., Brown D. G. & Tanksley S. D. The origins of genomic duplications in Arabidopsis. Science 290, 2114–7 (2000). [DOI] [PubMed] [Google Scholar]
- Yin G. et al. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol 13, 148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. et al. Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol 14, 93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V. K. & Jain M. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front Plant Sci 6, 918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010). [DOI] [PubMed] [Google Scholar]
- Schapire A. L., Valpuesta V. & Botella M. A. TPR Proteins in Plant Hormone Signaling. Plant Signal Behav 1, 229–30 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea L. D. & Regan L. TPR proteins: the versatile helix. Trends Biochem Sci 28, 655–62 (2003). [DOI] [PubMed] [Google Scholar]
- Blatch G. L. & Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–9 (1999). [DOI] [PubMed] [Google Scholar]
- Lakhssassi N. et al. The Arabidopsis tetratricopeptide thioredoxin-like gene family is required for osmotic stress tolerance and male sporogenesis. Plant Physiol 158, 1252–66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceserani T., Trofka A., Gandotra N. & Nelson T. VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J 57, 1000–14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N. W. & Chakrabarti S. There’s more to life than neurotransmission: the regulation of exocytosis by synaptotagmin VII. Trends Cell Biol 15, 626–31 (2005). [DOI] [PubMed] [Google Scholar]
- El Kasmi F. et al. SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol Biol Cell 24, 1593–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof T. C. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron 80, 675–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapire A. L. et al. Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20, 3374–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof T. C. A molecular machine for neurotransmitter release: synaptotagmin and beyond. Nat Med 19, 1227–31 (2013). [DOI] [PubMed] [Google Scholar]
- Matsye P. D. et al. The expression of a naturally occurring, truncated allele of an alpha-SNAP gene suppresses plant parasitic nematode infection. Plant Mol Biol 80, 131–55 (2012). [DOI] [PubMed] [Google Scholar]
- Cook D. E. et al. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–9 (2012). [DOI] [PubMed] [Google Scholar]
- Hirano T., Kinoshita N., Morikawa K. & Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+. Cell 60, 319–28 (1990). [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Boguski M. S., Goebl M. & Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60, 307–17 (1990). [DOI] [PubMed] [Google Scholar]
- Main E. R., Xiong Y., Cocco M. J., D’Andrea L. & Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure 11, 497–508 (2003). [DOI] [PubMed] [Google Scholar]
- Koenning S. R. & Wrather J. A. Suppression of Soybean Yield Potential in the Continental United States by Plant Diseases from 2006 to 2009. Plant Health Progress (2010). [Google Scholar]
- Liu S. et al. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492, 256–60 (2012). [DOI] [PubMed] [Google Scholar]
- Liu S. et al. The soybean GmSNAP18 underlies two types of resistance to soybean cyst nematode. Nature communications (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless A. M. et al. Disease resistance through impairment of α-SNAP–NSF interaction and vesicular trafficking by soybean Rhg1. Proceedings of the National Academy of Sciences 201610150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekal S. et al. A SNARE-Like Protein and Biotin Are Implicated in Soybean Cyst Nematode Virulence. PLoS One 10, e0145601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. et al. Synteny and Collinearity in Plant Genomes. Science 320, 486–488 (2008). [DOI] [PubMed] [Google Scholar]
- Tang H. et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res 18, 1944–54 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.-H., Tang H., Wang X. & Paterson A. H. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Research 41, D1152–D1158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B. D., Goel S. & Krishna P. In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS One 5, e12761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meksem K. et al. ’Forrest’ resistance to the soybean cyst nematode is bigenic: saturation mapping of the Rhg1and Rhg4 loci. Theoretical and Applied Genetics 103, 710–717 (2001). [Google Scholar]
- Clarindo W. R., de Carvalho C. R. & Alves B. M. G. Mitotic evidence for the tetraploid nature of Glycine max provided by high quality karyograms. Plant Systematics and Evolution 265, 101–107 (2007). [Google Scholar]
- Hanada K., Zou C., Lehti-Shiu M. D., Shinozaki K. & Shiu S. H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148, 993–1003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M. et al. The genome of the extremophile crucifer Thellungiella parvula. Nature Genetics 43, 913–918 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S. & Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38, 615–43 (2004). [DOI] [PubMed] [Google Scholar]
- Yue P., Arelli R. P. & Sleper A. D. Molecular characterization of resistance to Heterodera glycines in soybean PI 438489B. Theoretical and Applied Genetics 102, 921–928 (2001). [Google Scholar]
- Guo B., Sleper D. A., Nguyen H. T., Arelli P. R. & Shannon J. G. Quantitative Trait Loci underlying Resistance to Three Soybean Cyst Nematode Populations in Soybean PI 404198A. Crop Science 46, 224 (2006). [Google Scholar]
- Han Y. et al. Mapping QTL tolerance to Phytophthora root rot in soybean using microsatellite and RAPD/SCAR derived markers. Euphytica 162, 231–239 (2008). [Google Scholar]
- Liu X. et al. Soybean cyst nematode resistance in soybean is independent of the Rhg4 locus LRR-RLK gene. Funct Integr Genomics 11, 539–49 (2011). [DOI] [PubMed] [Google Scholar]
- Faghihi J. & Ferris J. M. An Efficient New Device to Release Eggs From Heterodera glycines. J Nematol 32, 411–3 (2000). [PMC free article] [PubMed] [Google Scholar]
- Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–25 (1987). [DOI] [PubMed] [Google Scholar]
- Lightfoot D. A., Njiti V. N., Gibson P. T., Kassem M. A., Iqbal J. M. & Meksem K. Registration of the Essex × Forrest Recombinant Inbred Line Mapping Population. Crop Science 45, 1678 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The developed cross between the resistant Forrest (+) and the susceptible Essex (−) using both recombinant inbred lines (RILs) and near-isogenic lines (NILs) seeds, are property of Southern Illinois University (SIU). Access to the germplasm is subjected to Transfer Material Agreement Form.