Abstract
Background
Human cytomegalovirus (HCMV) is the most common cause of intrauterine infections worldwide. The toll-like receptors (TLRs) have been reported as important factors in immune response against HCMV. Particularly, TLR2, TLR4 and TLR9 have been shown to be involved in antiviral immunity. Evaluation of the role of single nucleotide polymorphisms (SNPs), located within TLR2, TLR4 and TLR9 genes, in the development of human cytomegalovirus (HCMV) infection in pregnant women and their fetuses and neonates, was performed.
Methods
The study was performed for 131 pregnant women, including 66 patients infected with HCMV during pregnancy, and 65 age-matched control pregnant individuals. The patients were selected to the study, based on serological status of anti-HCMV IgG and IgM antibodies and on the presence of viral DNA in their body fluids. Genotypes in TLR2 2258 A > G, TLR4 896 G > A and 1196 C > T and TLR9 2848 G > A SNPs were determined by self-designed nested PCR-RFLP assays. Randomly selected PCR products, representative for distinct genotypes in TLR SNPs, were confirmed by sequencing. A relationship between the genotypes, alleles, haplotypes and multiple variants in the studied polymorphisms, and the occurrence of HCMV infection in pregnant women and their offsprings, was determined, using a logistic regression model.
Results
Genotypes in all the analyzed polymorphisms preserved the Hardy-Weinberg equilibrium in pregnant women, both infected and uninfected with HCMV (P > 0.050). GG homozygotic and GA heterozygotic status in TLR9 2848 G > A SNP decreased significantly the occurrence of HCMV infection (OR 0.44 95% CI 0.21–0.94 in the dominant model, P ≤ 0.050). The G allele in TLR9 SNP was significantly more frequent among the uninfected pregnant women than among the infected ones (χ2 = 4.14, P ≤ 0.050). Considering other polymorphisms, similar frequencies of distinct genotypes, haplotypes and multiple-SNP variants were observed between the studied groups of patients.
Conclusions
TLR9 2848 G > A SNP may be associated with HCMV infection in pregnant women.
Electronic supplementary material
The online version of this article (doi:10.1186/s12985-017-0730-8) contains supplementary material, which is available to authorized users.
Keywords: Human cytomegalovirus (HCMV), Toll-like receptors (TLRs), Single nucleotide polymorphism (SNP), Infection, Pregnant women
Background
Human cytomegalovirus (HCMV) is the most common cause of intrauterine infections worldwide, with seroprevalence rates at the range from 40 to 100% among pregnant women [1–4]. Our recent study, performed among Polish pregnant women between 2010 and 2011, showed seroprevalence of anti-HCMV IgG and IgM antibodies to have been 62.4 and 2.2%, respectively [4]. Compared to other European populations of pregnant women from the Netherlands (41%), France (46%) and the United Kingdom (49%), the prevalence of IgG anti-HCMV in the Polish pregnancy cohort was still high [5–7]. In case of primary infections, diagnosed during pregnancy, the transplacental transmissions of the virus from mother to fetus occur with the incidence rate of 30–40%, while the recurrent infections cause congenital cytomegaly within the range 0.2–2.2% [8–12]. Among fetuses and neonates, congenitally infected with HCMV, cytomegaly may have both asymptomatic and symptomatic course with severe symptoms, including microcephaly, ventriculomegaly, increased periventricular echogenicity and calcifications [8, 12].
Taking into account the immune response to HCMV, the Toll-like receptors (TLRs) have been reported to play important role [13–15]. Particularly, TLR2, TLR4 and TLR9 have been shown to be involved in antiviral immunity [14, 16–18]. In the most recent in vitro study, TLR2 was found as a target of HCMV miR-UL112-3p [19]. Previously, TLR2 was also determined to be involved in the functional sensing of HCMV through direct interactions with viral glycoproteins (gp, g) gB and gH [20]. In turn, TLR4 was reported to be correlated with inhibition of HCMV infection [21]. In human monocytoid THP1 cells and foreskin fibroblasts, TLR9 was determined to induce the expression of TNF-α at 1 h after HCMV infection [22]. A study performed for neonatal human fibroblasts, showed some involvement of TLR9 in the development of HCMV infection as well [23].
Previously, the role of single nucleotide polymorphisms (SNPs, variations of single nucleotides at specific positions in sequences of the genes), residing within TLR genes, was also reported [24–26]. In case of TLR2 + 1350 T > C polymorphic site, the CC genotype (homozygotic status with two minor C alleles) was correlated with congenital HCMV infection [27]. In turn, our recent study showed no genotypic variability within TLR2 + 1350 T > C as well as 2029 C > T SNPs among the analyzed Polish fetuses and neonates, who were both congenitally infected and uninfected with HCMV [28]. However, the study reported the GA heterozygotic status and A allele located within TLR2 2258 G > A SNP to be significantly more frequent among the infected offsprings than among uninfected individuals [28]. In an in vitro study of the transfected human embryonic kidney (HEK) 293 cells exposed to HCMV gB, the TLR2 2258 SNP was shown to be associated with TLR2 signaling impairment [25]. Considering TLR4, both 896 A > G and 1196 C > T polymorphisms were reported to impair TLR4/MD2 dimerization necessary to activate downstream signaling, involved in HCMV-induced immune response [29, 30]. TLR4 896 A > G and TLR4 1196 C > T SNPs were also determined to be significantly associated with more frequent opportunistic infections and cytomegaly, diagnosed among renal transplant recipients (RTRs) [14, 31]. Another study performed for RTRs and their unrelated donors, showed both TLR4 SNPs to be possibly associated with the risk factors of invasive aspergillosis that included HCMV seropositivity [24]. Considering TLR9, the -1237 T > C SNP was marginally correlated with recurrent urinary infections in RTRs [32]. The study performed for Polish fetuses and neonates congenitally infected with HCMV, showed the CC genotype in TLR4 1196 polymorphism, as well as the GA variant in TLR9 2848 G > A SNP to be correlated with the infection, and the heterozygotic status in TLR9 SNP increased the risk of congenital cytomegaly by 4.81 times [18]. Moreover, complex AA variants for both TLR2 2258 and TLR9 2848 G > A polymorphisms, were found to be associated with an increased risk of congenital HCMV infection [28]. Regarding TLR9 -1486 T > C and 2848 C > T SNPs, the heterozygous and homozygous recessive genotypes within the reported polymorphisms, were associated with an increased risk of HCMV disease among infants [33].
Considering reported data, the association between the presence of genetic changes within TLR2 2258 G > A, TLR4 896 A > G and TLR4 1196 C > T, as well as TLR9 2848 G > A SNPs and the occurrence of HCMV infection among pregnant women seems to be really possible, although there have been no such reports. Therefore, the current paper was aimed to describe the role of TLR2, TLR4 and TLR9 SNPs (see Fig. 1) in the occurrence of HCMV infection among pregnant women, acquired within the gestation period [18, 29, 34–39]. The estimation of the possible relationship between the genetic status within the analyzed polymorphic sites and the appearance of HCMV infection might be significant to provide new genetic alterations possibly associated with the infection occurring among pregnant women during pregnancy.
Fig. 1.
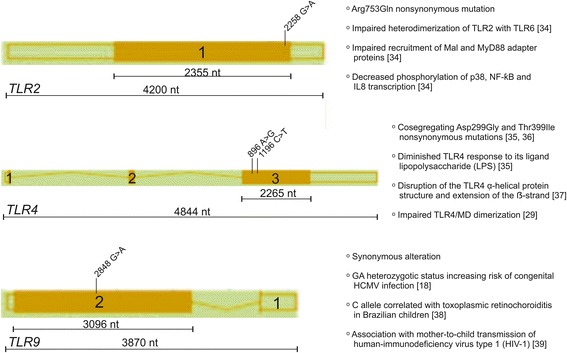
The primary structures of TLR2, TLR4 and TLR9 genes, loci of the polymorphic sites analyzed in the study, and role of the polymorphisms in immune response. Structures of the genes were developed using the Ensembl genome browser and NCBI dbSNP database. Numbered squares represent the following exons in the genes. Lengths of the exons containing analyzed polymorphisms, and of the whole genes, are shown by numbers of nucleotides (nt) over black lines. Diagonal subtitles show the names of studied SNPs. Role of the polymorphisms is indicated on the right side of genes
Methods
The study was performed with 131 pregnant women, including 66 patients infected with HCMV during pregnancy, and 65 age-matched control individuals uninfected with the virus, at the age between 18 and 41 years (mean 28.52 years). The mean age among HCMV-infected pregnant women was 28.29 ± 5.29 years, and among uninfected pregnant women was 28.75 ± 4.79 years. The samples were obtained from pregnant women, admitted to the Department of Feto-Maternal Medicine and Gynecology at the Polish Mother’s Memorial Hospital—Research Institute, between the years 2002 and 2014. Clinical samples, used in the TLRs’ SNPs genotyping, consisted of whole blood and serum specimens. HCMV infection, that occurred within the pregnancy period in pregnant women, was determined by serological status for anti-HCMV antibodies, HCMV DNA detection, as well as by ultrasound markers related to congenital cytomegaly in their fetuses. Symptoms associated with congenital disease included microcephaly (determined in 1 of 66 tested fetuses and neonates, 1.52%), ventriculomegaly (1/66, 1.52%), respiratory failure (5/66, 7.58%), heart defects (4/66, 6.06%), hepatitis (1/66, 1.52%), ascites (2/66, 3.03%), intrauterine growth restriction (IUGR, 6/66, 9.09%), as well as fetal/neonatal death (7/66, 10.61%), and were observed among the 28.8% (19/66) offsprings of the infected pregnant women. Active HCMV infections were confirmed in 15 pregnant women (15/66, 22.7%) and their 10 offsprings (10/66, 15.2%), based on the presence of the viral DNA in body fluids, including maternal whole blood, plasma, and urine samples, as well as fetal amniotic fluids and umbilical cord blood specimens. Considering all the studied parameters related to HCMV infection, the overall rate of congenital viral transmission from mother to the fetus was 33.3% (22/66). The study was approved by the Research Ethics Committee at the Polish Mother’s Memorial Hospital—Research Institute. All the samples, previously collected for diagnostic purposes, were anonymized in this report. Informed consent forms were signed by all the enrolled pregnant women.
Serological tests
Blood specimens were gained from the pregnant women by venipuncture on their first visit to the Hospital, between the 5th and 38th week of gestation (mean 21.96 weeks). The mean gestational age among HCMV-infected pregnant women was 22.50 ± 9.21 weeks, and among uninfected pregnant women was 21.63 ± 9.02 weeks. Serum samples were obtained by centrifugation and then stored at 4oC until analysis, on the day of blood collection. Serological status was determined at the Hospital’s Department of Clinical Microbiology.
Screening for anti-HCMV IgG and IgM antibodies was performed with VIDAS CMV IgG and IgM tests (bioMérieux, France), used between the years 2002 and 2006. Both IgG and IgM antibodies titers ≥ 6 IU/ml were determined as positive. Between 2006 and 2011, the antibodies levels and IgG avidity, were evaluated by the chemiluminescence immunoassays (CLIA), using anti-CMV IgG and IgM tests (Diasorin/Biomedica, Italy). Samples were considered as IgG- or IgM-positive for antibody levels ≥ 0.6 IU/ml and ≥ 30 IU/ml, respectively. The IgG avidity with indexes < 0.200 was interpreted as low, 0.200–0.300 as borderline, and ≥ 0.300 as high. From the year 2012, the CLIA method was replaced by the enzyme-linked fluorescence assays (ELFA), used to determine antibodies levels. IgG antibody titers ≥ 6 IU/ml and IgM indexes ≥ 0.9 were considered positive.
DNA extraction
Genomic and viral DNA was extracted from studied body fluids, using a QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). The isolated DNA was diluted in 100 μL of elution buffer and stored at -20oC until further genetic analyses, according to the manufacturer’s guidelines.
Quantification of HCMV DNA
Among HCMV infected patients, the amount of viral DNA in the study specimens, was determined by the quantitative real-time PCR assay for the detection of UL55 gene fragment, as described previously [40–42]. The reactions were performed in triplicate and the PCR conditions were as follows: initial activation for 10 min at 95°C and 50 cycles of repeated denaturation at 95°C for 15 s and annealing at 60°C for 1 min. The standard curves were obtained from serial 10-fold dilutions from 105 to 1 plasmid DNA, containing the entire HCMV UL55 open reading frame [42, 43]. The amplification was performed by a 7900HT Fast Real-Time PCR System (Applied Biosystems, USA).
Genotyping of TLR2, TLR4 and TLR9 SNPs
TLR2 2258 G > A, TLR4 896 A > G and 1196 C > T SNPs, as well as TLR9 2848 G > A SNP (see Fig. 1), were genotyped, using the self-designed nested PCR-RFLP assays. The sequences of the external and internal primers, the lengths of PCR products and annealing temperatures, used in the PCR assays for TLR SNPs, were previously shown [18, 28, 44]. The sequences, annealing temperatures, and the sizes of amplicons for TLR SNPs are presented in Table 1. The sequences of external primers were developed, using the Vector NTI Suite 5.5 software, whereas the internal primers were taken from the literature [45–49]. Nested PCR products were digested overnight with AciI, NcoI, HinfI or BstUI endonucleases, in order to study the genotypes residing within the analyzed TLR2 2258 G > A, TLR4 896 A > G, TLR4 1196 C > T or TLR9 2848 G > A SNPs, respectively. The genotypes and alleles were determined by the length of restriction fragments, resolved on 2% agarose gels, as previously described [45–49]; see Additional file 1: Figure S1. Randomly selected amplicons for the studied TLR SNPs were then sequenced by the Sanger method, at the Genomed Joint-Stock Company (Warsaw, Poland), to verify the genotypes, determined by the PCR-RFLP assay. Sequencing was performed for five GG homozygotes and seven GA heterozygotes in TLR2 SNP, for six AA homozygotes and four AG heterozygotes in TLR4 896 A > G SNP, for five CC homozygotes and five CT heterozygotes in TLR4 1196 C > T SNP, as well as for five GG homozygotes, five GA heterozygotes and five AA homozygotes in TLR9 SNP. The exemplary chromatograms for DNA fragments of sequenced PCR products encompassing different TLR SNPs are presented in Additional file 2: Figure S2. In order to determine the analyzed genotypes, the sequenced and the reference fragments of the analyzed TLR genes were compared, using the BLASTN program for the alignment of two (or more) sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=blast2seq&LINK_LOC=align2seq). Chromatograms were read, using the Chromas Lite 2.1.1 program.
Table 1.
Primer sequences, annealing temperatures and amplicon lengths, obtained in nested PCR assays for SNPs in the TLR genes
| Gene | GenBank Accession No.a | SNPb name | Primer sequences (5’-3’) | Annealing temperature [oC] | Amplicon length (bps) c | |
|---|---|---|---|---|---|---|
| TLR2 | NC_000004.12 | 2258 G > A (rs5743708) | External | For: CGGAATGTCACAGGACAGC Rev: GGACTTTATCGCAGCTCTCAG |
52 | 605 |
| Internal | For: GCCTACTGGGTGGAGAACCT Rev: GGCCACTCCAGGTAGGTCTT |
59 | 340 | |||
| TLR4 | NG_011475 | 896 A > G | External | For: AAAACTTGTATTCAAGGTCTGGC | 52 | 355 |
| (rs4986790) | Rev: TGTTGGAAGTGAAAGTAAGCCT | |||||
| Internal | For: AGCATACTTAGACTACTACCTCCATG | 61 | 188 | |||
| Rev: AGAAGATTTGAGTTTCAATGTGGG | ||||||
| 1196 C > T | External | For: AGTTGATCTACCAAGCCTTGAGT | 52 | 510 | ||
| (rs4986791) | Rev: GGAAACGTATCCAATGAAAAGA | |||||
| Internal | For: GGTTGCTGTTCTCAAAGTGATTTTGGGAGAA | 59 | 407 | |||
| Rev: ACCTGAAGACTGGAGAGTGAGTTAAATGCT | ||||||
| TLR9 | EU170539 | 2848 G > A | External | For: GTCAATGGCTCCCAGTTCC | 52 | 292 |
| (rs352140) | Rev: CATTGCCGCTGAAGTCCA | |||||
| Internal | For: AAGCTGGACCTCTACCACGA | 59 | 177 | |||
| Rev: TTGGCTGTGGATGTTGTT | ||||||
a No., number
b SNP, single nucleotide polymorphism
c bps, base pairs
Statistical analysis
Distribution of genotypes and alleles in the analyzed TLR2, TLR4 and TLR9 SNPs among HCMV-infected and uninfected pregnant women was estimated by means of descriptive statistics. The studied groups of patients were tested for the Hardy-Weinberg (H-W) equilibrium, the linkage disequilibrium (LD) and haplotypes, using the SNPStats software (http://bioinfo.iconcologia.net/en/SNPStats_web). Genotypes in all the analyzed polymorphisms preserved the H-W equilibrium in pregnant women, both infected with HCMV and control uninfected individuals (P > 0.050). The TLR4 896 A > G and 1196 C > T SNPs were in linkage disequilibrium among the studied groups of pregnant women (P ≤ 0.050). The relationships between the genotypes, alleles or haplotypes in TLR SNPs and the occurrence of the viral infections were determined by cross-tabulation, Pearson’s Chi-squared test and by the logistic regression model. The multiple-SNP analysis for the haplotypes in TLR4 SNPs, as well as for the complex genotypic variants within the range of all the analyzed TLR SNPs was performed by the Expectation Maximization (EM) algorithm. The outcomes of the analyses were determined statistically significant when the significance level of P ≤ 0.050 was obtained. The statistical analysis was in part supported by the NCSS 2004 software.
Results
Prevalence of anti-HCMV IgG and IgM antibodies
In pregnant women, HCMV infection during pregnancy was estimated on the basis of IgG seroconversion within gestation, on the presence of IgG and IgM specific antibodies or on a low IgG avidity index. IgG seropositivity was confirmed in 96.55% (56/58) of the infected pregnant women, while data on IgM antibodies suggestive of the recent infection, were obtained for 86.21% (50/58) of infected individuals. Control group in the study, consisted of pregnant women seronegative for both IgG and IgM antibodies against HCMV, who were classified as uninfected individuals.
HCMV DNA loads in body fluids
The median load of HCMV DNA in whole blood specimens of the infected pregnant women was 3.8 × 102 copies/ml and ranged from 1.3 × 102 to 1.1 × 103 copies/ml, while the mean viral load was 5.2 × 102 copies/ml. In case of plasma specimens, the median viral load was 5.6 × 102 copies/ml, ranging from 1.8 × 102 to 3.6 × 103 copies/ml, whereas the mean viral load was 1.4 × 103 copies/ml. Regarding urine samples, the median HCMV DNA load was 4.4 × 102 copies/ml, ranging from 1.5 × 102 to 2.4 × 103 copies/ml, and the mean viral load was 3.2 × 103 copies/ml. Considering fetal amniotic fluids, the median HCMV DNA load was 9.9 × 102 copies/ml, and ranged from 2.2 × 102 to 1.5 × 103 copies/ml, while the mean viral load was 9.2 × 102 copies/ml. In umbilical cord blood samples, the median HCMV DNA load was 3.2 × 103 copies/ml, and ranged from 1.6 × 102 to 6.6 × 103 copies/ml, and the mean viral load was 3.3 × 103 copies/ml.
Frequencies of the genotypes in TLR2, TLR4 and TLR9 SNPs
In the pregnant women, infected with HCMV, the frequencies of GG and GA genotypes found at TLR2 2258 G > A polymorphic site were 93.9% (62/66) and 6.1% (4/66), respectively (see Table 2). In cases of TLR4 SNPs, AA and AG genotypes in 896 A > G SNP were carried by 91.8% (56/61) and 8.2% (5/61) of the pregnant women, respectively, while CC and CT genotypes in 1196 C > T polymorphism—by 90.9% (60/66) and 9.1% (6/66), respectively. Considering TLR9 2848 G > A SNP, the GG, GA and AA genotypes were determined in 13.3% (8/60), 41.7% (25/60), and 45% (27/60) of patients, respectively. Among uninfected pregnant women, the frequencies of GG and GA genotypes in TLR2 SNP were 92.2% (59/64) and 7.8% (5/64), respectively. The frequencies of AA and AG genotypes in TLR4 896 A > G were 86.9% (53/61) and 13.1% (8/61), respectively, while the rates of CC and CT genotypes in TLR4 1196 C > T were 87.3% (55/63) and 12.7% (8/63), respectively. In cases of TLR9 2848 G > A polymorphism, the GG, GA and AA genotypes were observed in 20.3% (13/64), 53.1% (34/64), and 26.6% (17/64), respectively. Taking that into account, the GG homozygotic and GA heterozygotic status decreased significantly the occurrence of HCMV infection (OR 0.44, 95% CI 0.21–0.94 in the dominant model; P ≤ 0.050; see Table 2). In turn, the frequencies of the genotypes, located in TLR2 and TLR4 SNPs, were similar between the studied groups of the infected and uninfected pregnant women. Moreover, both the haplotypes in TLR4 polymorphisms and the complex variants within the range of all the analyzed SNPs, were observed in similar frequencies among the studied groups of patients. Considering congenital transmission of HCMV from the infected pregnant women to their fetuses, similar frequencies of various genetic variants of the analyzed polymorphisms, were observed between mothers of congenitally infected and uninfected offsprings.
Table 2.
Relationship between TLR2, TLR4 and TLR9 SNPs and the occurrence of HCMV infection among pregnant women
| Gene polymorphism | Genetic model | Genotype | Genotype prevalence rates; n (%) a | ORb (95% CI) c | P-valued | |
|---|---|---|---|---|---|---|
| Infected cases | Uninfected controls | |||||
| TLR2 2258 G > A | - | GG | 62 (93.9%) | 59 (92.2%) | 1.00 | |
| GA | 4 (6.1%) | 5 (7.8%) | 0.76 (0.19–2.97) | 0.690 | ||
| TLR4 896 A > G | - | AA | 56 (91.8%) | 53 (86.9%) | 1.00 | |
| AG | 5 (8.2%) | 8 (13.1%) | 0.59 (0.18–1.92) | 0.380 | ||
| TLR4 1196 C > T | - | CC | 60 (90.9%) | 55 (87.3%) | 1.00 | |
| CT | 6 (9.1%) | 8 (12.7%) | 0.69 (0.22–2.11) | 0.510 | ||
| TLR9 2848 G > A | Codominant | GG | 8 (13.3%) | 13 (20.3%) | 0.39 (0.13–1.13) | 0.093 |
| GA | 25 (41.7%) | 34 (53.1%) | 0.46 (0.21–1.03) | |||
| AA | 27 (45.0%) | 17 (26.6%) | 1.00 | |||
| Dominant | AA | 27 (45.0%) | 17 (26.6%) | 1.00 | ||
| GA-GG | 33 (55.0%) | 47 (73.4%) | 0.44 (0.21–0.94) | 0.032 | ||
| Recessive | AA-GA | 52 (86.7%) | 51 (79.7%) | 1.00 | ||
| GG | 8 (13.3%) | 13 (20.3%) | 0.60 (0.23–1.58) | 0.300 | ||
| Overdominant | GG-AA | 35 (58.3%) | 30 (46.9%) | 1.00 | ||
| GA | 25 (41.7%) | 34 (53.1%) | 0.63 (0.31–1.28) | 0.200 | ||
a n, number of tested pregnant women
b OR, odds ratio
c 95% CI, confidence interval
d logistic regression model; P ≤ 0.050 is considered as significant
Distribution of the alleles in TLR2, TLR4 and TLR9 SNPs
In the infected pregnant women, the frequencies of G and A alleles in TLR2 2258 G > A SNP were 97.0% (128/132) and 3.0% (4/132), respectively (see Table 3). In case of TLR4 polymorphisms, A and G alleles in 896 A > G SNP were determined in 96.0% (117/122) and 4.0% (5/122) of the patients, respectively, while C and T alleles in 1196 C > T SNP—in 95.0% (126/132) and 5.0% (6/132), respectively. Regarding TLR9 2848 G > A SNP, G and A alleles were carried by 34.0% (41/120) and 66.0% (79/120) of the infected pregnant women, respectively. Among uninfected pregnant women, the frequencies of G and A alleles in TLR2 2258 G > A SNP were 96.0% (123/128) and 4.0% (5/128), respectively. The frequencies of A and G alleles in TLR4 896 A > G SNP were 93.0% (114/122) and 7.0% (8/122), respectively, and the rates of C and T alleles in TLR4 1196 C > T SNP were 94.0% (118/126) and 6.0% (8/126), respectively. Considering TLR9 SNP, the G and A alleles were found in frequencies of 47.0% (60/128) and 53.0% (68/128), respectively. The G allele in TLR9 SNP was significantly more frequent among the uninfected pregnant women, as compared to the infected ones (χ2 = 4.14, P ≤ 0.050; Pearson’s Chi-squared test; see Table 3). The distribution of the alleles at other analyzed polymorphic sites was similar between the studied groups of patients.
Table 3.
Distribution of the alleles, located in TLR2, TLR4 and TLR9 SNPs
| Gene polymorphism and allele | No.a of carriers with TLR alleles (%) | P-valueb | ||
|---|---|---|---|---|
| Cases | Controls | |||
| TLR2 2258 G > A | ||||
| G | 128 (97%) | 123 (96%) | 0.699 | |
| A | 4 (3%) | 5 (4%) | ||
| TLR4 896 A > G | ||||
| A | 117 (96%) | 114 (93%) | 0.392 | |
| G | 5 (4%) | 8 (7%) | ||
| TLR4 1196 C > T | ||||
| C | 126 (95%) | 118 (94%) | 0.523 | |
| T | 6 (5%) | 8 (5%) | ||
| TLR9 2848 G > A | ||||
| G | 41 (34%) | 60 (47%) | 0.042 | |
| A | 79 (66%) | 68 (53%) | ||
a No. - number
b Pearson’s Chi-squared test; P ≤ 0.050 is considered significant
Discussion
Both GG homozygous and GA heterozygous variants in TLR9 2848 G > A SNP were found in our study to have been significantly associated with a decreased occurrence of HCMV infection among pregnant women. Additionally, the G allele in TLR9 SNP was observed significantly more frequently among the uninfected pregnant women than among the infected ones. The frequencies of the genotypes and alleles within TLR9 2848 G > A SNP determined among uninfected pregnant women, but not among HCMV-infected patients, were similar to the frequencies reported for European populations (see https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=352140).
Differences in distribution of both the genotypes and alleles within TLR9 2848 G > A SNP between the infected pregnant women analyzed in the current study, and the reported European populations might be due to a selection bias. Similarly, significantly higher prevalence rates of anti-HCMV IgG and IgM antibodies determined among the infected pregnant women studied in this paper, as compared to the previously reported cohort of Polish pregnant women between 2010 and 2011, as well as to other European populations, are possibly also caused by a classification of the patients to the presented study [4–7]. So far, it has been the first study to reveal a really significant role of TLR9 2848 G > A SNP in the occurrence of HCMV infection in pregnant women. Another research reported before that the GA heterozygotic status in the analyzed TLR9 SNP, carried by fetuses and neonates, congenitally infected with HCMV, had significantly increased the risk of the infection [18]. In turn, the G allele in TLR9 SNP was more frequent among the infected offsprings than among the uninfected ones, although that difference was statistically non-significant [18]. The distinct contribution of the analyzed TLR9 SNP in the occurrence of HCMV infection may have been age-related, as well as may have resulted from the classification bias. The qualification criteria of the pregnant women to the study included their serological status, the presence of HCMV DNA in their body fluids and were also based on confirmed congenital infection with the virus in their fetuses. In turn, the fetuses were diagnosed as congenitally infected with HCMV only on the basis of ultrasound markers and on the presence of HCMV DNA in their body fluids, since the diagnostics towards the infection is not routinely performed in pregnant women. Considering the role of TLR9 SNP in HCMV infection, the TT homozygotic status in TLR9 -1237 T > C SNP was reported to have been correlated with a decreased risk of the infection in HCMV-seropositive kidney transplant recipients [50]. In recipients of allogeneic hematopoietic stem cell transplants, the HCMV infection occurred significantly more frequently among carriers of minor C allele in TLR9 -1237 T > C, as compared to the patients, carrying the T allele [51]. On the other hand, the TLR9 2848 G > A polymorphism was not involved in the incidence of HCMV infection among the examined patients [51]. However, TLR9 -1486 T > C and 2848 C > T SNPs were associated with an increased risk of HCMV disease among infants [33]. Therefore, it seems possible that different SNPs, residing within TLR9 gene, may have been correlated with various disease types. Regarding the HCMV infection, previous papers showed some contribution of TLR9 to its occurrence [14, 22, 23]. In case of plasmacytoid dendritic cells (pDCs), the infection with HCMV was significantly associated with increased levels of TLR9 [52]. The inhibited cytokine expression, observed after treatment of pDCs with CpG, agonists for TLR7 and TLR9, suggested an involvement of the reported TLRs in the development of HCMV infection [52]. What is more, a significant role in the occurrence of HCMV infection was previously confirmed for TLR2 and TLR4 molecules as well [30, 53, 54]. TLR2 was found to have been involved in the functional sensing of HCMV through a direct interaction with the viral glycoproteins gB and gH [20]. In case of TLR4, the molecule was involved in an inhibition of HCMV infection [21]. The expression levels of both TLR2 and TLR4 were correlated with the levels of HCMV IE1-72 protein [55]. Considering genetic alterations in the current study, no differences were found in the distribution of distinct genotypes within the range of both TLR2 and TLR4 SNPs. The frequencies of the genotypes in the analyzed TLR2 and TLR4 polymorphic sites, found among HCMV-infected pregnant women were similar to the frequencies observed among European populations (see https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=5743708; https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=4986790; https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=4986791). Taking into account the allelic variability within TLR2 2258 G > A and TLR4 1196 C > T SNPs, the frequencies observed among all pregnant women studied in our research, were similar to the frequencies reported for European populations. In case of TLR4 896 C > T SNP, similarity to the European populations was found for HCMV-infected pregnant women (see https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=4986790). So far, no other study has shown before any possible involvement of TLR2 2258 G > A, TLR4 896 A > G and TLR4 1196 C > T SNPs in the occurrence of HCMV infection during pregnancy. However, in case of congenital HCMV infection, GA heterozygotic status and A allele within TLR2 2258 G > A, as well as CC genotype in the other TLR2 + 1350 T > C SNP, was reported to be correlated with the infection [27, 28]. Regarding presented data, the age-dependent type of the involvement of TLR2 2258 G > A polymorphism to HCMV infection seems to be quite possible. Considering TLR4 SNPs, the CC genotype in TLR4 1196 C > T SNP, GC haplotype in both analyzed TLR4 SNPs, as well as GCA multiple variants within the range of TLR4 and TLR9 2848 G > A SNPs, were found to have been correlated with congenital HCMV infection in fetuses and neonates [18]. In pregnant women, the frequencies of distinct genotypes in TLR4 SNPs were similar to those, determined among congenitally infected fetuses and neonates, although no CT genotype at TLR4 1196 C > T polymorphic site was observed among the infected offsprings [18]. Among pregnant women, the low genotypic variability within TLR2 and TLR4 SNPs seems to be the important cause of the lack of any associations with HCMV infection. Similarly to our study, almost the same frequencies of distinct genotypes in both TLR2 2258 G > A and TLR4 896 G > A SNPs were determined in patients with transplants, with and without clinical signs of HCMV infection [50]. Regarding the outcomes presented in this report, TLR9 2848 G > A SNP seems to be the major polymorphism, contributing to HCMV infection in pregnant women during pregnancy. However, the studied genetic alteration is not involved, either in amino acid changes in TLR9 molecule or in regulatory site modifications within TLR9 gene [56]. Therefore, other molecular changes occurring simultaneously with the analyzed polymorphism, may contribute to the course of immune response after the infection with HCMV.
Conclusions
The outcomes presented in the current study show that TLR9 2848 G > A SNP seems to be involved in the occurrence of HCMV infection in pregnant women. Since both the GG homozygotic and GA heterozygotic statuses, as well as the G allele at TLR9 polymorphic site were significantly more frequent among the uninfected pregnant women, when compared with the infected ones, the genetic alterations in the studied polymorphism may plausibly be associated with the infection with HCMV. However, we suggest that further detailed studies would highly be justified to investigate the molecular mechanism of the TLR9 2848 G > A SNP function in HCMV infection.
Additional files
Exemplary PCR-RFLP products representative for various genotypes within TLR2 2258 G > A (A), TLR4 896 A > G (B), TLR4 1196 C > T (C) and TLR9 2848 G > A (D) SNPs. DNA fragments were resolved in 2% agarose gel, stained with ethidium bromide. Disparate lanes show restriction profiles for distinct genotypes in the range of studied TLR polymorphisms, determined in different pregnant women. The numbers on the right side of electropherograms show the size of separated DNA fragments. M—50 bp DNA marker; GG, GA, AA, AG, CT, CC—genotypes determined in studied TLR polymorphisms. (TIF 501 kb)
a and b Representative chromatograms of DNA fragments of various sequenced PCR products, encompassing TLR2 2258 G > A (A, B), TLR4 896 A > G (C, D), TLR4 1196 C > T (E, F) and TLR9 2848 G > A (G-I) SNPs. For TLR2 SNP, DNA forward strands were sequenced, and for TLR4 and TLR9—reverse strands were assayed. The numbers above some peaks of chromatograms indicate the following nucleotides determined in sequenced DNA fragments. Loci of the polymorphisms and genotypes analyzed in the study, are indicated with arrows. GG, GA, AA, AG, CT, CC—genotypes determined in studied TLR SNPs. (ZIP 1732 kb)
Acknowledgements
Not applicable.
Funding
This study was funded by the Polish Ministry of Science & Higher Education, Polish Mother’s Memorial Hospital-Research Institute (Young Researcher Internal Grant No. 2015/II/16-MN).
Availability of data and materials
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author’s contributions
WW and DN contributed to conception and designed the research. WW, EP, MS, JW and DN acquired data in the study. WW analyzed data, and all authors interpreted them. WW drafted the paper, and all authors revised it critically, read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was in accordance with the Helsinki Declaration and was approved by the Research Ethics Committee at the Polish Mother’s Memorial Hospital—Research Institute.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wioletta Wujcicka, Phone: +48 (42) 271 15 20, Email: wwujcicka@yahoo.com.
Edyta Paradowska, Email: eparadowska@cbm.pan.pl.
Mirosława Studzińska, Email: mstudzinska@cbm.pan.pl.
Jan Wilczyński, Email: janszczesnywilczynski@gmail.com.
Dorota Nowakowska, Email: dnowakowska@yahoo.com.
References
- 1.Cordier AG, Guitton S, Vauloup-Fellous C, Grangeot-Keros L, Benachi A, Picone O. Awareness and knowledge of congenital cytomegalovirus infection among health care providers in France. J Clin Virol. 2012;55:158–163. doi: 10.1016/j.jcv.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 2.Paradowska E, Studzinska M, Nowakowska D, Wilczyński J, Rycel M, Suski P, et al. Distribution of UL144, US28 and UL55 genotypes in Polish newborns with congenital cytomegalovirus infections. Eur J Clin Microbiol Infect Dis. 2012;31:1335–1345. doi: 10.1007/s10096-011-1447-z. [DOI] [PubMed] [Google Scholar]
- 3.Rycel M, Wujcicka W, Zawilinska B, Paradowska E, Suski P, Gaj Z, et al. Mixed infections with distinct cytomegalovirus glycoprotein B genotypes in Polish pregnant women, fetuses, and newborns. Eur J Clin Microbiol Infect Dis. 2014;34:585–591. doi: 10.1007/s10096-014-2266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wujcicka W, Gaj Z, Wilczynski J, Sobala W, Spiewak E, Nowakowska D. Impact of socioeconomic risk factors on the seroprevalence of cytomegalovirus infections in a cohort of pregnant Polish women between 2010 and 2011. Eur J Clin Microbiol Infect Dis. 2014;33:1951–1958. doi: 10.1007/s10096-014-2170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picone O, Vauloup-Fellous C, Cordier AG, Parent Du Châtelet I, Senat MV, Frydman R, et al. A 2-year study on cytomegalovirus infection during pregnancy in a French hospital. BJOG. 2009;116:818–823. doi: 10.1111/j.1471-0528.2009.02139.x. [DOI] [PubMed] [Google Scholar]
- 6.Gaytant MA, Galama JM, Semmekrot BA, Melchers WJ, Sporken JM, Oosterbaan HP, et al. The incidence of congenital cytomegalovirus infections in The Netherlands. J Med Virol. 2005;76:71–75. doi: 10.1002/jmv.20325. [DOI] [PubMed] [Google Scholar]
- 7.Pembrey L, Raynor P, Griffiths P, Chaytor S, Wright J, Hall AJ. Seroprevalence of cytomegalovirus, Epstein Barr virus and varicella zoster virus among pregnant women in Bradford: a cohort study. PLoS One. 2013;27:e81881. doi: 10.1371/journal.pone.0081881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benoist G, Leruez-Ville M, Magny JF, Jacquemard F, Salomon LJ, Ville Y. Management of pregnancies with confirmed cytomegalovirus fetal infection. Fetal Diagn Ther. 2013;33:203–214. doi: 10.1159/000342752. [DOI] [PubMed] [Google Scholar]
- 9.Hollier LM, Grissom H. Human herpes viruses in pregnancy: cytomegalovirus, Epstein-Barr virus, and varicella zoster virus. Clin Perinatol. 2005;32:671–696. doi: 10.1016/j.clp.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol. 2007;17:253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 11.Rycel M, Gaj Z, Wilczynski J, Paradowska E, Studzińska M, Suski P, et al. Evaluation of the association between maternal HCMV viremia and the course of pregnancy and neonatal outcome. Ginekol Pol. 2013;84:1005–1011. doi: 10.17772/gp/1672. [DOI] [PubMed] [Google Scholar]
- 12.Malinger G, Lev D, Lerman-Sagie T. Imaging of fetal cytomegalovirus infection. Fetal Diagn Ther. 2011;29:117–126. doi: 10.1159/000321346. [DOI] [PubMed] [Google Scholar]
- 13.Renneson J, Dutta B, Goriely S, Danis B, Lecomte S, Laes JF, et al. IL-12 and type I IFN response of neonatal myeloid DC to human CMV infection. Eur J Immunol. 2009;39:2789–2799. doi: 10.1002/eji.200939414. [DOI] [PubMed] [Google Scholar]
- 14.Wujcicka W, Wilczynski J, Nowakowska D. Alterations in TLRs as new molecular markers of congenital infections with Human cytomegalovirus? Pathog Dis. 2014;70:3–16. doi: 10.1111/2049-632X.12083. [DOI] [PubMed] [Google Scholar]
- 15.Yew KH, Harrison CJ. Blockade of Lyn kinase upregulates both canonical and non-canonical TLR-3 pathways in THP-1 monocytes exposed to human cytomegalovirus. Acta Virol. 2011;55:243–253. doi: 10.4149/av_2011_03_243. [DOI] [PubMed] [Google Scholar]
- 16.Jablonska A, Paradowska E, Studzinska M, Suski P, Nowakowska D, Wiśniewska-Ligier M, et al. Relationship between toll-like receptor 2 Arg677Trp and Arg753Gln and toll-like receptor 4 Asp299Gly polymorphisms and cytomegalovirus infection. Int J Infect Dis. 2014;25:11–15. doi: 10.1016/j.ijid.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Muntasell A, Costa-Garcia M, Vera A, Marina-Garcia N, Kirschning CJ, Lopez-Botet M. Priming of NK cell anti-viral effector mechanisms by direct recognition of human cytomegalovirus. Front Immunol. 2013;4:40. doi: 10.3389/fimmu.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wujcicka W, Paradowska E, Studzinska M, Gaj Z, Wilczyński J, Leśnikowski Z, et al. TLR9 2848 GA Heterozygotic Status Possibly Predisposes Fetuses and Newborns to Congenital Infection with Human Cytomegalovirus. PLoS One. 2015;10:e0122831. doi: 10.1371/journal.pone.0122831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landais I, Pelton C, Streblow D, DeFilippis V, McWeeney S, Nelson JA. Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFkappaB Signaling Pathway. PLoS Pathog. 2015;11:e1004881. doi: 10.1371/journal.ppat.1004881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehme KW, Guerrero M, Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol. 2006;177:7094–7102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- 21.Harwani SC, Lurain NS, Zariffard MR, Spear GT. Differential inhibition of human cytomegalovirus (HCMV) by toll-like receptor ligands mediated by interferon-beta in human foreskin fibroblasts and cervical tissue. Virol J. 2007;4:133. doi: 10.1186/1743-422X-4-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yew KH, Carsten B, Harrison C. Scavenger receptor A1 is required for sensing HCMV by endosomal TLR-3/-9 in monocytic THP-1 cells. Mol Immunol. 2010;47:883–893. doi: 10.1016/j.molimm.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Iversen AC, Steinkjer B, Nilsen N, Bohnhorst J, Moen SH, Vik R, et al. A proviral role for CpG in cytomegalovirus infection. J Immunol. 2009;182:5672–5681. doi: 10.4049/jimmunol.0801268. [DOI] [PubMed] [Google Scholar]
- 24.Bochud PY, Chien JW, Marr KA, Leisenring WM, Upton A, Janer M, et al. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med. 2008;359:1766–1777. doi: 10.1056/NEJMoa0802629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown RA, Gralewski JH, Razonable RR. The R753Q polymorphism abrogates toll-like receptor 2 signaling in response to human cytomegalovirus. Clin Infect Dis. 2009;49:e96–e99. doi: 10.1086/644501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang SH, Abdel-Massih RC, Brown RA, Dierkhising RA, Kremers WK, Razonable RR. Homozygosity for the toll-like receptor 2 R753Q single-nucleotide polymorphism is a risk factor for cytomegalovirus disease after liver transplantation. J Infect Dis. 2012;205:639–646. doi: 10.1093/infdis/jir819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi R, Koyano S, Suzutani T, Goishi K, Ito Y, Morioka I, et al. Polymorphisms in TLR-2 are associated with congenital cytomegalovirus (CMV) infection but not with congenital CMV disease. Int J Infect Dis. 2013;17:e1092–e1097. doi: 10.1016/j.ijid.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Wujcicka W, Paradowska E, Studzińska M, Wilczyński J, Nowakowska D. TLR2 2258 G > A single nucleotide polymorphism and the risk of congenital infection with human cytomegalovirus. Virol J. 2017;14:12. doi: 10.1186/s12985-016-0679-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamakawa N, Ohto U, Akashi-Takamura S, Takahashi K, Saitoh S, Tanimura N, et al. Human TLR4 polymorphism D299G/T399I alters TLR4/MD-2 conformation and response to a weak ligand monophosphoryl lipid A. Int Immunol. 2013;25:45–52. doi: 10.1093/intimm/dxs084. [DOI] [PubMed] [Google Scholar]
- 30.Yew KH, Carpenter C, Duncan RS, Harrison CJ. Human cytomegalovirus induces TLR4 signaling components in monocytes altering TIRAP, TRAM and downstream interferon-beta and TNF-alpha expression. PLoS One. 2012;7:e44500. doi: 10.1371/journal.pone.0044500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ducloux D, Deschamps M, Yannaraki M, Ferrand C, Bamoulid J, Saas P, et al. Relevance of Toll-like receptor-4 polymorphisms in renal transplantation. Kidney Int. 2005;67:2454–2461. doi: 10.1111/j.1523-1755.2005.00354.x. [DOI] [PubMed] [Google Scholar]
- 32.Kruger B, Banas MC, Walberer A, Böger CA, Farkas S, Hoffmann U, et al. A comprehensive genotype-phenotype interaction of different Toll-like receptor variations in a renal transplant cohort. Clin Sci (Lond) 2010;119:535–54430. doi: 10.1042/CS20100190. [DOI] [PubMed] [Google Scholar]
- 33.Paradowska E, Jabłońska A, Studzińska M, Skowrońska K, Suski P, Wiśniewska-Ligier M, et al. TLR9–1486T/C and 2848C/T SNPs Are Associated with Human Cytomegalovirus Infection in Infants. PLoS One. 2016;11:e0154100. doi: 10.1371/journal.pone.0154100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong Y, Song C, Snyder GA, Sundberg EJ, Medvedev AE. R753Q polymorphism inhibits Toll-like receptor (TLR) 2 tyrosine phosphorylation, dimerization with TLR6, and recruitment of myeloid differentiation primary response protein 88. J Biol Chem. 2012;287:38327–38337. doi: 10.1074/jbc.M112.375493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 36.Bell JK, Mullen GE, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/S1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 37.Gibrat JF, Garnier J, Robson B. Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 38.Peixoto-Rangel AL, Miller EN, Castellucci L, Jamieson SE, Peixe RG, Elias Lde S, et al. Candidate gene analysis of ocular toxoplasmosis in Brazil: evidence for a role for toll-like receptor 9 (TLR9) Mem Inst Oswaldo Cruz. 2009;104:1187–1190. doi: 10.1590/S0074-02762009000800019. [DOI] [PubMed] [Google Scholar]
- 39.Ricci E, Malacrida S, Zanchetta M, Mosconi I, Montagna M, Giaquinto C, et al. Toll-like receptor 9 polymorphisms influence mother-to-child transmission of human immunodeficiency virus type 1. J Transl Med. 2010;8:49. doi: 10.1186/1479-5876-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassan-Walker AF, Mattes FM, Griffiths PD, Emery VC. Quantity of cytomegalovirus DNA in different leukocyte populations during active infection in vivo and the presence of gB and UL18 transcripts. J Med Virol. 2001;64:283–289. doi: 10.1002/jmv.1048. [DOI] [PubMed] [Google Scholar]
- 41.Paradowska E, Przepiórkiewicz M, Nowakowska D, Studzińska M, Wilczyński J, Emery VC, et al. Detection of cytomegalovirus in human placental cells by polymerase chain reaction. APMIS. 2006;114:764–771. doi: 10.1111/j.1600-0463.2006.apm_31.x. [DOI] [PubMed] [Google Scholar]
- 42.Wujcicka W, Wilczyński J, Paradowska E, Studzińska M, Nowakowska D. The role of single nucleotide polymorphisms, contained in proinflammatory cytokine genes, in the development of congenital infection with human cytomegalovirus in fetuses and neonates. Microb Pathog. 2017;105:106–116. doi: 10.1016/j.micpath.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 43.Temperton NJ, Quenelle DC, Lawson KM, Zuckerman JN, Kern ER, Griffiths PD, et al. Enhancement of humoral immune responses to a human cytomegalovirus DNA vaccine: adjuvant effects of aluminum phosphate and CpG oligodeoxynucleotides. J Med Virol. 2003;70:86–90. doi: 10.1002/jmv.10357. [DOI] [PubMed] [Google Scholar]
- 44.Wujcicka W, Gaj Z, Wilczynski J, Nowakowska D. Possible role of TLR4 and TLR9 SNPs in protection against congenital toxoplasmosis. Eur J Clin Microbiol Infect Dis. 2015;34:2121–2129. doi: 10.1007/s10096-015-2461-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biswas D, Gupta SK, Sindhwani G, Patras A. TLR2 polymorphisms, Arg753Gln and Arg677Trp, are not associated with increased burden of tuberculosis in Indian patients. BMC Res Notes. 2009;2:162. doi: 10.1186/1756-0500-2-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirchner M, Sonnenschein A, Schoofs S, Schmidtke P, Umlauf VN, Mannhardt-Laakmann W. Surface expression and genotypes of Toll-like receptors 2 and 4 in patients with juvenile idiopathic arthritis and systemic lupus erythematosus. Pediatr Rheumatol Online J. 2013;11:9–11. doi: 10.1186/1546-0096-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meena NK, Verma R, Verma N, Ahuja V, Paul J. TLR4 D299G polymorphism modulates cytokine expression in ulcerative colitis. J Clin Gastroenterol. 2013;47:773–780. doi: 10.1097/MCG.0b013e31828a6e93. [DOI] [PubMed] [Google Scholar]
- 48.Pandey S, Mittal B, Srivastava M, et al. Evaluation of Toll-like receptors 3 (c.1377C/T) and 9 (G2848A) gene polymorphisms in cervical cancer susceptibility. Mol Biol Rep. 2011;38:4715–4721. doi: 10.1007/s11033-010-0607-z. [DOI] [PubMed] [Google Scholar]
- 49.Schroder NW, Diterich I, Zinke A, Eckert J, Draing C, von Baehr V, et al. Heterozygous Arg753Gln polymorphism of human TLR-2 impairs immune activation by Borrelia burgdorferi and protects from late stage Lyme disease. J Immunol. 2005;175:2534–2540. doi: 10.4049/jimmunol.175.4.2534. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Ruiz M, Corrales I, Arias M, Campistol JM, Giménez E, Crespo J, et al. Association between individual and combined SNPs in genes related to innate immunity and incidence of CMV infection in seropositive kidney transplant recipients. Am J Transplant. 2015;15:1323–1335. doi: 10.1111/ajt.13107. [DOI] [PubMed] [Google Scholar]
- 51.Carvalho A, Cunha C, Carotti A, Aloisi T, Guarrera O, Di Ianni M, et al. Polymorphisms in Toll-like receptor genes and susceptibility to infections in allogeneic stem cell transplantation. Exp Hematol. 2009;37:1022–1029. doi: 10.1016/j.exphem.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Varani S, Cederarv M, Feld S, Tammik C, Frascaroli G, Landini MP, et al. Human cytomegalovirus differentially controls B cell and T cell responses through effects on plasmacytoid dendritic cells. J Immunol. 2007;179:7767–7776. doi: 10.4049/jimmunol.179.11.7767. [DOI] [PubMed] [Google Scholar]
- 53.Assinger A, Kral JB, Yaiw KC, Schrottmaier WC, Kurzejamska E, Wang Y, et al. Human cytomegalovirus-platelet interaction triggers toll-like receptor 2-dependent proinflammatory and proangiogenic responses. Arterioscler Thromb Vasc Biol. 2014;34:801–809. doi: 10.1161/ATVBAHA.114.303287. [DOI] [PubMed] [Google Scholar]
- 54.Xie F, von Dadelszen P, Nadeau J. CMV infection, TLR-2 and -4 expression, and cytokine profiles in early-onset preeclampsia with HELLP syndrome. Am J Reprod Immunol. 2014;71:379–386. doi: 10.1111/aji.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Qian D, Ju F, Wang B. Upregulation of Toll-like receptor 2 expression in colorectal cancer infected by human cytomegalovirus. Oncol Lett. 2015;9:365–370. doi: 10.3892/ol.2014.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanders MS, van Well GT, Ouburg S, Morre SA, van Furth AM. Toll-like receptor 9 polymorphisms are associated with severity variables in a cohort of meningococcal meningitis survivors. BMC Infect Dis. 2012;12:112. doi: 10.1186/1471-2334-12-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exemplary PCR-RFLP products representative for various genotypes within TLR2 2258 G > A (A), TLR4 896 A > G (B), TLR4 1196 C > T (C) and TLR9 2848 G > A (D) SNPs. DNA fragments were resolved in 2% agarose gel, stained with ethidium bromide. Disparate lanes show restriction profiles for distinct genotypes in the range of studied TLR polymorphisms, determined in different pregnant women. The numbers on the right side of electropherograms show the size of separated DNA fragments. M—50 bp DNA marker; GG, GA, AA, AG, CT, CC—genotypes determined in studied TLR polymorphisms. (TIF 501 kb)
a and b Representative chromatograms of DNA fragments of various sequenced PCR products, encompassing TLR2 2258 G > A (A, B), TLR4 896 A > G (C, D), TLR4 1196 C > T (E, F) and TLR9 2848 G > A (G-I) SNPs. For TLR2 SNP, DNA forward strands were sequenced, and for TLR4 and TLR9—reverse strands were assayed. The numbers above some peaks of chromatograms indicate the following nucleotides determined in sequenced DNA fragments. Loci of the polymorphisms and genotypes analyzed in the study, are indicated with arrows. GG, GA, AA, AG, CT, CC—genotypes determined in studied TLR SNPs. (ZIP 1732 kb)
Data Availability Statement
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.


