Abstract
Background
Repeat breeding directly affects reproductive efficiency in cattle due to an increase in services per conception and calving interval. This study aimed to investigate whether changes in endometrial gene expression profile are involved in repeat breeding in cows. Differential gene expression profiles of the endometrium were investigated during the mid-luteal phase of the estrous cycle between repeat breeder (RB) and non-RB cows using microarray analysis.
Methods
The caruncular (CAR) and intercaruncular (ICAR) endometrium of both ipsilateral and contralateral uterine horns to the corpus luteum were collected from RB (inseminated at least three times but not pregnant) and non-RB cows on Day 15 of the estrous cycle (4 cows/group). Global gene expression profiles of these endometrial samples were analyzed with a 15 K custom-made oligo-microarray for cattle. Immunohistochemistry was performed to investigate the cellular localization of proteins of three identified transcripts in the endometrium.
Results
Microarray analysis revealed that 405 and 397 genes were differentially expressed in the CAR and ICAR of the ipsilateral uterine horn of RB, respectively when compared with non-RB cows. In the contralateral uterine horn, 443 and 257 differentially expressed genes were identified in the CAR and ICAR of RB, respectively when compared with non-RB cows. Gene ontology analysis revealed that genes involved in development and morphogenesis were mainly up-regulated in the CAR of RB cows. In the ICAR of both the ipsilateral and contralateral uterine horns, genes related to the metabolic process were predominantly enriched in the RB cows when compared with non-RB cows. In the analysis of the whole uterus (combining the data above four endometrial compartments), RB cows showed up-regulation of 37 genes including PRSS2, GSTA3 and PIPOX and down-regulation of 39 genes including CHGA, KRT35 and THBS4 when compared with non-RB cows. Immunohistochemistry revealed that CHGA, GSTA3 and PRSS2 proteins were localized in luminal and glandular epithelial cells and stroma of the endometrium.
Conclusion
The present study showed that endometrial gene expression profiles are different between RB and non-RB cows. The identified candidate endometrial genes and functions in each endometrial compartment may contribute to bovine reproductive performance.
Electronic supplementary material
The online version of this article (doi:10.1186/s12958-017-0237-6) contains supplementary material, which is available to authorized users.
Keywords: Repeat breeder, Endometrium, Caruncle, Intercaruncle, Microarray, Cow
Background
Repeat breeder (RB) is generally defined as any cow that has failed to conceive after at least three inseminations. In both dairy and beef cattle herds, the presence of RB cows can directly lead a large economic loss for producers due to an extension of the length of the open period and frequent artificial insemination (AI) [1]. In addition to management problems such as inadequate estrus detection and AI techniques, various physiological problems of individual cows are one of major causes of repeat breeding. For example, infections of uterus, cervix and/or vagina, dysfunctions of uterus or ovary, obstructed oviducts, defective oocytes and anatomical defects of the reproductive tracts are involved in conception failure, early embryonic death and endocrine disorders of RB animals. [1]. It has been reported that embryo transfer is effective to improve the fertility of RB cows and heifers [2, 3]. On the other hand, a study of reciprocal transfers of embryos between RB and virgin heifers showed that a higher proportion of embryos transferred from RB to virgin heifers than from virgin to RB heifers survived at day 16 to 17, suggesting that the uterine environment in RB heifers is less suitable than in the virgins for supporting a successful embryo development [4]. This became more evident by transfer of identical demi-embryos to RB and virgin recipient heifers resulted less number of morphologically normal and elongated embryos in the RB heifers than in the virgin heifers at day 15 [5]. About an association between alteration of uterine environment and repeat breeding, Katagiri et al. have demonstrated that there is a close relationship between the endometrial epidermal growth factor profile and diminished fertility of RB cows [6].
The molecular mechanisms underlying endometrial function may contribute to reproductive performance in cattle. Increasing evidence using global gene expression analysis has identified numerous differentially expressed genes and related functional pathways in bovine endometrium among highly fertile, subfertile and infertile animal strains during estrous cycle or early pregnancy [7–10]. Recent studies have also investigated gene expression profiles under various conditions of the bovine endometrium during the estrous cycle and/or during early pregnancy using DNA microarray or RNA sequencing [11–18]. In addition, microarray studies have revealed that heat stress and steroid hormones directly affect bovine endometrial gene expression profiles [19, 20].
In ruminants, the endometrium shows structural and physiological differences depending on the uterine compartments. The caruncular (CAR) areas are aglandular and a limited area that forms placentomes by fusing with the fetal extraembryonic membrane [21, 22]. On the other hand, the intercaruncular (ICAR) areas contain endometrial glands that synthesize and secrete substances or factors that are essential for survival and development of the conceptus [23, 24]. A study that directly compared the gene expression profiles of CAR and ICAR during implantation in cows showed 1177 and 453 differentially expressed genes (DEG) were found for cyclic and pregnant animals, respectively [13]. In addition, it has been reported that tissues of the ipsilateral uterine horn to the ovary with the corpus luteum (CL) contain greater quantities of progesterone (P4) and are more sensitive to P4 as compared with tissues on the contralateral side [25]. Although a previous study demonstrated that a few genes show differences in expression between ipsilateral and contralateral uterine horns during the bovine estrous cycle [11], we consider that it is important to analyze each compartment of the bovine endometrium separately in order to understand enodometrial function more comprehensively.
These previous studies suggest that alteration of the endometrial function due to changes in gene expression may contribute to their lower reproductive performance in RB cows, whereas details of the molecular mechanisms and biological pathways of their endometria still need to be elucidated. Thus, we hypothesized that there is a characteristic gene expression profile in the endometrium of the RB cows. This study aimed to investigate differences in gene expression profiles of the endometrium between RB and non-RB cows during the mid-luteal phase of the estrous cycle. In pregnant cattle, maternal recognition of pregnancy occurs around Day 14–15 [26]. In addition, it has been reported that the majority of early embryo losses in cattle have occurred within 16 days of gestation (i.e. during the mid-luteal phase) [27, 28]. Therefore, the basal gene expression profiles of endometrium at mid luteal phase would have the most important association with reproductive performance.
Methods
Animals and sample collection
This study was carried out using non-lactating Japanese Black cows at the institute’s ranch (age: 7.8 ± 0.9 years, parity: 3.3 ± 0.8, open period from last parturition to first AI in this study: 104 ± 9.6 month). Repeat breeder cows (n = 4) were defined based on a previous study by Dochi et al. [3]. Briefly, the RB cows had three characteristics as follows: (1) detectable estrous behavior, but not always normal estrous cycles; (2) not conceiving after three or more inseminations following normal estrous behavior; and (3) healthy uterus and ovaries, as determined by transrectal palpation. Non-RB cows (n = 4) conceived within three inseminations. The non-RB cows were confirmed to be pregnant by transrectal ultrasonography (HS-1500V; Honda Electronics. Co., Aichi, Japan) at 40 days after insemination, then abortion was induced by a single intramuscular injection of 500 μg of prostaglandin F2α (cloprostenol [Dalmazin]; Kyoritsu Seiyaku. Co., Tokyo, Japan) followed by repeated normal estrous cycles at least twice. Both RB and non-RB cows were slaughtered on Day 15 of the estrous cycle (the day of estrus was designated as Day 0) and the uterus and both ovaries together were collected. Uterine horns were identified as ipsilateral to the ovary containing the CL or contralateral. We collected CAR and ICAR in the endometrium from the middle area of each uterine horns. The uterine horns were cut opened longitudinally using scissors and CAR were carefully dissected first not to include ICAR, subsequently, ICAR areas were cut off. Collected samples were snap-frozen in liquid nitrogen and stored at −80 °C until RNA extraction. Whole cross section of the uterus for immunohistochemistry were collected from the middle area of ipsilateral uterine horn of all cows and fixed in 10% formalin (v/v), embedded in paraffin wax, and then stored at 4 °C until use. All procedures in animal experiments were carried out in accordance with guidelines approved by the Animal Ethics Committee of the National Institute of Agrobiological Sciences for the use of animals (permission number: H18-036).
Microarray analysis
Total RNA was extracted from each sample by acid guanidinium thiocyanate-phenol-chloroform with ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instructions. All RNA samples were then treated with TURBO DNase (TURBO DNA-free™ Kit, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions to remove contaminating genomic DNA. The quantity and quality of the total RNA samples were assessed using a NanoDrop spectrophotometer (ND-1000; NanoDrop Technology Inc., Wilmington, DE, USA) and an Experion automated electrophoresis system with an Experion RNA StdSens kit (Bio-Rad Laboratories, Hercules, CA, USA), respectively. A custom-made bovine oligonucleotide microarray with 15,000 unique genes (GPL9284) fabricated by Agilent Technologies (Santa Clara, CA, USA) was used in this study, which was performed as described previously [29]. Sixty-mer nucleotide probes for the customized microarray were synthesized on a glass slide. We performed one-color microarray analysis. cDNA synthesis, Cy3-labeled cRNA preparation, hybridization, and the washing and scanning of array slides were performed according to the Agilent one color microarray-based gene expression analysis protocol. Briefly, 400 ng of total RNA from each sample were reverse-transcribed into cDNA using the Quick Amp Labeling Kit (Agilent Technologies) with an oligo dT-based primer, and then Cy3-labelled cRNA was prepared by in vitro transcription. Labeled cRNA was purified with an RNeasy Mini Kit (Qiagen, Hilden, Germany), and the concentration and Cy3 dye incorporation (pmol Cy3/μg cRNA) were measured with a spectrophotometer. Labeled cRNA (600 ng) was fragmented and hybridized using the Gene Expression Hybridization Kit (Agilent Technologies), according to the manufacturer’s instructions. The arrays were washed using a Gene Expression Wash Pack Kit (Agilent Technologies) and scanned using an Agilent Microarray Scanner. Feature Extraction ver. 9.5 was used for image analysis and data extraction. Microarray data from each sample were imported into GeneSpring 12 (Agilent Technologies) for further data characterization. The GEO accession numbers are as follows. Platform: GPL9284; samples: GSM2093338 to GSM2093369; series: GSE79367. To identify putative biological functions of DEG between RB and non-RB cows in each endometrial compartment, we performed functional annotation chart analysis of the lists of DEG using the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) based on Genebank Accession IDs [30]. Gene Ontology (GO) Biological Process was selected as the functional annotation category for the analysis with the threshold for minimum gene counts belonging to an annotation term set to 5 and an EASE score set to 0.05. The GO terms were ranked according to their P-values describing the significance of gene-term enrichment.
Quantitative real-time RT-PCR analysis
To validate the results of microarray analysis, we confirmed mRNA expression of the following representative genes using quantitative real-time RT-PCR (qPCR) analysis: (1) top two up- or down-regulated known genes in each endometrial compartment; and (2) top five up- or down-regulated known genes in the whole uterus. Details of the procedures for single-strand cDNA synthesis and qPCR were previously described [31]. Briefly, 50 ng of total RNA from the same sample used for the microarray were reverse-transcribed into cDNA for 30 min at 48 °C using MultiScribeTM Reverse Transcriptase (Applied Biosystems, Foster City, CA, USA) with a random primer, dNTP mixture, MgCl2 and RNase inhibitor. After heat inactivation of the reverse transcriptase for 5 min at 95 °C, PCR and resulting relative increase in reporter fluorescent dye emission were monitored in real time using an Mx3000P qPCR system (Agilent Technologies). Primers were designed using Primer Express computer software program (Applied Biosystems) or Primer3 Plus software (www.bioinformatics.nl/primer3plus/) based on the bovine sequences. The primer sequences for each gene are listed in Table 1. Thermal-cycling conditions included an initial sample incubation at 50 °C for 2 min and at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 1 min. The cycle threshold value (CT) indicate the quantity of the target gene in each sample. The relative difference in initial amount of each mRNA species (or cDNA) was determined by comparing the CT values. The standard curves for each gene were generated by serially diluting plasmids containing cDNA of each individual gene to quantify the mRNA concentrations. We confirmed the utility of the dissociation curve for detecting the SYBR Green-based objective amplicon because SYBR Green also detects double-stranded DNA including Primer dimers, contaminating DNA and PCR products from misannealed primers. Non-specific amplicons appear as a peak separate from the desired amplicon peak. The expression ratio of each gene to SUZ12 mRNA, which has been demonstrated to be suitable for normalization in bovine endometrial tissue [32], was calculated to adjust for any variations in the qPCR reaction.
Table 1.
Details of the primers used for quantitative real-time RT-PCR analysis
| Gene (GenBank accession number) | Primer | Sequence | Position |
|---|---|---|---|
| CHGA | Forward | 5′-GCCGAAAGAGGTGACAGAAGA-3′ | 538-558 |
| (NM_181005) | Reverse | 5′-GTCTCCGTCCGAGTCTTCATC-3′ | 637-617 |
| CNGA1 | Forward | 5′-AGCAGAGATCGCCATCAATGT-3′ | 1574-1594 |
| (NM_174278) | Reverse | 5′-ACCAACTCCACCAACAGACCA-3′ | 1663-1643 |
| CPXM2 | Forward | 5′- ACCAGTGGATTGAAGTGGACG-3′ | 581-601 |
| (NM_001206057) | Reverse | 5′- TCACTCAGCCAGAGTGAGTTCCT-3′ | 665-643 |
| FAM83D | Forward | 5′- GGCTCCTACAGTTTTACATGGACAG-3′ | 788-812 |
| (NM_001083393) | Reverse | 5′-CAACCACTTGGCCAGACAGAA-3′ | 863-843 |
| FMO2 | Forward | 5′- AAGCCAGACATCCTTTCTCTCTTG -3′ | 1459-1482 |
| (NM_001163274) | Reverse | 5′- CCCAACCAGGCGATACTGATA-3′ | 1554-1532 |
| GSTA3 | Forward | 5′-AGAGCCATCCTCAGCTACCTTG-3′ | 254-275 |
| (NM_001077112) | Reverse | 5′-TCGATCCTGACTGTCTCCTTCA-3′ | 327-306 |
| IFIH1 | Forward | 5′-GGGACTAACAGCTTCACCAGGT-3′ | 1764-1785 |
| (XM_002685338) | Reverse | 5′-GGTAACTGCATCAAGATTGGCA-3′ | 1860-1839 |
| IGG1C | Forward | 5′-ACCAAGGTGGACAAGGCTGTT-3′ | 274-294 |
| (S82409) | Reverse | 5′-GGAAGATGAAGACAGAGGGTCCT-3′ | 370-348 |
| KCNA2 | Forward | 5′-TGGGTTCCCTATGTGCAATTG-3′ | 1644-1664 |
| (NM_001101195) | Reverse | 5′-TCCCGGTGGTAGAAGTAGTTGAA-3′ | 1734-1712 |
| KLHL24 | Forward | 5′- TTATTGGCAAGGAGGAGATGGT-3′ | 901-922 |
| (NM_001206196) | Reverse | 5′- TCTCAGATCAACAGCGCGAT-3′ | 968-949 |
| KRT35 | Forward | 5′- GAGACCGAGGTATCCATGCG-3′ | 587-606 |
| (NM_001076073) | Reverse | 5′- TTCTTGAGGCAGAGCAGCTC -3′ | 726-707 |
| LPLUNC1 | Forward | 5′- TCGGTGTGTTCAACCCTAAGC-3′ | 1280-1300 |
| (NM_174697) | Reverse | 5′- TTCTCGTTTGGCAGCAGGAT -3′ | 1355-1336 |
| PIPOX | Forward | 5′- ACAGCATTAACACCGAGTCGG-3′ | 2140-2160 |
| (NM_001014878) | Reverse | 5′- GGCAGTTATGAGCCTGTTTCCT-3′ | 2210-2189 |
| PLEKHA5 | Forward | 5′- GATGGATTCAAGAACGGAACG-3′ | 2655-2675 |
| (XM_002687754) | Reverse | 5′- TTCCACAGTCATCCTAGGTCGA-3′ | 2739-2718 |
| PRF1 | Forward | 5′-CAAGCCAAATGCTAATGTCCGT-3′ | 408-429 |
| (NM_001143735) | Reverse | 5′-AAAGCGACACTCCACTAAGTCCAT-3′ | 531-508 |
| PRSS2 | Forward | 5′-GTGAGGCTGGGAGAATACAACA-3′ | 211-232 |
| (NM_174690) | Reverse | 5′-ATGATCTTGGACGCATCGATGA-3′ | 281-260 |
| SLC39A2 | Forward | 5′- TTGGCTGCCTATTTGCCCT-3′ | 355-373 |
| (NM_001205648) | Reverse | 5′- CTGGAACCACTTGAAGCAGATG-3′ | 428-407 |
| THBS4 | Forward | 5′- CACTCTGAACGAGCTCTACGTGAT 3′ | 331-354 |
| (NM_001034728) | Reverse | 5′- GAAGAGTAAAGGCCGAAGATGGT-3′ | 411-389 |
| SUZ12 | Forward | 5′-GAACACCTATCACACACATTCTTGT-3′ | 1565-1589 |
| (NM_001205587) | Reverse | 5′-TAGAGGCGGTTGTGTCCACT-3′ | 1694-1675 |
Immunohistochemistry
Immunohistochemistry for chromogranin A (CHGA), glutathione S-transferase A3 (GSTA3) and trypsin 2 (PRSS2) was performed in the endometrium of both RB and non-RB cows on Day 15 of the estrous cycle using the automated Ventana HX System Discovery with a DabMapKit (Roche Diagnostics, Basel, Switzerland) as described previously in detail by our laboratory [33]. Uterine cross sections 7-μm-thick were incubated at room temperature with rabbit polyclonal anti-human CHGA antibody (1.0 mg/ml, 20085, ImmunoStar Inc., Hudson, WI, USA), rabbit polyclonal anti-human GSTA3 antibody (0.5 mg/ml, orb5362, Biorbyt LLC, San Francisco, CA, USA) or rabbit polyclonal anti-bovine PRSS2 antibody (10 mg/ml, OASA07087, Aviva Systems Biology, San Diego, CA, USA) diluted 1:100 (anti-CHGA), 1:20 (anti-GSTA3) or 1:200 (anti-PRSS2) in Discovery Ab diluents (Roche) for 12 h. The signals were detected using anti-rabbit IgG-Biotin conjugate (Sigma) diluted 1:500 for 1 h. Negative controls were performed using normal rabbit IgG (0.5 mg/ml, 20304, Imgenex, San Diego, CA, USA) diluted at concentrations equivalent to the primary antibodies. The sections were observed with a Leica DMRE HC microscope (Leica Microsystems, Wetzlar, Germany) and a Nikon Digital Sight DS-Fi1-L2 (Nikon Instruments Co., Tokyo, Japan).
Statistical analysis
Microarray data were analyzed statistically with an unpaired Student’s t-test and summarized using GeneSpring 12 (Agilent Technologies). The analysis of each uterine compartment was performed by comparing the gene datasets which composed by microarray data of four cows in each RB and non-RB group (n = 4/group). The analysis of whole uterus was performed by comparing the gene datasets which composed by microarray data of all four compartments of four cows in each RB and non-RB group (n = 16/group). The qPCR results were analyzed using a Mann–Whitney U test. Results are presented as the mean ± SEM. Statistical significance is considered to be at P < 0.05.
Results
Gene expression profiles of CAR and ICAR in ipsilateral uterine horns
A total of 405 and 397 genes were differentially expressed in CAR and ICAR of the ipsilateral uterine horn of RB cows, respectively when compared with non-RB cows (adjusted P-value <0.05, fold-change >1.0). All data of individual gene changes in CAR and ICAR are available in Additional file 1: Tables S1 and S2, respectively. Out of these, 128 genes were up-regulated and 277 genes were down-regulated in CAR, whereas 169 genes were up-regulated and 228 genes were down-regulated in ICAR. The top 10 up- and down-regulated known genes in CAR are shown in Table 2. The most pronounced up- and down-regulation of gene expression in RB cows was observed for GSTA3 (Glutathione S-transferase, alpha 3; 19.2-fold) and CPXM2 (Carboxypeptidase X (M14 family), member 2; 5.3-fold), respectively. The top five functional annotations of DEG in the CAR of ipsilateral uterine horns between RB and non-RB cows are listed in Table 3. The GO terms involved in anatomical structure development, developmental process, cellular process, multicellular organismal development and biosynthetic process were highly enriched in up-regulated genes, whereas the GO terms involved in cellular process, cytoskeleton organization, biological adhesion, cell adhesion and cellular component organization were highly enriched in down-regulated genes.
Table 2.
Top 10 up- and down-regulated known genes in CAR of ipsilateral uterine horns of RB cows
| GenBank accession ID | Gene symbol | Gene description | Fold change | P-value |
|---|---|---|---|---|
| Up-regulated genes | ||||
| NM_001077112 | GSTA3 | Glutathione S-transferase, alpha 3 | 19.2 | 0.0016 |
| NM_001206196 | KLHL24 | Kelch-like 24 (Drosophila) | 3.0 | 0.0273 |
| XM_588022 | SPOPL | Speckle-type POZ protein-like | 2.8 | 0.0239 |
| NM_001103317 | ERCC2 | Excision repair cross-complementing rodent repair deficiency, complementation group 2 | 2.5 | 0.0437 |
| XM_002696037 | CD300LG | CD300 molecule-like family member g | 2.2 | 0.0378 |
| NM_001075908 | STK33 | Serine/threonine kinase 33 | 2.1 | 0.0351 |
| NM_174607 | SLC5A3 | Solute carrier family 5 (inositol transporters), member 3 | 2.0 | 0.0126 |
| NM_001192523 | KCNMB4 | Potassium large conductance calcium-activated channel, subfamily M, beta member 4 | 2.0 | 0.0307 |
| NM_001083638 | MEF2A | Myocyte enhancer factor 2A | 2.0 | 0.0290 |
| XM_002695445 | ZNF211 | Zinc finger protein 211 | 2.0 | 0.0063 |
| Down-regulated genes | ||||
| NM_001206057 | CPXM2 | Carboxypeptidase X (M14 family), member 2 | 5.3 | 0.0496 |
| NM_001076073 | KRT35 | Keratin 35 | 4.1 | 0.0279 |
| NM_001101239 | GRP | Gastrin-releasing peptide | 3.6 | 0.0319 |
| NM_001245926 | FGF9 | Fibroblast growth factor 9 | 3.5 | 0.0066 |
| NM_174145 | PKP1 | Plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) | 2.9 | 0.0021 |
| NM_001076864 | TMEM129 | Transmembrane protein 129 | 2.6 | 0.0087 |
| NM_001105478 | SSLP1 | Secreted seminal-vesicle Ly-6 protein 1 | 2.5 | 0.0474 |
| NM_001077962 | STAC | SH3 and cysteine rich domain | 2.4 | 0.0157 |
| NM_001077945 | PFN3 | Profilin 3 | 2.4 | 0.0106 |
| NM_001012685 | FCAR | Fc fragment of IgA, receptor for | 2.3 | 0.0322 |
Table 3.
Top 5 functional annotations of up- and down-regulated genes in CAR of ipsilateral uterine horns
| Term | Count | P-value |
|---|---|---|
| Up-regulated genes | ||
| GO:0048856 ~ anatomical structure development | 11 | 0.0029 |
| GO:0032502 ~ developmental process | 11 | 0.0161 |
| GO:0009987 ~ cellular process | 31 | 0.0186 |
| GO:0007275 ~ multicellular organismal development | 10 | 0.0230 |
| GO:0009888 ~ tissue development | 5 | 0.0246 |
| Down-regulated genes | ||
| GO:0009987 ~ cellular process | 95 | <0.0001 |
| GO:0007010 ~ cytoskeleton organization | 8 | 0.0061 |
| GO:0022610 ~ biological adhesion | 11 | 0.0065 |
| GO:0007155 ~ cell adhesion | 11 | 0.0065 |
| GO:0016043 ~ cellular component organization | 23 | 0.0099 |
The top 10 up- and down-regulated known genes in ICAR are shown in Table 4. The highest increase and decrease in gene expression in RB cows were observed in LPLUNC1 (Von Ebner minor salivary gland protein; 3.7-fold) and THBS4 (Thrombospondin 4; 3.4-fold), respectively. Table 5 summarizes the top five functional annotations of DEG in ICAR between RB and non-RB cows. As a result of DAVID analysis, only four GO terms related to metabolic process, cellular metabolic process, cellular biosynthetic process and chemical homeostasis were identified in up-regulated genes. In down-regulated genes, the GO terms involved in metabolic process, cellular metabolic process, cellular process, primary metabolic process and protein metabolic process were highly enriched.
Table 4.
Top 10 up- and down-regulated known genes in ICAR of ipsilateral uterine horns of RB cows
| GenBank accession ID | Gene symbol | Gene description | Fold change | P-value |
|---|---|---|---|---|
| Up-regulated genes | ||||
| NM_174697 | LPLUNC1 | Von Ebner minor salivary gland protein | 3.7 | 0.0214 |
| NM_001075162 | FMO2 | Flavin containing monooxygenase 2 (non-functional) | 3.3 | 0.0348 |
| NM_001166616 | C5 | Complement component 5 | 3.2 | 0.0429 |
| XM_002692160 | FOXA2 | Forkhead box A2 | 3.0 | 0.0350 |
| NM_181027 | AKR1C4 | Aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4) | 2.9 | 0.0104 |
| NM_001045878 | GATM | Glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 2.8 | 0.0472 |
| NM_001206196 | KLHL24 | Kelch-like 24 (Drosophila) | 2.6 | 0.0301 |
| NM_001034419 | HPGD | Hydroxyprostaglandin dehydrogenase 15-(NAD) | 2.6 | 0.0293 |
| XM_001254052 | ZNED1 | DNA-directed RNA polymerase I subunit RPA12-like | 2.4 | 0.0476 |
| NM_001038096 | CFI | Complement factor I | 2.4 | 0.0096 |
| Down-regulated genes | ||||
| NM_001034728 | THBS4 | Thrombospondin 4 | 3.4 | 0.0106 |
| NM_001083393 | FAM83D | Protein FAM83D | 2.6 | 0.0011 |
| NM_001105411 | GFRA1 | GDNF family receptor alpha 1 | 2.4 | 0.0391 |
| NM_001206057 | CPXM2 | Carboxypeptidase X (M14 family), member 2 | 2.3 | 0.0231 |
| NM_178572 | CA2 | Carbonic anhydrase II | 2.3 | 0.0474 |
| NM_001099381 | GALK1 | Galactokinase 1 | 2.1 | 0.0466 |
| NM_001035050 | VTN | Vitronectin | 2.0 | 0.0464 |
| NM_174745 | MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72 kDa gelatinase, 72 kDa type IV collagenase) | 1.9 | 0.0387 |
| NM_001075730 | STRA6 | Stimulated by retinoic acid gene 6 | 1.9 | 0.0405 |
| NM_174558 | KCNK17 | Potassium channel, subfamily K, member 17 | 1.9 | 0.0496 |
Table 5.
Top 5 functional annotations of up- and down-regulated genes in ICAR of ipsilateral uterine horns
| Term | Count | P-value |
|---|---|---|
| Up-regulated genes | ||
| GO:0008152 ~ metabolic process | 38 | 0.0033 |
| GO:0044237 ~ cellular metabolic process | 29 | 0.0242 |
| GO:0044249 ~ cellular biosynthetic process | 15 | 0.0345 |
| GO:0048878 ~ chemical homeostasis | 5 | 0.0423 |
| Down-regulated genes | ||
| GO:0008152 ~ metabolic process | 66 | <0.0001 |
| GO:0044237 ~ cellular metabolic process | 8 | <0.0001 |
| GO:0009987 ~ cellular process | 11 | 0.0001 |
| GO:0044238 ~ primary metabolic process | 11 | 0.0009 |
| GO:0019538 ~ protein metabolic process | 23 | 0.0023 |
Gene expression profiles of CAR and ICAR in contralateral uterine horns
A total of 443 and 257 genes were differentially expressed in CAR and ICAR of the contralateral uterine horn of RB cows, respectively when compared with non-RB cows (adjusted P-value <0.05, fold-change >1.0). All data of individual gene changes in CAR and ICAR are available in Additional file 1: Tables S3 and S4, respectively. Out of these, 333 genes were up-regulated and 110 genes were down-regulated in CAR, whereas 121 genes were up-regulated and 136 genes were down-regulated in ICAR. The top 10 up- and down-regulated known genes in CAR are shown in Table 6. Similar to CAR of the ipsilateral side, the most pronounced up-regulated gene in RB cows was GSTA3 (Glutathione S-transferase, alpha 3; 12.7-fold). The most down-regulated gene in RB cows was SLC39A2 (Solute carrier family 39 (zinc transporter), member 2; 2.7-fold). Table 7 shows the top five functional annotations of DEG in CAR between RB and non-RB cows. Biological functions of positive regulation of biological process, positive regulation of cellular process, organ morphogenesis, anatomical structure morphogenesis, and anatomical structure development were highly enriched in up-regulated genes, whereas biological functions of regulation of protein kinase activity, regulation of kinase activity, regulation of transferase activity and carboxylic acid metabolic process were highly enriched in down-regulated genes.
Table 6.
Top 10 up- and down-regulated known genes in CAR of contralateral uterine horns of RB cows
| GenBank accession ID | Gene symbol | Gene description | Fold change | P-value |
|---|---|---|---|---|
| Up-regulated genes | ||||
| NM_001077112 | GSTA3 | Glutathione S-transferase, alpha 3 | 12.7 | 0.0080 |
| NM_001014878 | PIPOX | Pipecolic acid oxidase | 8.4 | 0.0261 |
| NM_001024569 | ELF5 | E74-like factor 5 (ets domain transcription factor) | 4.3 | 0.0173 |
| NM_173981 | ACAN | Aggrecan | 3.0 | 0.0420 |
| NM_174404 | NRXN1 | Neurexin 1 | 3.0 | 0.0065 |
| NM_001079771 | SMOC1 | SPARC related modular calcium binding 1 | 2.7 | 0.0104 |
| NM_001034351 | TNNC1 | Troponin C type 1 (slow) | 2.6 | 0.0142 |
| NM_173945 | NTS | Neurotensin | 2.6 | 0.0289 |
| NM_001206196 | KLHL24 | Kelch-like 24 (Drosophila) | 2.4 | 0.0345 |
| NM_001046585 | CCL14 | Chemokine (C-C motif) ligand 14 | 2.4 | 0.0358 |
| Down-regulated genes | ||||
| NM_001205648 | SLC39A2 | Solute carrier family 39 (zinc transporter), member 2 | 2.7 | 0.0110 |
| XM_002687754 | PLEKHA5 | Pleckstrin homology domain containing, family A member 5 | 2.2 | 0.0181 |
| NM_001077962 | STAC | SH3 and cysteine rich domain | 2.0 | 0.0456 |
| NM_001098061 | SQLE | Squalene epoxidase | 2.0 | 0.0268 |
| NM_174145 | PKP1 | Plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) | 1.9 | 0.0304 |
| NM_001098938 | CYP39A1 | Cytochrome P450, family 39, subfamily A, polypeptide 1 | 1.9 | 0.0262 |
| NM_174489 | VLDLR | Very low density lipoprotein receptor | 1.9 | 0.0063 |
| NM_001034660 | SLC5A11 | Solute carrier family 5 (sodium/glucose cotransporter), member 11 | 1.8 | 0.0061 |
| NM_001075803 | FH | Fumarate hydratase | 1.8 | 0.0009 |
| NM_001099399 | CMTM3 | CKLF-like MARVEL transmembrane domain containing 3 | 1.8 | 0.0434 |
Table 7.
Top 5 functional annotations of up- and down-regulated genes in CAR of contralateral uterine horns
| Term | Count | P-value |
|---|---|---|
| Up-regulated genes | ||
| GO:0048518 ~ positive regulation of biological process | 25 | <0.0001 |
| GO:0048522 ~ positive regulation of cellular process | 22 | <0.0001 |
| GO:0009887 ~ organ morphogenesis | 12 | <0.0001 |
| GO:0009653 ~ anatomical structure morphogenesis | 16 | 0.0001 |
| GO:0048856 ~ anatomical structure development | 24 | 0.0002 |
| Down-regulated genes | ||
| GO:0045859 ~ regulation of protein kinase activity | 5 | 0.0029 |
| GO:0043549 ~ regulation of kinase activity | 5 | 0.0035 |
| GO:0051338 ~ regulation of transferase activity | 5 | 0.0040 |
| GO:0043436 ~ oxoacid metabolic process | 7 | 0.0075 |
| GO:0019752 ~ carboxylic acid metabolic process | 7 | 0.0075 |
Table 8 shows the top 10 up- and down-regulated known genes in ICAR. The highest increase and decrease in gene expression in RB cows were found for PIPOX (Pipecolic acid oxidase; 8.8-fold) and IFIH1 (Interferon induced with helicase C domain 1; 4.0-fold), respectively. The top five functional annotations of DEG in the ICAR of contralateral uterine horns between RB and non-RB cows are listed in Table 9. The GOs containing genes regulating gene expression, regulation of primary metabolic process, regulation of macromolecule metabolic process, metabolic process and regulation of metabolic process were highly enriched in up-regulated genes. In down-regulated genes, the GO terms involved in primary metabolic process, transport, establishment of localization, localization and metabolic process were highly enriched.
Table 8.
Top 10 up- and down-regulated known genes in ICAR of contralateral uterine horns of RB cows
| GenBank accession ID | Gene symbol | Gene description | Fold change | P-value |
|---|---|---|---|---|
| Up-regulated genes | ||||
| NM_001014878 | PIPOX | Pipecolic acid oxidase | 8.8 | 0.0156 |
| NM_174278 | CNGA1 | Cyclic nucleotide gated channel alpha 1 | 6.8 | 0.0390 |
| NM_001033608 | GSTA3 | Glutathione S-transferase, alpha 3 | 6.6 | 0.0340 |
| NM_001046400 | MIF | Macrophage migration inhibitory factor (glycosylation-inhibiting factor) | 3.1 | 0.0118 |
| NM_001046400 | ZNRD1 | Zinc ribbon domain containing 1 | 2.8 | 0.0400 |
| NM_001206196 | KLHL24 | Kelch-like 24 (Drosophila) | 2.6 | 0.0212 |
| NM_001076517 | LY6D | Lymphocyte antigen 6 complex, locus D | 2.5 | 0.0414 |
| NM_001035473 | GK5 | Glycerol kinase 5 | 2.2 | 0.0210 |
| NM_001075890 | KLK10 | Kallikrein-related peptidase 10 | 2.1 | 0.0445 |
| NM_001083791 | SH3BGRL2 | SH3 domain binding glutamic acid-rich protein like 2 | 1.9 | 0.0030 |
| Down-regulated genes | ||||
| XM_002685338 | IFIH1 | Interferon induced with helicase C domain 1 | 4.0 | 0.0485 |
| NM_001101195 | KCNA2 | Potassium voltage-gated channel, shaker-related subfamily, member 2 | 3.5 | 0.0204 |
| NM_180998 | LTF | Lactotransferrin | 2.9 | 0.0286 |
| NM_001076843 | SLC30A3 | Solute carrier family 30 (zinc transporter), member 3 | 2.6 | 0.0289 |
| NM_001076494 | C8H8orf13 | Chromosome 8 open reading frame 13 ortholog | 2.5 | 0.0406 |
| NM_001105411 | GFRA1 | GDNF family receptor alpha 1 | 2.5 | 0.0383 |
| NM_174018 | CFTR | Cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | 2.5 | 0.0316 |
| NM_001077941 | MARCH3 | Membrane-associated ring finger (C3HC4) 3 | 2.5 | 0.0158 |
| NM_173959 | SCD | Stearoyl-CoA desaturase (delta-9-desaturase) | 2.0 | 0.0096 |
| NM_174602 | SLC2A1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | 1.9 | 0.0057 |
Table 9.
Top 5 functional annotations of up- and down-regulated genes in ICAR of contralateral uterine horns
| Term | Count | P-value |
|---|---|---|
| Up-regulated genes | ||
| GO:0010467 ~ gene expression | 17 | 0.0004 |
| GO:0080090 ~ regulation of primary metabolic process | 19 | 0.0013 |
| GO:0060255 ~ regulation of macromolecule metabolic process | 19 | 0.0015 |
| GO:0008152 ~ metabolic process | 38 | 0.0033 |
| GO:0019222 ~ regulation of metabolic process | 19 | 0.0040 |
| Down-regulated genes | ||
| GO:0044238 ~ primary metabolic process | 34 | 0.0023 |
| GO:0006810 ~ transport | 17 | 0.0025 |
| GO:0051234 ~ establishment of localization | 17 | 0.0026 |
| GO:0051179 ~ localization | 18 | 0.0027 |
| GO:0008152 ~ metabolic process | 35 | 0.0028 |
Gene expression profiles of whole uterus
To characterize differential global gene expression profiles in the endometrium of RB and non-RB cows not only locally in each endometrial compartment but also globally in the uterus, we also performed bioinformatics analysis by combining the microarray gene data sets of four endometrial compartments in each cow as whole uterus. A total of 76 genes were found to be differentially expressed in the whole uterus of RB cows when compared with non-RB cows (adjusted P-value <0.05, fold-change >2.0). Among these, 37 genes were up-regulated and 39 genes were down-regulated. All up- and down-regulated known genes in the whole uterus are shown in Table 10. The most pronounced up- and down-regulated gene expression in RB cows was found for PRSS2 (Protease, serine, 2 (trypsin 2); 12.3-fold) and CHGA (Chromogranin A (parathyroid secretory protein 1); 3.9-fold), respectively.
Table 10.
Up- and down-regulated known genes in whole uterus of RB cows as compared with non-RB cows
| GenBank accession ID | Gene symbol | Gene description | Fold change | P-value |
|---|---|---|---|---|
| Up-regulated genes | ||||
| NM_174690 | PRSS2 | Protease, serine, 2 (trypsin 2) | 12.3 | 0.0018 |
| NM_001077112 | GSTA3 | Glutathione S-transferase, alpha 3 | 6.7 | 0.0002 |
| NM_001014878 | PIPOX | Pipecolic acid oxidase | 6.4 | <0.0001 |
| NM_174278 | CNGA1 | Cyclic nucleotide gated channel alpha 1 | 4.3 | 0.0024 |
| S82409 | IGG1C | IgG1 heavy chain constant region | 3.7 | 0.0081 |
| BC112657 | Vl1a | Immunoglobulin lambda light chain variable region | 3.7 | 0.0076 |
| S82407 | IgCgamma | IgG2a heavy chain constant region | 3.4 | 0.0347 |
| NM_001025346 | DAPL1 | death associated protein-like 1 | 3.4 | 0.0075 |
| NM_001080353 | PI3 | Peptidase inhibitor 3, skin-derived (SKALP) | 3.2 | 0.0022 |
| NM_001166616 | C5 | Complement component 5 | 2.8 | 0.0044 |
| NM_001024569 | ELF5 | E74-like factor 5 (ets domain transcription factor) | 2.8 | 0.0047 |
| NM_001075910 | CCDC113 | Coiled-coil domain containing 113 | 2.7 | 0.0432 |
| NM_173945 | NTS | Neurotensin | 2.6 | <0.0001 |
| NM_001034351 | TNNC1 | Troponin C type 1 (slow) | 2.5 | 0.0004 |
| NM_001206196 | KLHL24 | Kelch-like 24 (Drosophila) | 2.5 | <0.0001 |
| NM_001046400 | ZNRD1 | zinc ribbon domain containing 1 | 2.3 | <0.0001 |
| NM_001193109 | SDCCAG8 | Serologically defined colon cancer antigen 8 | 2.2 | 0.0001 |
| NM_174010 | CD36 | CD36 molecule (thrombospondin receptor) | 2.2 | 0.0073 |
| XM_588022 | SPOPL | Speckle-type POZ protein-like | 2.2 | <0.0001 |
| NM_173880 | H4 | Histone H4 | 2.1 | 0.0033 |
| NM_001098155 | ZNF322A | Zinc finger protein 322A | 2.1 | 0.0005 |
| NM_001035380 | GC | Group-specific component (vitamin D binding protein) | 2.0 | 0.0269 |
| NM_001035473 | GK5 | Glycerol kinase 5 | 2.0 | 0.0003 |
| Down-regulated genes | ||||
| NM_181005 | CHGA | Chromogranin A (parathyroid secretory protein 1) | 3.9 | 0.0005 |
| NM_001076073 | KRT35 | Keratin 35 | 3.3 | 0.0011 |
| NM_001034728 | THBS4 | Thrombospondin 4 | 3.2 | <0.0001 |
| NM_001206057 | CPXM2 | Carboxypeptidase X (M14 family), member 2 | 3.1 | <0.0001 |
| NM_001143735 | PRF1 | Perforin 1 (pore forming protein) | 3.0 | 0.0090 |
| NM_001002763 | CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 2.9 | 0.0097 |
| NM_176851 | FUT5 | Fucosyltransferase 5 (alpha (1,3) fucosyltransferase) | 2.7 | 0.0038 |
| XM_002685338 | IFIH1 | Interferon induced with helicase C domain 1 | 2.5 | 0.0040 |
| NM_001081734 | MOCS3 | Molybdenum cofactor synthesis 3 | 2.5 | 0.0465 |
| NM_174039 | DPP4 | Dipeptidyl-peptidase 4 | 2.4 | 0.0158 |
| NM_001102080 | CSNK1D | Casein kinase 1, delta | 2.3 | 0.0144 |
| NM_001102060 | TBC1D10C | TBC1 domain family, member 10C | 2.3 | 0.0391 |
| NM_001081539 | C11H2orf49 | Chromosome 11 open reading frame, human C2orf49 | 2.3 | 0.0354 |
| AF068848 | VpreB | Surrogate light chain | 2.3 | 0.0204 |
| NM_001127317 | MIC1 | Major histocompatibility class I related protein | 2.2 | 0.0135 |
| NM_205801 | CLDN3 | Claudin 3 | 2.2 | 0.0196 |
| NM_001077887 | CLASRP | CLK4-associating serine/arginine rich protein | 2.2 | 0.0245 |
| NM_174513 | ADAP1 | ArfGAP with dual PH domains 1 | 2.1 | 0.0169 |
| NM_001105478 | SSLP1 | Secreted seminal-vesicle Ly-6 protein 1 | 2.1 | 0.0004 |
| NM_001077962 | STAC | SH3 and cysteine rich domain | 2.1 | <0.0001 |
| XM_002687754 | PLEKHA5 | Pleckstrin homology domain containing, family A member 5 | 2.1 | 0.0003 |
| NM_001101239 | GRP | Gastrin-releasing peptide | 2.1 | 0.0059 |
| NM_001205648 | SLC39A2 | Solute carrier family 39 (zinc transporter), member 2 | 2.0 | 0.0001 |
Validation of gene expression by qPCR
We selected the top two and top five up- and down-regulated known genes in each endometrial compartment and whole uterus between RB and non-RB cows, respectively to validate the changes in gene expression obtained from microarray analysis by qPCR. qPCR analysis clearly confirmed the microarray results in each endometrial compartment except for FAM83D (Fig. 1h), SLC39A2 (Fig. 2c), PLEKHA5 (Fig. 2d) and IFIH1 (Fig. 2g). In the whole uterus, the microarray results were confirmed except for PRF1 (Fig. 3j).
Fig. 1.
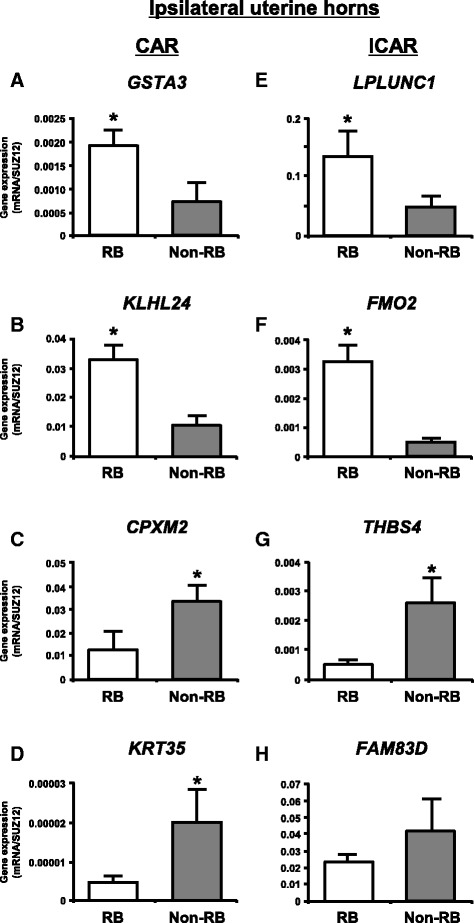
qPCR analysis of top two up- and down-regulated known genes in ipsilateral uterine horns between RB and non-RB cows for validation of the gene expression changes obtained from microarray analysis. a, b, c and d CAR and e, f, g and h ICAR. a, b, e and f up-regulated known genes in RB cows when compared with non-RB cows. c, d, g and h) down-regulated known genes in RB cows when compared with non-RB cows. The expression of mRNA was normalized to the expression of SUZ12 measured in the same RNA preparation. Data are shown as the mean ± SEM. Asterisks show significant differences (P < 0.05)
Fig. 2.
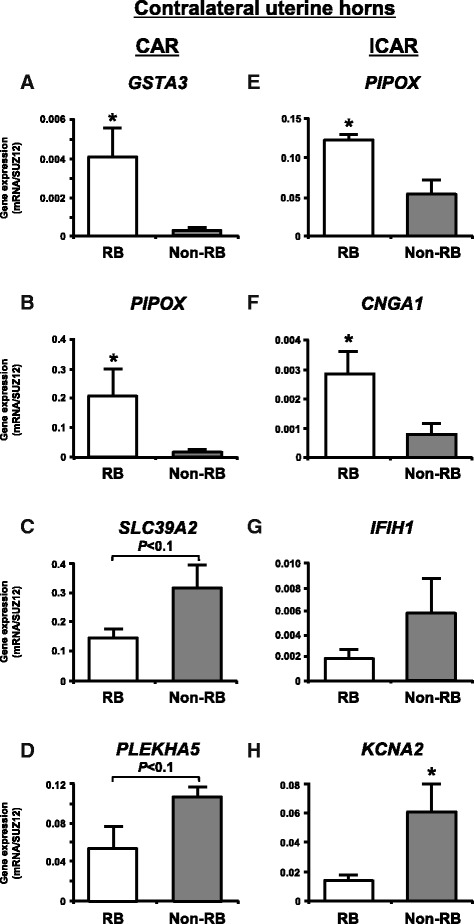
qPCR analysis of top two up- and down-regulated known genes in contralateral uterine horns between RB and non-RB cows for validation of the gene expression changes obtained from microarray analysis. a, b, c and d CAR and e, f, g and h ICAR. a, b, e and f up-regulated known genes in RB cows when compared with non-RB cows. c, d, g and h down-regulated known genes in RB cows when compared with non-RB cows. The expression of mRNA was normalized to the expression of SUZ12 measured in the same RNA preparation. Data are shown as the mean ± SEM. Asterisks show significant differences (P < 0.05)
Fig. 3.
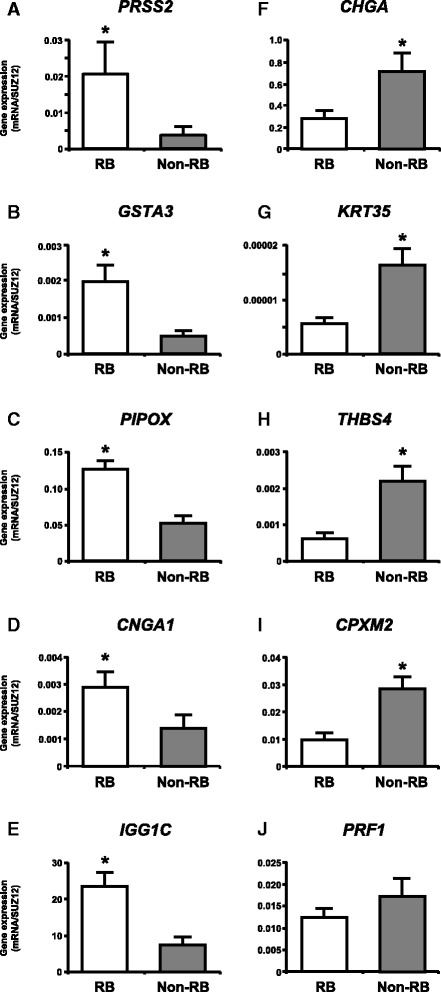
qPCR analysis of top five up- and down-regulated known genes in whole uterus between RB and non-RB cows for validation of the gene expression changes obtained from microarray analysis. a, b, c, d, e up-regulated known genes in RB cows when compared with non-RB cows. f, g, h, i, j down-regulated known genes in RB cows when compared with non-RB cows. The expression of mRNA was normalized to the expression of SUZ12 measured in the same RNA preparation. Data are shown as the mean ± SEM. Asterisks show significant differences (P < 0.05)
Protein localization of CHGA, GSTA3 and PRSS2 in the endometrium of RB and non-RB cows
Figure 4 shows the results of immunohistochemistry for CHGA, GSTA3 and PRSS2 in the endometrial tissues of ipsilateral uterine horns of RB and non-RB cows on Day 15 of the estrous cycle. In both RB and non-RB cows, a distinct CHGA signal was found in the uterine luminal epithelium and a part of uterine stroma under the epithelium (Fig. 4a and c). CHGA protein was also detected moderately in the glandular epithelium in both RB and non-RB cows and in the uterine stroma in RB cows (Fig. 4b and d). A positive GSTA3 signal was detected in the uterine luminal, uterine stroma and glandular epithelium in RB cows (Fig. 4e and f), whereas positive staining was not observed in non-RB cows (Fig. 4g and h). PRSS2 protein was moderately detected in the uterine luminal epithelium and glandular epithelium, and partially intense staining was observed in the uterine stroma under the epithelium in both RB and non-RB cows (Fig. 4i,j,k and l).
Fig. 4.
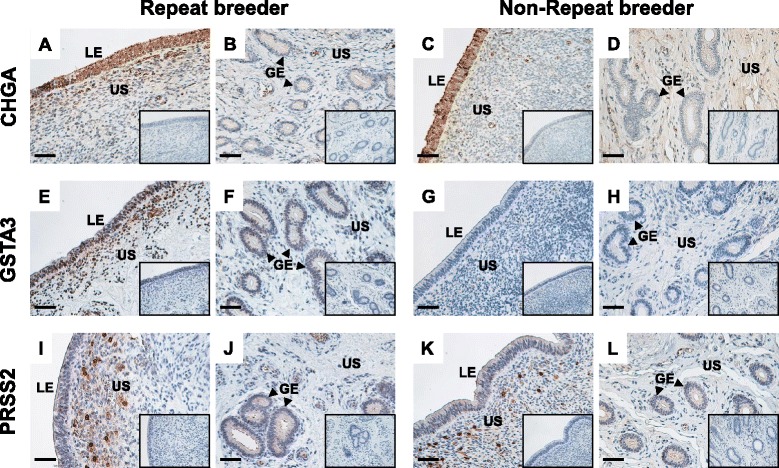
Representative photomicrographs of protein localization of CHGA, GSTA3 and PRSS2 in endometrial tissue from RB and non-RB cows on Day 15 of estrous cycle. Protein localization of (a, b, c and d) CHGA, (e, f, g and h) GSTA3 and (i, j, k and l) PRSS2 in endometrial tissue from RB (a, b, e, f, i and j) and non-RB (c, d, g, h, k and l) cows was detected by immunohistochemistry. Seven-micrometer sections of bovine endometrial tissues of ipsilateral uterine horns on Day 15 of estrous cycle were immunostained with anti-human CHGA, anti-human GSTA3 and anti-bovine PRSS2 polyclonal antibodies. Positive staining of CHGA and PRSS2 were found in the uterine luminal epithelium, uterine stroma and glandular epithelium of both RB and non-RB cows. GSTA3 was detected in the uterine luminal, uterine stroma and glandular epithelium in RB cows, whereas positive staining was not observed in non-RB cows. No signal was detected in the negative control sections using normal rabbit IgG (inserted panels). LE, luminal epithelium; US, uterine stroma; GE, glandular epithelium. Scale bars = 50 μm
Discussion
This is the first study to investigate global gene expression profiles of endometrium between RB and non-RB cows in both each endometrial compartments and the whole uterus. As we hypothesized, the microarray analysis identified a number of characteristic up- and down-regulated genes specific to each of four endometrial compartments of RB cows. The RB cows used in this study had experienced pregnancy and then became infertile. Thus, long-term infertility in the RB cows may be associated with alteration of endometrial function. Our results support that alteration of uterine environment, which may be induced by changes in the endometrial gene expression, could be a possible involvement of low fertility in the RB cattle.
Even though the endometrial gene expression profiles were regionally different in the endometrial compartments, GSTA3 was identified as the most pronounced up-regulated gene in the CAR of both ipsilateral and contralateral uterine horn. GSTA3 is a member of the class Alpha GST isoenzymes which exert a critical role in the detoxification of electrophilic decomposition products generated by reactive oxygen species (ROS) and metabolism of xenobiotics through glutathione conjugation with electrophilic compounds [34–37]. Similar to our results, a recent study has demonstrated that cows with low endometrial receptivity of the embryo show a higher expression of several oxidative stress-response genes in the endometrium compared with highly receptive cows at Day 7 of the estrous cycle [7]. Both oxidative stress and xenobiotics are directly responsible for not only an increase in embryonic mortality but also an alteration of uterine function inducing severe gynecological diseases such as endometriosis and preeclampsia [38–42]. We suppose that the CAR of RB cows may be accompanied by enhanced detoxification and elimination of ROS and xenobiotics. Another important contribution of GSTA3 isomerase is in the biosynthesis of steroids, especially testosterone and P4 in active steroidogenic tissues [43]. Progesterone inhibits endometrial epithelial cell proliferation, adenogenesis and uterine gland development [44, 45]. A previous study showed that RB cows had higher concentrations of P4 receptor in the endometrium than non-RB cows, implying the existence of a local hormonal imbalance in RB cows [46]. In the present study, the GSTA3 was also highly expressed in the ICAR of RB cows compared with non-RB cows. In addition, immunohistochemistry revealed that a strong signal of GSTA3 protein was detected in the uterine luminal and glandular epithelium and stroma in RB cows. GSTA3 may also be involved in ICAR functions in RB cows by mediating steroidogenesis.
Gene ontology analysis using DAVID revealed that a number of biological processes and functions were different between RB and non-RB cows in both CAR and ICAR. In the CAR of RB cows, genes involved in development and morphogenesis were mainly up-regulated. These genes included 14 and 9 genes regulating embryo development and vasculature development, respectively. The CAR eventually attaches with the trophoblast to give rise to the maternal side of the placentome in pregnant animals [22, 23]. Up-regulation of the genes involved in embryo and vasculature development in the CAR may contribute to the success of implantation and following placental formation at the maternal-fetal interface. An increase in the regulation of these genes in the CAR may be one of the characteristics of the RB uterus. In the ICAR of both the ipsilateral and contralateral uterine horns, genes related to metabolic processes were predominantly enriched in both up- and down-regulated genes in RB cows compared with non-RB cows. The ICAR is a specific compartment containing the uterine glands, which synthesize and secrete various metabolites and histotroph required for estrous cyclicity or development of the conceptus [24]. Alterations of endometrial metabolic processes in RB cows may seriously affect maintenance of uterine function.
The DAVID analysis also revealed that the CAR of the ipsilateral uterine horn of RB cows is characterized by down-regulation of a number of genes associated with cytoskeleton organization, cell adhesion and cellular component organization compared with non-RB cows. Previous global gene expression studies in bovine endometrium showed that profiles of the genes assigned to these functional categories changes during estrous cycle and peri-implantation [11–13], suggesting that these biological functions may be responsible for the regulation of uterine environment. Additionally, the endometrial cell adhesion molecules play a role in conceptus-endometrium attachment at implantation. A direct comparison of cyclic and pregnant endometrium found cell adhesion and cytoskeleton organization molecules affected by pregnancy in both CAR and ICAR [13]. Around the implantation period, the ipsilateral uterine horn is the site of first occurrence of conceptus-endometrial contact and modification of cytological character was seen exclusively on the CAR [47, 48]. Therefore, the lower expression of genes regulating cytoskeleton organization and cell adhesion in CAR of RB cows may be associated with inadequate endometrial responsiveness resulting in implantation failure.
CPXM2 was included in the top 10 down-regulated genes in both CAR and ICAR of the ipsilateral uterine horn. Previous microarray studies found no differences in CPXM2 expression in the bovine endometrium between highly fertile and poor fertile, and between highly fertile and subfertile cows at Day 14 of the estrous cycle [9], while expression decreasing at Day 7 compared to Day 3 of estrus in cows with low embryo receptivity [7]. CPXM2 is assumed to be more sensitive to P4 or some CL factors in a poorly fertile endometrium that includes the RB. Although the specific roles of CPXM2 remain unknown, DAVID analysis has assigned it belongs to the biological process of proteolysis and cell adhesion. Thus, CPXM2 may be related to alteration of endometrial cell adhesion in RB cows, as well as to the above described cell adhesion related genes that are down-regulated in the CAR of the ipsilateral uterine horn of RB cows.
KLHL24 (Kelch-like 24) was the only gene included in the top 10 up-regulated genes in all four endometrial compartments. A member of the KLHL family including KLHL24 is known to be involved in ubiquitination [49, 50]. It has been reported that lower expression of genes associated with ubiquitination in high fertile as compared with subfertile cows [9]. Although the specific roles of KLHL24 have not yet been elucidated, an increase in oxidative stress stimulated KLHL24 expression in human fibroblast cells [51], leading us to speculate that this gene is up-regulated to counteract cytoskeleton destruction by ROS- induced cell damage and/or to degrade proteins in cells exposed to ROS by ubiquitination reaction. Therefore, high expression of KLHL24 in RB cows compared with non-RB cows support the possibility that the endometrium of RB cows is under oxidative stress. However, it has been reported that the level of KLHL24 gene expression at Day 14 of the estrous cycle shows no significant difference among high fertile, low fertile and infertile cows [9]. The functional contribution of endometrial KLHL24 in bovine fertility remains unclear.
Analysis of the combined gene data sets of the four endometrial compartments revealed gene expression profiles of the whole uterus. PRSS2 and CHGA were the most pronounced up- and down-regulated genes, respectively. PRSS2 is a member of the trypsin family of serine proteases and degrades type I collagen directly or indirectly by activating several procollagenolytic matrix metalloproteinases (MMPs) [52, 53]. CHGA works as a pro-hormone for pancreastatin, vasostatin and catestatin [54–56]. Full-length CHGA and vasostatin act as anti-angiogenic factors to inhibit two potent angiogenic factors, basic fibroblast growth factor (bFGF) and vascular endothelial growth factor, while CHGA cleaved by thrombin and catestatin promote angiogenesis by inducing the release of bFGF from vascular endothelial cells [57]. In the present study, we found that both PRSS2 and CHGA proteins were localized in the luminal and glandular epithelium and in the stroma of the endometrium. These localizations coincide with the tissue site of gelatinase activity of MMP-2 and the localization of MMPs and bFGF in the bovine endometrium [58–60], suggesting paracrine and autocrine actions of PRSS2 and CHGA with MMPs and bFGF in the bovine endometrium. In addition, genes involved in cell death (DAPL1 and PRF1) or cell attachment (CD36, CDH1, CPXM2, KRT35 and THBS4) were also differentially expressed between RB and non-RB cows. Although further studies are needed to clarify, the endometrium of RB cows might not only be involved in the promotion of tissue remodeling and imbalance of angiogenesis but also in the degradation of cell renewal and tissue structure.
In cattle, around Day 15 of pregnancy is a stage of the beginning of conceptus elongation and maternal recognition of pregnancy [26]. A recent RNA-seq study identified numerous conceptus-expressed ligands that interact with corresponding receptors expressed on the endometrium and vice versa at Day 16 of pregnancy in cattle [61]. In the present study, some genes of endometrium expressed ligands (CCL4, CCL14, COL1A2, EDN1, F2, MMP2, THBS4 and TIMP3) and receptors (ACVR2B, BMPR2, CD4, CD36, IGF2R, IL10RB, KDR, TNFRSF25 and VLDLR) that interact with conceptus reported by Mamo et al. were differentially expressed between RB and non-RB cows. In addition, other genes encoding growth factors (FGF9 and GDF7) and cytokines (CCL8, CD14 and CD53) were down-regulated in the RB cows as compared with non-RB cows. Although the functional role of these two growth factors in bovine endometrium remains to be elucidated, FGF9 induces endometrial stromal cell proliferation [62]. Up-regulation of FGF9 and GDF7 expressions were detected in equine and/or swine pregnant endometrium and may be implicated in embryo-maternal communication at early pregnancy [63, 64]. The receptors of these growth factors were expressed in not only endometrium but also conceptus at Day 16 of pregnancy in cattle [61]. Therefore, alteration of the expression of these ligands and receptors in the RB cows may affect conceptus development and maternal recognition of pregnancy if a conceptus presents in the RB cows.
Conclusion
The results of the present study support the hypothesis that endometrial gene expression profiles are different between RB and non-RB cows. In RB cows, characteristic gene expression was identified in both the CAR and ICAR of both ipsilateral and contralateral uterine horns. The enriched GO terms of these genes were related to cell adhesion and morphogenesis in the CAR and metabolism in the ICAR. These results suggest that local regulation of molecular mechanisms in each endometrial compartment may contribute to normal uterine physiology. Therefore, the identified candidate endometrial genes and functions are likely to be involved in bovine reproductive performance. The present study could provide an information base for understanding underlying molecular pathogenesis and developing a treatment of repeat breeding in cattle from the point of view of endometrial function.
Acknowledgement
The authors thank the staff of Livestock Research Support Center of the National Agriculture and Food Research Organization for animal management and their technical assistance for sample collection. This manuscript was proofread by a professional service (SciRevision, Kagawa, Japan) prior to submission.
Funding
This study was supported by a Grant-in-Aid for Research Program on Innovative Technologies for Animal Breeding, Reproduction, and Vaccine Development (REP1001) from the Ministry of Agriculture, Forestry and Fisheries of Japan.
Availability of data and materials
All microarray data are available at the Gene Expression Omnibus (GEO) database at NCBI (http://www.ncbi.nlm.nih.gov/geo), under accession numbers GSE79367. All datasets on which the conclusions of the paper rely are available to readers.
Authors’ contributions
KGH participated in the design of the study, collected the materials, carried out all experiments and drafted the manuscript. MH collected the materials and helped to carry out qPCR and immunohistochemistry. KK carried out microarray and microarray data analysis. KH and SF carried out immunohistochemistry. TT participated in the design of the study, collected the materials and carried out microarray experiments. RS supervised the study, collected the materials and helped to carry out all experiments. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All procedures in animal experiments were carried out in accordance with guidelines approved by the Animal Ethics Committee of the National Institute of Agrobiological Sciences for the use of animals (permission number: H18-036).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ACVR2B
Activin A receptor type 2B
- AI
Artificial insemination
- bFGF
Basic fibroblast growth factor
- BMPR2
Bone morphogenetic protein receptor type 2
- CAR
Caruncular
- CCL
Chemokine (C-C motif) ligand
- CD4
CD4 molecule
- CD14
CD14 molecule
- CD36
CD36 molecule
- CD53
CD53 molecule
- CDH1
Cadherin 1, type 1
- CHGA
Chromogranin A
- CL
Corpus luteum
- COL1A2
Collagen type I alpha 2 chain
- CPXM2
Carboxypeptidase X (M14 family), member 2
- DAPL1
Death associated protein-like 1
- DAVID
Database for annotation, visualization and integrated discovery
- DEG
Differentially expressed genes
- EDN1
Endothelin 1
- F2
Coagulation factor II, thrombin
- FAM83D
Protein FAM83D
- FGF9
Fibroblast growth factor 9
- GDF7
Growth differentiation factor 7
- GE
Glandular epithelium
- GEO
Gene expression omnibus
- GO
Gene ontology
- GSTA3
Glutathione S-transferase A3
- ICAR
Intercaruncular
- IFIH1
Interferon induced with helicase C domain 1
- IGF2R
Insulin like growth factor 2 receptor
- IL10RB
Interleukin 10 receptor subunit beta
- KDR
Kinase insert domain receptor
- KLHL24
Kelch-like 24
- KRT35
Keratin 35
- LE
Luminal epithelium
- LPLUNC1
Von Ebner minor salivary gland protein
- MMP
Matrix metalloproteinase
- P4
Progesterone
- PIPOX
Pipecolic acid oxidase
- PLEKHA5
Pleckstrin homology domain containing, family A member 5
- PRF1
Perforin 1
- PRSS2
Trypsin 2
- qPCR
Quantitative real-time RT-PCR
- RB
Repeat breeder
- ROS
Reactive oxygen species
- SLC39A2
Solute carrier family 39 (zinc transporter), member 2
- SUZ12
Suppressor of zeste 12
- THBS4
Thrombospondin 4
- TIMP
Tissue inhibitor of metalloproteinase
- TNFRSF25
Tumor necrosis factor receptor superfamily member 25
- US
Uterine stroma
- VLDLR
Very low density lipoprotein receptor
Additional file
Table S1. List of up- and down-regulated genes in CAR of ipsilateral uterine horns of RB cows (n = 4) on Day 15 of the estrous cycle as compared with non-RB cows (n = 4). Table S2. List of up- and down-regulated genes in ICAR of ipsilateral uterine horns of RB cows on Day 15 of the estrous cycle as compared with non-RB cows. Table S3. List of up- and down-regulated genes in CAR of contralateral uterine horns of RB cows on Day 15 of the estrous cycle as compared with non-RB cows. Table S4. List of up- and down-regulated genes in ICAR of contralateral uterine horns of RB cows on Day 15 of the estrous cycle as compared with non-RB cows. (XLSX 254 kb)
Contributor Information
Ken-Go Hayashi, Email: hayaken@affrc.go.jp.
Misa Hosoe, Email: hosoe@affrc.go.jp.
Keiichiro Kizaki, Email: kizaki@iwate-u.ac.jp.
Shiori Fujii, Email: s1010590@affrc.go.jp.
Hiroko Kanahara, Email: kanahara@affrc.go.jp.
Toru Takahashi, Email: tatoru@iwate-u.ac.jp.
Ryosuke Sakumoto, Email: sakumoto@affrc.go.jp.
References
- 1.Perez-Marin CC, Calero GV, Moreno LM. Clinical Approach to the Repeat Breeder Cow Syndrome. In: Perez-Marin CC, editor. A Bird’s-Eye View of Veterinary Medicine. INTECH Open Access Publisher; 2012. doi:10.5772/31374.
- 2.Tanabe TY, Hawk HW, Hasler JF. Comparative fertility of normal and repeat-breeding cows as embryo recipients. Theriogenology. 1985;23:687–696. doi: 10.1016/0093-691X(85)90203-1. [DOI] [PubMed] [Google Scholar]
- 3.Dochi O, Takahashi K, Hirai T, Hayakawa H, Tanisawa M, Yamamoto Y, Koyama H. The use of embryo transfer to produce pregnancies in repeat-breeding dairy cattle. Theriogenology. 2008;69:124–128. doi: 10.1016/j.theriogenology.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Gustafsson H, Larsson K. Embryonic mortality in heifers after artificial insemination and embryo transfer: differences between virgin and repeat breeder heifers. Res Vet Sci. 1985;39:271–274. [PubMed] [Google Scholar]
- 5.Albihn A, Gustafsson H, Rodriguez-Martinez H, Larsson K. Development of day 7 bovine demi-embryos transferred into virgin and repeat-breeder heifers. Anim Reprod Sci. 1989;21:161–176. doi: 10.1016/0378-4320(89)90025-0. [DOI] [Google Scholar]
- 6.Katagiri S, Moriyoshi M. Alteration of the endometrial EGF profile as a potential mechanism connecting the alterations in the ovarian steroid hormone profile to embryonic loss in repeat breeders and high-producing cows. J Reprod Dev. 2013;59:415–420. doi: 10.1262/jrd.2013-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponsuksili S, Murani E, Schwerin M, Schellander K, Tesfaye D, Wimmers K. Gene expression and DNA-methylation of bovine pretransfer endometrium depending on its receptivity after in vitro-produced embryo transfer. PLoS One. 2012;7:e42402. doi: 10.1371/journal.pone.0042402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker CG, Littlejohn MD, Mitchell MD, Roche JR, Meier S. Endometrial gene expression during early pregnancy differs between fertile and subfertile dairy cow strains. Physiol Genomics. 2012;44:47–58. doi: 10.1152/physiolgenomics.00254.2010. [DOI] [PubMed] [Google Scholar]
- 9.Minten MA, Bilby TR, Bruno RG, Allen CC, Madsen CA, Wang Z, Sawyer JE, Tibary A, Neibergs HL, Geary TW, et al. Effects of fertility on gene expression and function of the bovine endometrium. PLoS One. 2013;8:e69444. doi: 10.1371/journal.pone.0069444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Killeen AP, Morris DG, Kenny DA, Mullen MP, Diskin MG, Waters SM. Global gene expression in endometrium of high and low fertility heifers during the mid-luteal phase of the estrous cycle. BMC Genomics. 2014;15:234. doi: 10.1186/1471-2164-15-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauersachs S, Ulbrich SE, Gross K, Schmidt SE, Meyer HH, Einspanier R, Wenigerkind H, Vermehren M, Blum H, Sinowatz F, Wolf E. Gene expression profiling of bovine endometrium during the oestrous cycle: detection of molecular pathways involved in functional changes. J Mol Endocrinol. 2005;34:889–908. doi: 10.1677/jme.1.01799. [DOI] [PubMed] [Google Scholar]
- 12.Mitko K, Ulbrich SE, Wenigerkind H, Sinowatz F, Blum H, Wolf E, Bauersachs S. Dynamic changes in messenger RNA profiles of bovine endometrium during the oestrous cycle. Reproduction. 2008;135:225–240. doi: 10.1530/REP-07-0415. [DOI] [PubMed] [Google Scholar]
- 13.Mansouri-Attia N, Aubert J, Reinaud P, Giraud-Delville C, Taghouti G, Galio L, Everts RE, Degrelle S, Richard C, Hue I, et al. Gene expression profiles of bovine caruncular and intercaruncular endometrium at implantation. Physiol Genomics. 2009;39:14–27. doi: 10.1152/physiolgenomics.90404.2008. [DOI] [PubMed] [Google Scholar]
- 14.Forde N, Beltman ME, Duffy GB, Duffy P, Mehta JP, O’Gaora P, Roche JF, Lonergan P, Crowe MA. Changes in the endometrial transcriptome during the bovine estrous cycle: effect of low circulating progesterone and consequences for conceptus elongation. Biol Reprod. 2011;84:266–278. doi: 10.1095/biolreprod.110.085910. [DOI] [PubMed] [Google Scholar]
- 15.Forde N, Carter F, Spencer TE, Bazer FW, Sandra O, Mansouri-Attia N, Okumu LA, McGettigan PA, Mehta JP, McBride R, et al. Conceptus-induced changes in the endometrial transcriptome: how soon does the cow know she is pregnant? Biol Reprod. 2011;85:144–156. doi: 10.1095/biolreprod.110.090019. [DOI] [PubMed] [Google Scholar]
- 16.Bauersachs S, Ulbrich SE, Reichenbach HD, Reichenbach M, Buttner M, Meyer HH, Spencer TE, Minten M, Sax G, Winter G, Wolf E. Comparison of the effects of early pregnancy with human interferon, alpha 2 (IFNA2), on gene expression in bovine endometrium. Biol Reprod. 2012;86:46. doi: 10.1095/biolreprod.111.094771. [DOI] [PubMed] [Google Scholar]
- 17.Forde N, Duffy GB, McGettigan PA, Browne JA, Mehta JP, Kelly AK, Mansouri-Attia N, Sandra O, Loftus BJ, Crowe MA, et al. Evidence for an early endometrial response to pregnancy in cattle: both dependent upon and independent of interferon tau. Physiol Genomics. 2012;44:799–810. doi: 10.1152/physiolgenomics.00067.2012. [DOI] [PubMed] [Google Scholar]
- 18.Spencer TE, Forde N, Dorniak P, Hansen TR, Romero JJ, Lonergan P. Conceptus-derived prostaglandins regulate gene expression in the endometrium prior to pregnancy recognition in ruminants. Reproduction. 2013;146:377–387. doi: 10.1530/REP-13-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu T, Krebs S, Bauersachs S, Blum H, Wolf E, Miyamoto A. Actions and interactions of progesterone and estrogen on transcriptome profiles of the bovine endometrium. Physiol Genomics. 2010;42A:290–300. doi: 10.1152/physiolgenomics.00107.2010. [DOI] [PubMed] [Google Scholar]
- 20.Sakumoto R, Hayashi KG, Saito S, Kanahara H, Kizaki K, Iga K. Comparison of the global gene expression profiles in the bovine endometrium between summer and autumn. J Reprod Dev. 2015;61:297–303. doi: 10.1262/jrd.2015-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King GJ, Atkinson BA, Robertson HA. Development of the bovine placentome during the second month of gestation. J Reprod Fertil. 1979;55:173–180. doi: 10.1530/jrf.0.0550173. [DOI] [PubMed] [Google Scholar]
- 22.King GJ, Atkinson BA, Robertson HA. Development of the bovine placentome from days 20 to 29 of gestation. J Reprod Fertil. 1980;59:95–100. doi: 10.1530/jrf.0.0590095. [DOI] [PubMed] [Google Scholar]
- 23.King GJ, Atkinson BA, Robertson HA. Development of the intercaruncular areas during early gestation and establishment of the bovine placenta. J Reprod Fertil. 1981;61:469–474. doi: 10.1530/jrf.0.0610469. [DOI] [PubMed] [Google Scholar]
- 24.Filant J, Spencer TE. Uterine glands: biological roles in conceptus implantation, uterine receptivity and decidualization. Int J Dev Biol. 2014;58:107–116. doi: 10.1387/ijdb.130344ts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pope WF, Maurer RR, Stormshak F. Distribution of progesterone in the uterus, broad ligament, and uterine arteries of beef cows. Anat Rec. 1982;203:245–250. doi: 10.1002/ar.1092030206. [DOI] [PubMed] [Google Scholar]
- 26.Ealy AD, Yang QE. Control of interferon-tau expression during early pregnancy in ruminants. Am J Reprod Immunol. 2009;61:95–106. doi: 10.1111/j.1600-0897.2008.00673.x. [DOI] [PubMed] [Google Scholar]
- 27.Dunne LD, Diskin MG, Sreenan JM. Embryo and foetal loss in beef heifers between day 14 of gestation and full term. Anim Reprod Sci. 2000;58:39–44. doi: 10.1016/S0378-4320(99)00088-3. [DOI] [PubMed] [Google Scholar]
- 28.Berg DK, van Leeuwen J, Beaumont S, Berg M, Pfeffer PL. Embryo loss in cattle between Days 7 and 16 of pregnancy. Theriogenology. 2010;73:250–260. doi: 10.1016/j.theriogenology.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Kizaki K, Shichijo-Kizaki A, Furusawa T, Takahashi T, Hosoe M, Hashizume K. Differential neutrophil gene expression in early bovine pregnancy. Reprod Biol Endocrinol. 2013;11:6. doi: 10.1186/1477-7827-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Ushizawa K, Takahashi T, Hosoe M, Ishiwata H, Kaneyama K, Kizaki K, Hashizume K. Global gene expression analysis and regulation of the principal genes expressed in bovine placenta in relation to the transcription factor AP-2 family. Reprod Biol Endocrinol. 2007;5:17. doi: 10.1186/1477-7827-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker CG, Meier S, Mitchell MD, Roche JR, Littlejohn M. Evaluation of real-time PCR endogenous control genes for analysis of gene expression in bovine endometrium. BMC Mol Biol. 2009;10:100. doi: 10.1186/1471-2199-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ushizawa K, Takahashi T, Hosoe M, Kizaki K, Hashizume K. Characterization and expression analysis of SOLD1, a novel member of the retrotransposon-derived Ly-6 superfamily, in bovine placental villi. PLoS One. 2009;4:e5814. doi: 10.1371/journal.pone.0005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayes JD, McLellan LI. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic Res. 1999;31:273–300. doi: 10.1080/10715769900300851. [DOI] [PubMed] [Google Scholar]
- 35.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 36.Ilic Z, Crawford D, Vakharia D, Egner PA, Sell S. Glutathione-S-transferase A3 knockout mice are sensitive to acute cytotoxic and genotoxic effects of aflatoxin B1. Toxicol Appl Pharmacol. 2010;242:241–246. doi: 10.1016/j.taap.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kensler KH, Slocum SL, Chartoumpekis DV, Dolan PM, Johnson NM, Ilic Z, Crawford DR, Sell S, Groopman JD, Kensler TW, Egner PA. Genetic or pharmacologic activation of Nrf2 signaling fails to protect against aflatoxin genotoxicity in hypersensitive GSTA3 knockout mice. Toxicol Sci. 2014;139:293–300. doi: 10.1093/toxsci/kfu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baranova H, Canis M, Ivaschenko T, Albuisson E, Bothorishvilli R, Baranov V, Malet P, Bruhat MA. Possible involvement of arylamine N-acetyltransferase 2, glutathione S-transferases M1 and T1 genes in the development of endometriosis. Mol Hum Reprod. 1999;5:636–641. doi: 10.1093/molehr/5.7.636. [DOI] [PubMed] [Google Scholar]
- 39.Jackson LW, Schisterman EF, Dey-Rao R, Browne R, Armstrong D. Oxidative stress and endometriosis. Hum Reprod. 2005;20:2014–2020. doi: 10.1093/humrep/dei001. [DOI] [PubMed] [Google Scholar]
- 40.Yokoi R, Hayashi M, Tamura T, Kobayashi K, Kuroda J, Kusama H, Kagami H, Ono T. Embryonic mortality and intrauterine growth retardation (IUGR) associated with placental alterations in pregnant rats treated with methyl methanesulfonate (MMS) at the peri-implantation stage. J Toxicol Sci. 2008;33:585–598. doi: 10.2131/jts.33.585. [DOI] [PubMed] [Google Scholar]
- 41.Celi P, Merlo M, Da Dalt L, Stefani A, Barbato O, Gabai G. Relationship between late embryonic mortality and the increase in plasma advanced oxidised protein products (AOPP) in dairy cows. Reprod Fertil Dev. 2011;23:527–533. doi: 10.1071/RD10268. [DOI] [PubMed] [Google Scholar]
- 42.Loset M, Mundal SB, Johnson MP, Fenstad MH, Freed KA, Lian IA, Eide IP, Bjorge L, Blangero J, Moses EK, Austgulen R. A transcriptional profile of the decidua in preeclampsia. Am J Obstet Gynecol. 2011;204:84. doi: 10.1016/j.ajog.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson AS, Mannervik B. Human glutathione transferase A3-3, a highly efficient catalyst of double-bond isomerization in the biosynthetic pathway of steroid hormones. J Biol Chem. 2001;276:33061–33065. doi: 10.1074/jbc.M104539200. [DOI] [PubMed] [Google Scholar]
- 44.Gray CA, Bazer FW, Spencer TE. Effects of neonatal progestin exposure on female reproductive tract structure and function in the adult ewe. Biol Reprod. 2001;64:797–804. doi: 10.1095/biolreprod64.3.797. [DOI] [PubMed] [Google Scholar]
- 45.Filant J, Zhou H, Spencer TE. Progesterone inhibits uterine gland development in the neonatal mouse uterus. Biol Reprod. 2012;86:146. doi: 10.1095/biolreprod.111.097089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Almeida Fo AP, Ayalon N, Bartoov B. Progesterone receptors in the endometrium of normal and repeat-breeder cows. Anim Reprod Sci. 1987;14:11–19. doi: 10.1016/0378-4320(87)90072-8. [DOI] [Google Scholar]
- 47.Boshier DP. A histological and histochemical examination of implantation and early placentome formation in sheep. J Reprod Fertil. 1969;19:51–61. doi: 10.1530/jrf.0.0190051. [DOI] [PubMed] [Google Scholar]
- 48.Spencer TE, Johnson GA, Bazer FW, Burghardt RC. Fetal-maternal interactions during the establishment of pregnancy in ruminants. Soc Reprod Fertil Suppl. 2007;64:379–396. doi: 10.5661/rdr-vi-379. [DOI] [PubMed] [Google Scholar]
- 49.Dhanoa BS, Cogliati T, Satish AG, Bruford EA, Friedman JS. Update on the Kelch-like (KLHL) gene family. Hum Genomics. 2013;7:13. doi: 10.1186/1479-7364-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Z, Li S, Feng C, Yang S, Wang H, Ma D, Zhang J, Gou M, Bu D, Zhang T, et al. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat Genet. 2016;48:1508–1516. doi: 10.1038/ng.3701. [DOI] [PubMed] [Google Scholar]
- 51.Schweikl H, Hiller KA, Eckhardt A, Bolay C, Spagnuolo G, Stempfl T, Schmalz G. Differential gene expression involved in oxidative stress response caused by triethylene glycol dimethacrylate. Biomaterials. 2008;29:1377–1387. doi: 10.1016/j.biomaterials.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 52.Sorsa T, Salo T, Koivunen E, Tyynela J, Konttinen YT, Bergmann U, Tuuttila A, Niemi E, Teronen O, Heikkila P, et al. Activation of type IV procollagenases by human tumor-associated trypsin-2. J Biol Chem. 1997;272:21067–21074. doi: 10.1074/jbc.272.34.21067. [DOI] [PubMed] [Google Scholar]
- 53.Moilanen M, Sorsa T, Stenman M, Nyberg P, Lindy O, Vesterinen J, Paju A, Konttinen YT, Stenman UH, Salo T. Tumor-associated trypsinogen-2 (trypsinogen-2) activates procollagenases (MMP-1, −8, −13) and stromelysin-1 (MMP-3) and degrades type I collagen. Biochemistry. 2003;42:5414–5420. doi: 10.1021/bi020582s. [DOI] [PubMed] [Google Scholar]
- 54.Tatemoto K, Efendic S, Mutt V, Makk G, Feistner GJ, Barchas JD. Pancreastatin, a novel pancreatic peptide that inhibits insulin secretion. Nature. 1986;324:476–478. doi: 10.1038/324476a0. [DOI] [PubMed] [Google Scholar]
- 55.Aardal S, Helle KB, Elsayed S, Reed RK, Serck-Hanssen G. Vasostatins, comprising the N-terminal domain of chromogranin A, suppress tension in isolated human blood vessel segments. J Neuroendocrinol. 1993;5:405–412. doi: 10.1111/j.1365-2826.1993.tb00501.x. [DOI] [PubMed] [Google Scholar]
- 56.Mahata SK, O’Connor DT, Mahata M, Yoo SH, Taupenot L, Wu H, Gill BM, Parmer RJ. Novel autocrine feedback control of catecholamine release. A discrete chromogranin a fragment is a noncompetitive nicotinic cholinergic antagonist. J Clin Invest. 1997;100:1623–1633. doi: 10.1172/JCI119686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helle KB, Corti A. Chromogranin A: a paradoxical player in angiogenesis and vascular biology. Cell Mol Life Sci. 2015;72:339–348. doi: 10.1007/s00018-014-1750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michael DD, Alvarez IM, Ocon OM, Powell AM, Talbot NC, Johnson SE, Ealy AD. Fibroblast growth factor-2 is expressed by the bovine uterus and stimulates interferon-tau production in bovine trophectoderm. Endocrinology. 2006;147:3571–3579. doi: 10.1210/en.2006-0234. [DOI] [PubMed] [Google Scholar]
- 59.Kizaki K, Ushizawa K, Takahashi T, Yamada O, Todoroki J, Sato T, Ito A, Hashizume K. Gelatinase (MMP-2 and −9) expression profiles during gestation in the bovine endometrium. Reprod Biol Endocrinol. 2008;6:66. doi: 10.1186/1477-7827-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ulbrich SE, Meyer SU, Zitta K, Hiendleder S, Sinowatz F, Bauersachs S, Buttner M, Frohlich T, Arnold GJ, Reichenbach HD, et al. Bovine endometrial metallopeptidases MMP14 and MMP2 and the metallopeptidase inhibitor TIMP2 participate in maternal preparation of pregnancy. Mol Cell Endocrinol. 2011;332:48–57. doi: 10.1016/j.mce.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 61.Mamo S, Mehta JP, Forde N, McGettigan P, Lonergan P. Conceptus-endometrium crosstalk during maternal recognition of pregnancy in cattle. Biol Reprod. 2012;87:6. doi: 10.1095/biolreprod.112.099945. [DOI] [PubMed] [Google Scholar]
- 62.Tsai SJ, Wu MH, Chen HM, Chuang PC, Wing LY. Fibroblast growth factor-9 is an endometrial stromal growth factor. Endocrinology. 2002;143:2715–2721. doi: 10.1210/endo.143.7.8900. [DOI] [PubMed] [Google Scholar]
- 63.Ostrup E, Bauersachs S, Blum H, Wolf E, Hyttel P. Differential endometrial gene expression in pregnant and nonpregnant sows. Biol Reprod. 2010;83:277–285. doi: 10.1095/biolreprod.109.082321. [DOI] [PubMed] [Google Scholar]
- 64.Klein C. Novel equine conceptus-endometrial interactions on day 16 of pregnancy based on RNA sequencing. Reprod Fertil Dev. 2016;28:1712–1720. doi: 10.1071/RD14489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All microarray data are available at the Gene Expression Omnibus (GEO) database at NCBI (http://www.ncbi.nlm.nih.gov/geo), under accession numbers GSE79367. All datasets on which the conclusions of the paper rely are available to readers.


