Abstract
Introduction
Both reverse-phase and HILIC chemistries are deployed for liquid-chromatography mass spectrometry (LC-MS) metabolomics analyses, however HILIC methods lag behind reverse-phase methods in reproducibility and versatility. Comprehensive metabolomics analysis is additionally complicated by the physiochemical diversity of metabolites and array of tunable analytical parameters.
Objective
Our aim was to rationally and efficiently design complementary HILIC-based polar metabolomics methods on multiple instruments using Design of Experiments (DoE).
Methods
We iteratively tuned LC and MS conditions on ion-switching triple quadrupole (QqQ) and quadrupole-time-of-flight (qTOF) mass spectrometers through multiple rounds of a workflow we term COLMeD (Comprehensive optimization of LC-MS metabolomics methods using design of experiments). Multivariate statistical analysis guided our decision process in the method optimizations.
Results
LC-MS/MS tuning for the QqQ method on serum metabolites yielded a median response increase of 161.5% (p<0.0001) over initial conditions with a 13.3% increase in metabolite coverage. The COLMeD output was benchmarked against two widely used polar metabolomics methods, demonstrating total ion current increases of 105.8% and 57.3%, with median metabolite response increases of 106.1% and 10.3% (p<0.0001 and p<0.05 respectively). For our optimized qTOF method, 22 solvent systems were compared on a standard mix of physiochemically diverse metabolites, followed by COLMeD optimization, yielding a median 29.8% response increase (p<0.0001) over initial conditions.
Conclusions
The COLMeD process elucidated response tradeoffs, facilitating improved chromatography and MS response without compromising separation of isobars. COLMeD is efficient, requiring no more than 20 injections in a given DoE round, and flexible, capable of class-specific optimization as demonstrated through acylcarnitine optimization within the QqQ method.
Keywords: Design of Experiments, HILIC LC-MS, Method Development, Multivariate Statistical Analysis
1 Introduction
An ideal metabolomics platform would profile all the metabolites in a living system. Complementary approaches such as gas-chromatography mass spectrometry (GC-MS), liquid chromatography mass spectrometry (LC-MS), or nuclear magnetic resonance (NMR) can be employed to enhance analytical coverage of the metabolome, however the high physiochemical diversity in metabolites and technological limitations confine any individual analyses to a relatively small subset of the metabolome. Within LC-MS, multiple column chemistries are also increasingly incorporated into metabolomics workflows to further enhance coverage (Want et al. 2010). Hydrophilic interaction chromatography (HILIC) methods hold significant promise for comprehensive analysis of polar metabolites, however reverse-phase methods are routinely used for small polar molecules (New et al. 2008), in part due to a longer history of reproducible chromatography. The choice of MS detection is also critical for augmenting metabolome coverage. For instance, untargeted high-resolution instruments can provide good coverage and sensitivity (Want et al. 2010), while modern triple quadrupole (QqQ) or ion-trap instruments provide ion-switching and fast-scanning capabilities for targeted metabolite identification and quantification (Yuan et al. 2012; Gika et al. 2012; Lv et al. 2011). Holistic improvement of both chromatography and detection parameters requires bespoke methods to address a large multivariate problem space (Gika et al. 2014). This problem has been previously addressed using genetic algorithms and large-scale Bayesian networks (Napoles et al. 2014; Correa et al. 2011), however these approaches do not concurrently optimize numerous parameters inherent in the comprehensive LC-MS methodology, nor have they been applied to HILIC, which is often sensitive to small LC parameter adjustments (Nguyen et al. 2008; Hao et al. 2008). An alternative approach for complex method optimization is Design of Experiments (DoE), which incorporates multivariate modeling of many response variables simultaneously (Eriksson et al. 2006). DoE allows for the manipulation of several factors concurrently and efficiently searches for interaction effects, as opposed to simply changing one factor at a time. Typical DoE workflows start with screening objectives, where the most important factors and their appropriate ranges are chosen and subsequently optimized iteratively. DoE has been used in optimizing other steps of the typical metabolomics workflow, including sample preparation and data processing (A et al. 2005; Eliasson et al. 2012; Zheng et al. 2013). LC-MS methods have also been improved in this manner, however the response of interest has typically been targeted to one metabolite or a single class of compounds (Zhou et al. 2009; Székely et al. 2012; Kostić et al. 2013; Riter et al. 2005).
Here we demonstrate that a DoE-driven approach has potential for large-scale metabolomics method development by improving metabolome coverage without overtly sacrificing individual metabolite chromatography and MS response. Our main objective was to design and optimize a polar metabolomics platform, while addressing the idiosyncrasies of targeted and untargeted LC-MS metabolomics. We coin this method comprehensive optimization of LC-MS methods through DoE (COLMeD) as a workflow procedure and assess the capability of DoE to improve responses on a diverse set of polar metabolites. We find this workflow is robust to method development across MS detection methods, tailoring the COLMeD approach to an LC-MS electrospray positive ionization (ESI+) method using high-resolution quadrupole time-of-flight (qTOF) detection subsequent to the initial polarity-switching QqQ optimization. We show that the results are robust to multiple sample types and can be tailored in a class-specific manner by specifically optimizing acylcarnitines from the comprehensive QqQ method. We note improvements over commonly used methods (Yuan et al. 2012; Paglia et al. 2012), and our workflow informed parameter decisions to limit response tradeoffs. Moreover, we describe a generalized procedure, bearing in mind the utility of this approach for efficiently optimizing other facets of analytical method development.
2 Methods
2.1 Chemicals
All chemical standards used in this study were minimally analytical grade and obtained from commercial sources (Table S1). Optima grade acetonitrile and methanol were purchased from Fisher Scientific (Fair Lawn, NJ) for the mobile phase and standard solutions. Optima LC/MS ammonium acetate and formic acid and TraceMetal grade ammonium hydroxide were used as mobile phase additives and also obtained from Fisher Scientific. All water used in this study was deionized and filtered (18.2MΩ, 0.22μm).
2.2 Preparation of Standard Solutions and Biological Samples
Standard solutions for positive mode qTOF DoE were prepared as 1mg/mL stocks in 100% methanol and diluted to 1μg/mL in 3:1 acetonitrile:methanol. All samples were centrifuged at 18787g for 5 minutes before LC-MS injection. Gibco horse serum (Invitrogen, Grand Island, NY) and homogenized Drosophila melanogaster samples were prepared using a modified Bligh-dyer extraction (Bligh et al. 1959). Briefly, 120μL of 2:1 methanol:chloroform was added to 20μL of serum or 40mg of fly tissue, followed by a brief vortex and 15 minute sonication. 40μL of both chloroform and water were added to the solution, followed by centrifugation at 18787g for 7 minutes to form the bilayer. The top layer, containing the aqueous fraction, was isolated and dried down overnight. The dried pellet was resuspended in either 100μL or 400μL of 50:50 water:acetonitrile for serum and fly respectively before LC-MS injection.
2.3 LC-MS Conditions
Chromatographic separations for the ion-switching DoE were performed on an XBridge BEH Amide column (2.1x100mm, 2.5μm, Waters Corporation, Milford, MA) with a 2.1x5mm Vanguard pre-column. DoE chromatography for untargeted qTOF analysis was performed on an ACQUITY UPLC BEH Amide column (2.1x150mm, 1.7μm), with a 0.2μm in-line filter. Both methods utilized an ACQUITY H-Class UPLC (Waters Corporation). The mobile phases for the ion-switching analysis were initially taken from Yuan et al., where the aqueous mobile phase consisted of 95:5 water:acetonitrile with 20mM ammonium acetate and ammonium hydroxide, pH 9, with the organic mobile phase as 100% acetonitrile. Mobile phase A for the qTOF LC-MS method was comprised of 95:5 water:acetonitrile with 2mM ammonium acetate and 0.2% formic acid, while mobile phase B consisted of 90:10 acetonitrile:water with 2mM ammonium acetate and 0.2% formic acid, which was determined through experimental testing as described later in the text. Mass spectrometry was performed on either a Waters TQD or Waters G2-S qTOF in positive ion mode using electrospray ionization (ESI+), using Leucine-enkephalin for the lock-mass calibration. As a basis of comparison for our approach, the LC-MS methods described by Yuan et al. (Method 1) and from a Waters HILIC Application Note (Paglia et al. 2012, Method 2) were followed as published. Chromatograms were processed using TargetLynx under MassLynx version 4.1. Statistical analyses and plotting was performed in R version 3.2, and comparisons between DoE rounds were made via paired Wilcoxon signed-rank tests.
2.4 Model Generation, Design, and Optimization in the COLMeD Process
DoE serves to discover important predictor variables which contribute to one or many desired responses to determine optimal factor tuning (Eriksson et al. 2006). To deal with the complexity of tuning multiple factors to manipulate the many responses in our metabolomics methods, our DoE-driven COLMeD approach employs a partial least squares (PLS) fitting algorithm. Specifically, PLS fits a model to the variation of all responses with the variation of the factors by accounting for their covariance. This method of fitting is more efficient than multiple linear regression (MLR), which is also common in multivariate optimization problems, since MLR fits separate regression models for each response. MLR also suffers when handling missing data points, which we had encountered in our response matrix, given some chromatographic peaks were not always present depending on the LC-MS factor settings. In our case, dependent variables are the original analytical responses (e.g. metabolite peak area and chromatographic values), which are tuned to independent LC-MS factors by transformation into latent variables. The number of latent variables, or PLS components, were determined through the default mechanism in MODDE v11 (Umetrics, Umeå, Sweden), whereby components were added to improve goodness of fit (R2) until the goodness of prediction (Q2) was compromised by overfitting the model. The predictive performance of the model was computed via 7-fold cross validation. A major advantage of this approach is the ability to weigh one response more than another across DoE rounds, which we utilize heavily in our COLMeD process. For example, responses with significant peak area under the curve (AUC) after the first round were downweighted in the predictive modeling to favor LC-MS settings which improve other features with lower responses. AUC responses were set to be maximized in the modeling, while the peak widths were tailored such that peak width objectives were either 4 seconds for the untargeted qTOF method or 15 seconds for the QqQ method, which benefits from slightly wider peaks due to tradeoffs of scanning over hundreds of MRMs. Additionally, the response objectives can be adjusted round over round to achieve iterative improvement. After modeling, a new set of experiments are generated, in the form of LC-MS settings. This process constitutes one round of DoE, which would be repeated until optimal conditions are met (specific COLMeD processes listed in Table 1).
Table 1.
COLMeD Workflow for both QqQ and qTOF Methods
| Workflow | QqQ | qTOF |
|---|---|---|
| Initial factor range and response selection |
Factors: LC only (round 1), followed by LC- MS Responses: Metabolites from horse serum (Table S2) |
Factors LC and MS optimization separate Responses: Standard mix, both LC and MS response (Tables S5 and S6) |
| Pilot LC-MS batch |
D-Optimal linear screening design: 13 runs |
D-Optimal quadratic design for LC: 20 runs D-Optimal linear for MS: 13 runs |
| Data processing and fitting to PLS models |
Main effect plots and PLS loadings for data assessment |
|
| Model analysis and optimum predictions |
Predictive design space plots, and optimizer function for optimum predictions, VIP > 1 for important factors |
|
| Update factor settings and constrain design space for next round; change design as needed |
1 round for LC (13 runs) 3 rounds for LC- MS (13, 17, 9 runs) |
3 rounds for LC (20, 12, 12 runs) 2 rounds for MS (13, 12 runs) |
In a regularly shaped design region, central composite or full factorial designs are typically chosen to explore the edges of the design space. However, D-Optimal designs generated in MODDE can accommodate experiments with irregular design regions (Eriksson et al. 2006), which allowed us to impose constraints on our LC-MS settings which were not feasible or desirable, for example long LC gradients coupled with high flow rates. We tested the edges of the irregular design space in addition to replicate LC-MS injections in the center of the space to gauge reproducibility and model validity. In addition, we performed a conserved triplicate injection at the end of each DoE round as a quality control measure across batches. To rationalize the LC-MS parameters for the next round of experiments, we utilized both visual representations of optimal regions within the design space and a quantitative optimizer function which generated a list of parameters to yield an optimized solution using the PLS model. In addition to the model statistics and predictive functions of the PLS model, we also evaluated the VIP value, which is a multivariate metric used to identify the relative importance of an original predictor variable (i.e. before transformation) to the model (Eriksson et al. 2006). These values identify non-significant contributions of LC-MS parameters to the metabolite responses, which allowed us to assign fixed values and simplify the design space for the next DoE round. We chose to use MODDE software due to integrated cross-validated model fitting, model fit visualizations, and predictive capabilities. Open-source platforms for each of the steps in the COLMeD process could be alternatively used to build an in-house workflow.
2.5 Ion-Switching COLMeD
While the ultimate goal was to optimize both LC and MS conditions, the initial experimental design space including LC gradients was too large to combine with MS parameters and therefore required a two-stage approach, outlined in Table 1. A screening linear objective design of LC-only tuning was chosen for this initial round, omitting interaction effects and requiring only 13 LC-MS injections. Response measurements were taken on multiple sample types, horse serum and homogenized fly samples, which provided an added measure of confidence in designing the next round of experiments. The total initial response optimization consisted of a set of 33 responses in horse serum (Table S2), as well as unique responses found in fly tissue but uncommon in serum in order to enhance overall coverage in the initial screen (Table S3). Measured response variables were chosen based on criteria designed to elucidate a broad physiochemical range of metabolites and the presence of marginally detectable metabolites, in addition to measures of peak quality. We tailored the responses to reflect particular considerations of MRM-based analysis. For example, glutamine and lysine have overlapping MRMs, thus we fit the PLS model to predict maximal retention time separation. MRM transitions and voltages were optimized by using pure standards or from the METLIN and HMDB mass spectrometry databases (Smith et al. 2005; Wishart et al. 2013). For AUC response optimization, the objective defined in the PLS fitting was set by using the mean AUC from the LC-MS injections of that DoE round as a threshold, from which the optimizer and design space plots were used to find conditions predicted to increase AUC for the subsequent DoE round.
Factors considered for the LC optimization included the initial LC flow rates (0.1–0.25mL/min, continuous variable) and gradient types (1–4, Figure S1, discrete). The gradients were rationalized from both published (Yuan et al. 2012) and unpublished work. The LC-MS parameters proposed by Yuan et al. (Gradient 1) served as a starting point to build our metabolomics platform, however we felt the COLMeD process could improve response within the QqQ method as well as and LC solvents as Yuan et al. for the QqQ method, we relaxed the LC parameters in an exploratory manner before refined tuning in later rounds. The MS parameters were initially set as close to the published settings as possible while aligned with vendor-specific voltage parameters. Initial gradient times (10–16min) were purposefully imposed to achieve reasonably high throughput. The gradient time refers to the time of injection until the wash step.
2.6 Modular Workflow Optimization
After Rounds 2 and 3, we sought to further optimize the method for a specific class of compounds with shared chemical properties, in which case the parent method could be used to tune the response for specific compounds, such as carnitines. The analytical factors were analogous to other rounds of QqQ DoE, but the responses were limited to peak AUCs and peak widths for carnitine, acetylcarnitine, propionylcarnitine, and butrylcarnitine. The data from DoE rounds 2 and 3 were combined and used as inclusions in a D-Optimal quadratic design, whereby only an additional six test runs were needed to finish out the model design. The experimental space in which the method optimum predictions from Rounds 2 and 3 overlapped with predictions based off of these additional test runs with good model statistics confirmed the optimized method and thus completed the class-specific DoE.
2.7 Untargeted qTOF COLMeD
Optimizing chromatography for an untargeted method requires additional considerations due to the large number of unknown responses. Rather than optimizing responses on a serum sample, which would contain many unknown features, we initially developed the qTOF method on a standard mix of 48 diverse polar metabolites injected at 1μg/mL (Table S1). Prior to DoE, 22 LC solvent systems were compared using the standard mix. These solvents were based on a literature search and are listed in Table S4 (Want et al. 2010; Kivilompolo et al. 2013; Ivanisevic et al. 2013; Zhou et al. 2013). Mass spectrometry settings were based on data from the ion-switching method development, where desolvation temperature and gas flow were set to 500°C and 1000L/Hr respectively. Unless specified by a particular published method, the gradients were aligned across each solvent system, with 45°C column temperature. Given the solvents used in the QqQ method are pH 9, new solvents were required for a qTOF method in positive ionization. Aggregate measures of peak capacity, peak skew, peak resolution between any two pairs of peaks, number of peaks, MS response, and peak widths were all compared to choose the initial LC solvents (Table S5). These metrics were used in addition to inspection of chromatography to choose the best solvents manually. We found that some solvents yielded split peaks and prohibited detection of all the metabolites in our mix, thus we felt the need to inspect these results and choose accordingly before proceeding to strictly quantitative optimization of the method using DoE. After selecting the LC solvents, the COLMeD approach was divided into two parts to optimize LC and MS settings separately, which was in large part guided by our QqQ COLMeD findings. Three rounds of DoE were performed for the LC factors (Table S6), which included responses for isobar separations of leucine/isoleucine and alanine/sarcosine, with fixed MS parameters. For ease of comparison, peak response (AUC), peak width, and peak skew were converted to rank-based values, whereby each injection was ranked in each of these metrics across every injection from a given DoE round. The holistic peak metrics used as responses in the qTOF COLMeD are more amenable to peak-picking methods used in untargeted data processing algorithms. Our objective with the initial LC DoE was to heavily favor optimization of chromatography, which naturally derives from the variety of peak quality metrics (peak shape, width, separation, etc..) chosen compared to a singular readout of pure metabolite response on the MS. In addition, automated integration of peaks can be difficult when flow rates and gradients are tuned, which does not change during MS factor tuning. AUC was thus one of several responses optimized in the LC DoE, but subsequently the sole response variable used in the MS DoE after chromatography was fixed.
Thus we maintained a similar workflow to the QqQ COLMeD procedure, with additional fit-for-purpose modifications to the developmental process. After LC optimization, two rounds of DoE were performed for the MS parameters in ESI+. Similar to the polarity switching design, the LC factors consisted of flow rate (0.2–0.5mL/min), gradient slope (4–9, which was calculated by percent change in solvent B divided by gradient time), and column temperature (30–60°C). By having a simpler design space with only three factors, we could employ a more complex a D-Optimal quadratic model (20 runs). We were able to further simplify the design by eliminating non-significant factors and perform rounds 2 and 3 as full factorial designs (12 runs each, in a 2x2x3 design). A full factorial design allows for simultaneously testing three levels of each factor and support a quadratic model. While typically experimentally costly, with only two factors, this design space can be tested with only 12 injections, including center point replicate injections. This design is thus identical to the central composite face-centered (CCF) analysis, which is recommended for full scale investigations and optimization after elimination of less important factors from earlier DoE rounds (Eriksson et al. 2006). Our criteria for removing factors included both analysis of coefficient plots and displaying a VIP score below 1. The detailed models informed tradeoffs in analyte response, while also considering conditions providing sufficient chromatographic resolution between isobars. The MS factors consisted of sampling cone voltage (20–40V), desolvation temperature (400–550°C), source temperature (90–150°C), cone gas flow (20–80L/Hr), and source offset (60–100V). The sole response optimized for the MS DoE was average AUC rank for each injection on the 48 standards. DoE was completed when we were able to identify the LC-MS parameters that met our response thresholds, the elucidation of tradeoffs in the method, and weak PLS model statistics, which indicated a tightly constrained design space with minimal gains for further improvement.
3 Results and Discussion
3.1 Round 1: Initial LC Screening for Polarity Switching Method
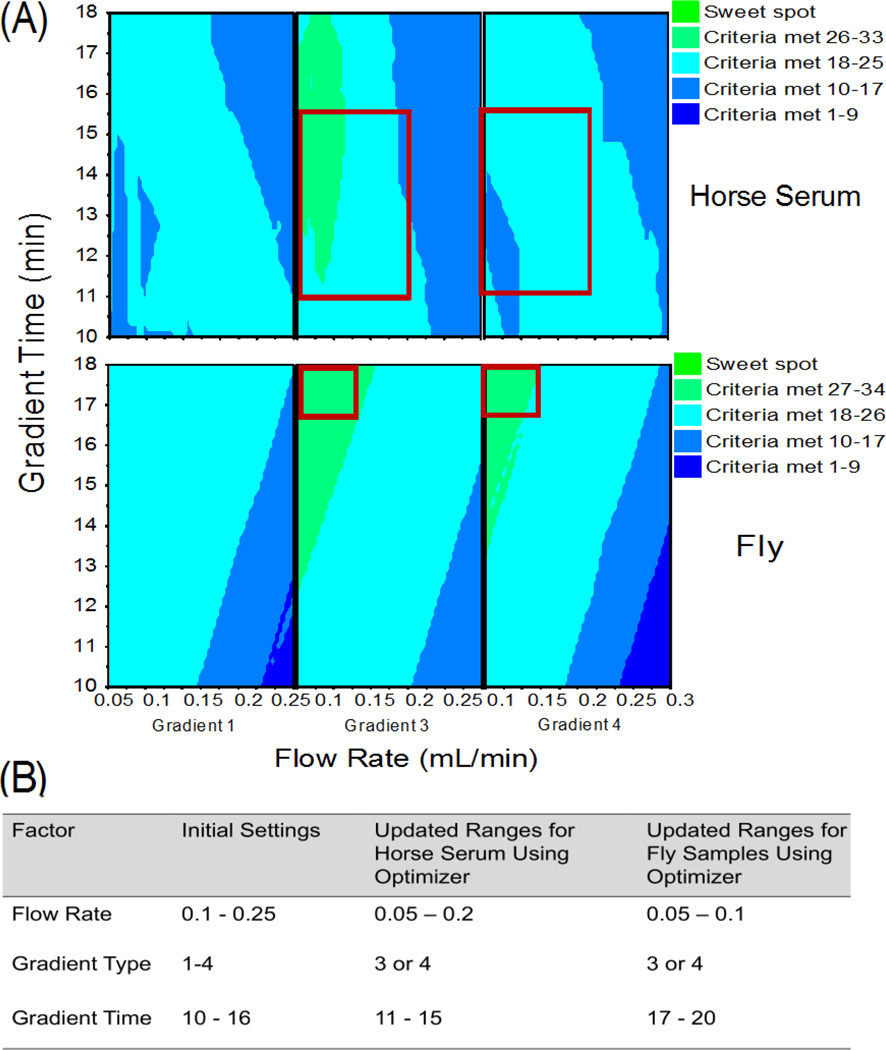
The initial screening batch for the comprehensive quadrupole method LC conditions consisted of 13 injections, repeated for both horse serum and fly samples. Predictive design space plots and optimizer analysis of both data sets yielded similar trends for all three factors (Figure 1). Notably, lower flow rates and/or longer gradients were predicted to improve response, with gradients 3 and 4 yielding predicted design space regions with the most response criteria met. Some differences in the predicted gradient time optimum likely result from slightly different response lists across sample types. However, given both flow rate and gradient time had significant impact (VIPs of 1.43 and 1.08 respectively), also supported by analysis of PLS loadings plot (Figure S2), our initial estimates of factor ranges based on a priori rationale and desirability of run time and flow rates needed to be expanded. The model statistics from Round 1 serum analysis gave us confidence in obtaining the method optimum via expanding these factor ranges (Table S7).
Figure 1.
(a) Predictive plots displaying design space regions with predicted optimal response, based on initial LC screening (round 1) with horse serum and fly samples. Green indicates design space where a maximal number of endpoint response thresholds are predicted to be met. Red boxes indicate regions predicted to improve responses for the next round. (b) Factor settings before and after analysis of serum and fly samples us-ing the optimizer function. These complementary approaches rationalize LC-MS parameters for the next round of experiments
3.2 Continued DoE with LC and MS Factors
After adjusting the LC factors from Round 1, the MS factors and column temperature were added to the design. While displaying a relatively weak effect compared to other factors, gradient 4 slightly outperformed gradient 3 (nonsignificantly), and was arbitrarily selected and fixed in future designs. Running multiple optimizations predicted flow rates above 0.15mL/min to improve results. To some degree this prediction contradicts the predictions from Round 1, which may be resolved with increased sampling. Given that flow rate maintained an important contribution to the PLS model (VIP = 1.09), the flow rate was restricted to regions of the design space where the predictive plots and optimization functions overlapped in optimum predictions (0.15–0.3mL/min). Desolvation temperature also had a large effect on response as demonstrated by the largest VIP value (1.19) amongst LC-MS factors. Consequently, this factor was restricted to 300–500°C for Round 3 based on the predictive plots and optimizer function. The predicted optimal gradient times were variable, however given the desire to increase throughput, 12–18 minute gradients were set for Round 3. Column temperature had less effect on the model compared to the other factors (VIP=0.89), and these temperatures were restricted to 40–55°C for Round 3 based on the predictions. Desolvation gas flow was retained as a factor in the subsequent model despite a minimal contribution (VIP=0.8), albeit restricted to maintain compatibility with the desolvation temperatures according to the manufacturer’s recommendations.
3.3 Further Refinement of Design Space
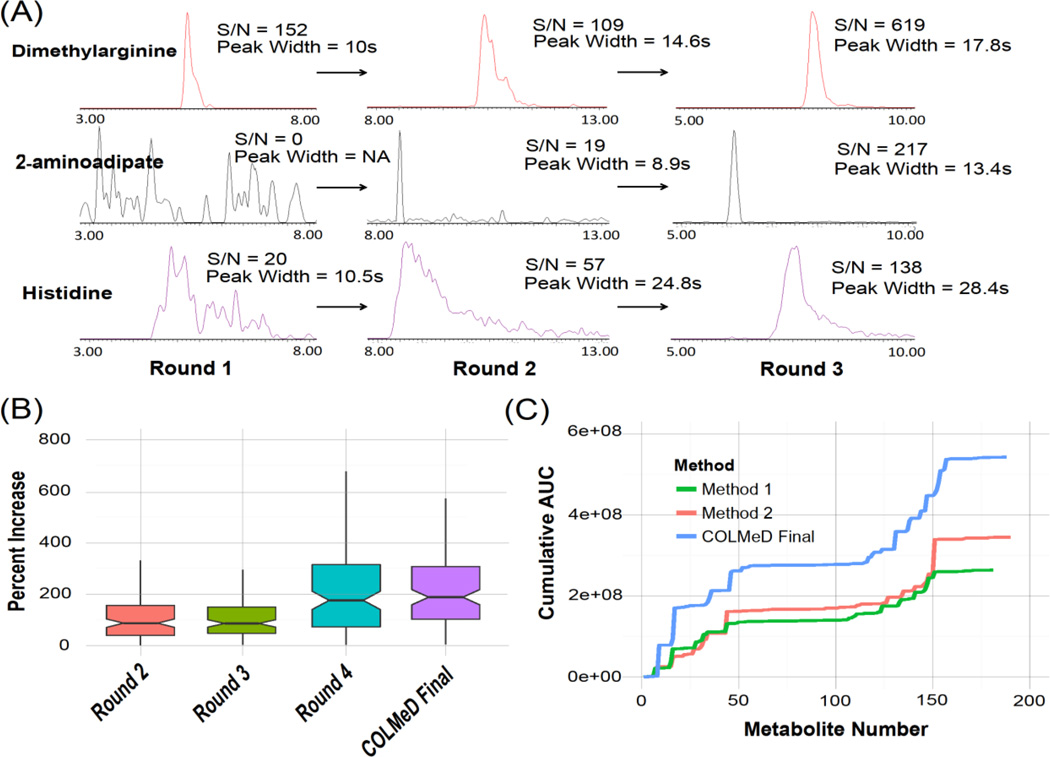
The updated factor ranges were used for the fractional factorial design in Round 3, necessitating 17 runs. Predictions from the model yielded slightly shifted but generally consistent results with Round 2. Flow rate and desolvation temperature again were the most significant factors, (VIP of 1.88 and 1.37, respectively). Optimal flow rates gravitated towards the low end of the 0.15–0.3mL/min range. Conversely, higher desolvation temperatures were predicted to perform better. Gradient times were consistent from predictions from Round 2, likewise, column temperature and desolvation gas flow were largely irrelevant, with the factor ranges being unaffected by the updated predictions. Over the course of the three rounds we noticed marked improvement in multiple endpoints, including MS response, peak width (towards our goal of 15sec widths at half height), and number of metabolites (Figure 2a).
Figure 2.
(a) Chromatographic improvement in selected metabolites after three rounds of DoE in the QqQ COLMeD process. Signal-to-noise (S/N) and peak width (full width half height) measurements were generated from vendor software after limited peak smoothing and integration. (b) Percent response increase by metabolite across each round. (c) Cumulative AUC plots for QqQ COLMeD benchmarking
3.4 Acylcarnitine-specific DoE
To explore the notion of targeting subsets of metabolites from the comprehensive parent method, data from rounds 2 and 3 of the polarity switching DoE were combined to generate predictions for increased AUC and optimized peak width of carnitine, acetylcarnitine, propionylcarnitine, and butrylcarnitine. Based on this data, lower flow and longer gradients were expected to improve response and peak width. Analysis of the acylcarnitine-specific LC-MS runs generated from modeling the data in rounds 2 and 3 revealed consistencies in the predictions and improved AUC response and peak width with strong PLS fitting (Table S7), demonstrating successful confirmation of the predictions and optimization of a compound class specific method (Figure S4).
3.5 Refinement and Validation of Comprehensive Method
We primarily attributed improvements in the acylcarnitines across Rounds 2 and 3 to adjustments in desolvation temperature. Given this information, we then evaluated the original 33 responses chosen in the comprehensive method with the acylcarnitine-specific experimental design to observe tradeoffs between response improvement of the carnitines, which exhibit positive ionization, versus other metabolites in both ESI+ and ESI- modes (labeled as Round 4 in Figure 2b). While not a true round 4 DoE design for the comprehensive method, we can leverage this extra information to refine our final COLMeD output. We found that between Round 4 and Round 1, which contained the maximal spread of desolvation temperatures, the median carnitine AUC increase was 82.2% compared to 54.9% for all other metabolites. We subsequently adjusted desolvation temperature to 450°C as a final tuning of the comprehensive method which contains both ESI+ and ESI- metabolites. The final parameters for the comprehensive QqQ method were 0.15mL/min flow rate, 20 minute gradient time (Mobile phase B changed from 85–30% over the first 5 minutes and held until the wash step at 20 minutes), 950L/Hr gas flow and 45°C column temperature. The overall COLMeD progression of LC-MS parameters is listed in Table 2. Manufacturer’s notes from Waters suggest 400°C and 800L/Hr desolvation settings for a 0.15mL/min flow rate, corroborating optimized and safe conditions. Although the flow rate is below the optimum efficiency for a 2.5μm particle size, the COLMeD approach optimized our methods to be fit-for-purpose. Additional injections of horse serum were analyzed at these conditions as a validation measure of the final method (labeled as ‘COLMeD Final’ in Figure 2b). Improvements were noted over Round 1 in both the percent AUC increase (median increase of 161.5%, p=7.76e-16, Figure 2b) and the number of metabolites detected, from 163 to 188.
Table 2.
COLMeD Factor Settings for Comprehensive Ion-Switching Analysis
| Factor | Round 1 | Round 2 | Round 3 | Final |
|---|---|---|---|---|
| Flow Rate (mL/min) |
0.1–0.25 | 0.05–0.3 | 0.15–0.3 | 0.15 |
| Gradient | 1–4 | 3–4 | 4 | 4 |
| Time (min) | 10–16 | 12–20 | 12–18 | 20 |
| Desolvation Temp (°C) |
Not tested |
200–650 | 300–500 | 450 |
| Desolvation Gas (L/Hr) |
Not tested |
750–1100 | 800–1100 | 950 |
| Column Temp (°C) |
Not tested |
30–60 | 40–55 | 45 |
3.6 Benchmarking the COLMeD Result
We compared our final method to two other well utilized methods: a polarity-switching method utilizing the same solvents and column from Yuan et al., (Method 1) as well as a vendor published method optimized for high-resolution untargeted qTOF analysis compatible with our instrumentation (Waters) used in several metabolomics studies (Want et al. 2010; Paglia et al. 2012; Bruce et al. 2009, Method 2). We observed a total ion current increase in measured metabolite channels of 105.8% and 57.3% over these two methods respectively, (Figure 2c), with a median 106.1% increase in metabolite response over Method 1 (paired comparison, p=1.46e-13) and a median 10.3% increase over Method 2 (p=0.042). We were also able to maintain broad metabolite coverage with the COLMeD output method, yielding 188 metabolites compared to 181 and 190 in Methods 1 and 2 respectively, meeting our overall objective of developing sensitive and deep polar metabolomics methods. The coefficient of variation across replicate injections also decreased for a given metabolite compared to Methods 1 and 2 (p=6.27e-03 and p=7.16e-04 respectively, Figure S5), suggesting improved method precision.
3.7 Untargeted qTOF DoE
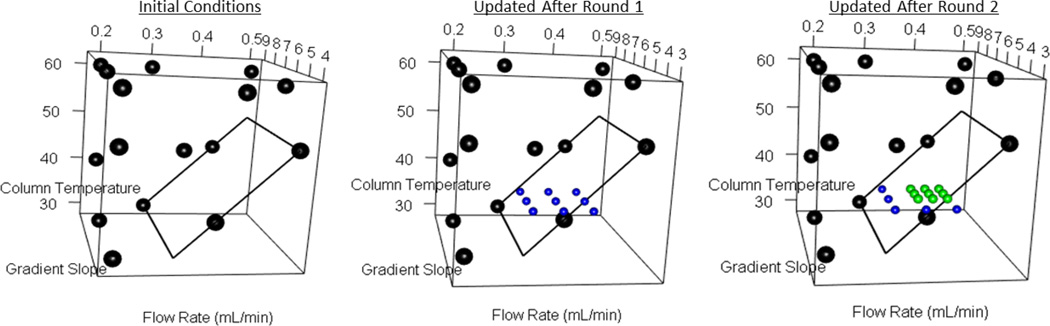
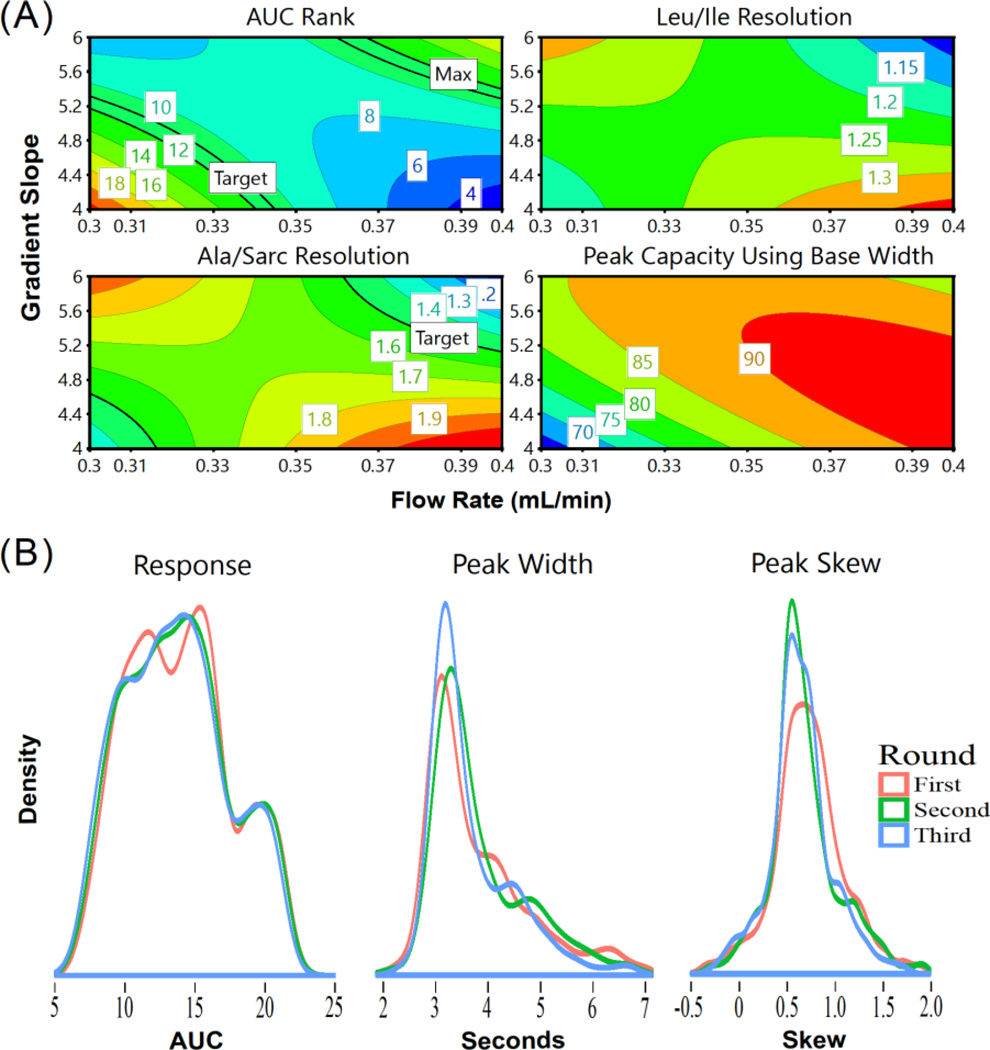
Given the difference between triple quadrupole and untargeted qTOF-based metabolomics, we employed a modified COLMeD approach. Ranked-based metrics (Table S5) and visual inspection of chromatography were used to choose the initial LC solvents between 22 solvent combinations in ESI+ mode before further DoE optimization of chromatography (Table S4, 2mM ammonium acetate with 0.2% formic acid chosen as the final additives). Of the six interaction variables for LC and MS factors in the PLS model for the Round 4 QqQ DoE, only gradient time with desolvation temperature yielded a VIP score over 1, while four of these six factors had a VIP score below 0.8. Thus we felt the interaction of LC and MS factors were small enough to optimize LC and MS on the qToF separately (Table S8). After the first round of DoE, optimizing LC parameters only, predicted response optimums resided around 0.3–0.4mL/min with 3–5 gradient slope. Both of these factors contributed significantly to the data (VIP values of 1.9 and 1.65), unlike column temperature (VIP=0.62). Consequently, column temperature was fixed at 40°C for Rounds 2 and 3, which facilitated full factorial designs for optimizing flow rate and gradient slope (Figure 3). Results were consistent after Round 2, and flow rates were focused to 0.35–0.4mL/min with a gradient slope between 4 and 5. In this process, important tradeoffs were noted. For example, 0.35mL/min was the minimum flow rate to maintain a Leucine/Isoleucine and Alanine/Sarcosine resolution of at least 1.2. Peak capacity also improved with higher flow rates, but AUC ranks improved towards 0.3mL/min, thus 0.35mL/min was considered a good compromise for these responses (with a 5 gradient slope as defined in the methods and 40°C column temperature chosen as the final conditions, Figure 4a). The PLS fitting to data from the Round 1 LC dropped significantly by Round 3, informing us of a highly constrained design space where there existed little room for improvement (Table S7). Chromatographic parameters improved with minimal sacrifice to response, particularly in regards to decreasing the spread of peak widths and peak skews (Figure 4b). These settings also closely follow the UPLC linear velocity for the 1.7μm column while maintaining safe backpressures. Thus for the final LC conditions, the gradient was changed from 100–20.6% B over 15 minutes at 0.35mL/min, followed by a wash of 100% A for 5 minutes. Mobile phase B was changed from 0–100% from 20–22 minutes and held for column equilibration until 30 minutes. For the MS parameters, analysis of the initial linear screening batch produced a compelling model (Table S7) with desolvation temperature as the dominant factor (VIP=2). Positive correlation of AUC and desolvation temperature is also in line with the QqQ method optimization. Desolvation temperature was then fixed for Round 2 to reduce design complexity. Source offset was the only other factor which weighed significantly in the PLS models (VIP=1.75), improving AUC rank at minimum voltages. The other factors had low VIP scores (all below 0.9) and nonsignificant coefficient values, though the trends in improving response were consistent in both rounds. Two rounds of DoE were considered sufficient to improve response to complete the LC-MS optimization of the untargeted method in a highly efficient manner. Round 2 yielded an average response increase of 29.8% (p=3.016e-05), while the chosen parameters within the Round 2 factor settings only yielded a 2.9% increase over the average AUC for the entire round, indicating minimal room for further improvement. Final conditions were 550°C desolvation temperature, 25V cone voltage, 60V source offset, 120°C source temperature, and 50L/Hr cone gas flow (Figure S6).
Figure 3.
Visualization of the design space changes across each LC qTOF DoE Round, including the reduction of 3-D to 2-D design spaces formed after fixing column temperature. The black plane within the box depicts an initial constraint on the space. The black, blue, and green spheres represent the parameter settings tested for each round. The final conditions were chosen within the plane of the green dots
Figure 4.
(a) Response contour plots after Round 3 of LC DoE for un-targeted qTOF metabolomics, demonstrating the tradeoffs to be considered in finding a method optimum. (b) Decreased spread of peak widths and peak skews (with a target objective of 4 seconds widths and minimized skews) across three rounds. These improvements are without compromising the MS response (depicted here as log-transformed AUC values)
4 Concluding Remarks
In this study, the COLMeD approach is demonstrated as an efficient and flexible tool to optimize multiple LC-MS metabolomics methods with different objectives. This may be particularly useful due to differences between results an individual lab compared to literature parameters. Improvements from the starting points of the LC-MS design space were noted for metabolite responses and their chromatography using a limited number of injections. We also noted improvements in these responses over other established polar metabolomics methods. We do acknowledge that while all columns tested in this study and the benchmarked methods have an amide chemistry, the column dimensions vary which may impact the data. A subset of metabolites with similar chemical properties were further optimized within the comprehensive method, which could allow for acylcarnitine-specific analyses without needing to switch solvents or columns. We feel this modular approach can efficiently optimize analyses of other particular metabolite groups of interest. Tailoring the COLMeD approach for untargeted metabolomics on the qTOF also yielded improved chromatography and response while maintaining sufficient isobar separation. Consequently, it is important to note that there are many ways to optimize these methods within the COLMeD framework. One could optimize only LC factors, using more thorough designs after the screening round to add confidence in obtaining the method optimum, followed by a similar workflow for the MS factors, which was the route was taken for the untargeted qTOF method development. This approach may require more injections and time, but has the advantage of being amenable to more automated methods for peak analysis once the chromatography is fixed. Conversely, we felt our combined LC-MS linear model approach for the polarity-switching method could yield our desired output while minimizing injections, given most of the coefficients for the interaction variables to be among the lowest in both rounds 3 and 4 of the QqQ COLMeD process (Table S8). A combination of more detailed model designs, along with a smaller initial design space, will likely yield a stronger predictive model, as we had found with the separated LC and MS optimizations (Table S7). We generally recommend separating LC and MS optimizations if time allows and additional rigor is required, however without parallel analysis of combined versus sequential LC-MS optimizations, we cannot definitively say if the response gains would be significant. In addition, broader assessments of peak quality and response are more suitable for optimizing untargeted metabolomics methods. In our case, optimizing these aggregated metrics met our objective of developing comprehensive metabolomics methods, as opposed to more focused and quantitative metrics such as limits of detection and response variance. Others have recently used a Derringer function approach to comprehensively assess peak characteristics in a multi-analyte mixture (Sampsonidis et al. 2015), similar to what we employed here. We stress that during optimization of complex methods over many responses, not every response can be maximally improved and thus requires analysis of tradeoffs. However, one benefit in utilizing these statistical models is the ability to stress the optimization of more important metabolites or responses. We also note that in method development for LC-MS metabolomics, the method optimum is largely defined by the user, and thus the response selection must be fit for the experimental purpose. Future studies will further validate these methods with different sample types and responses, such as the compounds found in our homogenized fly samples but not horse serum. More importantly, we have laid out a thorough description of the COLMeD workflow from which we hope can be useful in not only LC-MS metabolomics but other complex method types which require adjusting multiple factors to optimize multiple responses.
Supplementary Material
Acknowledgments
The authors would also like to thank Saikumari Krishnaiah for assistance with qTOF data acquisition and Barry Slaff for fruitful discussions regarding qTOF data analysis. S.D.R. is supported through a Pharmacology T32 Training Grant (T32 GM008076). Supported in part by the Institute for Translational Medicine and Therapeutics (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics.
Footnotes
Compliance with ethical standard
Conflict of interest The authors declare no competing financial interest.
Ethical approval This article does not contain any experiments with human participants or animals as performed by the authors.
References
- A J, Trygg J, Gullberg J, Johansson AI, Jonsson P, Antti H, Marklund SL, Moritz T. Extraction and GC/MS analysis of the human blood plasma metabolome. Anal. Chem. 2005;77(24):8086–8094. doi: 10.1021/ac051211v. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJCanJ. A rapid method of total lipid extraction and purification. Biochem. Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bruce SJ, Tavazzi I, Parisod V, Rezzi S, Kockhar S, Guy PA. Investigation of human blood plasma sample preparation for performing metabolomics using ultrahigh performance liquid chromatography/mass spectrometry. Anal Chem. 2009;81(9):3285–3296. doi: 10.1021/ac8024569. [DOI] [PubMed] [Google Scholar]
- Correa E, Goodacre R. A genetic algorithm-Bayesian network approach for the analysis of metabolomics and spectroscopic data: application to the rapid identification of Bacillus spores and classification of Bacillus species. BMC Informatics. 2011;12(33):1–17. doi: 10.1186/1471-2105-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson M, Rännar S, Madsen R, Donten MA, Marsden-Edwards E, Moritz T, Shockcor JP, Johansson E, Trygg J. Strategy for optimizing LC-MS data processing in metabolomics: a design of experiments approach. Anal. Chem. 2012;84(15):6869–6876. doi: 10.1021/ac301482k. [DOI] [PubMed] [Google Scholar]
- Eriksson L, Johansson E, Kettaneh-Wold N, Wikström C, Wold S. Design of Experiments, Principles and Applications. 2nd. Umeå: Umetrics Academy; 2006. [Google Scholar]
- Gika HG, Theodoridis GA, Vrhovsek U, Mattivi FJ. Quantitative profiling of polar primary metabolites using hydrophilic interaction ultrahigh performance liquid chromatography-tandem mass spectrometry. Chromatogr A. 2012;1259:121–127. doi: 10.1016/j.chroma.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Gika HG, Wilson ID, Theodoridis GA. LC-MS-based holistic metabolic profiling. Problems, limitations, advantages, and future perspectives. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;966:1–6. doi: 10.1016/j.jchromb.2014.01.054. [DOI] [PubMed] [Google Scholar]
- Hao Z, Xiao B, Weng NJ. Impact of column temperature and mobile phase components on selectivity of hydrophilic interaction chromatography (HILIC) Sep. Sci. 2008;31(9):1449–1464. doi: 10.1002/jssc.200700624. [DOI] [PubMed] [Google Scholar]
- Ivanisevic J, Zhu Z, Plate L, Tautenhahn R, Chen S, O’Brien PJ, Johnson CH, Marletta MA, Patti GJ, Siuzdak G. Toward 'omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal. Chem. 2013;85(14):6876–6884. doi: 10.1021/ac401140h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivilompolo M, Ӧhnrberg L, Orešič M, Hyӧtylӓinen TJ. Rapid quantitative analysis of carnitine and acylcarnitines by ultra-high performance-hydrophilic interaction liquid chromatography-tandem mass spectrometry. Chromatogr. A. 2013;1292:189–194. doi: 10.1016/j.chroma.2012.12.073. [DOI] [PubMed] [Google Scholar]
- Kostić N, Dotsikas Y, Malenović A, Stojanović JB, Rakić T, Ivanović D, Medenica MJ. Stepwise optimization approach for improving LC-MS/MS analysis of zwitterionic antiepileptic drugs with implementation of experimental design. Mass Spectrom. 2013;48(7):875–884. doi: 10.1002/jms.3236. [DOI] [PubMed] [Google Scholar]
- Lv H, Palacios G, Hartil K, Kurland IJ. Advantages of tandem LC-MS for the rapid assessment of tissue-specific metabolic complexity using a pentafluorophenylpropyl stationary phase. J Proteome Res. 2011;10(4):2104–2112. doi: 10.1021/pr1011119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoles MO, Steenbergen RDJM. Analysis of axle and vehicle load properties through Bayesian Networks based on Weigh-in-Motion data. Reliab. Eng. Syst. Safe. 2014;125:153–164. [Google Scholar]
- New L, Chan ECY. Evaluation of BEH C18, BEH HILIC, and HSS T3 (C18) column chemistries for the UPLC-MS-MS analysis of glutathione, glutathione disulfide, and ophthalmic acid in mouse liver and human plasma. J Chromatogr. Sci. 2008;46:209–214. doi: 10.1093/chromsci/46.3.209. [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Schug KA. The advantages of ESI-MS detection in conjunction with HILIC mode separations: Fundamentals and applications. J. Sep. Sci. 2008;31(9):1465–1480. doi: 10.1002/jssc.200700630. [DOI] [PubMed] [Google Scholar]
- Paglia G, Hrafnsdóttir S, Magnúsdóttir M, Fleming RM, Thorlacious S, Palsson BØ, Thiele I. Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS) Anal Bioanal Chem. 2012;402(3):1183–1198. doi: 10.1007/s00216-011-5556-4. [DOI] [PubMed] [Google Scholar]
- Riter LS, Vitek O, Gooding KM, Hodge BD, Julian RK. Statistical design of experiments as a tool in mass spectrometry. J. Mass Spectrom. 2005;40(5):565–579. doi: 10.1002/jms.871. [DOI] [PubMed] [Google Scholar]
- Sampsonidis I, Witting M, Koch W, Virgillou C, Gika HG, Schmitt-Kopplin P, Theodoridis GA. Computational analysis and ratiometric comparison approaches aimed to assist column selection in hydrophilic interaction liquid chromatography-tandem mass spectrometry targeted metabolomics. J Chromatogr A. 2015;1406:145–155. doi: 10.1016/j.chroma.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Suizdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Székely GY, Henriques B, Gil M, Ramos A, Alvarez CJ. Design of experiments as a tool for LC-MS/MS method development for the trace analysis of the potentially genotoxic 4-dimethylaminopyridine impurity in glucocorticoids. Pharm. Biomed. Anal. 2012;70:251–258. doi: 10.1016/j.jpba.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockor J, Holmes E, Nicholson JK. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. 2010;5(6):1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelni-kov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012;7(5):872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Clausen MR, Dalsgaard KT, Mortensen G, Bertram CH. Time-saving design of experiment protocol for optimization of LC-MS data processing in metabolomic approaches. Anal. Chem. 2013;85(15):7109–7116. doi: 10.1021/ac4020325. [DOI] [PubMed] [Google Scholar]
- Zhou G, Pang H, Tang Y, Yao X, Mo X, Zhu S, Guo S, Qian D, Qian Y, Su S, Zhang L, Jin C, Qin Y, Duan JA. Hydrophilic interaction ultra-performance liquid chromatography coupled with triple-quadrupole tandem mass spectrometry for highly rapid and sensitive analysis of underivatized amino acids in functional foods. Amino Acids. 2013;44(5):1293–1305. doi: 10.1007/s00726-013-1463-7. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Song J, Choi FF, Wu H, Qiao C, Ding L, Gesang S, Xu H. An experimental design approach using response surface techniques to obtain optimal liquid chromatography and mass spectrometry conditions to determine the alkaloids in Meconopsi species. J. Chromatogr., A. 2009;1216(42):7013–7023. doi: 10.1016/j.chroma.2009.08.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.