Abstract
Osteoarthritis (OA) is a common chronic joint disease, the etiology of which is complex. Disturbance to proinflammatory and anti-inflammatory signaling pathways is a major cause of OA. MicroRNAs (miRNAs/miR) are a group of endogenous, short, non-coding RNAs, the expression profile of which is disturbed in the cartilage of patients with OA. To determine the function of miRNAs during the progression of OA, the present study detected the expression levels of nine candidate miRNAs in cartilage samples from 33 patients with OA. The results demonstrated that miR-26a, miR-26b, miR-138 and miR-140 were downregulated in patients with OA. As predicted by a bioinformatics analysis and confirmed by luciferase assay and western blotting, the present study revealed that miR-26a and miR-26b are able to suppress karyopherin subunit alpha 3 (KPNA3) expression by targeting its 3′-untranslated region. Since KPNA3 is an important mediator that modulates nuclear factor (NF)-κB p65 translocation, the present study examined the impact of miR-26a and miR-26b on NF-κB signaling. The results indicated that transfection of cells with a miR-26a or miR-26b inhibitor may promote NF-κB p65 translocation from the cytoplasm to the nucleus via the upregulation of KPNA3. Furthermore, the expression levels of matrix metalloproteinase-3, −9, −13 and cyclooxygenase-2 were upregulated following transfection with a miR-26a or miR-26b inhibitor. These results indicate that downregulation of miR-26a and miR-26b may contribute to the pathogenesis of OA via promotion of the NF-κB signaling pathway. The present study sheds light on the pathogenesis of OA and may provide a target for the development of therapeutic methods for the treatment of OA.
Keywords: osteoarthritis, microRNA, KPNA3, nuclear factor-κB
Introduction
Osteoarthritis (OA) is a multifactorial disease, which is characterized by degeneration of articular cartilage, limited intra-articular inflammation with synovitis, and alterations in periarticular and subchondral bone (1). The majority of individuals >65 years old exhibit radiographic and/or clinical evidence of OA (2,3). It is widely accepted that the pathogenesis of OA is complex, and is associated with mechanical influences, genetic disposition and epigenetic factors.
MicroRNAs (miRNAs/miR) are a group of endogenous, short, non-coding RNAs, which regulate gene expression by targeting the 3′-untranslated region (3′UTR) of mRNA. miRNAs have been found in various organisms, and several of them are evolutionary conserved. Furthermore, it is estimated that >50% of all human protein-coding genes are potentially regulated by miRNAs (4). miRNAs have previously been demonstrated to exist in cartilage and have essential roles during cartilage development (5). Appropriate miRNA expression is important for maintaining cartilage homeostasis, and abnormal miRNA profiles have been shown to be associated with cartilage diseases (5,6).
A previous study demonstrated that several aberrantly expressed miRNAs in cartilage are associated with the pathogenesis of OA (7). In addition, numerous studies have indicated that miR-140 exhibits a chondrocyte differentiation-related expression pattern, the downregulation of which is associated with an enhanced interleukin (IL)-1β response, which may contribute to the pathogenesis of OA (6,8,9). Furthermore, miR-146a is an IL-1β-responsive miRNA, which contributes to OA pathogenesis by increasing vascular endothelial growth factor levels (10).
To explore the function of miRNAs during the pathogenesis of OA and find a target for clinical treatment, the present study detected the expression levels of nine miRNAs in cartilage tissue samples from patients with OA, and investigated the role of candidate miRNAs during the progression of OA.
Materials and methods
Tissue sample collection
Human cartilage was obtained from femoral condyles and tibial plateaus. OA cartilage samples were obtained from female patients with OA that underwent total knee arthroplasty (n=33; age, 62±11 years), and normal human cartilage samples were obtained from individuals within 12 h of death (n=15; age, 60±16 years old). The normal individuals had no history of joint disease and succumbed to causes unrelated to arthritic diseases. The cartilage was examined macroscopically and microscopically to ensure that only normal tissue was used. All patients with OA were evaluated by a certified rheumatologist and were diagnosed as having OA according to the American College of Rheumatology criteria (11). The Ethics Committee Board of the People's Hospital of Dongying (Dongying, China) approved the use of human articular tissues. Patients with OA provided written informed consent, and post-mortem tissues were obtained with the consent of a family member or authorized individual.
Chondrocyte isolation and culture
Human knee articular chondrocytes were isolated from 9 patients (age, 55–67 years) that underwent joint replacement surgery for OA. Briefly, cartilage slices (~0.5 cm2) were initially digested with 0.25% trypsin for 1 h at 37°C, and were subsequently incubated with 0.04% collagenase type II overnight in a 37°C water bath and filtered a 20 µm-pore strainer. Primary cell cultures were maintained at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (DMEM)/F12 (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences), 100 U/ml penicillin and 100 U/ml streptomycin. Chondrocytes at the third passage were used for subsequent experiments.
The SW1353 chondrosarcoma cell line was purchased from the China Infrastructure of Cell Line Resources (Beijing, China). The cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin at 37°C in a humidified atmosphere.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Single-stranded cDNA was synthesized by using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and then amplified by using TaqMan Universal PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) together with miRNA-specific TaqMan MGB probes (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR cycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Primers were supplied by Applied Biosystems (Thermo Fisher Scientific, Inc.). The expression levels of candidate miRNAs were measured using stem-loop qPCR with U6 small nuclear RNA as an internal control. The primers used were purchased from Thermo Fisher Scientific, Inc. (miR-26a, cat. no. 000405; miR-26b, cat. no. 000407; miR-27b, cat. no. 002174; miR-125a, cat. no. 002198; miR-138, cat. no. 02284; miR-140, cat. no. 002234; miR-146a, cat. no. 000468; miR-181a, cat. no. 000480; miR-199a, cat. no. 002304; and U6, cat. no. 4451372). Each sample was measured in triplicate and the experiment was repeated at least three times. The relative expression levels of the candidate genes were calculated using the 2−ΔΔCq method (12).
For mRNA quantification, first strand cDNA synthesis was achieved using a Primescript RT reagent kit (Takara Bio, Inc., Otsu, Japan). Amplification of cDNA was performed using SYBR Premix Ex Taq (Takara Bio, Inc.). The primers were synthesized by BGI (Shenzhen, China) and sequences were as follows: Matrix metalloproteinase (MMP)-3, forward 5′-CTCGTTGCTGCTCATGAAATTG-3′, reverse 5′-TCAGGTCTGTGAGTGAGTGATA-3′; MMP-9, forward 5′-GAACTTTGACAGCGACAAGAAG-3′, reverse 5′-CGGCACTGAGGAATGATCTAA-3′; MMP-13, forward 5′-AGCATCTGGAGTAACCGTATTG-3′, reverse 5′-CCCGCACTTCTGGAAGTATT-3′; cyclooxygenase (COX)2, forward 5′-TCAGTAGGTGCATTGGAATCAA-3′, reverse 5′-GGAGAAACGAAGTGATGAGAAGA-3′; and GAPDH, forward 5′-GACCACTTTGTCAAGCTCATTTC-3′, reverse 5′-CTCTCTTCCTCTTGTGCTCTTG-3′. The PCR cycling condition were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The expression levels of GAPDH were used to normalize the levels of target mRNA, and the relative expression levels of genes were calculated using the 2−ΔΔCq method (12).
Dual luciferase assay
miRNA mimics, inhibitors and controls were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences for miRNAs mimics were as follows: miR-26a, 5′-UUCAAGUAAUCCAGGAUAGGCU-3′; miR-26b, 5′-UUCAAGUAAUUCAGGAUAGGU-3′; and miR-138, 5′-AGCUGGUGUUGUGAAUCAGGCCG-3′. The sequences for miRNAs inhibitors were as follows: miR-26a, 5′-AGCCUAUCCUGGAUUACUUGAA-3′; miR-26b, 5′-ACCUAUCCUGAAUUACUUGAA-3′; and miR-138, 5′-CGGCCUGAUUCACAACACCAGCU-3′. The full-length karyopherin subunit α 3 (KPNA3) 3′UTR was amplified from cDNA of HEK293T cells and cloned downstream of the firefly luciferase coding region in the pmirGLO vector (Promega Corporation, Madison, WI, USA) in order to generate a luciferase reporter vector.
For the luciferase reporter assays, 1×105 SW1353 cells were seeded in 48-well plates. Subsequently, 20 µM miRNA mimics or inhibitors were transfected into the cells alongside 100 ng of the reporter vector, using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Scrambled short single-stranded RNA or double-stranded RNA sequences were used as a control for the miRNA mimics or inhibitors. Briefly, miRNA mimics or inhibitors were diluted in DMEM, and Lipofectamine 2000 was also diluted in DMEM. The mimics, inhibitors and Lipofectamine 2000 dilutions were then incubated at room temperature for 5 min, before being combined for 20 min at room temperature. The complex (200 µl) was added to each well of the cell-containing plates, and the plates were incubated for 4 h. A total of 48 h post-transfection, the cells were harvested, lysed and assessed using the Dual-Luciferase Assay kit (Promega Corporation). Each treatment was performed in triplicate in three independent experiments. The results are expressed as relative luciferase activity (firefly luciferase activity/Renilla luciferase activity).
Western blotting
Protein was extracted from the cells using M-PER™ Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, Inc.); protein samples were quantified using the bicinchoninic acid method. Protein extracts were subsequently boiled in SDS/β-mercaptoethanol sample buffer, and 10 µg samples were loaded into each lane of 10% polyacrylamide gels. The proteins were then separated by electrophoresis and were blotted onto polyvinylidene fluoride membranes (Amersham; GE Healthcare Life Sciences, Little Chalfont, UK) by electrophoretic transfer. The membranes were blocked in 5% bovine serum albumin in TBS-Tween 20 for 1 h at room temperature. The membranes were incubated with rabbit anti-KPNA3 polyclonal antibody (1:500; cat. no. ab117578; Abcam, Cambridge, MA, USA), rabbit anti-NF-κB p65 monoclonal antibody (1:500; cat. no. ab32536; Abcam), mouse anti-GAPDH monoclonal antibody (1:5,000; cat. no. sc-32233; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), rabbit anti-α-tubulin polyclonal antibody (1:1,000; cat. no. ab18251; Abcam), rabbit anti-lamin B1 monoclonal antibody (1:1,000; cat. no. ab13374; Abcam) and mouse anti-β-actin monoclonal antibody (1:5,000; cat. no. sc-47778; Santa Cruz Biotechnology Inc.) at 4°C overnight. The specific protein-antibody complexes were subsequently detected following incubation with horseradish peroxidase-conjugated goat anti-rabbit (1:5,000) or rabbit anti-mouse (1:5,000) immunoglobulin G antibodies (cat. nos ab6721 and ab6728, respectively; Abcam) for 2 h at 37°C. The blots were visualized using an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.). The β-actin or GAPDH signals were used as loading controls for the total cell lysate. The α-tubulin and lamin B1 signals were used as loading controls for the cytosolic and nuclear lysates, respectively. The band density was analyzed using Quantity One software (version 4.6.2; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The experiment was conducted in triplicate and results were analyzed using Student's t-test.
miRNA transfection and IL-1β treatment
Chondrocytes were transfected with miRNA mimics, inhibitors or scrambled control RNA using Lipofectamine 2000. A total of 48 h post-transfection, the cells were washed three times with PBS and were incubated for 1 h in serum-starved media (0.5% FBS). Serum-starved chondrocytes were then stimulated with 10 ng/ml IL-1β (R&D Systems, Inc., Minneapolis, MN, USA) for 1 h, and were collected for mRNA and protein expression detection.
NF-κB nuclear translocation
Following various transfections and treatments, cells were harvested for preparation of the cytoplasmic and nuclear extracts using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Inc.). The NF-κB p65 expression levels in the extracts were then examined by western blotting.
Statistical analysis
Data were analyzed using SPSS Statistical Package version 16 (SPSS Inc., Chicago, IL, USA) and are presented as the mean ± standard deviation. Analyses of two independent groups were conducted using Welch's unpaired t-test. For multiple comparisons, one-way analysis of variance was conducted with the Newman-Keuls method. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-26a, miR-26b, miR-138 and miR-140 are downregulated in patients with OA
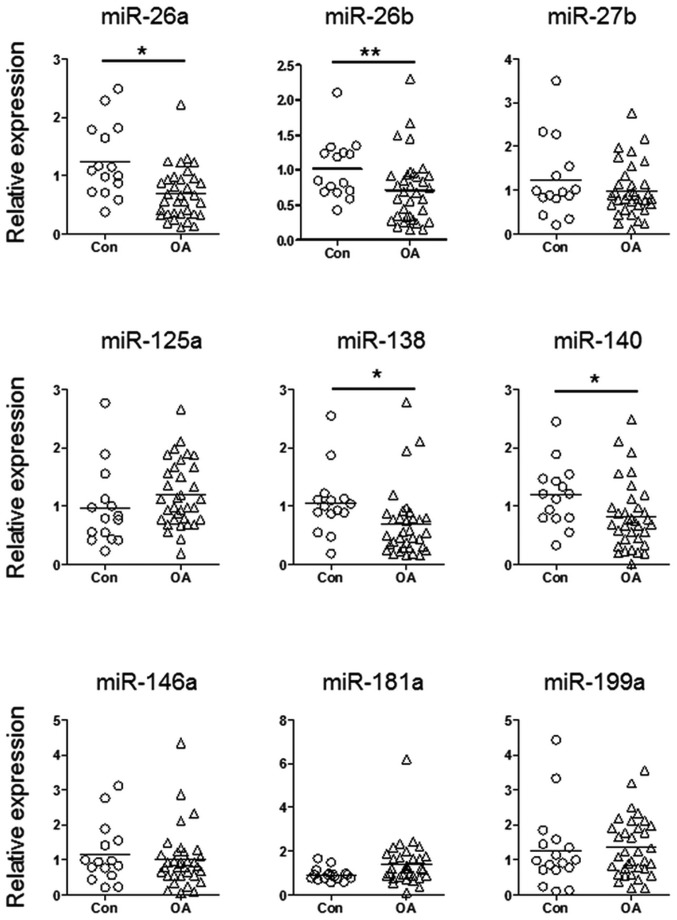
To explore the function of aberrantly expressed miRNAs during the pathogenesis of OA, the present study detected the expression levels of nine candidate miRNAs in cartilage samples from patients with OA and normal controls. These candidate miRNAs have previously been reported to be aberrantly expressed in cartilage samples from patients with OA, or have been shown to have a role during chondrogenesis (7,13). As shown in Fig. 1, miR-26a, miR-26b, miR-138 and miR-140 exhibited significantly reduced expression in cartilage samples from patients with OA. Since the function of miR-140 during the pathogenesis of OA is well understood (6,14–16), the present study focused on investigating the biological functions of miR-26a, miR-26b and miR-138.
Figure 1.
Aberrant expression of miRNAs detected in cartilage samples from patients with OA. The expression levels of nine candidate miRNAs were detected in cartilage samples from patients with OA and normal controls using reverse transcription-quantitative polymerase chain reaction. Results were analyzed using a t-test. *P<0.05, **P<0.01. miRNA/miR, microRNA; OA, osteoarthritis; Con, controls.
miR-26a and miR-26b suppress KPNA3 expression by targeting its 3′UTR
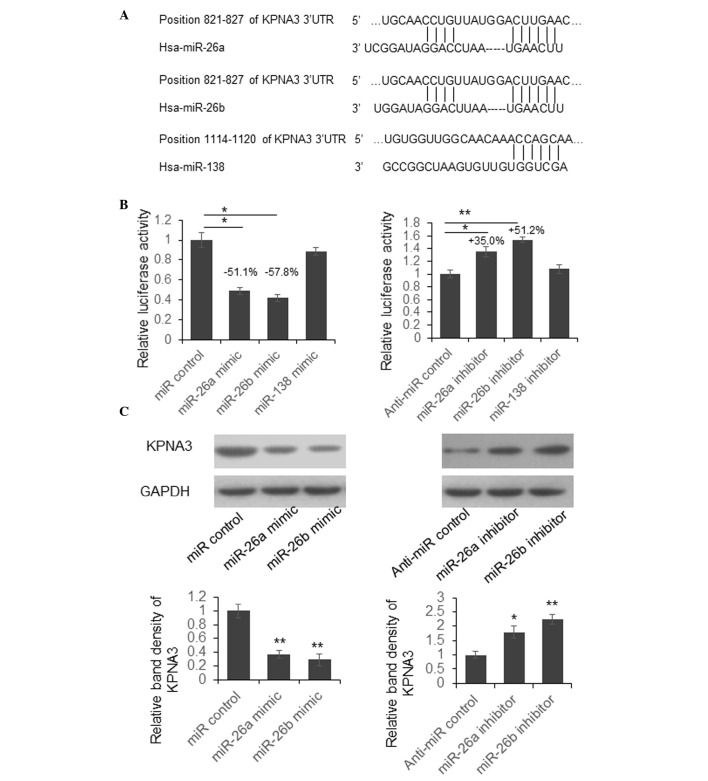
To investigate the biological function of aberrant miRNAs during the pathogenesis of OA, the present study predicted the target genes of miR-26a, miR-26b and miR-138 using the online bioinformatics tool TargetScan (http://www.targetscan.org/). The predicted interactions between miRNAs and the 3′UTR of KPNA3 are presented in Fig. 2A.
Figure 2.
miR-26a and miR-26b suppress KPNA3 expression by targeting its 3′UTR. (A) Schematic diagram of the predicted interaction between miRNAs and the 3′UTR of KPNA3. (B) Dual luciferase assay. A reporter vector containing the 3′UTR of KPNA3 was transfected into SW1353 cells alongside miRNA mimics or inhibitors. A total of 48 h post-transfection, the cells were lysed and luciferase activity was detected. Results were analyzed by Student's t-test. *P<0.05, **P<0.01. (C) miRNA mimics or inhibitors were transfected into SW1353 cells. A total of 48 h post-transfection, cells were lysed and the protein expression levels of KPNA3 were determined by western blotting. GAPDH was used as a loading control. *P<0.05, **P<0.01. miR/miRNA, microRNA; 3′UTR, 3′-untranslated region; KPNA3, karyopherin subunit alpha 3.
To examine whether miR-26a, miR-26b and miR-138 are able to suppress KPNA3 expression by targeting its 3′UTR, the present study constructed a luciferase reporter vector by cloning the full-length KPNA3 3′UTR into the pmirGLO plasmid, downstream of the firefly luciferase coding region. Dual luciferase assays were then conducted in SW1353 cells. As presented in Fig. 2B, luciferase activity was significantly reduced by 51.1 or 57.8% in cells overexpressing miR-26a or miR-26b. Conversely, luciferase activity was increased by 35.0 or 51.2% in cells in which miR-26a or miR-26b expression was suppressed. There was no significant change in luciferase activity in cells with miR-138 overexpression or downregulation. These results indicate that miR-26a and miR-26b may suppress luciferase activity by targeting KPNA3 3′UTR.
To further examine whether endogenous KPNA3 expression was suppressed by miR-26a and miR-26b, SW1353 cells were transfected with miR-26a and miR-26b mimics or inhibitors. A total of 48 h post-transfection, the cells were lysed and KPNA3 expression was detected by western blotting. As shown in Fig. 2C, the protein expression levels of KPNA3 were downregulated following transfection with miR-26a or miR-26b mimics, whereas KPNA3 expression was upregulated in cells transfected with miR-26a or miR-26b inhibitors. These results suggest that endogenous KPNA3 expression may be directly regulated by miR-26a and miR-26b.
miR-26a and miR-26b modulate NF-κB p65 translocation, and the expression of MMP-3, −9, −13 and COX-2
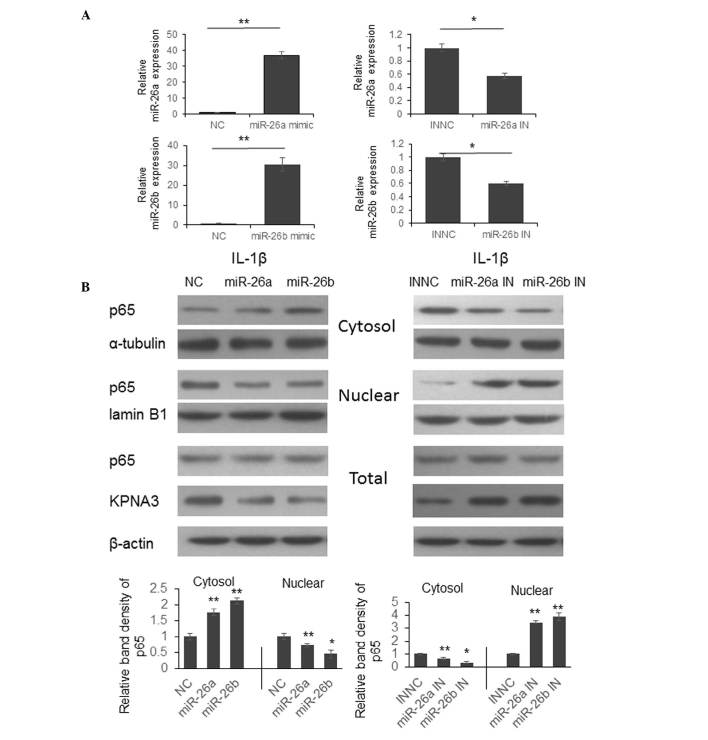
Previous studies have indicated that KPNA3 is able to bind to the nuclear localization sequence region of NF-κB p65 and mediate translocation between the cytoplasm and nucleus (17,18). To determine whether miR-26a and miR-26b are able to regulate NF-κB signaling by modulating p65 translocation, chondrocytes were transfected with miR-26a and miR-26b mimics or inhibitors. A total of 48 h post-transfection, the cells were stimulated with 10 ng/ml IL-1β for 1 h and were collected for protein expression detection.
As shown in Fig. 3A, the expression levels of miR-26a or miR-26b were significantly upregulated by miR-26a or miR-26b mimics, and were reduced by miR-26a or miR-26b inhibitors. Following IL-1β stimulation, the process of p65 cytoplasmic-nuclear translocation was arrested following transfection with miR-26a or miR-26b mimics, and p65 was detained in the cytoplasm (Fig. 3B, left panel). However, the p65 translocation process was promoted following transfection with miR-26a or miR-26b inhibitors, and increased p65 expression was detected in the nucleus (Fig. 3B, right panel).
Figure 3.
Downregulation of miR-26a or miR-26b promotes p65 cytoplasmic-nuclear translocation by upregulating KPNA3 expression. (A) Chondrocytes were isolated from cartilage samples from patients with osteoarthritis and were transfected with miR-26a and miR-26b mimics or inhibitors. A total of 48 h post-transfection, miR-26a and miR-26b expression levels were detected. The results were analyzed by Student's t-test. *P<0.05, **P<0.01. (B) Chondrocytes were transfected with miRNA mimics or inhibitors for 48 h. The cells were then treated with IL-1β for 1 h, and the expression levels of p65 were detected in the cytoplasm, nucleus and total lysate by western blotting.*P<0.05, **P<0.01. miR/miRNA, microRNA; KPNA3, KPNA3, karyopherin subunit alpha 3; IL-1β, interleukin-1β; IN, inhibitor; NC, negative control.
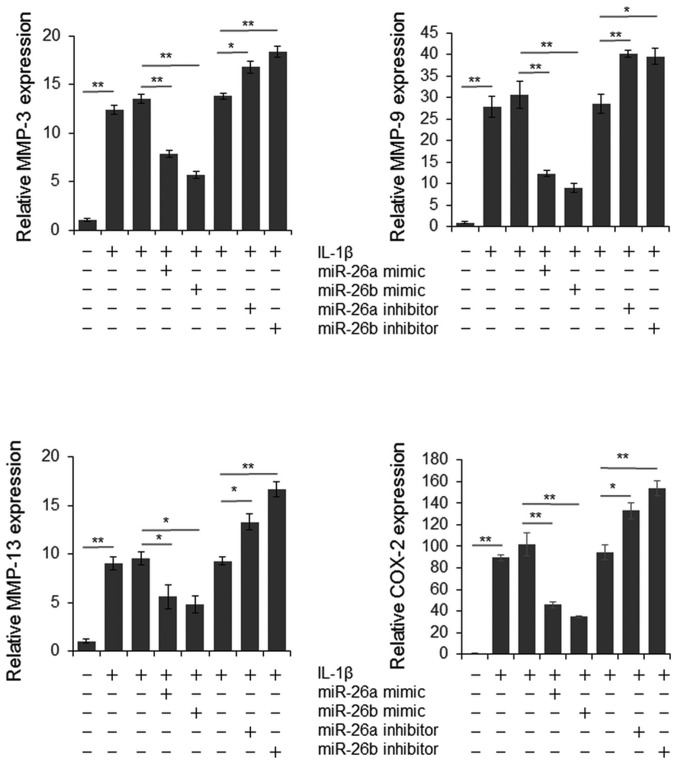
To explore the function of miR-26a and miR-26b on modulating NF-κB signaling, the present study examined the mRNA expression levels of MMP-3, −9, −13 and COX-2. As presented in Fig. 4, the expression levels of MMP-3, −9, −13 and COX-2 were significantly downregulated by miR-26a or miR-26b mimics, and were upregulated by miR-26a or miR-26b inhibitors. These results indicate that reduced miR-26a and miR-26b expression may contribute to the pathogenesis and progression of OA by upregulating NF-κB signaling.
Figure 4.
MMPs and COX-2 expression was modulated by miR-26a and miR-26b. Chondrocytes were transfected with miRNA mimics or inhibitors for 48 h and were then treated with IL-1β for 1 h. Total RNA was extracted and the expression levels of MMP-3, −9, −13 and COX-2 were detected by reverse transcription-quantitative polymerase chain reaction. The experiment was carried out in triplicate and the results between multiple groups were analyzed by one-way analysis of variance. *P<0.05, **P<0.01. miR/miRNA, microRNA; MMP, matrix metalloproteinase; COX-2, cyclooxygenase-2.
Discussion
miRNAs are a group of endogenous, short, non-coding RNAs, the function of which was demonstrated to be associated with the development and homeostasis of articular cartilage. Recently, increasing reports indicated that an altered miRNA expression profile is associated with the pathogenesis of OA, however, the mechanisms were not well understood. The current study examined the level of nine selected miRNAs in the cartilage samples of 33 patients with OA. In contrast with other reports, upregulation of miR-146a in cartilage samples from patients with OA was not detected (19). The expression of miR-27b and miR-125a was also not altered, in contrast with what was reported by other researchers, which may be caused by the differences of samples collection and the genetic background of the patients in the study (14,20). However, the current study observed that the levels of miR-26a, miR-26b, miR-138 and miR-140 were downregulated in the patients with OA. It was also confirmed that miR-26a and miR-26a function as an immune response modulators that regulate NF-κB p65 translocation by targeting KPNA3. To the best of our knowledge, this is the first report to demonstrate the role of miR-26a and miR-26b as NF-κB signaling regulators during OA pathogenesis.
The nucleocytoplasmic traffic of large molecules (>25 nm in diameter) in eukaryotic cells is regulated by specific nuclear import and export systems. Proteins that contain classical nuclear localization sequences are imported into the nucleus by importin α/β heterodimers (21). Although eight importin α members were found in human cells KPNA3 was confirmed to be the most important mediator of p50/p65 heterodimer and p50 homodimer translocation (17). The present study confirmed that KPNA3 is a direct target of miR-26a and miR-26b for the first time. Furthermore, upregulation of MMPs and COX-2 induced by IL-1β in chondrocytes are predominantly modulated by NF-κB signaling (22,23). The reduction of MMP-3, −9, −13 and COX-2 mRNA level by miR-26a/b mimics and upregulation by miR-26a/b inhibitors indicates the functional role of miR-26a/b in regulating NF-κB signaling. Thus, the current study successfully demonstrated an association between miR-26a/b reduction and OA pathogenesis, which may shed light on the mechanism of why altered miRNA expression is associated with OA.
In conclusion, the present study demonstrated that miR-26a, miR-26b, miR-138 and miR-140 are downregulated in cartilage samples from patients with OA. Furthermore, reduced miR-26a and miR-26b expression induced upregulation of MMPs and COX-2 by upregulating KPNA3 expression and promoting p65 cytoplasmic-nuclear translocation.
References
- 1.Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213:626–634. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence RC, Helmick CG, Arnett FC, Deyo RA, Felson DT, Giannini EH, Heyse SP, Hirsch R, Hochberg MC, Hunder GG, et al. Estimates of the prevalence of arthritis and selected musculoskeletal disorders in the United States. Arthritis Rheum. 1998;41:778–799. doi: 10.1002/1529-0131(199805)41:5<778::AID-ART4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 3.Shane Anderson A, Loeser RF. Why is osteoarthritis an age-related disease? Best Pract Res Clin Rheumatol. 2010;24:15–26. doi: 10.1016/j.berh.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirzamohammadi F, Papaioannou G, Kobayashi T. MicroRNAs in cartilage development, homeostasis and disease. Curr Osteoporos Rep. 2014;12:410–419. doi: 10.1007/s11914-014-0229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK, Asahara H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barter MJ, Young DA. Epigenetic mechanisms and non-coding RNAs in osteoarthritis. Curr Rheumatol Rep. 2013;15:353. doi: 10.1007/s11926-013-0353-z. [DOI] [PubMed] [Google Scholar]
- 8.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura Y, Inloes JB, Katagiri T, Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31:3019–3028. doi: 10.1128/MCB.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Huang J, Dai L, Yu D, Chen Q, Zhang X, Dai K. miR-146a, an IL-1beta responsive miRNA, induces vascular endothelial growth factor and chondrocyte apoptosis by targeting Smad4. Arthritis Res Ther. 2012;14:R75. doi: 10.1186/ar3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke TD, Greenwald R, Hochberg M, et al. Development of criteria for the classification and reporting of osteoarthritis. classification of osteoarthritis of the knee. diagnostic and therapeutic criteria committee of the american rheumatism association. Arthritis Rheum. 1986;29:1039–1049. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Le LT, Swingler TE, Clark IM. Review: The role of microRNAs in osteoarthritis and chondrogenesis. Arthritis Rheum. 2013;65:1963–1974. doi: 10.1002/art.37990. [DOI] [PubMed] [Google Scholar]
- 14.Tardif G, Hum D, Pelletier JP, Duval N, Martel-Pelletier J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord. 2009;10:148. doi: 10.1186/1471-2474-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araldi E, Schipani E. MicroRNA-140 and the silencing of osteoarthritis. Genes Dev. 2010;24:1075–1080. doi: 10.1101/gad.1939310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tardif G, Pelletier JP, Fahmi H, Hum D, Zhang Y, Kapoor M, Martel-Pelletier J. NFAT3 and TGF-beta/SMAD3 regulate the expression of miR-140 in osteoarthritis. Arthritis Res Ther. 2013;15:R197. doi: 10.1186/ar4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fagerlund R, Kinnunen L, Köhler M, Julkunén I, Melen K. NF-(kappa)B is transported into the nucleus by importin (alpha)3 and importin (alpha)4. J Biol Chem. 2005;280:15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- 18.Agrawal T, Gupta GK, Agrawal DK. Calcitriol decreases expression of importin alpha3 and attenuates RelA translocation in human bronchial smooth muscle cells. J Clin Immunol. 2012;32:1093–1103. doi: 10.1007/s10875-012-9696-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamasaki K, Nakasa T, Miyaki S, Ishikawa M, Deie M, Adachi N, Yasunaga Y, Asahara H, Ochi M. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60:1035–1041. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nugent M. MicroRNAs: exploring new horizons in osteoarthritis. Osteoarthritis Cartilage. 2016;24:573–580. doi: 10.1016/j.joca.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Pumroy RA, Cingolani G. Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem J. 2015;466:13–28. doi: 10.1042/BJ20141186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh R, Ahmed S, Islam N, Goldberg VM, Haqqi TM. Epigallocatechin-3-gallate inhibits interleukin-1beta-induced expression of nitric oxide synthase and production of nitric oxide in human chondrocytes: suppression of nuclear factor kappaB activation by degradation of the inhibitor of nuclear factor kappaB. Arthritis Rheum. 2002;46:2079–2086. doi: 10.1002/art.10443. [DOI] [PubMed] [Google Scholar]
- 23.Liacini A, Sylvester J, Li WQ, Huang W, Dehnade F, Ahmad M, Zafarullah M. Induction of matrix metalloproteinase-13 gene expression by TNF-alpha is mediated by MAP kinases, AP-1, and NF-kappaB transcription factors in articular chondrocytes. Exp Cell Res. 2003;288:208–217. doi: 10.1016/S0014-4827(03)00180-0. [DOI] [PubMed] [Google Scholar]