Abstract
The aim of the present study was to investigate the possible pathogenesis of osteosarcoma using bioinformatics analysis to examine gene-gene interactions. A total of three datasets associated with osteosarcoma were downloaded from the Gene Expression Omnibus. The differentially expressed genes (DEGs) were identified using the significance analysis of microarrays method, which then were subjected to the Human Protein Reference Database to identify the protein-protein interaction (PPI) pairs and to construct a PPI network of the DEGs. Subsequent multilevel community analysis was applied to mine the modules in the network, followed by screening of the differential expression module using the GlobalAncova package. The genes in the differential expression modules were verified in the valid datasets. The verified genes underwent functional and pathway enrichment analysis. A total of 616 DEGs were selected to construct the PPI network, which included 5,808 osteosarcoma-specific interaction pairs and 8,012 normal-specific pairs. Tumor protein p53 (TP53), mitogen-activated protein kinase 1 (MAPK1) and estrogen receptor 1 (ESR1) were identified the most important osteosarcoma-associated genes, with the highest levels of topological properties. Neurogenic locus notch homolog protein 3 (NOTCH3) and caspase 1 (CASP1) were identified as the osteosarcoma-specific interaction pairs. Among all 23 mined modules, three were identified as differential expression modules, which were verified in the other two datasets. The genes in these modules were predominantly enriched in the FGFR, MAPK and Notch signaling pathways. Therefore, TP53, MAPK1, ESR1, NOTCH3 and CASP1 may be important in the development of osteosarcoma, and provides valuable clues to investigate the pathogenesis of osteosarcoma using the three differential expression modules.
Keywords: osteosarcoma, differential expression, interaction pairs, module, classification
Introduction
Osteosarcoma, as the most common primary bone sarcoma, has a relatively high morbidity rate in young individuals (1). In the first two decades of life, ~60% of cases of bone cancer are osteosarcoma (2). Despite intensive investigation, effective therapies for osteosarcoma have remained unchanged for the last 30 years, with a five-year survival rate of ≤20% (3). Metastasis is considered to be one of the primary reasons for the unsatisfactory improvements in survival rates (4). To develop effective treatments for osteosarcoma, further investigations are required to understand the pathogenesis of this malignant tumor.
There is evidence that molecular interactions are important in the mechanism of tumorigenesis and as a risk factor (5). Gene-gene interaction analysis is an essential tool to elucidate the nature of human traits (6). At present, it is clear that investigating the interactions of genes can provide increased understanding of disease risk factors, compared with the independent effects of gene variations (7,8). Park et al (9) identified a novel genetic risk factor for colorectal cancer using gene-gene interaction analysis. Shen et al (10) found that the interaction between DAB1 and RELN potentially increases the risk of autism in the Han Chinese population. Network analysis is an effective method to examine interactions and enables the potential identification of communities corresponding to genes with associated functions by module mining (11).
The present study aimed to investigate the differentially expressed genes (DEGs) and the interaction network of osteosarcoma samples, compared with normal samples. The network was furthered divided into different interaction modules, and genes in the differential expression modules were verified in the other two datasets.
Materials and methods
Gene expression profiles
The gene expression profiles of osteosarcoma samples and matched normal samples were downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). For profile retrieval, four keywords (osteosarcoma, normal, human and array express) were used. In addition, eligible profiles were those including at least three samples. In total, three datasets were obtained (GSE36001, GSE19276 and GSE16088). A total of 21 osteosarcoma samples and four normal bone tissues were included in GSE36001 (12), which was based on the Illumina human-6 v2.0 expression beadchip platform. GSE19276 (13) was comprised of 44 osteosarcoma samples and five normal bone tissue samples, based on the Agilent-012391 Whole Human Genome Oligo Microarray G4112A platform. For the GSE16088 (14) dataset, the platform used was the Affymetrix Human Genome U133A Array, which included 17 osteosarcoma samples and six normal bone tissue samples. The probe symbols in these profiles were converted into gene symbols, and the average values of genes with multiple corresponding probes were used as the expression values.
Screening of DEGs
To identify which genes were differentially expressed in the osteosarcoma samples and normal tissue samples in GSE16088, the significance analysis of microarrays (SAM) (15) method was applied rather than a t-test to control the rate of false positives using the false discovery rate (FDR). The cut-off level was FDR<0.05. Subsequently, principle component analysis (PCA) (16) was performed to determine whether the selected DEGs were able to distinguish the osteosarcoma samples from the normal samples in this dataset.
Protein-protein interaction (PPI) network
The Human Protein Reference Database, which stores the most information on PPIs, was examined to determine the interactions of the genes in GSE16088. Briefly, Pearson coefficients between each interacting gene pair in osteosarcoma and normal conditions were calculated using R version 3.1.3 (www.r-project.org), and pairs with coefficients >0.5 were retained for the construction of two condition-specific interaction networks. The two networks were then integrated to remove the common interaction between the normal and osteosarcoma PPI networks, leaving the interactions of the DEGs only. Finally, the topological properties (17) of this differential interaction network were examined, including the degree, betweenness and hub score.
Differential expression module mining
The modules, which comprise a group of specific genes or proteins in the PPI network, exert important effects. The multilevel community (18) function of the igraph package (R/igraph 1.0.0; www.igraph.org) was utilized for network module mining. Screening of the modules containing >15 genes was used to perform differential expression analysis in the GSE16088 profile using the GlobalAncova package in R language (version 3.42.0; https://www.bioconductor.org/packages/release/bioc/html/GlobalAncova.html). P<0.01 was considered to indicate a differential expression module.
Enrichment analysis
Genes in the differential expression modules were subjected to Database for Annotation, Visualization and Integrated Discovery for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment (david.ncifcrf.gov). The GO and KEGG terms with FDR<0.05 were regarded as significant functions and pathways.
Differential expression module verification
To verify the differential expression modules of the osteosarcoma samples, the GSE36001 and GSE19276 datasets were used. The expression levels of genes from modules 117, 135 and 152 in these two datasets were calculated, based on which a hierarchical clustering method was applied to confirm whether it was possible to confirm the differential expression profile of these genes between the osteosarcoma and normal samples.
Results
Screening of DEGs
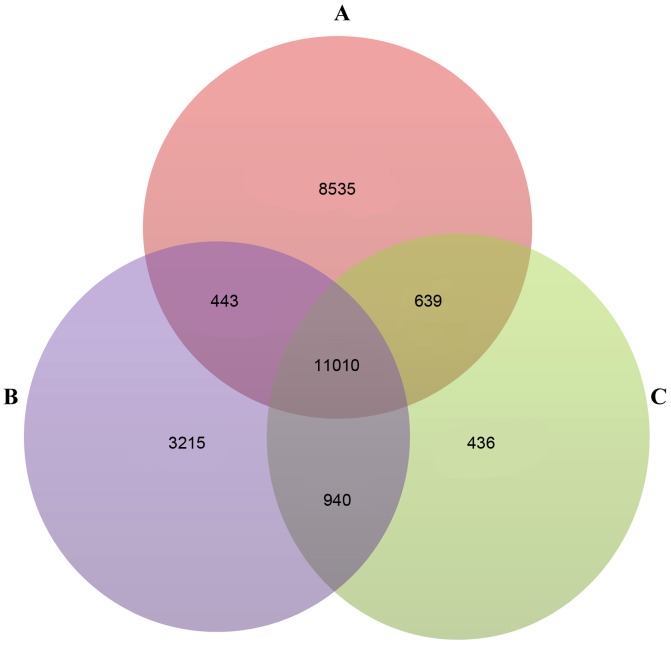
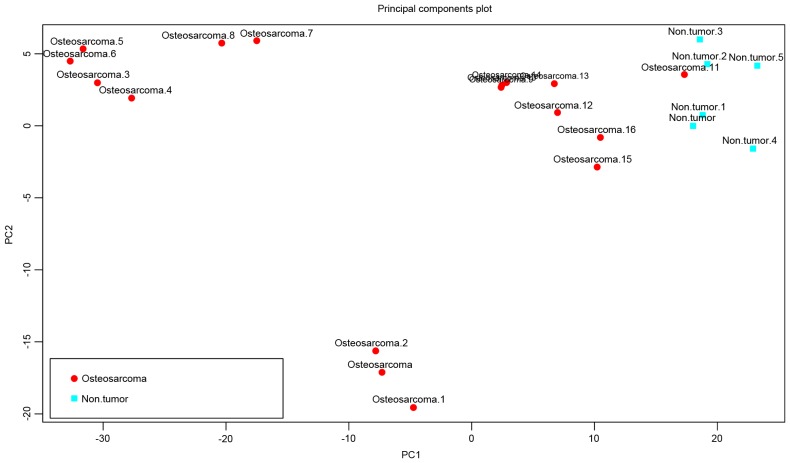
Following preprocessing, 24,614, 19,595 and 13,025 genes were obtained in the GSE36001, GSE19276 and GSE16088 datasets, respectively. A total of 11,010 genes were identified as genes common genes to the three datasets (Fig. 1A-C). Using the SAM method, 616 DEGs were screened out between the osteosarcoma and normal samples in the GSE16088 dataset. The results of the following PCA analysis showed that these DEGs enabled isolation of the osteosarcoma samples from the normal samples (Fig. 2).
Figure 1.
Numbers of common genes in the expression profiles of (A) GSE36001, (B) GSE19276 and (C) GSE16088.
Figure 2.
Sample classification of differentially expressed genes in GSE16088. Principle component analysis revealed that these differentially expressed genes were able to partially isolate osteosarcoma samples from normal samples.
PPI network of the DEGs
Proteins are the components, which perform the majority of the biological functions in the human body and usually function in interactive pairs during these processes. The PPI network of the DEGs in the GSE16088 dataset comprised 13,820 protein interaction pairs and 5,780 genes. The degree represents the importance of a node in the network, whereas the betweenness is the distance between a node and other nodes in the network. The 10 genes with the highest degree, betweenness and hub score are listed in Table I, and included tumor protein p53 (TP53), sarcoma (SRC), mitogen-activated protein kinase 1 (MAPK1) and estrogen receptor 1 (ESR1). SRC was the gene with the highest degree, betweenness and hub score.
Table I.
Top 10 gene nodes with highest degrees, hub scores and betweenness in the differentially expressed gene network.
| Gene | Degree | Gene | Hub score | Gene | Betweenness |
|---|---|---|---|---|---|
| SRC | 109 | SRC | 1 | SRC | 0.046616 |
| GRB2 | 97 | ESR1 | 0.777419 | TP53 | 0.042007 |
| EP300 | 96 | SHC1 | 0.76465 | SMAD2 | 0.032582 |
| TP53 | 96 | GRB2 | 0.763542 | GRB2 | 0.031228 |
| ESR1 | 87 | MAPK1 | 0.741441 | CTNNB1 | 0.029823 |
| MAPK1 | 79 | EGFR | 0.696561 | ESR1 | 0.02954 |
| FYN | 76 | FYN | 0.620895 | EP300 | 0.028269 |
| CSNK2A1 | 76 | PRKCD | 0.585712 | SMAD3 | 0.02601 |
| SMAD2 | 75 | EP300 | 0.582316 | EGFR | 0.024847 |
| SMAD3 | 75 | LCK | 0.579908 | MAPK1 | 0.023829 |
SRC, sarcoma; TP53 tumor protein p53; GRB2, growth factor receptor-bound protein 2; EP300, E1A binding protein p300; ESR1, estrogen receptor 1; MAPK1, mitogen-activated protein kinase 1; CSNK2A1 casein kinase 2 α1; SMAD, small mothers against decapentaplegic; PRKCD, protein kinase Cδ; LCK, lymphocyte-specific protein tyrosine kinase; CTNNB1, catenin-β1; EGFR, epidermal growth factor receptor.
Differential expression module mining
The multilevel community method identified 23 modules in the PPI network of the DEGs, and each of these modules had >15 genes (Table II). Following screening using the GlobalAncova package, a total of three modules (modules 117, 135 and 152) were selected as the differential expression modules (P<0.01; Table II).
Table II.
Differential expression modules.
| Module | F-value | P.perm |
|---|---|---|
| 117 | 3.27 | 0 |
| 152 | 2.87 | 0 |
| 135 | 3.15 | 0 |
| 86 | 2.64 | 0.01 |
| 123 | 3.02 | 0.01 |
| 101 | 2.14 | 0.02 |
| 124 | 2.53 | 0.02 |
| 130 | 3.10 | 0.02 |
| 103 | 2.82 | 0.02 |
| 96 | 2.63 | 0.02 |
| 147 | 2.73 | 0.03 |
| 85 | 2.54 | 0.03 |
| 120 | 2.66 | 0.03 |
| 106 | 2.76 | 0.04 |
| 121 | 2.53 | 0.04 |
| 127 | 2.93 | 0.04 |
| 61 | 2.65 | 0.05 |
| 159 | 2.55 | 0.05 |
| 122 | 2.70 | 0.05 |
| 153 | 3.11 | 0.05 |
| 137 | 2.77 | 0.06 |
| 50 | 2.49 | 0.06 |
| 114 | 2.60 | 0.12 |
Enrichment analysis
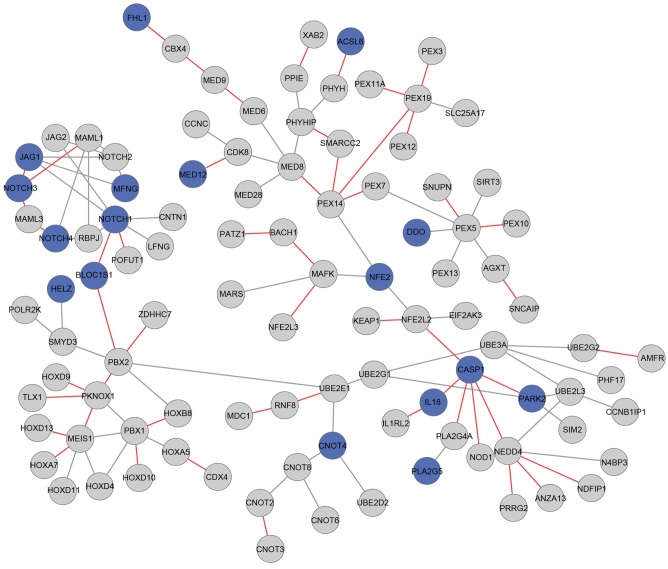
The genes in the differential expression modules underwent GO and pathway enrichment analyses (Table III). The genes in module 117 were primarily associated with the autophagy function, including intracellular membrane-bounded organelle and intracellular organelle, whereas module 135 was primarily enriched in the fibroblast growth factor receptor (FGFR) signaling pathway and MAPK signaling pathway. Module 152 was most closely involved with tumor-associated terms, including the Notch signaling pathway. Furthermore, in module 152, the neurogenic locus notch homolog protein 3 (NOTCH3) and caspase 1 (CASP1) genes only interacted with other genes in the osteosarcoma status (Fig. 3).
Table III.
Top 10 enriched function and pathway terms of the differential expression modules.
| Module | GO term | P-value | Bonferroni |
|---|---|---|---|
| 117 | GO:0030118~clathrin coat | 1.21E-15 | 2.23E-13 |
| 117 | GO:0030135~coated vesicle | 1.90E-13 | 3.47E-11 |
| 117 | GO:0030117~membrane coat | 6.94E-13 | 1.27E-10 |
| 117 | GO:0044431~Golgi apparatus part | 7.05E-13 | 1.29E-10 |
| 117 | GO:0043231~intracellular membrane-bounded organelle | 2.41E-11 | 4.41E-09 |
| 117 | GO:0005794~Golgi apparatus | 1.64E-10 | 3.00E-08 |
| 117 | GO:0030125~clathrin vesicle coat | 3.03E-10 | 5.54E-08 |
| 117 | hsa04142:Lysosome | 5.38E-10 | 2.37E-08 |
| 117 | GO:0043229~intracellular organelle | 8.78E-10 | 1.61E-07 |
| 117 | GO:0006897~endocytosis | 2.02E-09 | 1.22E-06 |
| 135 | GO:0044459~plasma membrane part | 1.46E-16 | 1.73E-14 |
| 135 | REACT_9470:Signaling by FGFR | 1.30E-13 | 2.33E-12 |
| 135 | REACT_1505:Integration of energy metabolism | 7.89E-13 | 1.42E-11 |
| 135 | GO:0005834~heterotrimeric G-protein complex | 1.79E-11 | 2.80E-09 |
| 135 | hsa04130:SNARE interactions in vesicular transport | 5.08E-11 | 2.95E-09 |
| 135 | GO:0034703~cation channel complex | 1.39E-10 | 2.17E-08 |
| 135 | REACT_15380:Diabetes pathways | 8.10E-09 | 1.46E-07 |
| 135 | GO:0019897~extrinsic to plasma membrane | 1.99E-09 | 3.10E-07 |
| 135 | GO:0031201~SNARE complex | 2.71E-09 | 4.23E-07 |
| 135 | hsa04010:MAPK signaling pathway | 1.31E-08 | 7.58E-07 |
| 152 | GO:0006625~protein targeting to peroxisome | 1.74E-14 | 1.07E-11 |
| 152 | GO:0042579~microbody | 1.07E-13 | 1.10E-11 |
| 152 | GO:0005777~peroxisome | 1.07E-13 | 1.10E-11 |
| 152 | GO:0043574~peroxisomal transport | 3.71E-14 | 2.28E-11 |
| 152 | GO:0005778~peroxisomal membrane | 2.56E-13 | 2.61E-11 |
| 152 | GO:0031903~microbody membrane | 2.56E-13 | 2.61E-11 |
| 152 | hsa04330:Notch signaling pathway | 8.59E-13 | 3.27E-11 |
| 152 | GO:0044438~microbody part | 6.73E-12 | 6.86E-10 |
| 152 | GO:0044439~peroxisomal part | 6.73E-12 | 6.86E-10 |
| 152 | GO:0007219~Notch signaling pathway | 3.26E-12 | 2.01E-09 |
GO, Gene Ontology; FGFR, fibroblast growth factor receptor; MAPK, mitogen-activated protein kinase.
Figure 3.
Interaction network of genes in the differential expression module 152. Blue nodes represent differentially expressed genes. Grey edges represent normal status. Red edges represent osteosarcoma status.
Differential expression module verification
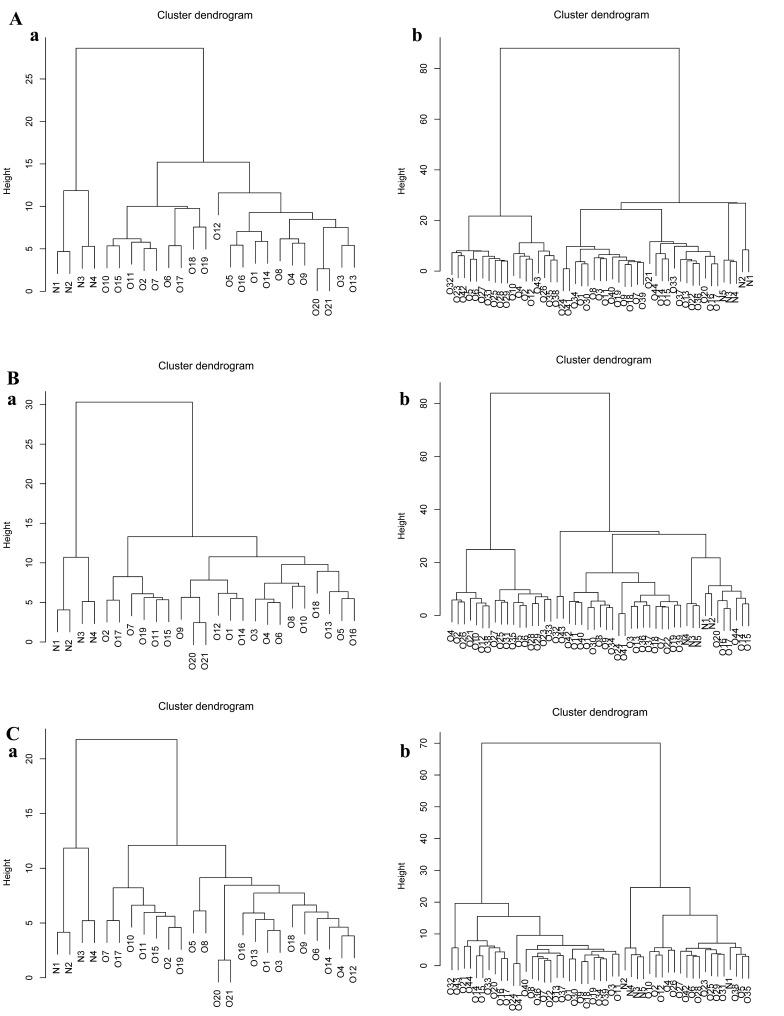
The genes in the differential expression modules were assessed for their ability to classify different samples in the other two expression profiles to verify the results from the GSE16088 dataset. The 151 genes in differential expression module 117 (Fig. 4A) were able to distinguish osteosarcoma genes from normal genes. The genes in modules 135 and 152 also enabled satisfactory distinguishing of osteosarcoma samples (Fig. 4B and C).
Figure 4.
Classification of genes in the differential expression modules of GSE36001 and GSE19276. (A) Effect of genes in module 117 in (a) GSE36001 and (b) GSE19276. (B) Effect of genes in module 135 in (a) GSE36001 and (b) GSE19276. (C) Effect of genes in module 152 in (a) GSE36001 and (b) GSE19276; O, osteosarcoma samples; N, normal samples.
Discussion
In order to investigate the possible molecular pathogenesis of osteosarcoma, the expression profiles and the interactions between DEGs were examined in the present study. A total of 616 DEGs were obtained in the osteosarcoma samples using the GSE16088 microarray data. A tumor-specific PPI network was then constructed using 13,820 interaction pairs and 5,780 genes, including normal specific-interaction pairs and disease-specific interaction pairs. Subsequently, the significantly connected sub-network modules were mined in this PPI network, from which three differential expression modules were identified. Functional and pathway enrichment analyses were used to analyze the enriched terms of the genes in these three modules. Finally, the genes in these modules were confirmed to be able to distinguish osteosarcoma samples from normal samples in the GSE36001 and GSE19276 datasets.
The topological properties of the degree, betweenness and hub score of the PPI network were examined. DEGs, including SRC, TP53 and ESR1 were found to be altered significantly in the osteosarcoma status, compared with the normal status. Thus, it was inferred that their roles were markedly altered in the osteosarcoma samples, and they may have specific contributions to the occurrence and development of this disease.
SRC showed the highest degree, betweenness and hub score levels, suggesting that SRC is the central gene during the development of osteosarcoma, and all the other connections were initiated by SRC. SRC is a known proto-oncogene (19), and the vital roles of SRC in colon cancer (20) and breast cancer (21) have been reported. SRC is involved in regulating cellular signaling, which results in the migration and invasion of cancer cells (22). In addition, evidence shows that SRC is a prognostic gene for tumors in osteosarcoma (23). In the present study, genes in the PPI network with a high rank in terms of their topological properties were considered to be causal genes for osteosarcoma. TP53, MAPK1 and ESR1 were also in the list of top 10 genes with important roles in the PPI network. It has been reported that these are all important oncogenes and osteosarcoma-associated genes (24). In a previous study on sporadic osteosarcoma reported ~3–7% had TP53 mutations (25). TP53 suppresses the proliferation, metastasis and angiogenesis of osteosarcoma cells through inhibition of the phosphoinositide 3-kinase/AKT/mammalian target of rapamycin pathway (26). ESR1 can initiate or enhance the transcription of genes responding to estrogen stimulation (27) and the DNA methylation of ESR1 is reported to be a useful prognostic indicator in osteosarcoma (24).
By examining the expression levels of genes in the differential expression modules, it was found that not all of these genes were DEGs in the osteosarcoma samples, suggesting it is not sufficient to investigate the mechanism of osteosarcoma by analyzing the differential expression of DEGs alone. The present study found that the interaction between NOTCH3 and CASP1 only existed under the osteosarcoma status in module 152, indicating that they were potential osteosarcoma-associated genes. The involvement of Notch signaling is important in cancer, and NOTCH3 is reported to be involved in the growth and survival of lung cancer (28). The expression of NOTCH3 is upregulated in osteosarcoma cells (29). The activation of caspase is involved in the execution of apoptosis in humans, and activation of caspase 1 has been shown to affect apoptosis in human prostate cancer (30). The overexpression of CASP1 has been confirmed as an inducer of apoptosis in mammalian cells (31).
The PCA analysis performed in the present study demonstrated that the differential expression module was verified in the GSE36001 and GSE19276 datasets. Therefore, the differential expression modules were able to isolate osteosarcoma samples from normal samples. The reliable molecular modules identified based on the analysis of gene expression profiles and interaction mining may be provide valuable clues to elucidating the molecular pathogenesis of tumors.
In conclusion, TP53, MAPK1, ESR1, NOTCH3 and CASP1 may be important in the development of osteosarcoma, which provides valuable clues to investigate the pathogenesis of osteosarcoma together with three differential expression modules.
References
- 1.Mirabello LJ, Yeager M, Mai PL, Gastier-Foster JM, Gorlick R, Khanna C, Patiño-Garcia A, Sierrasesúmaga L, Lecanda F, Andrulis IL, et al. Abstract 5574: High prevalence of germline TP53 mutations in young osteosarcoma cases. Cancer Res. 2015;75(Suppl):S5574. doi: 10.1158/1538-7445.AM2015-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lussier DM, Johnson JL, Hingorani P, Blattman JN. Combination immunotherapy with α-CTLA-4 and α-PD-L1 antibody blockade prevents immune escape and leads to complete control of metastatic osteosarcoma. J Immunother Cancer. 2015;3:21. doi: 10.1186/s40425-015-0067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrle D, Bielack S. Pediatric and Adolescent Osteosarcoma. Springer; New York, NY: 2010. Osteosarcoma lung metastases detection and principles of multimodal therapy; pp. 165–184. [DOI] [PubMed] [Google Scholar]
- 5.Betts JA, French JD, Brown MA, Edwards SL. Long-range transcriptional regulation of breast cancer genes. Genes Chromosomes Cancer. 2013;52:113–125. doi: 10.1002/gcc.22020. [DOI] [PubMed] [Google Scholar]
- 6.Hall MA, Verma SS, Wallace J, Lucas A, Berg RL, Connolly J, Crawford DC, Crosslin DR, De Andrade M, Doheny KF, et al. Biology-driven gene-gene interaction analysis of age-related cataract in the eMERGE network. Genet Epidemiol. 2015;39:376–384. doi: 10.1002/gepi.21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallberg H, Padyukov L, Plenge RM, Ronnelid J, Gregersen PK, van der Helm-van Mil AH, Toes RE, Huizinga TW, Klareskog L, Alfredsson L. Epidemiological Investigation of Rheumatoid Arthritis study group: Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22 and smoking in two subsets of rheumatoid arthritis. Am J Hum Genet. 2007;80:867–875. doi: 10.1086/516736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seddighzadeh M, Korotkova M, Källberg H, Ding B, Daha N, Kurreeman FA, Toes RE, Huizinga TW, Catrina AI, Alfredsson L, et al. Evidence for interaction between 5-hydroxytryptamine (serotonin) receptor 2A and MHC type II molecules in the development of rheumatoid arthritis. Eur J Hum Genet. 2010;18:821–826. doi: 10.1038/ejhg.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J, Kim I, Jung KJ, Kim S, Jee SH, Yoon SK. Gene-gene interaction analysis identifies a new genetic risk factor for colorectal cancer. J Biomed Sci. 2015;22:73. doi: 10.1186/s12929-015-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Y, Xun G, Guo H, He Y, Ou J, Dong H, Xia K, Zhao J. Association and gene-gene interactions study of reelin signaling pathway related genes with autism in the Han Chinese population. Autism Res. 2016;9:436–442. doi: 10.1002/aur.1540. [DOI] [PubMed] [Google Scholar]
- 11.Wang YR, Jiang K, Feldman LJ, Bickel PJ, Huang H. Inferring gene-gene interactions and functional modules using sparse canonical correlation analysis. Ann Appl Stat. 2015;9:300–323. doi: 10.1214/14-AOAS792. [DOI] [Google Scholar]
- 12.Kresse SH, Rydbeck H, Skårn M, Namløs HM, Barragan-Polania AH, Cleton-Jansen AM, Serra M, Liestøl K, Hogendoorn PC, Hovig E, et al. Integrative analysis reveals relationships of genetic and epigenetic alterations in osteosarcoma. PLoS One. 2012;7:e48262. doi: 10.1371/journal.pone.0048262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endo-Munoz L, Cumming A, Sommerville S, Dickinson I, Saunders NA. Osteosarcoma is characterised by reduced expression of markers of osteoclastogenesis and antigen presentation compared with normal bone. Br J Cancer. 2010;103:73–81. doi: 10.1038/sj.bjc.6605723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paoloni M, Davis S, Lana S, Withrow S, Sangiorgi L, Picci P, Hewitt S, Triche T, Meltzer P, Khanna C. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics. 2009;10:625. doi: 10.1186/1471-2164-10-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grace C, Nacheva EP. Significance analysis of microarrays (SAM) offers clues to differences between the genomes of adult Philadelphia positive ALL and the lymphoid blast transformation of CML. Cancer Inform. 2012;11:173–183. doi: 10.4137/CIN.S9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDermott JE, Diamond DL, Corley C, Rasmussen AL, Katze MG, Waters KM. Topological analysis of protein co-abundance networks identifies novel host targets important for HCV infection and pathogenesis. BMC Syst Biol. 2012;6:28. doi: 10.1186/1752-0509-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subelj L, Bajec M. Unfolding communities in large complex networks: Combining defensive and offensive label propagation for core extraction. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:036103. doi: 10.1103/PhysRevE.83.036103. [DOI] [PubMed] [Google Scholar]
- 19.Liu W, Yue F, Zheng M, Merlot A, Bae DH, Huang M, Lane D, Jansson P, Lui GY, Richardson V, et al. The proto-oncogene c-Src and its downstream signaling pathways are inhibited by the metastasis suppressor, NDRG1. Oncotarget. 2015;6:8851–8874. doi: 10.18632/oncotarget.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartolomé RA, García-Palmero I, Torres S, López-Lucendo M, Balyasnikova IV, Casal JI. IL-13 receptor α2 signaling requires a scaffold protein, FAM120A, to activate the FAK and PI3K pathways in colon cancer metastasis. Cancer Res. 2015;75:2434–2444. doi: 10.1158/0008-5472.CAN-14-3650. [DOI] [PubMed] [Google Scholar]
- 21.Correction: Associations and Interactions between Ets-1 and Ets-2 and Coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clin Cancer Res. 2015;21:2190–2191. doi: 10.1158/1078-0432.CCR-15-0405. [DOI] [PubMed] [Google Scholar]
- 22.Guarino M. Src signaling in cancer invasion. J Cell Physiol. 2010;223:14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 23.Hu C, Deng Z, Zhang Y, Yan L, Cai L, Lei J, Xie Y. The prognostic significance of Src and p-Src expression in patients with osteosarcoma. Med Sci Monit. 2015;21:638–645. doi: 10.12659/MSM.892803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonaglio V, De Carvalho AC, Toledo SR, Salinas-Souza C, Carvalho AL, Petrilli AS, de Camargo B, Vettore AL. Aberrant DNA methylation of ESR1 and p14ARF genes could be useful as prognostic indicators in osteosarcoma. Onco Targets Ther. 2013;6:713–723. doi: 10.2147/OTT.S44918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McIntyre JF, Smith-Sorensen B, Friend SH, Kassell J, Borresen AL, Yan YX, Russo C, Sato J, Barbier N, Miser J, et al. Germline mutations of the p53 tumor suppressor gene in children with osteosarcoma. J Clin Oncol. 1994;12:925–930. doi: 10.1200/JCO.1994.12.5.925. [DOI] [PubMed] [Google Scholar]
- 26.Song R, Tian K, Wang W, Wang L. P53 suppresses cell proliferation, metastasis, and angiogenesis of osteosarcoma through inhibition of the PI3K/AKT/mTOR pathway. Int J Surg. 2015;20:80–87. doi: 10.1016/j.ijsu.2015.04.050. [DOI] [PubMed] [Google Scholar]
- 27.Stossi F, Barnett DH, Frasor J, Komm B, Lyttle CR, Katzenellenbogen BS. Transcriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER) alpha or ERbeta in human osteosarcoma cells: Distinct and common target genes for these receptors. Endocrinology. 2004;145:3473–3486. doi: 10.1210/en.2003-1682. [DOI] [PubMed] [Google Scholar]
- 28.Yi F, Amarasinghe B, Dang TP. Manic fringe inhibits tumor growth by suppressing Notch3 degradation in lung cancer. Am J Cancer Res. 2013;3:490–499. [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, Zhang W, Cui J, Shui W, Yin L, Wang Y, Zhang H, Wang N, Wu N, Nan G, et al. Targeting BMP9-promoted human osteosarcoma growth by inactivation of notch signaling. Curr Cancer Drug Targets. 2014;14:274–285. doi: 10.2174/1568009614666140305105805. [DOI] [PubMed] [Google Scholar]
- 30.Guo Y, Kyprianou N. Restoration of transforming growth factor beta signaling pathway in human prostate cancer cells suppresses tumorigenicity via induction of caspase-1-mediated apoptosis. Cancer Res. 1999;59:1366–1371. [PubMed] [Google Scholar]
- 31.Whyte M. ICE/CED-3 proteasesin apoptosis. Trends Cell Biol. 1996;6:245–248. doi: 10.1016/0962-8924(96)20025-X. [DOI] [PubMed] [Google Scholar]