Abstract
The present study aimed to investigate the effect of Dickkopf-related protein 3 (DKK3) on osteogenic differentiation of rat dental follicle cells (DFCs). A PCR array analysis of Wnt pathway activation in DFCs identified genes dysregulated by mineral induction. Among them, DKK3expression levels were decreased, and further experiments were conducted to investigate its role in DFC osteogenesis. By comparing DFCs grown in normal growth and mineral-induction media for 4 weeks, the present study confirmed that DKK3 was a potential target gene of osteogenesis through reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting (WB). A short hairpin RNA (shRNA) was introduced into DFCs using a lentiviral vector to inhibit DKK3 expression. An alkaline phosphatase (ALP) activity assay and Alizarin Red staining were performed to observe the DKK3-shRNA DFCs. In addition, the osteogenic differentiation of DKK3-shRNA DFCs was analyzed by RT-qPCR and WB. In vivo, DKK3-shRNA DFCs seeded on hydroxyapatite/β-tricalcium phosphate (HA/TCP) scaffolds were transplanted into the subcutaneous tissue of mice with severe combined immunodeficiency, followed by hematoxylin-eosin and Masson staining. The results confirmed that DKK3 expression was downregulated during mineral induction in rat DFCs. Lentivirus-mediated expression of DKK3 shRNA in DFCs promoted calcified-nodule formation, ALP activity and the expression of β-catenin, runt-related transcription factor 2 and osteocalcin, compared with control cells. In vivo, the implanted section presented the majority of newly formed osteoid matrices and collagen, with limited space between the HA/TCP scaffolds and matrices. In conclusion, DKK3 expression negatively regulates the osteogenic differentiation of DFCs and, conversely, downregulation of DKK3 may enhance DFC osteogenesis.
Keywords: Dickkopf-related protein 3, dental follicle, osteogenic, Wnt
Introduction
As loose connective tissues surrounding the developing tooth germ, dental follicles not only control tooth eruption, but also contain progenitor cells that give rise to the cementum, periodontal ligaments and alveolar bones (1,2). Progenitor cells in dental follicles expressing Notch-1 and nestin proteins are believed to possess mesenchymal stem cell-like properties (3). Dental follicle cells (DFCs) can differentiate into osteoblasts and cementoblasts through the regulation of specific signaling pathways (4,5). Using suitable biocompatible scaffolds, DFCs form dentin and cementum-like matrices, indicating that DFCs may potentially serve as seed cells in tooth regeneration engineering (6–8).
Previous studies have identified several effectors that have been implicated in the osteogenic differentiation of DFCs, including bone morphogenetic protein (BMP), Wnt, Sonic hedgehog and the Notch signaling pathway (4,9–11). Previously, the canonical and non-canonical Wnt pathways have been demonstrated to promote DFC differentiation into osteoblasts/cementoblasts (9,12). Results have indicated that the Wnt signaling pathway branches into at least three distinct pathways: The canonical, non-canonical and planar-cell polarity pathways (13,14). The canonical Wnt pathway is the most extensively studied, and refers to Wnt ligands causing β-catenin accumulation in the cytoplasm, leading to eventual nuclear translocation, where it acts as a transcriptional co-activator of transcription factors belonging to the T-cell factor/lymphoid enhancer factor family (15).
The canonical Wnt/β-catenin pathway serves a critical function in the cell cycle and cell growth, which mediate oral tissue development (16). Previous research has suggested that Wnt/β-catenin signaling inhibits dental pulp stem cell differentiation into odontoblasts (17), and enhances the mineralization capacity of ameloblasts (18). However, the biological role of the canonical Wnt pathway in dental follicles has not been fully elucidated. To investigate the effects of Wnt pathway activation on DFCs, a polymerase chain reaction (PCR) array was implemented to compare the expression levels of 84 Wnt-associated genes maintained in routine or mineral-induction media for 4 weeks. The results indicated that the Dishevelled 3 (DVL3), cyclin D2 (CCND) and Dickkopf-related protein 3 (DKK3) genes were downregulated upon mineral induction, thus providing novel areas to study the osteogenic differentiation of DFCs.
The DVL protein family function as scaffold proteins that bridge receptors and distinct downstream signaling components (19). CCND proteins are members of the cyclin protein family, which regulate cell cycle progression (20). In the present study, there was a particular focus on the role of DKK3, since DKK family members are secreted glycoproteins known to antagonize Wnt signaling (21,22). Specifically, DKK3 is downregulated in various tumor cells, including hepatocellular carcinoma, lymphoblastic leukemia, prostate cancer, renal cell carcinoma, and melanoma cells (23–28). It has also previously been reported that DKK-family members participate in tooth development (29) DKK1 has previously been reported to be markedly expressed in the distal, incisor-bearing mesenchyme area of mouse mandibular processes during the initial stages of tooth formation. However, during molar morphogenesis, DKK1 was detected in the dental mesenchyme, DKK2 was expressed in the dental papilla, and DKK3 was specifically expressed in the primary and secondary enamel knots. In addition, DKK3 was transiently expressed postnatally in pre-ameloblasts, prior to the onset of enamel matrix secretion (29). However, the supportive roles of DKK family members in tooth development or dental-related cells remain elusive. Considering the evidence provided by the Wnt PCR array in the present study, the role of DKK3 in rat DFCs during osteogenesis was investigated.
Materials and methods
Ethical approval
All procedures performed in the present study involving animals were approved by the Ethics Committee of the Guanghua College of Stomatology at Sun Yat-sen University (Guangzhou, China).
DKK3 expression assay of DFCs with/without mineral induction
DFCs were cultured from 6–7-day-old Sprague Dawley rats, as described previously (2). A total of 5 rats [2 female/3 male, ~10 to 15 g, bred at 24°C on a 12 h light/dark cycle with ad libitum access to food and water, supplied by the Animal Research Center of Sun Yat-sen University (Guangzhou, China)] were sacrificed by cervical dislocation. The mandibles were dissected to separate the dental follicles. Following this, the dental follicles were digested by 0.1% collagenase type I (cat. no. C0130; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and 10 U/ml dispase (cat. no. D4818; Sigma-Aldrich; Merck Millipore) for 30 min at 37°C to obtain DFCs. The DFCs were identified by Alizarin red staining and Oil Red O staining. For the Oil Red O Staining, DFCs were treated with Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS, 0.5 mmol/l 3-isobutyl-1-methylxanthine, 200 µm/l indometacin, 10 µg/ml insulin, and 1 µm/l dexamethasone (Sigma-Aldrich; Merck-Millipore). The cells were stained with Oil Red O (Sigma-Aldrich; Merck-Millipore) 2 weeks subsequent to this. For the DKK3 assay, DFCs were cultured in 10% DMEM or mineral-induction medium (10% DMEM, 10 µl β-glycerophosphate, 50 µM ascorbate-2 phosphate and 0.1 µM dexamethasone; Sigma-Aldrich; Merck-Millipore) for 4 weeks at 37°C. Subsequently, the rat Wnt signaling pathway profile (cat. no. PARN-043Z; Qiagen, Inc., Valencia, CA, USA) was used to detect the expression of 84 genes associated with Wnt-mediated signal transduction on DFCs with/without mineral induction for 4 weeks. Briefly, the experiment was assayed on 96-well plates by running the reverse transcription-quantitative PCR (RT-qPCR) cycling program, according to the manufacturer's protocol.
Based on the results of the RT-qPCR, DKK3 was selected for further study. DFCs were cultured using DMEM or mineral-induction medium for 1, 2 or 4 weeks at 37°C. DKK3 gene expression in each group was measured by RT-qPCR and western blot (WB) analysis.
RT-qPCR analysis
Cells were collected and total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Reverse transcription was performed using the 1st Strand cDNA Synthesis kit with random hexamer primers (Invitrogen; Thermo Fisher Scientific, Inc.) as follows: The RNA/primer mixture was incubated at 65°C for 5 min, then placed on ice for 1 min. 2X reaction mix was added to the prepared RNA/primer mixture followed with incubation at room temperature (~25°C) for 2 min. The mixture with 1 µl SuperScript II RT was incubated at 42°C for 50 min and the reaction was terminated at 70°C for 15 min. The obtained cDNA was collected by brief centrifugation (4,989.5 × g, 30 sec, room temperature) and the quantification of DKK3 expression was analyzed by RT-qPCR using the TaqMan Gene Expression Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the following parameters: 95°C for 3 min for initial denaturation, 40 cycles at 95°C for 3 sec, 57°C for 30 sec, 68°C for 1 min. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control and the primers were as follows: Forward 5′-GCAAGAGAGAGGCCCTCAG-3′ and reverse 5′-TGTGAGGGAGATGCTCAGTG-3′. The primer sequences of the DKK3 gene were as follows: Forward 5′-TATACATGTGCAAGCCAGCC-3′ and reverse 5′-TCCTCAAATGCCATCTCCTG-3′. Threshold cycle values (Cq) were determined and data were analyzed with the 2−∆∆Cq method (30).
Western blotting analysis
DFCs from each group were obtained and protein was extracted using a NE-PER Extraction kit (Pierce; Thermo Fisher Scientific, Inc.). Protein concentrations were measured with a bicinchoninic acid (BCA) protein assay (Beyotime Institute of Biotechnology, Haimen, China). Proteins (20 µg) were separated by 10% sodium dodecylsulfate-polyacrylamide gel electrophoresis and electrophoretically transferred onto a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). The membrane was incubated for 1 h at room temperature in TBST containing 5% skim milk to block nonspecific protein binding and incubated at 4°C overnight with the primary antibodies. The following primary antibodies were used: Rabbit anti-DKK3 polyclonal antibody (dilution, 1:1,000; cat. no. ABIN411466; Biorbyt, Ltd., Cambridge, UK) and rabbit anti-GAPDH polyclonal antibody (dilution, 1:5,000; cat. no. ab9485; Abcam, Cambridge, UK), which was used as internal control. Following washing for 20 min, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (dilution, 1:2,000; cat. no. ab6721; Abcam) for 1 h at room temperature. Blots were visualized using an enhanced chemiluminescence system (Millipore ECL Western Blotting Detection System; EMD Millipore), and band densities were obtained and normalized to GAPDH and the background using ImageJ software version 2 (National Institutes of Health, Bethesda, MD, USA).
Construction of DFCs expressing a lentiviral vector containing short hairpin RNA (shRNA) against DKK3
A lentiviral vector expressing shRNA against DKK3 mRNA was constructed. The DKK3-targeting shRNA sequence (forward, 5′-TGCCACAGTCTGGTATACATCTTCCTGTCAATGTATACCAGACTGTGGCTTTTTTC-3′ and reverse, 5′-TCGAGAAAAAAGCCACAGTCTGGTATACATTGACAGGAAGATGTATACCAGACTGTGGCA-3′) was designed and cloned into the PLL3.79 lentiviral vector (Addgene, Inc., Cambridge, MA, USA), which encodes a green fluorescence protein (GFP) tag, and was validated by sequence analysis. The packaging 293T cell line (cat. no. CMH010; Shanghai Gaining Biotechnology Co., Ltd., Shanghai China) was transfected with the lentiviral vector using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The viral supernatant was harvested and the lentiviral particle titer was determined. The DFCs were seeded into 75 cm2 culture flasks at a density of 1×106 cells/ml. Following overnight culture, the cells were infected for 12 h at 37°C with the lentiviral vector in the presence of polybrene (final concentration, 8 mg/ml). Subsequently, the cells were washed and cultured in fresh medium for 3 days. Infected DFCs were selected by fluorescence activated cell sorting (FACS). Cells were trypsinized with 0.05% Trypsin-EDTA (cat. no. 25300-062; Gibco; Thermo Fisher Scientific, Inc.) for 40 sec at 37°C, harvested, and resuspended in PBS. FACS analyses were performed with the BD Influx Cell Sorter (BD Biosciences, Franklin Lakes, NJ, USA) and samples were analyzed by collecting 10,000 events using FlowJo software version 1.0 (BD Biosciences). Cells transfected with control vectors served as negative controls. The GFP+ transduced cells were also isolated by FACS with the BD Influx Cell Sorter (BD Biosciences).
Alkaline phosphatase (ALP) activity assays and Alizarin Red staining of DKK3-shRNA DFCs
DKK3-shRNA DFCs and control vector DFCs were placed in 24-well plates at a density of 3×104 cells/well. After 24 h, the cells were grown in mineral-induction medium for 3, 7, 14 or 21 days at 37°C. Each experimental group of cells was washed with phosphate-buffered saline at the appointed time and lysed by the addition of 200 µl/well 1% Triton X-100. Cellular ALP activities and total protein concentrations were determined with an ALP kit (Nanjing Jiancheng Chemical Industrial Co., Ltd., Nanjing, China) and the Bicinchoninic Acid Protein Assay kit (Boshide, Wuhan, China), respectively.
During the Alizarin Red staining experiments, the DKK3-shRNA DFCs and control vector DFCs were seeded in 6-well plates at a density of 1×105 cells/well. Following 24 h, the cells were grown in mineral-induction medium for 3 weeks at 37°C. The DFCs were subsequently fixed in formaldehyde for 15 min and stained with 0.5% Alizarin Red. Mineral deposits were visualized under a light microscope (Zeiss GmbH, Jena, Germany).
Mineral capability assays of DKK3-shRNA DFCs
RT-qPCR and WB analysis were employed to compare the respective expression levels of osteogenic markers in DKK3-shRNA DFCs and control DFCs in vitro. Following mineral induction for 1 or 3 weeks in shRNA DFCs and control vector DFCs, the mRNA expression levels of ALP (forward 5′-GGAAGCTAGATGCGGACAAG-3′ and reverse 5′-TCCCTGACATCGAAGTACCC-3′) and collagen-type-I (Col-I; forward, 5′-AGAGCATGACCGATGGATTCC-3′ and reverse, 5′-TTGCCAGTCTGCTGGTCCATG-3′) were measured by RT-qPCR.
In addition, the protein expression levels of DKK3 (dilution, 1:1,000; cat. no. ABIN411466; Biorbyt, Ltd.), β-catenin (dilution, 1:500; cat. no. ab16051; Abcam), runt-related transcription factor 2 (RUNX2; dilution, 1:1,000; cat. no. ab23981; Abcam), and osteocalcin (OCN; dilution, 1:1,000; cat. no. ab93876; Abcam) were investigated using WB analysis.
To detect the osteogenic differentiation of DKK3-shRNA DFCs or vector DFCs in vivo, cells at a density of 4×106 cells were mixed with 40 mg hydroxyapatite/b-tricalcium phosphate powder (HA/TCP; Engineering Research Centre in Biomaterials, Sichuan University, Chengdu, China) Each mixture (1 ml) was centrifuged (148.8 × g, 3 min, room temperature) and transplanted into the subcutaneous tissue of 4 severe combined immunodeficiency (SCID) mice (6 weeks old; ~30 g; 2 female/2 male, supplied by Animal Research Center of Sun Yat-sen University, Guangzhou, China). The mice were maintained in a clean animal room under a temperature of ~24°C, humidity of 40 to 70%, on a 12 h light/dark cycle. At 5 weeks post-implantation, the mice were sacrificed by cervical dislocation and the transplants were separated, then fixed with 4% paraformaldehyde for 24 h, and decalcified in 5% methanoic acid for 5 days. The sections were embedded in paraffin and cut longitudinally at 5-µm intervals. Initially, hematoxylin and eosin (H&E) staining was performed to observe the general view of transplants. The paraffin sections were deparaffinized, rehydrated and incubated with hematoxylin to stain the nuclei. After washing with tap water, the sections were stained with eosin. Finally, the samples were dehydrated, cleared and mounted. To further explore the collagen fiber of the transplants, Masson staining was applied with a similar protocol to the H&E staining. A total of three sections from each group were selected randomly, and three independent visual fields were captured to observe the newly formed collagen and mineralized matrices. The images captured by light microscope (Zeiss GmbH) were analyzed by Image-Pro Plus software (version, 6.0; Media Cybernetics, Rockville, MD, USA) to calculate the integrated optical density (IOD) and area of new collagen and mineralized matrices. The mineralized capability was obtained using the following formula: IOD/area × 100.
Statistical analysis
Results were presented as the mean ± standard deviation of at least 3 independent experiments. Data analysis was performed using SPSS software (version 20.0; IBM SPSS, Armonk, NY, USA). A Student's t-test was used to compare two means. One-way analysis of variance was applied to compare two or more means, followed by Student-Newman-Keuls test. P<0.05 was considered to indicate a statistically significant difference.
Results
Decreased DKK3 expression in DFCs upon mineral induction
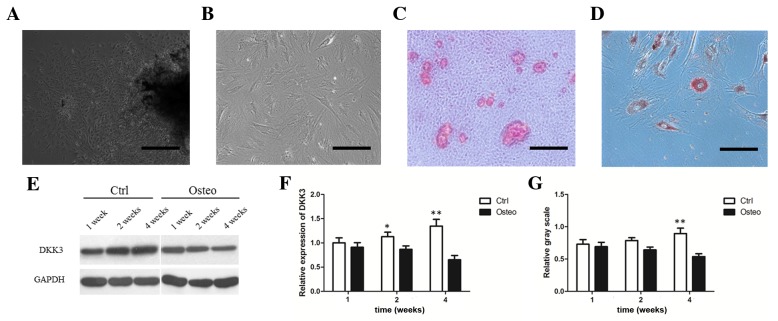
DFCs were successfully cultured and induced to form mineral deposits and adipocytes (Fig. 1A-D). The Wnt RT-qPCR array detected the expression levels of 84 Wnt-associated genes in DFCs maintained in routine or mineral-induction media for 4 weeks. The results indicated that DVL3, CCND2 and DKK3 genes were downregulated upon mineral induction (Table I; P<0.05).
Figure 1.
DKK3 expression in DFCs during osteogenic differentiation. (A) Primary culture of DFCs after 3 days. Scale bar, 200 µm. (B) Most DFCs at passage 3 presented fibroblast-like features. Scale bar, 100 µm. (C) Mineral deposits on DFCs grown in osteogenic medium for 3 weeks, as detected by Alizarin Red staining. Scale bar, 100 µm. (D) Adipocytes stained with Oil Red O following adipocyte induction for 4 weeks. Scale bar, 100 µm. (E) WB analysis demonstrating DKK3 protein downregulation following osteogenic differentiation. (F) DKK3 mRNA downregulation following osteogenic differentiation. (G) Relative gray scale of DKK3 expression determined following WB analysis. *P<0.05, **P<0.01 vs. Osteo. DKK3, Dickkopf-related protein 3; DFCs, dental follicle cells; WB, western blot; Control, DFCs grown in 10% Dulbecco's modified Eagle's medium; Osteo, DFCs grown in mineral induction medium.
Table I.
Wnt polymerase chain reaction array results of dental follicle cells cultured in mineral induction medium or 10% Dulbecco's modified Eagle's medium for 4 weeks.
| Average ∆∆Cq | 2−∆∆Cq | t-test | Fold up or downregulation | |||
|---|---|---|---|---|---|---|
| Gene | Mineral 4 weeks | 10% DMEM 4 weeks | Mineral 4 weeks | 10% DMEM 4 weeks | P-value | Mineral 4 weeks/10% DMEM 4 weeks |
| Apc | 5.88 | 5.68 | 1.7E-02 | 2.0E-02 | 0.716282 | −1.15 |
| Apc2 | 11.35 | 10.39 | 3.8E-04 | 7.5E-04 | 0.715525 | −1.95 |
| Axin1 | 6.93 | 6.93 | 8.2E-03 | 8.2E-03 | 0.854303 | 1.00 |
| Axin2 | 8.82 | 9.12 | 2.2E-03 | 1.8E-03 | 0.570925 | 1.23 |
| Bcl9 | 6.56 | 5.89 | 1.1E-02 | 1.7E-02 | 0.273815 | −1.59 |
| Btrc | 6.53 | 6.17 | 1.1E-02 | 1.4E-02 | 0.198065 | −1.28 |
| Ctnnb1 | 2.80 | 2.48 | 1.4E-01 | 1.8E-01 | 0.794771 | −1.25 |
| Ccnd1 | 2.20 | 2.38 | 2.2E-01 | 1.9E-01 | 0.520418 | 1.13 |
| Ccnd2 | 4.18a | 3.07a | 5.5E-02a | 1.2E-01a | 0.031100 | −2.15a |
| Ccnd3 | 4.22 | 3.92 | 5.3E-02 | 6.6E-02 | 0.719887 | −1.24 |
| Csnk1a1 | 3.29 | 2.99 | 1.0E-01 | 1.3E-01 | 0.476922 | −1.23 |
| Csnk1d | 4.32 | 4.50 | 5.0E-02 | 4.4E-02 | 0.475343 | 1.13 |
| Csnk2a1 | 4.52 | 4.48 | 4.3E-02 | 4.5E-02 | 0.940791 | −1.03 |
| Csnk2b | 3.71 | 3.69 | 7.6E-02 | 7.7E-02 | 0.940995 | −1.02 |
| Daam1 | 6.20 | 6.04 | 1.4E-02 | 1.5E-02 | 0.632151 | −1.12 |
| Dixdc1 | 8.45 | 7.99 | 2.9E-03 | 3.9E-03 | 0.971372 | −1.38 |
| Dkk1 | 11.79 | 12.24 | 2.8E-04 | 2.1E-04 | 0.414675 | 1.37 |
| Dkk3 | 3.15a | 2.21a | 1.1E-01a | 2.2E-01a | 0.049257a | −1.91a |
| Dkk4 | 7.09 | 6.59 | 7.3E-03 | 1.0E-02 | 0.299526 | −1.42 |
| Dvl1 | 6.15 | 6.02 | 1.4E-02 | 1.5E-02 | 0.955741 | −1.10 |
| Dvl2 | 4.48 | 4.26 | 4.5E-02 | 5.2E-02 | 0.426044 | −1.17 |
| Dvl3 | 7.08a | 6.09a | 7.4E-03a | 1.5E-02a | 0.011969a | −1.99a |
| Ep300 | 6.05 | 5.82 | 1.5E-02 | 1.8E-02 | 0.976422 | −1.17 |
| Fbxw11 | 4.56 | 4.29 | 4.2E-02 | 5.1E-02 | 0.585973 | −1.21 |
| Fbxw2 | 3.24 | 3.16 | 1.1E-01 | 1.1E-01 | 0.902830 | −1.06 |
| Fgf4 | 13.88 | 13.19 | 6.6E-05 | 1.1E-04 | 0.154326 | −1.61 |
| Frzb | 13.59 | 13.02 | 8.1E-05 | 1.2E-04 | 0.541255 | −1.48 |
| Fzd1 | 2.62 | 1.65 | 1.6E-01 | 3.2E-01 | 0.537557 | −1.96 |
| Fzd2 | 3.83 | 2.76 | 7.0E-02 | 1.5E-01 | 0.200846 | −2.09 |
| Fzd3 | 6.90 | 6.93 | 8.4E-03 | 8.2E-03 | 0.843733 | 1.02 |
| Fzd4 | 6.28 | 6.94 | 1.3E-02 | 8.2E-03 | 0.405254 | 1.58 |
| Fzd5 | 8.85 | 8.44 | 2.2E-03 | 2.9E-03 | 0.233498 | −1.32 |
| Fzd6 | 6.30 | 6.26 | 1.3E-02 | 1.3E-02 | 0.744854 | −1.03 |
| Fzd7 | 6.83 | 5.46 | 8.8E-03 | 2.3E-02 | 0.155697 | −2.59 |
| Fzd9 | 14.13 | 13.93 | 5.6E-05 | 6.4E-05 | 0.941213 | −1.15 |
| Gsk3a | 3.11 | 3.08 | 1.2E-01 | 1.2E-01 | 0.947278 | −1.02 |
| Gsk3b | 3.89 | 3.59 | 6.8E-02 | 8.3E-02 | 0.388341 | −1.23 |
| Jun | 5.31 | 4.74 | 2.5E-02 | 3.7E-02 | 0.579975 | −1.48 |
| Kremen1 | 5.85 | 5.63 | 1.7E-02 | 2.0E-02 | 0.540391 | −1.17 |
| Lef1 | 10.74 | 10.72 | 5.8E-04 | 5.9E-04 | 0.832187 | −1.02 |
| Lrp5 | 6.78 | 7.04 | 9.1E-03 | 7.6E-03 | 0.489102 | 1.19 |
| Lrp6 | 3.81 | 3.37 | 7.1E-02 | 9.7E-02 | 0.734114 | −1.36 |
| Mitf | 6.57 | 7.00 | 1.1E-02 | 7.8E-03 | 0.318318 | 1.35 |
| Myc | 4.65 | 5.27 | 4.0E-02 | 2.6E-02 | 0.311155 | 1.53 |
| Nkd1 | 7.24 | 6.07 | 6.6E-03 | 1.5E-02 | 0.145820 | −2.25 |
| Nkd2 | 4.78 | 4.29 | 3.6E-02 | 5.1E-02 | 0.451828 | −1.40 |
| RGD1561440 | 6.47 | 6.24 | 1.1E-02 | 1.3E-02 | 0.437063 | −1.17 |
| Pitx2 | 10.00 | 10.03 | 9.8E-04 | 9.6E-04 | 0.857792 | 1.02 |
| Porcn | 4.75 | 5.34 | 3.7E-02 | 2.5E-02 | 0.415860 | 1.50 |
| Ppp2ca | 3.52 | 3.35 | 8.7E-02 | 9.8E-02 | 0.652950 | −1.12 |
| Ppp2r1a | 2.99 | 3.10 | 1.3E-01 | 1.2E-01 | 0.611666 | 1.08 |
| Pygo2 | 6.82 | 6.55 | 8.9E-03 | 1.1E-02 | 0.917801 | −1.20 |
| Rhoa | 0.82 | 0.98 | 5.7E-01 | 5.1E-01 | 0.451892 | 1.12 |
| Senp2 | 5.13 | 4.95 | 2.9E-02 | 3.2E-02 | 0.606892 | −1.13 |
| Sfrp1 | 4.83 | 3.51 | 3.5E-02 | 8.8E-02 | 0.434107 | −2.49 |
| Sfrp2 | 6.31 | 6.01 | 1.3E-02 | 1.6E-02 | 0.470746 | −1.23 |
| Sfrp4 | 7.43 | 7.00 | 5.8E-03 | 7.8E-03 | 0.885189 | −1.34 |
| Sfrp5 | 11.47 | 11.58 | 3.5E-04 | 3.3E-04 | 0.786232 | 1.08 |
| Tcf3 | 6.38 | 6.24 | 1.2E-02 | 1.3E-02 | 0.618723 | −1.10 |
| Tcf4 | 4.21 | 4.13 | 5.4E-02 | 5.7E-02 | 0.699509 | −1.06 |
| Tcf7 | 11.27 | 10.70 | 4.1E-04 | 6.0E-04 | 0.216594 | −1.48 |
| Tcfe2a | 6.00 | 5.55 | 1.6E-02 | 2.1E-02 | 0.505454 | −1.36 |
| Tle1 | 7.21 | 6.69 | 6.8E-03 | 9.7E-03 | 0.770935 | −1.43 |
| Tle2 | 7.22 | 6.54 | 6.7E-03 | 1.1E-02 | 0.529451 | −1.60 |
| Wif1 | 7.16 | 7.56 | 7.0E-03 | 5.3E-03 | 0.328635 | 1.32 |
| Wisp1 | 2.52 | 1.95 | 1.7E-01 | 2.6E-01 | 0.586035 | −1.49 |
| Wnt1 | 13.16 | 12.49 | 1.1E-04 | 1.7E-04 | 0.374245 | −1.59 |
| Wnt10a | 13.75 | 13.27 | 7.2E-05 | 1.0E-04 | 0.948650 | −1.40 |
| Wnt10b | 13.09 | 12.90 | 1.1E-04 | 1.3E-04 | 0.748260 | −1.14 |
| Wnt11 | 8.51 | 7.61 | 2.8E-03 | 5.1E-03 | 0.287261 | −1.86 |
| Wnt2 | 11.62 | 10.83 | 3.2E-04 | 5.5E-04 | 0.373755 | −1.73 |
| Wnt2b | 10.97 | 10.77 | 5.0E-04 | 5.7E-04 | 0.634100 | −1.15 |
| Wnt3 | 10.48 | 10.23 | 7.0E-04 | 8.3E-04 | 0.564868 | −1.19 |
| Wnt3a | 6.61 | 6.06 | 1.0E-02 | 1.5E-02 | 0.540905 | −1.46 |
| Wnt4 | 8.59 | 8.88 | 2.6E-03 | 2.1E-03 | 0.869003 | 1.23 |
| Wnt5a | 3.42 | 3.69 | 9.4E-02 | 7.8E-02 | 0.588542 | 1.21 |
| Wnt5b | 5.54 | 5.31 | 2.1E-02 | 2.5E-02 | 0.402236 | −1.17 |
| Wnt6 | 15.60 | 16.84 | 2.0E-05 | 8.5E-06 | 0.332610 | 2.35 |
| Wnt7a | 17.12 | 17.92 | 7.0E-06 | 4.0E-06 | 0.415976 | 1.74 |
| Wnt7b | 7.46 | 6.70 | 5.7E-03 | 9.6E-03 | 0.243376 | −1.70 |
| Wnt8a | 14.74 | 13.88 | 3.6E-05 | 6.6E-05 | 0.276427 | −1.82 |
| Wnt8b | 16.05 | 16.15 | 1.5E-05 | 1.4E-05 | 0.502530 | 1.07 |
| Wnt9a | 8.94 | 8.93 | 2.0E-03 | 2.1E-03 | 0.497268 | −1.01 |
| Wnt9b | 9.38 | 9.38 | 1.5E-03 | 1.5E-03 | 0.642663 | 1.00 |
| Rplp1 | −2.04 | −2.43 | 4.1E+00 | 5.4E+00 | 0.058664 | −1.31 |
| Hprt1 | 4.43 | 4.61 | 4.6E-02 | 4.1E-02 | 0.526594 | 1.13 |
| Rpl13a | −2.39 | −2.18 | 5.2E+00 | 4.5E+00 | 0.372639 | 1.16 |
| Ldha | 2.88 | 2.93 | 1.4E-01 | 1.3E-01 | 0.431255 | 1.03 |
| Actb | −3.08 | −2.86 | 8.5E+00 | 7.3E+00 | 0.536666 | 1.16 |
P<0.05 vs. control.
WB and RT-qPCR analysis demonstrated that DKK3 expression was increased in a time-dependent manner in the control groups but decreased in a time-dependent manner in mineral-induction medium (Fig. 1E-G). After 2 weeks, DKK3 mRNA expression was decreased in DFCs grown in mineral-induction medium compared with those grown in routine medium (Fig. 1F; P<0.05). After 4 weeks, DKK3 mRNA and protein expression levels were significantly downregulated in the mineral-induction medium group (Fig. 1F-G; P<0.01).
Expression of osteogenic markers in DKK3-shRNA DFCs
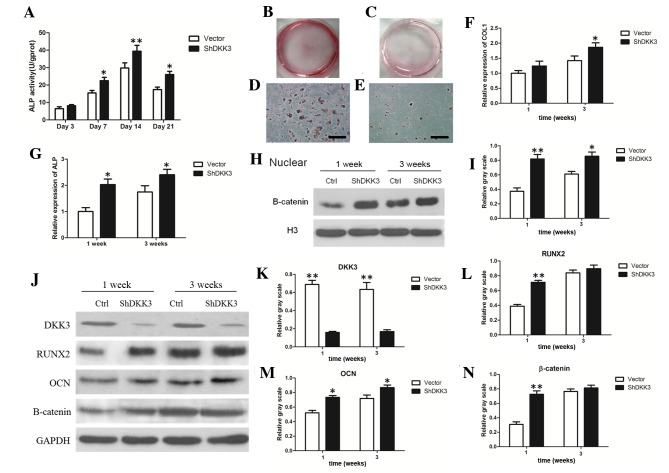
ALP activity, mineral deposits, RT-qPCR and WB analysis were used to analyze osteogenic markers. ALP activity was upregulated in DKK3-shRNA DFCs cultured for 7, 14 or 21 days, compared with in the vector-infected DFCs (Fig. 2A). In addition, following mineral induction for 3 weeks, DKK3-shRNA DFCs exhibited greater mineral deposition by Alizarin Red staining (Fig. 2B-E). RT-qPCR results demonstrated that Col-I and ALP levels were upregulated in DKK3-shRNA DFCs grown in mineral-induction medium for 1 or 3 weeks (Fig. 2F and G). The WB analysis results indicated that nuclear β-catenin, RUNX2, OCN and total β-catenin expression were increased, while DKK3 expression was inhibited in DKK3-shRNA DFCs following growth in mineral-induction medium for 1 or 3 weeks (Fig. 2H-N). These findings indicated that DKK3 suppression may promote osteogenic differentiation-marker expression in DFCs.
Figure 2.
Osteogenic differentiation markers analysis in DKK3-shRNA DFCs in vitro. (A) ALP activity of DKK3-shRNA DFCs was enhanced compared with in vector-infected DFCs between days 7 and 21. (B) General view of mineralized nodules of DKK3-shRNADFCs following growth in mineral-induction medium for 3 weeks. (C) General view of mineralized nodules of vector-infected DFCs following mineral induction for 3 weeks. (D) Calcified nodules of DKK3-shRNADFCs under microscopy. Scale bar, 200 µm. (E) Calcified nodules of vector-infected DFCs under microscopy. Scale bar, 200 µm. (F) mRNA expression of Col-I was upregulated in DKK3-shRNA DFCs grown in mineral-induction medium. (G) mRNA expression of ALP was upregulated in DKK3-shRNA DFCs grown in mineral-induction medium. (H) WB analysis demonstrating nuclear β-catenin expression inDKK3-shRNADFCs grown in mineral-induction medium. (I) Relative gray scale of WB analysis results in (H). (J) WB analysis of DKK3, RUNX2, OCN and β-catenin expression in DKK3-shRNADFCs grown in mineral-induction medium. (K-N) Relative gray scale of WB analysis results in (J). *P<0.05, **P<0.01 vs. vector-infected DFCs. ALP, alkaline phosphatase; DKK3, Dickkopf-related protein 3; ShRNA/sh, short hairpin RNA; DFCs, dental follicle cells; Col-I, collagen-type-I; WB, western blot; RUNX2, runt-related transcription factor 2; OCN, osteocalcin.
Mineralized capability of DKK3-shRNA DFCs in vivo
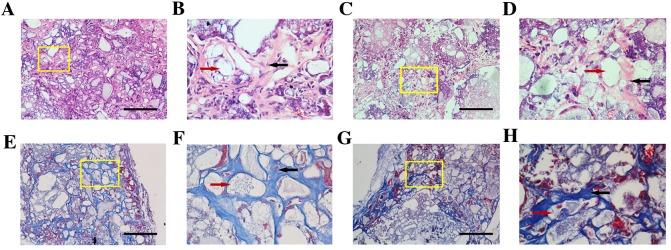
The DKK3-shRNA DFCs and vector-infected DFCs were successfully implanted in SCID mice. At 5 weeks post-implantation, H&E staining demonstrated that DKK3-shRNA DFCs with HA/TCP scaffolds formed new collagen fibers and vessels (Fig. 3A-D). Similarly, the Masson staining demonstrated that DKK3-shRNA DFCs with HA/TCP formed more mineralized matrices and collagen compared with the control group (Fig. 3E-H), indicating that low expression of DKK3 enhanced osteogenic differentiation in vivo. The quantitative analysis also indicated that the mineralization capabilities of the DKK3-shRNA DFCs with HA/TCP were higher than those of the control group (Fig. 4).
Figure 3.
H&E and Masson staining of DKK3-shRNA DFCs and vector-infected DFCs. Representative images obtained following (A-D) H&E and (E-H) Masson staining. (A) A marked amount of mineralized matrices were deposited on the implant of DKK3-shRNA DFCs. Scale bar, 100 µm. (B) Local magnification of yellow box in A indicating limited space occurring between the HA/TCP (red arrow) and matrices. Collagen fibers and vasculogenesis (black arrow) are apparent. (C) Vector-infected DFCs presented the formation of numerous semi-bone matrices. Scale bar, 100 µm. (D) Local magnification of yellow box in C demonstrated that vasculogenesis and collagen fiber (black arrow) formation around the cells and HA/TCP (red arrow) were weak. (E) Newly formed blue osteoid matrices and collagen in DKK3-shRNA DFCs. Scale bar, 100 µm. (F) Local magnification of yellow box in E indicating mineralized matrices, collagen, newly formed vessels (black arrow) and HA/TCP (red arrow) constituted a network in the implants. (G) Vector-infected DFCs formed loose osteoid matrices and collagen. Scale bar, 100 µm. (H) Local magnification of yellow box in G indicating numerous intervals were formed in the osteoid matrix (black arrow) around HA/TCP (red arrow). H&E, hematoxylin and eosin; DKK3, Dickkopf-related protein 3; shRNA, short hairpin RNA; DFCs, dental follicle cells; HA/TCP, hydroxyapatite/β-tricalcium phosphate.
Figure 4.
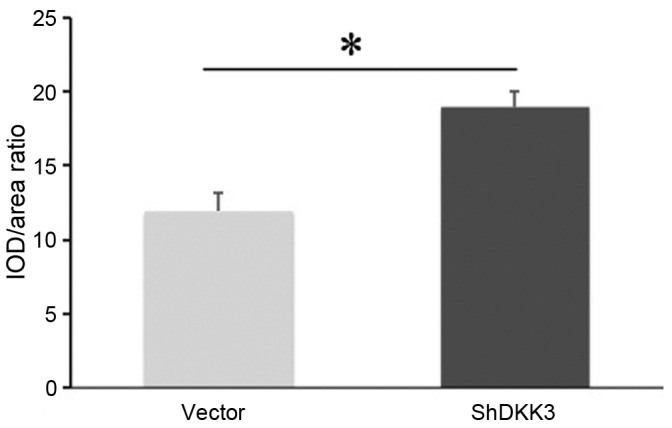
Quantitative comparison of mineral capabilities of DKK3-shRNA DFCs and vector-infected DFCs in vivo. The ratio of IOD/area in DKK3-shRNA DFCs was statistically higher than vector-infected DFCs. *P<0.05 vs. vector. DKK3, Dickkopf-related protein 3; shRNA/Sh, short hairpin RNA; DFCs, dental follicle cells; IOD, integrated optical density.
Discussion
Wnt pathway activation serves an essential function in tooth development and regeneration. The roles of specific Wnt genes have been studied by monitoring their expression levels and subcellular localization. Numerous Wnt-associated genes have been implicated in early tooth formation. For example, at the initiation stage, Wnt10b is expressed specifically in molar and incisor dental epithelial thickenings, whereasWnt5a is expressed in mesenchymal cells (31). Dental epithelial and mesenchymal Wnt/β-catenin signaling is suppressed by forced epithelial expression of the secreted Wnt inhibitor DKK1, followed by arrested tooth development at the early bud stage (32). At the early cap stage, lymphoid enhancer binding factor 1, Wnt3, Wnt6, Wnt10b and MFz6 are expressed in the primary enamel knot, whereas Wnt5a and MFrzb1 are detected in the dental papilla mesenchyme (31). Our previous studies demonstrated that, in DFCs, β-catenin expression increased in a time-dependent manner, andWnt5a is expressed in DFCs in postnatal rats (9,12). These observations suggested that the effects of Wnt family proteins on tooth formation should be focused on; however, the associated Wnt-associated gene has yet to be identified, therefore total genes or proteins should be screened in future studies.
At present, proteomics and DNA microarrays facilitate the study of the genome-wide transcriptome of human cells. Several studies have investigated signaling molecule activation during the osteogenic differentiation of dental-derived cells (33–36). A DNA microarray study previously reported that Wnt-associated genes are differentially expressed in differentiated DFCs (36). Since Wnt-associated gene expression may be limited during DFC differentiation, it suggests that the expression of certain genes may not be high enough to be detected in a total gene microarray assay. Therefore, a Wnt signaling pathway PCR array profile was used in the present study to investigate a panel of genes associated with Wnt-mediated signal transduction. It was demonstrated that the DKK3 gene was downregulated in DFCs upon mineral induction, supplying a novel target to control the osteogenic differentiation of DFCs. Notably, a recent microarray study of in vivo-implanted C3H10T1/2 cells expressing BMP2 also implicated DKK3 as a pivotal gene in endochondral bone formation (37). Consequently, the osteogenic differentiation of DFCs by controlling the expression of DKK3 was focused on in subsequent experiments.
As a secreted protein in the Dickkopf family, DKK3 serves an essential function in early embryonic development. Amphioxus DKK3 may regulate head formation by inhibiting Wnt/β-catenin and Nodal/Vg1 signaling, and these functions may have been partitioned among various vertebrate lineages during the evolution of DKK3 proteins (38). DKK3 expression has also been demonstrated o occur in the dental mesenchyme of embryonic day12 mouse embryos (39). Another report claimed that the DKK1-3 genes exhibited distinct spatiotemporal expression. During molar morphogenesis, DKK3 was specifically expressed in the primary and secondary enamel knots, whereas postnatal DKK3 mRNA expression was transiently observed in preameloblasts prior to the onset of enamel matrix secretion (29). In the present study, DKK3 expression in DFCs cultured in 10% DMEM or mineral-induction medium for 1, 2 or 4 weeks was explored. DKK3 mRNA expression was decreased at 2 weeks, whereasDKK3 protein decreased after 4 weeks in DFCs cultured in mineral-induction medium. In a previous study, DFCs were collected from postnatal rats, during which period DKK3 expression was not detected in the mesenchyme (29). Therefore, it may be concluded that low DKK3 expression may be associated with the osteogenic differentiation of DFCs in a time-dependent manner.
Based on the shRNA experiments in the present study, silencing DKK3 expression in DFCs led to enhanced formation of calcified nodules, ALP activity and osteogenic marker expression. WB analysis of DKK3-shRNA DFCs following mineral induction revealed that total β-catenin was upregulated at 1 week, whereas nuclear β-catenin was increased at 1 and 3 weeks. Since the Wnt canonical pathway is activated by nuclear β-catenin accumulation, it is possible that low DKK3 expression activated the canonical Wnt/β-catenin pathway, thereby regulating downstream target genes, including RUNX2 and OCN. In a previous study, osteoblast/cementoblast differentiation could be promoted by adding a canonical Wnt/β-catenin activator to DFCs (9). However, in murine SVF4 DFCs, Wnt3a-dependent β-catenin activation inhibited BMP2-mediated induction of cementoblast/osteoblast differentiation (40), which indicated that canonical Wnt signaling may inhibit the osteogenic differentiation of DFCs. It was hypothesized that this discrepancy may be caused by the source and type of DFCs, since the SVF4 cell line is an immortalized cell line, whereas rat DFCs were collected from primary cultures in the present study. Furthermore, DKK3 is an effective inhibitor of the canonical Wnt pathway; therefore, the positive effect of Wnt/β-catenin signaling on the osteogenic differentiation of DFCs was demonstrated.
To further confirm the aforementioned hypothesis, the osteogenic differentiation of DKK3-shRNA DFCs was completed in a SCID mouse model. Biphasic calcium phosphate bioceramics have been developed for tissue engineering applications, with various ratios of β-TCP and HA biphasic calcium phosphates (41). In the present study, the HA/TCP scaffold (HA/TCP=1:8; 1 mm) was loaded on DFCs in order to observe their osteogenic differentiation in vivo. Previous studies have demonstrated that DFCs acquired cementoblast or osteoblast-like features under BMP2 stimulation (4,5). However, DKK3 and BMP2 co-expression in implanted C3H10T1/2 cells significantly impaired bone formation, compared with cells expressing only BMP2 (37). These findings indicated that DKK3 may inhibit osteogenesis. In a similar manner, the shRNA-DKK3 DFCs treated with HA/TCP injected into the SCID mice presented increased osteoid matrices and collagen compared with control DFCs, also indicating that DKK3 is a negative regulator of osteogenesis. However, the small number of samples used was a limitation of the present study, since the expression of osteoblast markers, including RUNX2, could not be analyzed quantitatively.
In conclusion, the results presented in the current study indicated that DKK3 is a negative regulator during the osteogenic differentiation of DFCs and, conversely, that DKK3 downregulation may enhance DFC osteogenesis.
Acknowledgements
The present study was funded by the National Natural Science Foundation of China (grant nos. 81170932 and 81300846), the Science and Technology Research Fund of Guangdong Province (grant no. B2013153) and the Science and Technology Planning Project of Guangdong Province (grant no. 2013B021800058).
References
- 1.Ten Cate AR. The development of the periodontium-a largely ectomesenchymally derived unit. Periodontology 2000. 1997;13:9–19. doi: 10.1111/j.1600-0757.1997.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 2.Wise GE, Lin F, Fan W. Culture and characterization of dental follicle cells from rat molars. Cell Tissue Res. 1992;267:483–492. doi: 10.1007/BF00319370. [DOI] [PubMed] [Google Scholar]
- 3.Morsczeck C, Götz W, Schierholz J, Zeilhofer F, Kühn U, Möhl C, Sippel C, Hoffmann KH. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24:155–165. doi: 10.1016/j.matbio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Kemoun P, Laurencin-Dalicieux S, Rue J, Farges JC, Gennero I, Conte-Auriol F, Briand-Mesange F, Gadelorge M, Arzate H, Narayanan AS, et al. Human dental follicle cells acquire cementoblast features under stimulation by BMP-2/−7 and enamel matrix derivatives (EMD) in vitro. Cell Tissue Res. 2007;329:283–294. doi: 10.1007/s00441-007-0397-3. [DOI] [PubMed] [Google Scholar]
- 5.Zhao M, Xiao G, Berry JE, Franceschi RT, Reddi A, Somerman MJ. Bone morphogenetic protein 2 induces dental follicle cells to differentiate toward a cementoblast/osteoblast phenotype. J Bone Miner Res. 2002;17:1441–1451. doi: 10.1359/jbmr.2002.17.8.1441. [DOI] [PubMed] [Google Scholar]
- 6.Viale-Bouroncle S, Bey B, Reichert TE, Schmalz G, Morsczeck C. β-tricalcium-phosphate stimulates the differentiation of dental follicle cells. J Mater Sci Mater Med. 2011;22:1719–1724. doi: 10.1007/s10856-011-4345-0. [DOI] [PubMed] [Google Scholar]
- 7.Guo W, Gong K, Shi H, Zhu G, He Y, Ding B, Wen L, Jin Y. Dental follicle cells and treated dentin matrix scaffold for tissue engineering the tooth root. Biomaterials. 2012;33:1291–1302. doi: 10.1016/j.biomaterials.2011.09.068. [DOI] [PubMed] [Google Scholar]
- 8.Yang B, Chen G, Li J, Zou Q, Xie D, Chen Y, Wang H, Zheng X, Long J, Tang W, et al. Tooth root regeneration using dental follicle cell sheets in combination with a dentin matrix-based scaffold. Biomaterials. 2012;33:2449–2461. doi: 10.1016/j.biomaterials.2011.11.074. [DOI] [PubMed] [Google Scholar]
- 9.Du Y, Ling J, Wei X, Ning Y, Xie N, Gu H, Yang F. Wnt/β-catenin signaling participates in cementoblast/osteoblast differentiation of dental follicle cells. Connect Tissue Res. 2012;53:390–397. doi: 10.3109/03008207.2012.668980. [DOI] [PubMed] [Google Scholar]
- 10.Khan M, Seppala M, Zoupa M, Cobourne MT. Hedgehog pathway gene expression during early development of the molar tooth root in the mouse. Gene Expr Patterns. 2007;7:239–243. doi: 10.1016/j.modgep.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Viale-Bouroncle S, Gosau M, Morsczeck C. NOTCH1 signaling regulates the BMP2/DLX-3 directed osteogenic differentiation of dental follicle cells. Biochem Biophys Res Commun. 2014;443:500–504. doi: 10.1016/j.bbrc.2013.11.120. [DOI] [PubMed] [Google Scholar]
- 12.Xiang L, Chen M, He L, Cai B, Du Y, Zhang X, Zhou C, Wang C, Mao JJ, Ling J. Wnt5a regulates dental follicle stem/progenitor cells of the periodontium. Stem Cell Res Ther. 2014;5:135. doi: 10.1186/scrt525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones WM, Bejsovec A. Wingless signaling: An axin to grind. Curr Biol. 2003;13:R479–R481. doi: 10.1016/S0960-9822(03)00407-X. [DOI] [PubMed] [Google Scholar]
- 14.McEwen DG, Peifer M. Wnt signaling: Moving in a new direction. Curr Biol. 2000;10:R562–R564. doi: 10.1016/S0960-9822(00)00611-4. [DOI] [PubMed] [Google Scholar]
- 15.Kim JA, Choi HK, Kim TM, Leem SH, Oh IH. Regulation of mesenchymal stromal cells through fine tuning of canonical Wnt signaling. Stem Cell Res. 2015;14:356–368. doi: 10.1016/j.scr.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Liu F, Millar SE. Wnt/beta-catenin signaling in oral tissue development and disease. J Dent Res. 2010;89:318–330. doi: 10.1177/0022034510363373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheller EL, Chang J, Wang CY. Wnt/beta-catenin inhibits dental pulp stem cell differentiation. J Dent Res. 2008;87:126–130. doi: 10.1177/154405910808700206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian H, Lv P, Ma K, Zhou C, Gao X. Beta-catenin/LEF1 activated enamelin expression in ameloblast-like cells. Biochem Biophys Res Commun. 2010;398:519–524. doi: 10.1016/j.bbrc.2010.06.111. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama N, Golebiewska U, Wang HY, Malbon CC. Wnt-dependent assembly of supermolecular Dishevelled-3-based complexes. J Cell Sci. 2010;123:3693–3702. doi: 10.1242/jcs.075275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moschovi M, Alexiou GA, Patereli A, Siozos G, Sfakianos G, Prodromou N, Stefanaki K. Immunohistochemical expression of cell-cycle regulators in pediatric embryonal brain tumors. J Neurooncol. 2012;109:529–534. doi: 10.1007/s11060-012-0920-6. [DOI] [PubMed] [Google Scholar]
- 21.Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K, et al. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–313. doi: 10.1016/S0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- 22.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 23.Tsuji T, Miyazaki M, Sakaguchi M, Inoue Y, Namba M. A REIC gene shows down-regulation in human immortalized cells and human tumor-derived cell lines. Biochem Biophys Res Commun. 2000;268:20–24. doi: 10.1006/bbrc.1999.2067. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh SY, Hsieh PS, Chiu CT, Chen WY. Dickkopf-3/REIC functions as a suppressor gene of tumor growth. Oncogene. 2004;23:9183–9189. doi: 10.1038/sj.onc.1208138. [DOI] [PubMed] [Google Scholar]
- 25.Kurose K, Sakaguchi M, Nasu Y, Ebara S, Kaku H, Kariyama R, Arao Y, Miyazaki M, Tsushima T, Namba M, et al. Decreased expression of REIC/Dkk-3 in human renal clear cell carcinoma. J Urol. 2004;171:1314–1318. doi: 10.1097/01.ju.0000101047.64379.d4. [DOI] [PubMed] [Google Scholar]
- 26.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, Barrios M, Andreu EJ, Prosper F, Heiniger A, Torres A. Transcriptional silencing of the Dickkopfs-3 (Dkk-3) gene by CpGhypermethylation in acute lymphoblastic leukaemia. Br J Cancer. 2004;91:707–713. doi: 10.1038/sj.bjc.6602008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218–4227. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- 28.Kuphal S, Lodermeyer S, Bataille F, Schuierer M, Hoang BH, Bosserhoff AK. Expression of Dickkopf genes is strongly reduced in malignant melanoma. Oncogene. 2006;25:5027–5036. doi: 10.1038/sj.onc.1209508. [DOI] [PubMed] [Google Scholar]
- 29.Fjeld K, Kettunen P, Furmanek T, Kvinnsland IH, Luukko K. Dynamic expression of Wnt signaling-related Dickkopf1, −2 and −3 mRNAs in the developing mouse tooth. Dev Dyn. 2005;233:161–166. doi: 10.1002/dvdy.20285. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar L, Sharpe PT. Expression of Wnt signalling pathway genes during tooth development. Mech Dev. 1999;85:197–200. doi: 10.1016/S0925-4773(99)00095-7. [DOI] [PubMed] [Google Scholar]
- 32.Liu F, Chu EY, Watt B, Zhang Y, Gallant NM, Andl T, Yang SH, Lu MM, Piccolo S, Schmidt-Ullrich R, et al. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei X, Wu L, Ling J, Liu L, Liu S, Liu W, Li M, Xiao Y. Differentially expressed protein profile of human dental pulp cells in the early process of odontoblast-like differentiation in vitro. J Endod. 2008;34:1077–1084. doi: 10.1016/j.joen.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Wu L, Wei X, Ling J, Liu L, Liu S, Li M, Xiao Y. Early osteogenic differential protein profile detected by proteomic analysis in human periodontal ligament cells. J Periodontal Res. 2009;44:645–656. doi: 10.1111/j.1600-0765.2008.01174.x. [DOI] [PubMed] [Google Scholar]
- 35.Morsczeck C, Petersen J, Völlner F, Driemel O, Reichert T, Beck HC. Proteomic analysis of osteogenic differentiation of dental follicle precursor cells. Electrophoresis. 2009;30:1175–1184. doi: 10.1002/elps.200800796. [DOI] [PubMed] [Google Scholar]
- 36.Morsczeck C, Schmalz G, Reichert TE, Völlner F, Saugspier M, Viale-Bouroncle S, Driemel O. Gene expression profiles of dental follicle cells before and after osteogenic differentiation in vitro. Clin Oral Investig. 2009;13:383–391. doi: 10.1007/s00784-009-0260-x. [DOI] [PubMed] [Google Scholar]
- 37.Aslan H, Ravid-Amir O, Clancy BM, Rezvankhah S, Pittman D, Pelled G, Turgeman G, Zilberman Y, Gazit Z, Hoffmann A, et al. Advanced molecular profiling in vivo detects novel function of dickkopf-3 in the regulation of bone formation. J Bone Miner Res. 2006;21:1935–1945. doi: 10.1359/jbmr.060819. [DOI] [PubMed] [Google Scholar]
- 38.Onai T, Takai A, Setiamarga DH, Holland LZ. Essential role of Dkk3 for head formation by inhibiting Wnt/β-catenin and Nodal/Vg1 signaling pathways in the basal chordate amphioxus. Evol Dev. 2012;14:338–350. doi: 10.1111/j.1525-142X.2012.00552.x. [DOI] [PubMed] [Google Scholar]
- 39.Monaghan AP, Kioschis P, Wu W, Zuniga A, Bock D, Poustka A, Delius H, Niehrs C. Dickkopf genes are co-ordinately expressed in mesodermal lineages. Mech Dev. 1999;87:45–56. doi: 10.1016/S0925-4773(99)00138-0. [DOI] [PubMed] [Google Scholar]
- 40.Silvério KG, Davidson KC, James RG, Adams AM, Foster BL, Nociti FH, Jr, Somerman MJ, Moon RT. Wnt/β-catenin pathway regulates bone morphogenetic protein (BMP2)-mediated differentiation of dental follicle cells. J Periodontal Res. 2012;47:309–319. doi: 10.1111/j.1600-0765.2011.01433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen SS, Bornstein MM, Dard M, Bosshardt DD, Buser D. Comparative study of biphasic calcium phosphates with different HA/TCP ratios in mandibular bone defects. A long-term histomorphometric study in minipigs. J Biomed Mater Res B Appl Biomater. 2009;90:171–181. doi: 10.1002/jbm.b.31271. [DOI] [PubMed] [Google Scholar]