Abstract
The present study aimed to investigate the regulatory effect of probiotics on the expression of Toll-like receptors (TLRs) in an ulcerative colitis (UC) rat model, and to determine the role of probiotics in the underlying mechanisms through which UC develops and progresses in rat models. Rats were randomly allocated to one of the four following groups: i) The healthy control, ii) the model, iii) the Golden bifid treatment group, and iv) the TLR4 monoclonal antibody (TLR4mAb) intervention group. The UC rat model was established using 2,4,6-trinitro-benzene sulfonic acid. The general status and histological changes of rats were scored using the disease activity index and the histopathological scoring method, respectively. In these rats, the expression of TLR4 and TLR2 was measured using reverse transcription-quantitative polymerase chain reaction. The expression of TLR4 and TLR2 in the model group was significantly higher than that in the healthy control group. However, when compared with the model rats, those that received either Golden bifid treatment or TLR4mAb intervention exhibited significantly decreased mRNA expression levels of TLR4 and TLR2 (P<0.05). The development of UC is characterized by an abnormal immune response in the intestines. Probiotics alleviated inflammatory reactions in rats with UC. The underlying mechanism of UC may be associated with the expression of TLRs and the subsequent release of inflammatory cytokines.
Keywords: ulcerative colitis, intestinal flora, TLRs, probiotics, TNF-α, IL-1β
Introduction
Ulcerative colitis (UC) is a chronic non-specific inflammatory bowel disease (IBD) that most commonly afflicts people living in Western countries. Its clinical manifestations predominantly include repeated diarrhea, mucus, blood and pus in feces, and abdominal pain. In recent years, the incidence of UC has significantly increased in China (1). Numerous factors are involved in the pathogenesis of UC, leading to the World Health Organization (WHO) including UC in the list of modern diseases that are most difficult to treat (2). The etiology and pathogenesis of UC remain to be elucidated. Multiple factors, including intestinal flora disorders, autoimmune dysfunction, genetic mutations, infectious diseases and neuropsychological disorders, are involved in the development of UC (3). Among these, intestinal flora disorders and autoimmune dysfunction have been most investigated. As patients with UC develop intestinal flora disorders, probiotics maybe used to restore the balance of flora, thereby reducing intestinal inflammation (4). Bifidobacterium and Lactobacillus are two types of probiotics that are extensively observed in the human intestine. Previous studies have reported that these probiotics have anti-inflammatory and immunomodulatory activities (5), however, the exact mechanisms underlying the action of these probiotics remain to be elucidated. In normal flora, Bifidobacterium and Lactobacillus directly exert antibacterial effects on pathogenic Gram-positive and Gram-negative bacteria by producing bacteriocins and toxic acidic substances (6,7). In a previous study, it was demonstrated that VSL#3 probiotic capsules improved the function of the colonic barrier and protected the intestinal epithelial integrity of interleukin (IL)-10-deficient rats (8). Certain Lactobacillus strains also upregulate the expression of mucin 3, and enhance the intestinal mucus layer, thereby improving colonic barrier function in humans (9). Toll-like receptors (TLRs), which are type I transmembrane proteins homologous to the Drosophila Toll protein, are pivotal in eliciting an immune response to microbial infection (10). Increased expression of TLR4 has been demonstrated in UC animal models, while the inhibition of TLR activity alleviated UC-induced inflammation (11). Thus, the present study proposes that intestinal flora may be inducing an abnormal immune response and should be modulated to inhibit the development of abnormal immune responses via certain mechanisms. The present study examined the mRNA expression levels of TLR4/TLR2 and determined the concentration of tumor necrosis factor-α (TNF-α) and IL-1β in the intestinal mucosa of UC rats treated with Golden bifid. Results were then compared with the intervention group, which contained UC rats treated with TLR4 monoclonal antibody (TLR4mAb), and the model group, which contained untreated UC rats. Thus, in order to provide a further theoretical basis for the use of probiotics in UC treatment, the present study investigated the underlying mechanisms via which probiotics inhibit TLR expression.
Materials and methods
Experimental animal grouping
A total of 40 specific pathogen free-grade Sprague-Dawley rats (male and female; 2 months of age; weight, 280–320g; free access to food and water; temperature, 22–25°C; humidity, 50–70%; 12 h light/dark cycle), were purchased from the Laboratory Animal Center of Xinjiang Medical University (Xinjiang, China). The present study was approved by the Animal Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University. Rats were randomly assigned to one of the following four groups (n=10): i) Group I, normal control group, ii) group II, the model group, iii) group III, the Golden bifid treatment group, and iv) group IV, the TLR4mAb intervention group. The rats were maintained in a clean-grade room at constant temperature and fed ad libitum.
Reagents
2,4,6-trinitrobenzene sulfonic acid (TNBS) was purchased from Sigma-Aldrich (Merck Millipore, Darmstadt, Germany). Purified anti-mouse TLR4, CD284 (TLR4mAb; cat. no. 117602) was obtained from BioLegend, Inc. (San Diego, CA, USA). Golden bifid was purchased from Inner Mongolia Shuangqi Pharmaceutical Co., Ltd. (Hohhot, China). TRIzol reagent was obtained from Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA). TLR4 and TLR2 primers were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). First Strand cDNA Synthesis kit was purchased from Thermo Fisher Scientific, Inc. Synergy Brands SYBR Green PCR kit was purchased from Qiagen GmbH (Hilden, Germany). Taq PCR Master mix (2X) were purchased from Beijing Biomad Biological Gene Technology Co., Ltd. (Beijing, China). SYBR® Premix Ex Taq™ (2X) was purchased from Takara Bio, Inc. (Otsu, Japan). Loading buffer wash (6X) was purchased from Tiangen Biotech Co., Ltd. (Beijing, China). Agarose was purchased from Shanghai Generay Biotech Co., Ltd. (Shanghai, China) and Golden View nucleotide dye was purchased from Beijing Biomad Biological Gene Technology Co., Ltd.
UC model establishment and method of administration
Following acclimatization of the experimental rats in a clean-grade room for seven days, the animal model for the present study was developed. Prior to the development of the model, rats were not provided with food for 24 h. They were anesthetized by injection of a 1-fold diluted mixture of ketamine (100 mg/2 ml), diazepam (10 mg/2 ml), and atropine (0.5 mg/ml) in the abdomen; at a fixed dose of 0.75 ml/100 g. Experimental reagents were subsequently injected into the rats via an 8 Fr pediatric catheter, which was inserted at ~8 cm into the rat anus. Rats from groups II, III, and IV were injected with 1:1 mixture of 5% TNBS and 50% ethanol at a dose of 0.4 ml/100 g, while those from group I were injected with saline (0.4 ml/100 g). Following injection, the rats were lifted by their tails for ~10 min to prevent the outflow of reagents. Following establishment of the animal model, group III rats were fed Golden bifid via a gavage (2 ml/day, 1.0 ×109CFU) for seven days, while the other groups were administered saline via gavage. Group IV rats were abdominally injected with TLR4mAb (10 µg) (12); four times over 8 days. The rats from the other three groups were injected with saline. After 8 days, the rats were weighed and their abdominal cavity was opened following terminal anesthesia [a 1-fold diluted mixture of ketamine (100 mg/2 ml), diazepam (10 mg/2 ml), and atropine (0.5 mg/ml) in the abdomen; at a fixed dose of 0.75 ml/100 g]. The entire colon of each rat was exposed (excluding rats that has succumbed to natural death; or were close to death, i.e. inability to or extreme reluctance to stand up for 24 h, one in group II, two in group III and one in group IV), and 5 ml blood samples were taken from the abdominal aorta.
Symptom observation and scoring
Following establishment of the animal model, the rat feces were collected daily. The general status of the rats was also observed, including activity, fur, food intake, and weight, in addition to stool, blood and occult blood in stool. Furthermore, the number of incidence cases were recorded and scored using the disease activity index (DAI) proposed by Cooper et al (13).
Histopathological observation and scoring (HPS)
Approximately 1 cm of the colon exhibiting clear ulcers, congestion, and edema was sampled. The sample was first rinsed in pre-cooled saline and then fixed in 4% paraformaldehyde for 72 hat room temperature and embedded in paraffin. Subsequently, the sample was cut into 5 µm sections, which were de-waxed in xylene and dehydrated in ethanol: Xylene I (15 min), xylene II (10 min), and a graded series of 100, 95, and 80% ethanol (5 sec each). These sections were rinsed with water for 2 min. Subsequently, the sections were stained with hematoxylin (4 min) and rinsed in water, differentiated in HCl/ethanol (2 sec) and blued in water (2 min). These sections were stained with eosin (3 min) and rinsed in water, prior to finally being dehydrated in an ascending ethanol series (80, 95, 95 and 100% ethanol, 5 sec each). The dehydrated sections were made transparent using xylene I and xylene II (5 sec each). Finally, they were mounted with neutral gum in a hood. Following drying, the colonic mucosal tissue injuries and lesions were observed under a light microscope and scored using the scoring method proposed by Cooper et al (13). The remaining colon samples were frozen in liquid nitrogen and preserved at −80°C for further use.
Immunohistochemical staining and results interpretation
Subsequent to embedding and sectioning (4 µm) as described above, immunohistochemical examination was performed using a two-step method. Sections were de-waxed and dehydrated in a series of graded solvents, xylene I (15 min), xylene II (10 min), and 100, 95, 95, and 80% ethanol (5 sec each). The sections were then rinsed with water (2 min) and endogenous peroxidases were removed with 3% hydrogen peroxide at room temperature (10 min). Subsequently, the sections were washed with water (2 min) and phosphate-buffered saline (PBS; 2 min). Antigen retrieval was then conducted and the sections were placed in a 0.01 M citrate antigen retrieval solution and microwaved for 10 min (thawing temperature, 92–98°C). Following this, the sections were cooled to room temperature and rinsed in water and PBS, each for 2 min. The sections were incubated with rabbit anti-TLR4 polyclonal antibody (diluted in PBS, 1:150) at 37°C for 60 min. Subsequently, the sections were washed three times with PBS, for 2 min each time. These sections were incubated with goat anti-rabbit horseradish peroxidase-conjugated IgG secondary antibody (cat. no. PV-6001; OriGene Technologies, Inc. Beijing China), diluted in PBS (1:500), at 37°C for 25 min and subsequently washed with PBS. These sections were stained with a freshly prepared diaminobenzidine solution and observed under a light microscope (x100). The coloring process was conducted for 3–10 min, prior to the use of distilled water to terminate the reaction. The sections were re-stained with hematoxylin (4 min) and rinsed in water (2 min). They were again differentiated in HCL/ethanol (2 sec) and blued in water (1 min). Finally, the sections were dehydrated using an ascending ethanol series (80, 95, 95 and 100% ethanol, 5 sec each). They were made transparent with xylene I and xylene II (5 sec each), and mounted with neutral gum on a microscope for observation. PBS served as the negative control, while cells with a brown-stained membrane and/or cytoplasm were defined as positive. Based on the semi-quantitative scoring criteria proposed by Fromowitz et al (14), the positive staining percentages of <5, 5–25, 26–50, 51–75 and >75% were scored as 0, 1, 2, 3 and 4, respectively. Non-stained, light-yellow, brown, and dark-brown cells were scored as 0, 1, 2, and 3, respectively. The total score was calculated by adding the scores obtained using these two indicators. In each section, an average of five randomly selected fields were used for statistical analysis.
RNA extraction and purification
Colon tissue (50 mg) was ground in liquid nitrogen placed in a 1.5 ml Eppendorf (EP) tube and vortexed for 15 sec. This tissue was then maintained at room temperature for 15 min. The cells were then lysed and RNA was extracted in TRIzol/chloroform (1:0.2 by volume). Subsequently, the tube was kept at room temperature for 5 min and then centrifuged at 4°C, 4,834 × g for 15 min. The resulting supernatant was then transferred to another 1.5 ml EP tube containing isopropanol (TRIzol:isopropanol, 1:0.5). The tube was vortexed and placed at −20°C for 20–30 min, prior to centrifugation at 4°C, 4,834 × g for 10 min. Following centrifugation, the supernatant was aspirated and discarded, and the RNA precipitate was added to 1 ml of pre-cooled 75% diethylpyrocarbonate (DEPC)-treated ethanol. The solution was vortexed for 30 sec and centrifuged at 4°C, 4,834 × g for 10 min. The supernatant was discarded again, and the RNA precipitate was dried for 5–10 min. Subsequently, it was dissolved in DEPC-treated water (50–100 µl), followed by vortexing and centrifuging (4°C, 1,888 × g, 5 min). The concentration of the extracted RNA (2 µl) was then measured using a UV-spectrophotometer. If the concentration was too high, RNA was diluted with DEPC-treated water. The optical density (OD)260/OD280 was controlled between 1.8–2.0, and the RNA was stored at −80°C for further use.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) amplification
Primers for reverse transcription were designed and synthesized by Sangon Biotech Co. Ltd (Shanghai, China). β-actin served as the internal reference (Table I). cDNA was synthesized from the total RNA (1.5 g) according to the manufacturer's protocol of the First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). The cDNA was amplified by PCR for 35 cycles (the reaction system and conditions are listed in Tables II and III). The amplification product was electrophoresed using 2% agarose gel (110 V, 100 mA, 35 min). This was observed under a Bio-Rad Gel Doc 2000 gel imager (Bio-Rad Laboratories, Inc., Hercules, CA, USA) to check primer specificity.
Table I.
Primer sequences.
| Gene | PCR amplification fragment length (bp) | Primer sequence (5′→3′) |
|---|---|---|
| β-actin | 231 | F: 5′-ACTGCCCTGGCTCCTAGCA-3′ |
| R: 5′-GCCAGGATAGAGCCACCAATC-3′ | ||
| TLR4 | 100 | F: 5′-GCATCATCTTCATTGTCCTTGAGA-3′ |
| R: 5′-CTCCCACTCGAGGTAGGTGTTT-3′ | ||
| TLR2 | 285 | F: 5′-CGCTTCCTGAACTTGTCC-3′ |
| R: 5′-GGTTGTCACCTGCTTCCA-3′ |
PCR, polymerase chain reaction; TLR 4, Toll-like receptor 4; TLR 2, Toll-like receptor 2; F, forward; R, reverse.
Table II.
PCR amplification system.
| Agent | Volume (µl) |
|---|---|
| 2X Taq PCR Master Mix | 10 |
| P-f (10 pmol/µl) | 0.3 |
| P-r (10 pmol/µl) | 0.3 |
| DNA template | 2.0 |
| ddH2O | 7.4 |
PCR, polymerase chain reaction; ddH2O, double distilled water; P-f, forward primer; P-r, reverse primer.
Table III.
PCR reaction conditions.
| Procedure | Temperature (°C) | Duration |
|---|---|---|
| Pre-denaturation | 94 | 3 min |
| Denaturation 35 cycles | 94 | 30 sec |
| Annealing | 56.8, 62.3 and 57.5 for β-actin, TLR4 and TLR2, respectively | 30 sec |
| Elongation | 72 | 1 min |
| Renaturation | 72 | 5 min |
PCR, polymerase chain reaction; TLR 4, Toll-like receptor 4; TLR 2, Toll-like receptor 2.
Standard preparation
The brightest band observed under ultraviolet light was cut. The target gene and internal reference were purified and recovered using the manufacturer's protocols in the GeneJET DNA recovery kit (Thermo Fisher Scientific, Inc.). The concentration and absorbance value of recovered DNA were measured and the DNA was preserved at −20°C for further use. The standard was amplified by RT-PCR for 40 cycles (the reaction system and conditions are listed in Tables IV and V). The temperature was increased from 65 to 95°C at a temperature transition rate of 0.5°C/sec. The iCycler Optical System Interface software (version 2.0; Bio-Rad Laboratories, Inc.) was used to read the fluorescence data, which was then used to calculate the Cq value. Thus, the standard and amplified melting curves were obtained.
Table IV.
RT-qPCR reaction system.
| Agent | Volume (µl) |
|---|---|
| 2X SYBR® Premix Ex Taq™ | 10 |
| P-f (10 pmol/µl) | 0.3 |
| P-r (10 pmol/µl) | 0.3 |
| DNA template | 2.0 |
| ddH2O | 7.4 |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction; ddH2O, double distilled water; P-f, forward primer; P-r, reverse primer.
Table V.
RT-qPCR reaction conditions.
| Procedure | Temperature (°C) | Duration |
|---|---|---|
| Pre-denaturation 35 cycles | 95 | 3 min |
| Denaturation | 95 | 10 sec |
| Annealing | 56.8, 62.3 and 57.5 for β-actin, TLR4 and TLR2 respectively | 30 sec |
| Elongation | 65 | 1 min |
| Renaturation | 72 | 5 min |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TLR 4, Toll-like receptor 4; TLR 2, Toll-like receptor 2.
RT-qPCR of target genes
The β-actin, TLR4, and TLR2 genes of each sample were amplified by RT-qPCR. Each measurement was performed in triplicate. The standard and double distilled H2O was introduced as calibration and negative control for each gene. Details of the primers, reaction volume and thermocycling conditions are presented in Tables I, IV and V. The RT-qPCR product specificity was assessed by observing whether the melting curve had a single peak. Quantitative results of each sample were measured using the 2-∆∆Cq method by PCR amplifier (15).
ELISA examination and quantification
Blood samples were kept at room temperature for 2 h. These samples were then centrifuged at 33 × g for 20 min. Following centrifugation, the supernatant solution was collected. Serially diluted standard solutions (100 µl) were added to 7 wells (TNF-α successively diluted to 500, 250, 62.5, 31.2, 15.6 and 7.8 pg/ml; and IL-1β successively diluted to 400, 200, 100, 50, 25, 12.5 and 6.25 pg/ml. Water (100 µl) served as the blank solution, while samples were added to the other wells (100 µl). Each sample was replicated twice. The 96-well plate was covered and incubated at 37°C for 2 h after discarding the liquid. Following addition of 100 µl of detection reagent A, (working solution, prepared prior to use), the plate was again incubated at 37°C for 1 h. The liquid was discarded, and the wells were washed three times using 350 µl of washing solution. The wells were then dried by tapping them on absorbent paper. Subsequently, 100 µl of detection reagent B was added (working solution, prepared prior to use) to the wells, which were then incubated at 37°C for 30 min. Finally, the wells were washed and dried and the substrate solution (90 µl) was added to each well and the 96-well plate was developed in a dark room at 37°C. The development of the wells took place over 15–25 min. Notably, the first 3–4 standard wells exhibited a gradient of blue intensity, while the last 3–4 wells did not exhibit any such gradient, the development was terminated by adding 50 µl of termination solution (H2SO4). The color of the wells turned from blue to yellow following addition of this solution. Subsequent to ensuring there were no water droplets at the bottom of the ELISA plate or bubbles in the wells, the OD value was measured at 450 nm using a microplate reader. A 7-point plot was obtained by subtracting the OD value of blank solution from the OD values of the standard or each sample. The mean OD value of repeated wells was used for calculation. The standard curve was plotted using the standard concentrations as the ordinate (or logarithmic scale) and the corresponding OD values as the abscissa (or logarithmic scale). The actual concentration of each sample was calculated using the CurveExpert software 1.30 (Wuhan Boster Biological Technology, Ltd., Wuhan, China).
Statistical analysis
Statistical analysis was performed using the SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard deviation. Inter-group comparisons were made by one-way analysis of variance. P<0.05 was considered to indicate a statistically significant difference.
Results
General status and DAI scoring
DAI scoring is presented in Table VI. The rats from group I were active, exhibiting smooth fur, normal food intake, continuous weight gain, and normal stool. On the second day after the development of the animal model, rats from group II developed a sickly disposition exhibiting less vigorous movement, rough fur, decreased appetite, and increased stool frequency. Of the ten rats, two passed visibly bloody stool, whilst four tested positive for fecal occult blood. Only two rats passed loose stool, one rat died. All the rats of group II exhibited significant weight loss, and their DAI score was significantly higher than that of the group I rats (P=0.000). Rats of groups III and IV also demonstrated similar symptoms two days after establishment of the animal model. Prior to succumbing to the UC, one rat of group III and two rats of group IV passed bloody stool, while the remaining rats passed either pasty or loose stool, two rats of group III and one rat of group IV died. All the rats of groups III and IV had varying degrees of weight loss, however, their DAI scores were not significantly different from each other (P=0.631). However, compared with group II rats, the symptoms of rats belonging to groups III (P=0.016) and IV (P=0.028) were alleviated, and the difference between their symptoms was observed to be statistically significant.
Table VI.
DAI and HPS scores of each group.
| Group | DAI | HPS |
|---|---|---|
| Control | 0 | 0 |
| Model | 8.05±1.97a | 14.55±0.86a |
| Golden bifid | 4.73±1.04a,b | 9.97±1.58a,b |
| TLR4mAb | 5.08±2.18a,b | 10.11±2.11a,b |
P<0.05 vs. the control
P<0.05 vs. the model group. TLR4mAb, Toll-like receptor 4 monoclonal antibody; DAI, disease activity index; HPS, histopathological scoring.
Pathological observation and HPS scoring
Under a light microscope, it was observed that the intestinal epithelial cells of group I rats were intact; the sections indicated that closely arranged glands were enriched by goblet cells. Few neutrophils or lymphocytes were scattered in the lamina propria. In group II rats, the intestinal epithelial cells were either markedly damaged or were absent, the mucosa was eroded and the glands appeared to be damaged and disordered. The goblet cells were either notably reduced or absent. The neutrophils and lymphocytes had markedly infiltrated the lamina propria and crypt abscesses were clearly formed. Congestion was also observed, along with the expansion of capillaries in these sections (Fig. 1). The HPS score of the model rats was significantly higher than that of the control (P=0.000). Compared with the rats of groups I and II, the tissue damage in rats belonging to groups III and IV was less intense. The HPS scores of rats belonging to groups III (P=0.035) and IV (P=0.047) were significantly lower than that of the model rats however these HPS scores did not indicate a significant difference from each other (P=0.88; Table VI).
Figure 1.
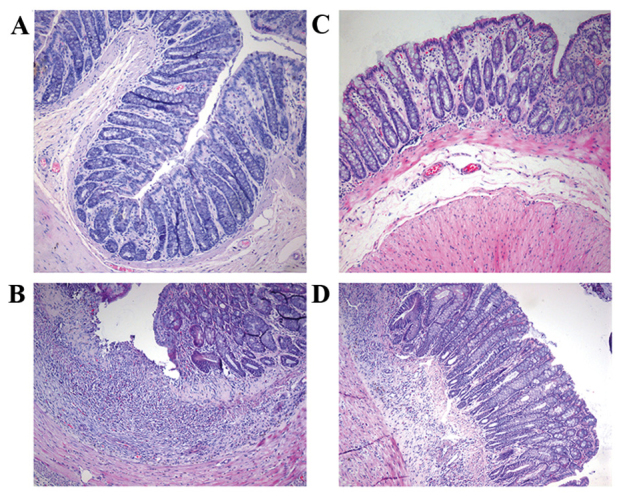
Hematoxylin and eosin staining of rat colon tissues (magnification, ×100). (A) Healthy control, (B) model group, (C) Golden bifid group and (D) Toll-like receptor 4 monoclonal antibody group.
TLR4 expression in each group
In group II rats, strong expression of TLR4 (indicated by brown granules) was exhibited in the cytoplasm and the mucosal and submucosal membrane of inflammatory cells, including neutrophils, plasma cells and monocytes. TLR4 was also strongly expressed in the lamina propria and the perivascular areas. However, in group I rats, TLR4 was weakly expressed in the mucosa and submucosa (Fig. 2). Compared with group I rats, TLR4 expression was significantly greater in group II rats (P=0.001). Rats of group III (P=0.042) and IV (P=0.048) indicated significantly decreased expression of TLR4 (Fig. 3).
Figure 2.
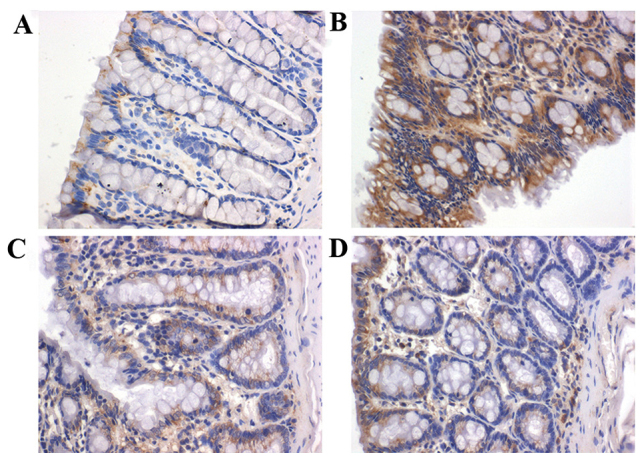
Immunohistological examination of TLR4 expression in rat colon (magnification, ×400). (A) Healthy control, (B) model group, (C) Golden bifid group and (D) TLR 4 monoclonal antibody group. TLR 4, Toll-like receptor 4.
Figure 3.
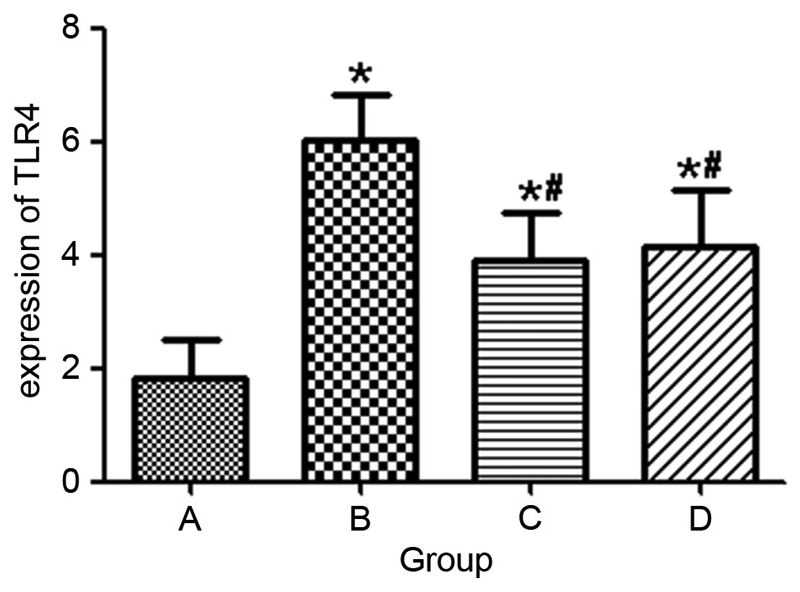
Histopathological scoring of TLR4 expression. A indicates the healthy control, B indicates the model group, C indicates the Golden bifid group and D indicates the TLR4 monoclonal antibody group. *P<0.05 vs. the control, #P<0.05 vs. the model group. TLR4, Toll-like receptor 4.
mRNA expression levels of TLR4 and TLR2
RT-PCR demonstrated that the mRNA expression levels ofTLR4 in groups I, II, III and IV were 0.00057±0.00023, 1.58±1.00, 0.018±0.015, and 0.037±0.027, respectively. Furthermore, the mRNA expression levels of TLR2 in groups I, II, III and IV were 0.00038±0.00018, 0.42±0.17, 0.027±0.019, and 0.020±0.007, respectively. When compared with rats of group I, rats of group II demonstrated significantly increased mRNA expression levels of TLR4 and TLR2 (P=0.000; Fig. 4). Compared with groups II, groups III (P=0.000) and IV (P=0.000) indicated significantly decreased mRNA expression levels of TLR4 and TLR2 (Fig. 4). However, the mRNA levels of TLR4 (P=0.898) and TLR2 (P=0.963) in groups III and IV were not significantly different from each other.
Figure 4.
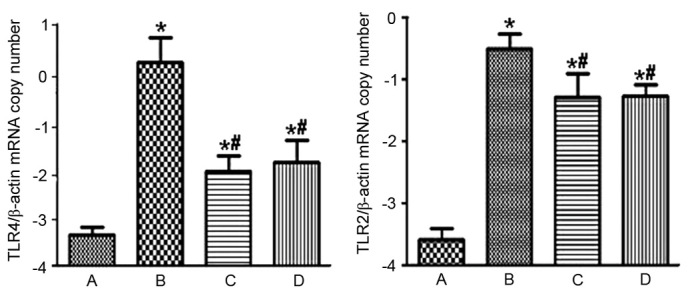
mRNA expression levels of TLR4 and TLR2 in each group. A indicates the healthy control, B indicates the model group, C indicates the Golden bifid group and D indicates the TLR4 monoclonal antibody group. *P<0.05 vs. the control, #P<0.05 vs. the model group. TLR4, Toll-like receptor 4; TLR2, Toll-like receptor 2.
Serum concentration of TNF-α and IL-1β
Compared with rats in group I, rats in group II exhibited a significantly higher concentration of serum TNF-α (P=0.032) and IL-1β (P=0.023) Compared to group II, rats of groups III (P=0.041, 0.048) and IV (P=0.045, 0.049) were observed to have varying levels of decrease in their serum TNF-α and IL-1β concentration (Table VII).
Table VII.
Serum concentration of TNF-α and IL-1β in each group.
| Group | TNF-α | IL-1β |
|---|---|---|
| Control | 17.36±3.85 | 11.64±3.50 |
| Model | 29.42±7.51a | 23.05±4.52a |
| Golden bifid | 22.98±5.22a,b | 16.37±4.22a,b |
| TLR4mAb | 24.18±4.62a,b | 16.78±5.89a,b |
P<0.05 vs. the control
P<0.05 vs. the model group. TLR4mAb, Toll-like receptor 4 monoclonal antibody; TNF-α, tumor necrosis factor-α; IL-1β, interleukin-1β.
Discussion
UC is a chronic and recurrent IBD that primarily affects the rectum and colon. The etiology and pathogenesis of UC remain to be elucidated, however, there have been several advancements in the study of intestinal microecology and immunology, suggesting that intestinal flora disorder and abnormal immune response are characteristic symptoms of UC. Bullock et al (16) reported that compared with UC patients in remission, patients with acute UC demonstrated continuous inflammation and intestinal flora disorder. These patients had significantly reduced amounts of beneficial bacteria, however, there was widespread proliferation of opportunistic and harmful bacteria in the gut. A recent study has demonstrated hat when probiotics are administered along with conventional treatment to patients with mild and moderate UC, there is a notable improvement in their condition (17). Under the combined effect of probiotics and conventional treatment, the proportion of patients with disease activity decreased by 50%, which was higher than that of patients receiving placebo and conventional treatment. In order to provide a theoretical basis for the clinical use of probiotics, the underlying mechanisms via which probiotics inhibit inflammation first require elucidation. Golden bifid is a preparation containing three prominent probiotics, Bifidobacteriumlongum, Lactobacillus bulgaricus and Streptococcus thermophilus. Golden bifid alleviates intestinal inflammation by releasing these live bacteria into the intestine, thereby increasing the number of beneficial or opportunistic bacteria that promote intestinal flora. Sang et al (18) used gavage to administer heat-inactivated probiotic VSL#3 to UC rats for seven days. The inactivated probiotic VSL#3 elicited anti-inflammatory activity in the dextran sodium sulfate-induced acute UC rat model. The probiotic VSL#3 downregulated the expression of IL-6, IL-23, and TGF-β in rat colonic tissues. In addition, a previous study described how Bifidobacterium and Lactobacillus were administered via gavage to UC rats (19). It was observed that these probiotic species reduced the expression of TLR4 in colonic rat tissues.
Presently, it has been suggested that TLRs are closely associated with the development of UC (20–22). TLR, a pattern recognition receptor, is a protein of the natural immune system. TLRs are widely expressed on cell membranes in cells that are involved in the innate and the specific immune system, indicating their importance as part of the defense mechanism of the body. TLRs first recognize the pathogen associated molecular patterns (PAMPs) and trigger a series of cascades to activate nuclear factor-κB and mitogen activated protein kinase P38, which subsequently induce the release of various cytokines, including IL-6, IL-1, IL-8, IL-10, IL-12, TNF-α, nitric oxide synthase, and B7 molecules. Thus, TLRs aggravate inflammatory reactions (23,24). Haller et al (25) has reported that peptidoglycans and lipopolysaccharides of bacteria may act as PAMPs to interact with TLR2 and TLR4, respectively. In addition to being inflammatory mediators, particularly in abnormal release of cytokines, TLRs also increase mucosal damage and disease progression (26). During the development of IBD, TNFs function beyond their pro-inflammatory activity to activate endothelial cells and induce the release of chemokines. Furthermore, they recruit neutrophils, resulting in mucosal lesions and edema. They activate coagulation and participate in the formation of granuloma (27). TNF-α is an important cytokine that causes mucosal inflammation during the development of UC. A previous study has reported that TNF-α enhances the expression of TLR-4-myeloid differentiation 2 complex in intestinal epithelial cells, thus increasing sensitivity of the host to LPS (28). This series of events aggravates the progression of UC. Canto et al (29) has demonstrated that compared with remission patients or healthy controls, 48% patients with active IBD exhibited significantly increased TNF-α expression. IL-1β is another inflammatory factor that is highly expressed in patients with UC. Rachmilewitz et al (30) has reported that in patients with acute UC, the expression of IL-1β in lesions was three-fold that observed in normal sites. As the intestinal barrier is severely damaged in patients with UC, bacterial products can easily pass from the intestine into the intestinal mucosa, triggering a series of inflammatory reactions by the immune cells.
The findings of the present study are in agreement with the results of Hausmann et al (31). In the present study, UC model rats exhibited significantly increased expression of TLR4 and TLR2, when compared with the healthy control group, indicating that TLR4 and TLR2 are important in the pathogenesis of UC. Compared with rats of group II, rats of groups III and IV exhibit significantly lower levels of pathological inflammation and expression of TLR4 and TLR2. However, the differences in the pathological inflammation or TLR4/TLR2 expression were not significant when comparing these parameters amongst rats of groups III and IV, indicating that Golden bifid had a strong inhibitory effect on the expression of TLR4 and TLR2. Furthermore, such an effect was comparable to that of TLR4mAb. Compared with the control group, rats of group II had a significantly greater concentration of TNF-α and IL-1β in their serum samples. This indicates that TNF-α and IL-1β are associated with the development of UC in rats. By contrast, compared to group II, the rats of groups III and IV exhibited a markedly lower concentration of TNF-α and IL-1β; however, there was no statistically significant difference in the concentration of TNF-α and IL-1β levels among groups III and IV. These results indicate that probiotics regulate the balance of intestinal flora, thus decreasing the occurrence of abnormal immune responses to bacterial antigens. Furthermore, probiotics either alleviate or inhibit further inflammation by suppressing the expression of TLR4. Thus, probiotics prevent the occurrence of TLR4-mediated cascades, which are otherwise induced by these inflammatory reactions.
In conclusion, the present study suggests that abnormal intestinal immune responses are associated with the development of UC. Thus, probiotics regulate the balance of intestinal flora and inhibit TLR-mediated immune response, thus alleviating the pathogenesis of intestinal inflammatory reactions.
Acknowledgements
The present study was supported by the Xinjiang Uyghur Autonomous Region Natural Science Foundation (grant no. 2013211A092).
References
- 1.Zeng Z, Zhu Z, Yang Y, Ruan W, Peng X, Su Y, Peng L, Chen J, Yin Q, Zhao C, et al. Incidence and clinical characteristics of inflammatory bowel disease in a developed region of Guangdong Province, China: A prospective population-based study. J Gastroenterol Hepatol. 2013;28:1148–1153. doi: 10.1111/jgh.12164. [DOI] [PubMed] [Google Scholar]
- 2.Sprong RC, Schonewille AJ, van der Meer R. Dietary cheese whey protein protects rats against mild dextran sulfate sodium-induced colitis: Role of mucin and microbiota. J Dairy Sci. 2010;93:1364–1371. doi: 10.3168/jds.2009-2397. [DOI] [PubMed] [Google Scholar]
- 3.Schirbel A, Fiocchi C. Inflammatory bowel disease: Established and evolving considerations on its etiopathogenesis and therapy. J Dig Dis. 2010;11:266–276. doi: 10.1111/j.1751-2980.2010.00449.x. [DOI] [PubMed] [Google Scholar]
- 4.Nagasaki A, Takahashi H, Iinuma M, Uchiyama T, Watanabe S, Koide T, Tokoro C, Inamori M, Abe Y, Nakajima A. Ulcerative colitis with multidrug-resistant Pseudomonas aeruginosa infection successfully treated with bifidobacterium. Digestion. 2010;81:204–205. doi: 10.1159/000236042. [DOI] [PubMed] [Google Scholar]
- 5.Wang LS, Li YX, Zhu HM, Zhu ZS, Ma XD. Influence of bifidobacterium DNA on PKC and NF-kappaB in murine macrophages. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2007;23:11–13. (In Chinese) [PubMed] [Google Scholar]
- 6.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Collado MC, Surono IS, Meriluoto J, Salminen S. Potential probiotic characteristics of Lactobacillus and Enterococcus strains isolated from traditional dadih fermented milk against pathogen intestinal colonization. J Food Prot. 2007;70:700–705. doi: 10.4315/0362-028X-70.3.700. [DOI] [PubMed] [Google Scholar]
- 8.Madsen KL. Inflammatory bowel disease: Lessons from the IL-10 gene-deficient mouse. Clin Invest Med. 2001;24:250–257. [PubMed] [Google Scholar]
- 9.Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut. 2003;52:827–833. doi: 10.1136/gut.52.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolova L, Drastich P, Rossmann P, Klimesova K, Tlaskalova-Hogenova H. Expression of Toll-like receptor 2 (TLR2), TLR4, and CD14 in biopsy samples of patients with inflammatory bowel diseases: Upregulated expression of TLR2 in terminal ileum of patients with ulcerative colitis. J Histochem Cytochem. 2008;56:267–274. doi: 10.1369/jhc.7A7303.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Zhang Z, Wang L, Li J, Dong L, Yue W, Chen J, Sun X, Zhong L, Sun D. TLR4 monoclonal antibody blockade suppresses dextran-sulfate-sodium-induced colitis in mice. J Gastroenterol Hepatol. 2010;25:209–214. doi: 10.1111/j.1440-1746.2009.06046.x. [DOI] [PubMed] [Google Scholar]
- 12.Fort MM, Mozaffarian A, Stöver AG, Jda S Correia, Johnson DA, Crane RT, Ulevitch RJ, Persing DH, Bielefeldt-Ohmann H, Probst P, et al. A synthetic TLR4 antagonist has anti-inflammatory effects in two murine models of inflammatory bowel disease. J Immunol. 2005;174:6416–6423. doi: 10.4049/jimmunol.174.10.6416. [DOI] [PubMed] [Google Scholar]
- 13.Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- 14.Fromowitz FB, Viola MV, Chao S, Oravez S, Mishriki Y, Finkel G, Grimson R, Lundy J. ras p21 expression in the progression of breast cancer. Hum Pathol. 1987;18:1268–1275. doi: 10.1016/S0046-8177(87)80412-4. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Method. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Bullock NR, Booth JC, Gibson GR. Comparative composition of bacteria in the human intestinal microflora during remission and active ulcerative colitis. Curr Issues Intest Microbiol. 2004;5:59–64. [PubMed] [Google Scholar]
- 17.Mardini HE, Grigorian AY. Probiotic mix VSL#3 is effective adjunctive therapy for mild to moderately active ulcerative colitis: A meta-analysis. Inflamm Bowel Dis. 2014;20:1562–1567. doi: 10.1097/MIB.0000000000000084. [DOI] [PubMed] [Google Scholar]
- 18.Sang LX, Chang B, Dai C, Gao N, Liu WX, Jiang M. Heat-killed VSL#3 ameliorates dextran sulfate sodium (DSS)-induced acute experimental colitis in rats. Int J Mol Sci. 2013;15:15–28. doi: 10.3390/ijms15010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Fu Y, Liu J, Ren HY. Impact of probiotics on toll-like receptor 4 expression in an experimental model of ulcerative colitis. J Huazhong Univ Sci Technolog Med Sci. 2013;33:661–665. doi: 10.1007/s11596-013-1177-9. [DOI] [PubMed] [Google Scholar]
- 20.Cario E. Toll-like receptors in inflammatory bowel diseases: A decade later. Inflamm Bowel Dis. 2010;16:1583–1597. doi: 10.1002/ibd.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Østvik AE, Granlund AV, Torp SH, Flatberg A, Beisvåg V, Waldum HL, Flo TH, Espevik T, Damås JK, Sandvik AK. Expression of Toll-like receptor-3 is enhanced in active inflammatory bowel disease and mediates the excessive release of lipocalin 2. Clin Exp Immunol. 2013;173:502–511. doi: 10.1111/cei.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrello S, Nicolo C, Delogu G, Pandolfi F, Ria F. TLR2: A crossroads between infections and autoimmunity? Int J Immunopathol Pharmacol. 2011;24:549–556. doi: 10.1177/039463201102400301. [DOI] [PubMed] [Google Scholar]
- 23.D'Incà R, Barollo M, Scarpa M, Grillo AR, Brun P, Vettorato MG, Castagliuolo I, Sturniolo GC. Rectal administration of Lactobacillus casei DG modifies flora composition and Toll-like receptor expression in colonic mucosa of patients with mild ulcerative colitis. Dig Dis Sci. 2011;56:1178–1187. doi: 10.1007/s10620-010-1384-1. [DOI] [PubMed] [Google Scholar]
- 24.Roberts-Thomson IC, Fon J, Uylaki W, Cummins AG, Barry S. Cells, cytokines and inflammatory bowel disease: A clinical perspective. Expert Rev Gastroenterol Hepatol. 2011;5:703–716. doi: 10.1586/egh.11.74. [DOI] [PubMed] [Google Scholar]
- 25.Haller D, Russo MP, Sartor RB, Jobin C. IKK beta and phosphatidylinositol 3-kinase/Akt participate in non-pathogenic Gram-negative enteric bacteria-induced RelA phosphorylation and NF-kappa B activation in both primary and intestinal epithelial cell lines. J Biol Chem. 2002;277:38168–38178. doi: 10.1074/jbc.M205737200. [DOI] [PubMed] [Google Scholar]
- 26.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Deventer SJ. Tumour necrosis factor and Crohn's disease. Gut. 1997;40:443–448. doi: 10.1136/gut.40.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki M, Hisamatsu T, Podolsky DK. Gamma interferon augments the intracellular pathway for lipopolysaccharide (LPS) recognition in human intestinal epithelial cells through coordinated up-regulation of LPS uptake and expression of the intracellular Toll-like receptor 4-MD-2 complex. Infect Immun. 2003;71:3503–3511. doi: 10.1128/IAI.71.6.3503-3511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantó E, Ricart E, Monfort D, González-Juan D, Balanzó J, Rodríguez-Sánchez JL, Vidal S. TNF alpha production to TLR2 ligands in active IBD patients. Clin Immunol. 2006;119:156–165. doi: 10.1016/j.clim.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Rachmilewitz D, Karmeli F, Shteingart S, Lee J, Takabayashi K, Raz E. Immunostimulatory oligonucleotides inhibit colonic proinflammatory cytokine production in ulcerative colitis. Inflamm Bowel Dis. 2006;12:339–345. doi: 10.1097/01.MIB.0000217335.30689.77. [DOI] [PubMed] [Google Scholar]
- 31.Hausmann M, Kiessling S, Mestermann S, Webb G, Spöttl T, Andus T, Schölmerich J, Herfarth H, Ray K, Falk W, Rogler G. Toll-like receptors 2 and 4 are up-regulated during intestinal inflammation. Gastroenterology. 2002;122:1987–2000. doi: 10.1053/gast.2002.33662. [DOI] [PubMed] [Google Scholar]


