Abstract
The metal-affinities of metal-sensing transcriptional regulators co-vary with cellular metal concentrations over more than 12 orders of magnitude. To understand the cause of this relationship, we determined the structure of the Ni(II)-sensor InrS then created cyanobacteria (Synechocystis PCC 6803) in which transcription of genes encoding a Ni(II)-exporter and a Ni(II)-importer were controlled by InrS variants with weaker Ni(II)-affinities. Variant strains were sensitive to elevated nickel and contained more nickel but the increase was small compared to the change in Ni(II)-affinity. All of the variant-sensors retained the allosteric mechanism which inhibits DNA binding upon metal binding but a response to nickel in vivo was only observed when the sensitivity was set to respond within a relatively narrow (less than 2 orders of magnitude) range of nickel-concentrations. The Ni(II)-affinity of InrS is attuned to cellular metal concentrations rather than the converse.
The available concentration of each metal at the locations where nascent proteins fold is important for correct protein-metalation1. Biological catalysis uses metals in almost a half of enzymes with each metal selected to match the needs of a reaction2,3. However, metals such as Ni(II) and Zn(II) form thermodynamically more stable complexes with proteins than do others such as Mg(II)4. This creates a challenge for cells to populate some proteins with tight-binding metals and others with weaker-binding ones. Crucially, metals in cells are maintained in an order of available concentrations which is the inverse of the order of stabilities of their metal-ligand complexes: Mg(II) at milli-, but Ni(II) and Zn(II) at sub nano-molar buffered concentrations1,5–7. These observations highlight the importance of the mechanisms that control these vital set-points for intracellular metal concentrations.
Bacterial metal-sensors (commonly DNA-binding, metal-responsive transcriptional regulators) modulate expression of genes encoding proteins involved in metal-import, metal-export, metal-storage or metal-usage when metal levels deviate from some optimal set-point6,8. The metal-affinities of bacterial metal-sensors correlate over multiple orders of magnitude with available intracellular metal concentrations7, with a long standing hypothesis that these affinities define the set-point for buffered metal concentrations6,8–10. This suggests that metal availability within a cell might be altered by adjusting the metal-affinity of a metal-sensor. The cyanobacterial (Synechocystis PCC 6803) Ni(II) sensor InrS is known to de-repress expression of a gene encoding a nickel exporter (nrsD) in response to elevated Ni(II)11. This sensor was chosen to examine the relationship between intracellular metal concentrations and metal-sensor metal-affinity.
InrS is a member of the CsoR-RcnR family of DNA-binding, metal-responsive transcriptional regulators. The founding members of this family respond to Cu(I) or Ni(II)/Co(II)12,13. Promoter-targets that have been characterised to date are repressed in response to protein binding11–21. Structures are known for CsoR Cu(I)-sensors12,17,20,21, but there are no structures for RcnR-like Ni(II) and Co(II) sensors.
Here we report the structure of Ni(II)-sensing InrS and discover that, in addition to repressing expression of nrsD, InrS also binds upstream of the nik operon where it acts positively on expression of genes encoding the nickel-import machinery (including nikM). The abundance of nikM and nrsD transcripts, along with nickel contents, were monitored in cells in which Ni(II)-homeostasis was controlled by InrS variants with weakened KNi(II). Competition for Ni(II) between either InrS or a variant with 20-fold weaker KNi(II), and a buffer matching the cellular concentrations of the most abundant free amino acids plus glutathione, was analysed. The purpose of this work was to understand why the metal-affinities of metal-sensors co-vary with cellular metal concentrations: Specifically we aimed to discover whether or not these metal-affinities are the reference points that govern available intracellular metal concentrations, and found that the opposite was true.
Results
Structure of InrS
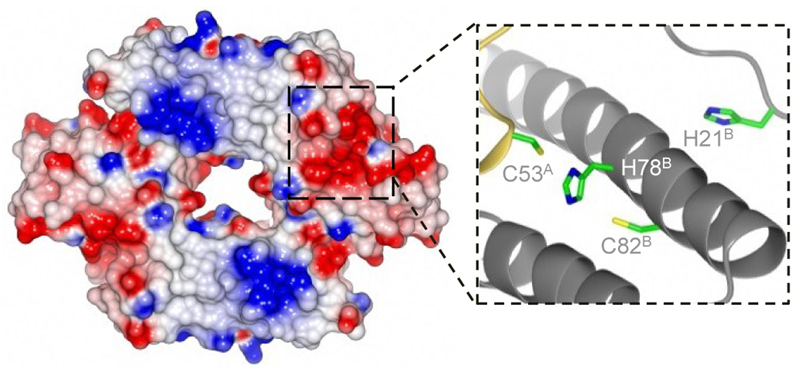
To aid the design of InrS variants with weaker KNi(II), diffraction quality crystals of apo-InrS were produced and a structure determined to 2.4 Å resolution (Fig. 1; Supplementary Results, Supplementary Table 1 and Supplementary Fig. 1a). C53, C82 and H78 were found to be proximal in the determined structure and are implicated in Ni(II)-binding by previous site directed mutagenesis22. InrS and Escherichia coli RcnR were aligned with cyanobacterial InrS homologues deduced to detect Ni(II) (Supplementary Fig. 1b). InrS H21, which due to its position on the flexible N-terminal extension could approach the Ni(II)-binding site comprising C53, H78 and C82 (Fig. 1), aligned with the previously assigned metal-ligand H3 of E. coli RcnR23,24. A histidine residue at this position was also found to be conserved across all cyanobacterial InrS-like sequences (Supplementary Fig. 1b). Additionally, there are multiple histidine residues located between the N-terminus and H21 which could also approach the Ni(II)-binding site. Folding of the N-terminus over the metal-coordination site is also observed for Cu(I)-sensing CsoR from Geobacillus thermodenitrificans21. H21 and H78 were selected with the aim of generating InrS variants of weaker Ni(II)-affinities.
Figure 1. DNA-binding Ni(II)-sensor InrS.
Electrostatic surface of InrS tetramer and candidate Ni(II)-binding site with superscript letters referring to chains A and B. A positive patch (necessary for interaction of CsoR with DNA41,42), runs diagonally across the surface of InrS (blue).
Variants with weaker KNi(II) retained allostery
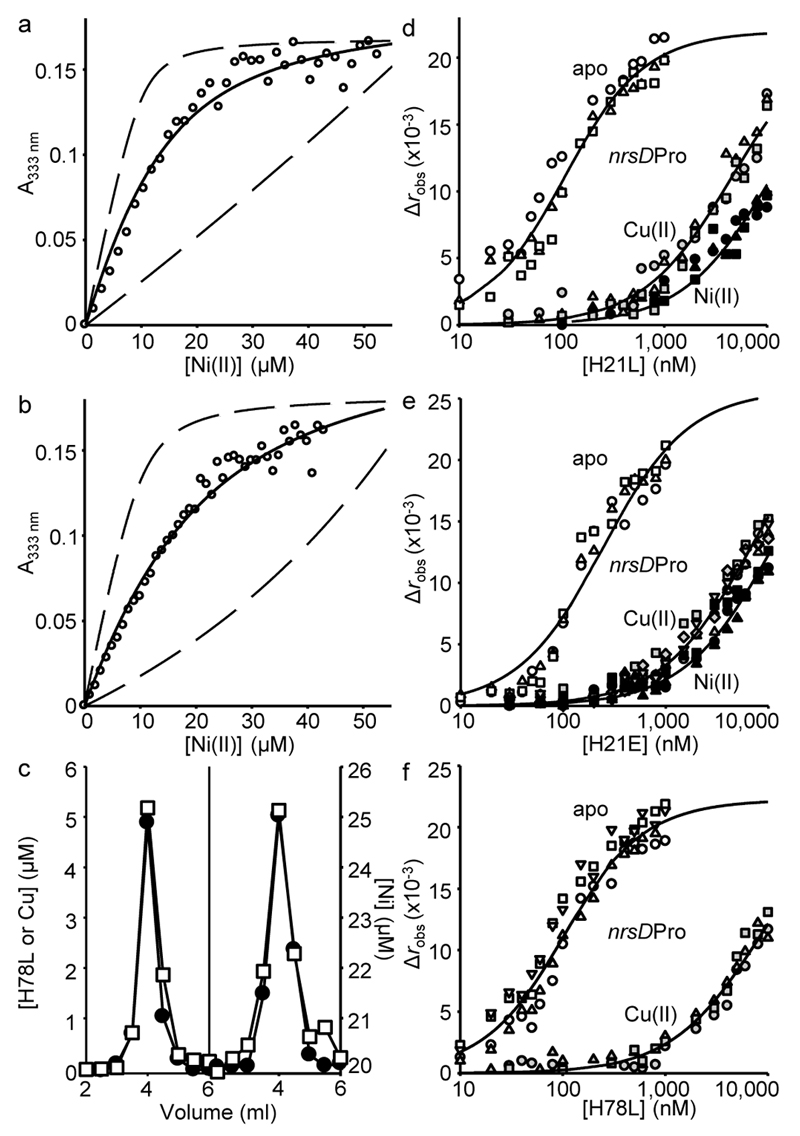
The Ni(II)-affinities of H21L, H21E and wild type InrS were determined by competition against ethylene glycol tetraacetic acid (EGTA)25. H21L and H21E exhibited Ni(II)-dependent UV-visible spectra equivalent to wild type InrS (Supplementary Fig. 2a,b)11, making it possible to monitor metal-partitioning between EGTA and protein via an intense Ni(II)-dependent feature at 333 nm (Fig. 2a,b and Supplementary Fig. 2c). Data were fit to a model describing competition for four Ni(II) ions per tetramer (Supplementary Table 2), and simulated curves with affinities ten times tighter or ten times weaker than the fitted values confirmed that the data were within the limits of the assay (Fig. 2a,b and Supplementary Fig. 2c). Retention of Ni(II) by variant H78L during gel filtration chromatography required 20 μM Ni(II) in the elution buffer (Fig. 2c)22, revealing a weak 10–5 M KNi(II) (Supplementary Table 2). The larger effect on KNi(II) of H78L relative to H21L or H21E, could be explained either by the recruitment of alternative amino-terminal ligands in the H21 variants or solely indirect (such as second coordination sphere) effects of the H21 substitutions on the primary coordination sphere.
Figure 2. Weak Ni(II)-affinity sensors with altered predicted sensitivities.
a. Representative (n = 3) H21L absorbance upon titration of H21L (10 μM protomer) and EGTA (50 μM) with NiCl2. Solid line is a fit to a model describing competition from H21L for one molar equivalent of Ni(II), revealing KNi(II)= 5.5(±1) × 10–11 M. Dashed lines are simulated curves with KNi(II) 10-fold tighter and 10-fold weaker than the fitted value. b. As in ‘a’ with H21E ([H21E] = 10 μM protomer, [EGTA] = 50 μM), KNi(II) = 8.6(±2) × 10–11 M (n = 3). c. Elution profile of H78L (10 μM protomer) incubated with CuCl2 (15 μM) before (left), or with NiCl2 (20 μM) during (right), fractionation by size exclusion chromatography. Protein (closed symbols), metal (open symbols). Note the 20 μM [Ni] baseline when NiCl2 was included in the buffer. d. Anisotropy change upon titration of nrsDProFA (10 nM) with either H21L in 5 mM EDTA (open symbols), Ni(II)-H21L (closed symbols) or Cu(II)-H21L (gray symbols). Symbol shapes represent individual experiments (n = 3). Data were fit to a model describing a 1:1 InrS tetramer (non-dissociable):DNA stoichiometry and lines are simulated curves using the mean KDNA across the experiments shown (Supplementary Table 2). e. As in ‘d’ with H21E (apo n = 3, Cu(II) n = 3, Ni(II) n = 5). f. As in ‘d’ with H78L (Cu(II) n = 3, apo n = 4).
Affinities of InrS variants for DNA, and effects of metal on DNA-binding, were determined by fluorescence anisotropy using a fluorescently labelled fragment of the nrsD promoter11, which confirmed that all three variants bind DNA and undergo an allosteric change upon metal-binding that weakens DNA binding (Fig. 2d–f and Supplementary Table 2)22. Due to the weak KNi(II) of H78L, Cu(II), with tighter affinity (Fig. 2c), was used as a surrogate. The free energy coupling metal-binding to DNA-binding, ΔGC, was calculated for each variant (Supplementary Table 2). KDNA was similar for the metal-bound forms of all three proteins showing that the mutations did not interfere with the allosteric mechanism. The apo-form of H21E had a significantly (~6 fold) weaker KDNA than InrS, perhaps resulting from the extra negative-charge. Mean DNA- and Ni(II)-affinities from (at least) triplicate analyses were used to model nrsD promoter occupancy with InrS as a function of Ni(II) concentration (Supplementary Fig. 3). The concentration of Ni(II) required to displace 95% of InrS from the nrsD promoter was estimated to be 3 × 10–10 M while equivalent promoter occupancy (and hence de-repression) by H21E and H21L would require 7 and 10 times more Ni(II), respectively (Supplementary Table 2 and Supplementary Fig. 3). Equivalent clearance of H78L from the nrsD promoter was estimated to require 2.3 × 106 times more Ni(II) if it is assumed that Ni(II), in common with Cu(II) (Fig. 2f), also triggers allostery in H78L.
InrS KNi(II) is near a limit for regulating export
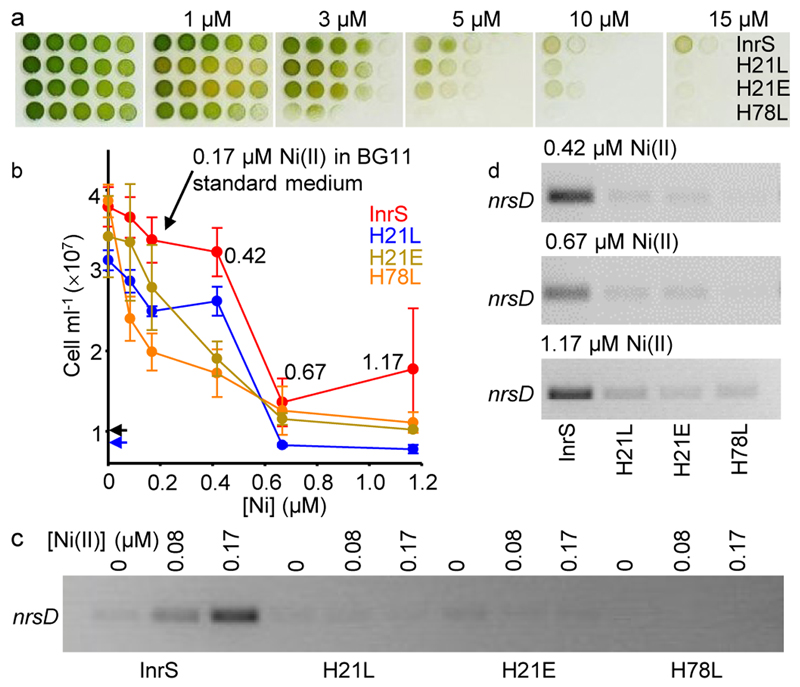
Cyanobacterial strains InrS, H21L, H21E and H78L were created in which genes encoding these sensors were integrated into ΔinrS11, which was also inactivated in a second Ni(II)-export system22,26 (Supplementary Fig. 4). All three variants (H21L, H21E and H78L) were more sensitive to Ni(II) relative to the InrS strain (Fig. 3a). In liquid culture, Ni(II) concentrations up to the level in standard BG11 cyanobacterial culture medium (0.17 μM) did not significantly inhibit the growth of strain InrS, while 1.17 μM Ni(II) was bacteriostatic to all strains expressing variants and significantly inhibited growth of the InrS strain (Fig. 3b).
Figure 3. The tight Ni(II)-affinity of InrS was needed to regulate nrsD.
a. Spotting assay showing growth of strains InrS, H21L, H21E and H78L on standard BG11 medium plus the indicated [NiSO4]. b. Growth after 48 h in liquid medium (BG11 without NiSO4) plus the indicated [NiSO4]. Starting inoculum for H21L (blue) and other strains (black) indicated by arrows. Values are means of triplicate biological replicates with error bars representing ± one standard deviation. c. nrsD transcript abundance in media containing Ni(II) concentrations non-inhibitory to InrS cells (48 h) (loading controls shown in supplementary figure 4, full gel shown in supplementary figure 5). d. nrsD transcript abundance in media containing inhibitory Ni(II) concentrations (48 h) (all loading controls shown in supplementary figure 4, full gels shown in supplementary figures 6-8).
In response to increasing [Ni(II)] (up to 0.17 μM), nrsD transcripts accumulated in the InrS strain but there was no Ni(II)-dependent increase in nrsD transcript abundance in strains containing InrS variants with weaker KNi(II) (Fig. 3c; Supplementary Fig. 5). Inhibitory Ni(II)-concentrations also failed to induce accumulation of nrsD transcripts in the variant strains (Fig. 3d; Supplementary Figs.6-8). Constitutive DNA-binding by H78L must be disadvantageous since a spontaneous mutation was selected after prolonged culture of this strain, introducing a premature stop codon and conferring basal expression of nrsD (Supplementary Figs. 9-11). Taken together, these data show that the weakest KNi(II) allowing regulation of nrsD by InrS was close to the 2.8 × 10–12 M KNi(II) of InrS, such that even the modest weakening of KNi(II) in H21L and H21E (Supplementary Table 2), did not allow Ni(II)-dependent nrsD-regulation in living cells (Fig. 3).
InrS also regulates Ni(II)-import
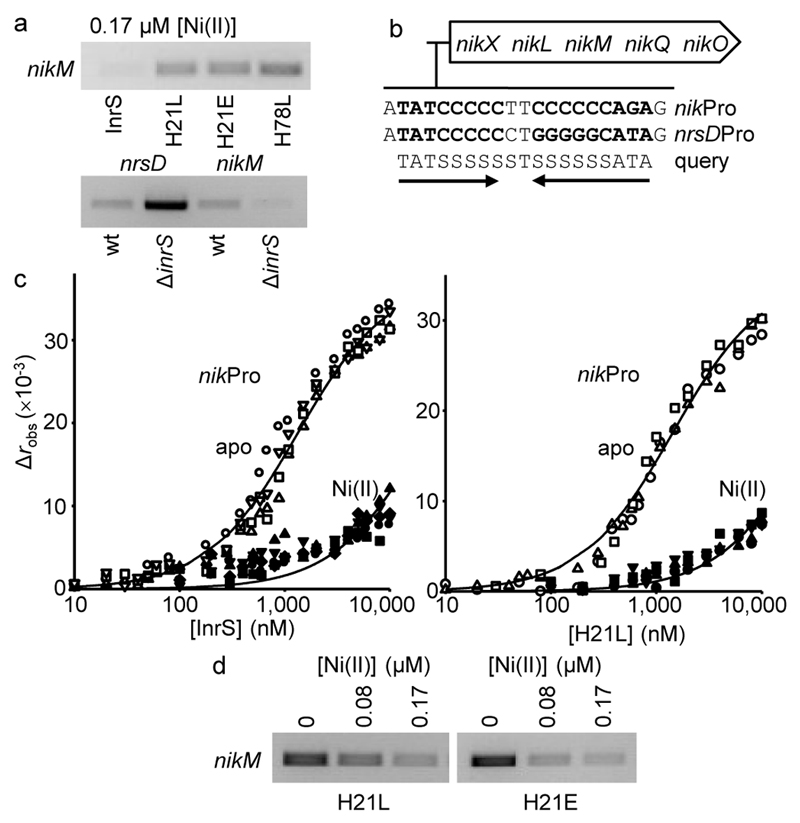
NikM is a component of the Nik-importer of nickel27. Surprisingly, strains expressing InrS variants with weaker KNi(II) contained more nikM transcripts (not less as expected due to greater repression of Ni(II)-export, see figure 3) relative to the InrS strain grown in standard BG11 medium (0.17 μM Ni(II)) (Fig. 4a). Conversely, nikM transcripts were less abundant in cells missing inrS (not more as expected due to de-repression of Ni(II)-export) (Fig. 4a). The Synechocystis genome does not encode NikR but regulation of nikM via some unknown second sensor of cellular Ni(II) would be expected to give the exact opposite patterns of nikM expression to those observed in Figure 4a. A candidate nucleotide binding-site for InrS was identified upstream of the nik operon (Fig. 4b), and binding of InrS, H21L and H21E, was confirmed by fluorescence anisotropy (Fig. 4c and Supplementary Fig. 12). Mechanisms by which a metal-sensing, DNA-binding protein can repress transcription on some promoters while directly activating others have been described28, but this is the first evidence of a member of this family of metal-sensors directly enhancing gene expression as well as repressing.
Figure 4. InrS also regulates nikM.
a. nikM transcript abundance in strains grown in standard BG11 medium (48 h). wt = wild type Synechocystis, ΔinrS = inrS deletion mutant11. nrsD transcript abundance was included to contrast with nikM transcript abundance in the wt versus ΔinrS comparison (all loading controls shown in supplementary figure 4, full gels shown in supplementary figures 10 and 11). b. The nik operon of Synechocystis with the InrS binding site of the nrsD promoter aligned with a binding site upstream of the nik operon (136 nt from the nikX start codon) plus a query sequence which retrieves only these two sequences (within 150 nucleotides of start codons allowing two mismatches), where S = G or C. c. Anisotropy change on titration of nikProFA (the identified InrS binding site plus 7 flanking base pairs) (10 nM) with InrS (apo n = 4, Ni(II) n = 5) or H21L (apo n = 3, Ni(II) n = 4) in 5 mM EDTA (open symbols) or Ni(II)-bound forms (closed symbols). Symbol shapes represent individual experiments. Simulated curves generated as in Fig. 2d–f. d. nikM transcript abundance in H21L and H21E variants in response to Ni(II) (48 h) (all loading controls shown in supplementary figure 4, full gels shown in supplementary figure 13).
Binding of InrS, H21L and H21E to the nik promoter was impaired by Ni(II) (Fig. 4c and Supplementary Fig 12). The abundance of nikM transcripts declined in H21L and H21E strains with the addition of non-lethal concentrations of Ni(II) (Fig. 4d and Supplementary Fig. 13), consistent with dissociation of a positive regulator. A weaker InrS KDNA on the nik promoter would lower the set point for regulation of nikM relative to nrsD (Supplementary Fig. 3), in this way import can be switched off at lower levels of Ni(II) than needed to trigger export. The theoretical [Ni(II)] required to displace InrS, H21E and H21L from the nikM promoter were calculated in the same manner as the set points for the nrsD promoter to be 4 × 10–12, 1.5 × 10–11 and 6 × 10–11 M respectively (Supplementary Table 2). Expression of nikM in cultures containing wild type InrS was variable, probably because the set point is sufficiently low that the presence of trace levels of Ni(II) in culture media make it challenging to observe Ni(II)-responsive regulation of the importer in cyanobacteria adapted to survive in fresh water.
Cellular nickel did not co-vary with altered KNi(II)
It is known that inactivation of metal-sensing transcriptional regulators can alter cell metal content11,13,29,30. If the metal-affinities of metal-sensors govern the buffered intracellular metal-concentrations, then variants in which Ni(II)-import and/or -export are controlled by InrS with weaker KNi(II) should contain proportionately more nickel. The number of nickel ions per cell increased by 1.2- and 1.5-fold in H21E and H21L variants grown for 24 h in 0.42 μM nickel (Table 1), but the [Ni(II)] set point for these variants on the nik promoter was 4- and 15-fold greater than for wild-type InrS, and 7- and 10-fold greater on the nrsD promoter (Supplementary Table 2). Any increase in nickel in the H78L strain should represent the maximum achievable (the hypothetical set point is 2.3 million fold higher than InrS, Supplementary Table 2) but in four growth conditions the increases were only 0-, 2.4-, 9.4- and 4.1-fold (Table 1). The fluorophore Newport Green was used to visualise exchangeable (rather than total) Ni(II). An approximate two-fold increase in signal was observed in the H78L strain grown in 0.67 μM Ni(II) relative to the InrS strain (Supplementary Fig. 14), noting that Newport Green is also influenced by pH. In other cyanobacterial species nickel supplementation enhances hydrogenase activity31, but there was no difference in hydrogenase activity in H78L variants and crucially nickel supplementation also had no effect on activity (Supplementary Fig. 15), hence in this strain there was no scope for engineering hydrogenase activity solely by increasing nickel content. In summary, weakening KNi(II) of InrS increased the number of Ni(II) ions per cell but the increase in total (or buffered) Ni(II) was less than one order of magnitude and ceased to correlate with KNi(II).
Table 1. Nickel contents of Synechocystis strains.
| 24 h exposure at early-log | ||
|---|---|---|
| [Ni]exogenous (μM) | Strain | Ni ions cell-1 (× 105) |
| 0.42* | wt | 5.6(±0.3) |
| ΔinrS | 4.5(±0.1) | |
| ΔPnrs | 6.0(±0.1) | |
| ΔinrSΔPnrs | 4.4(±0.3) | |
| InrS | 5.1(±0.2) | |
| H21E | 6.1(±0.0) | |
| H21L | 7.4(±0.1) | |
| H78L | 12.1(±0.3) | |
| 12 h exposure at mid-log | ||
| 0 | InrS | 0.3(±0.1) |
| H78L | 0.2(±0.0) | |
| 0.17 | InrS | 0.5(±0.2) |
| H78L | 4.7(±0.7) | |
| 0.67† | InrS | 1.8(±0.9) |
| H78L | 7.4(±0.8) | |
cells cultured in standard BG11 with addition of 0.25 μM NiSO4 24 h prior to harvest.
Cells cultured in standard BG11 with addition of 0.5 μM NiSO4 24 h prior to harvest.
InrS competes for Ni(II) with metabolites
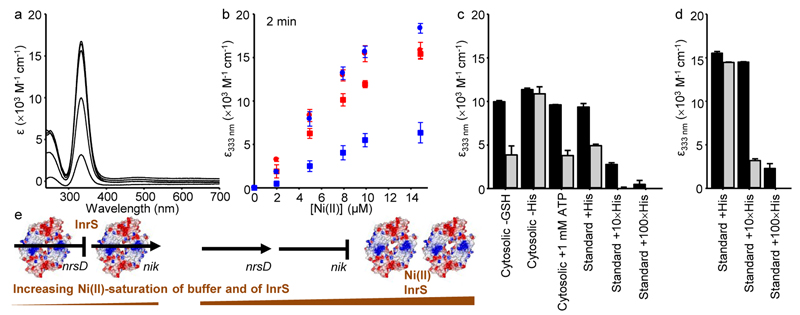
The cytoplasm enables Ni(II)-InrS to form, but the cytoplasm did not allow formation of sufficient Ni(II)-species of the InrS variants to de-repress nrsD, and these cells became hypersensitive to elevated nickel (Fig. 3 and Supplementary Fig. 9). To explore competition with cytosolic components, cyanobacterial extracts were analysed for amino acid and glutathione content (Supplementary Table 3), then equivalent solutions of the most abundant molecules were made from pure reagents (with 10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl). Ni(II)-dependent spectra of InrS (Fig. 5a), and H21L (the variant with the closest KNi(II) to wild type InrS), were monitored upon titration with Ni(II) in this (partial) cell-like buffer. At equilibrium Ni(II) partitioned to the proteins as observed in a standard buffer (10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl) (Supplementary Fig. 16a). Equilibration times were slower with H21L as evident at 2 min post-addition of protein to Ni(II) containing buffer (Fig. 5b). Exclusion of glutathione from the cell-like buffer had negligible effect on Ni(II)-binding by either protein, but exclusion of histidine led to loss of competition and standard buffer containing the same concentration of histidine alone restored competition (Fig. 5c). A known ATP-histidine complex has a high affinity for Ni(II)32, but competition was unaffected by addition of ATP at concentrations known to occur in vivo (Fig. 5c). A cellular concentration of 2.8 × 10–8 M InrS tetramers (assuming 100 InrS tetramers per 6 fl cell) is two orders of magnitude less than used in figure 5. Experiments were therefore repeated with 10 and 100 times more histidine, and these assays showed competition for Ni(II) at equilibrium and discerned H21L from InrS (Fig. 5d). A 10-fold increase in histidine largely out-competed H21L but not InrS for Ni(II), and a 100-fold increase fully out-competed H21L and largely out-competed InrS. Thus KNi(II) of InrS relative to histidine is finely poised for competition at the molar ratios which occur in vivo. Importantly, H21L was less able than InrS to compete with histidine at cellular concentrations (Fig. 5d). Figure 5e summarises InrS-mediated Ni(II) homeostasis.
Figure 5. InrS competes for Ni(II) with metabolites at cellular concentrations.
a. UV-vis apo-subtracted spectra of InrS (10 μM protomer) titrated with NiCl2 (up to 14.85 μM) in cytosolic-matched buffer (Supplementary Table 3, with 10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl). b. Extinction coefficient of InrS (red) or H21L (blue) at 333 nm 2 min post-addition of protein (10 μM protomer) to cytosolic-matched buffer (squares), or standard buffer (circles) containing NiCl2 (all values are means of triplicate analyses, error bars represent ± one standard deviation). c. Extinction coefficient of InrS (black bars) or H21L (gray bars) at 333 nm 2 min post-addition of protein (10 μM protomer) to cytosolic-like or standard buffer variants (where 1 × His = 45 μM, GSH = reduced glutathione) containing 7.92 μM NiCl2 (all values are means of triplicate analyses with error bars showing one standard deviation, note that the values for H21L in standard buffer with 100 × His are too small to be visible). d. As in ‘c’ but at equilibrium, note that the values for H21L in standard buffer with 100 × His are too small to be visible (all values are means of triplicate analyses with error bars showing one standard deviation). e. Occupancy of nrsD (regulating Ni(II)-export) and nik (regulating Ni(II)-import) promoters (black lines or arrows) by InrS as a function of Ni(II)-saturation of the buffer (including His) and hence of InrS. Arrows indicate transcription: InrS represses nrsD and acts positively on nik. A weaker KDNA on the nik promoter lowers the set point for regulation.
Discussion
The observations described here define a tight range of intracellular [Ni(II)] within which the sensitivity of InrS must be tuned to sense fluctuations in [Ni(II)] and modulate Ni(II)-homeostasis in live cells. The calculated set points for all three promoters that were observed to respond to Ni(II) in vivo (InrS on the nrsD promoter, H21L and H21E on the nik promoter) all cluster around 1 × 10–10 M. Weakening KNi(II) by twenty-fold in variant H21L (which combined with H21L KDNA and ΔGC, increased the [Ni(II)] needed for de-repression of nrsD by ten-fold) led to loss of Ni(II)-dependent accumulation of nrsD transcripts. Loss of nrsD expression in the H21E strain reduced this margin to seven-fold. Notably, both H21L and H21E variants still regulated nikM, where a weaker KDNA lowered the set point for sensing. All of the variant strains were more sensitive to elevated [Ni(II)] consistent with the accumulation of Ni(II) above the tuneable range inhibiting growth. Importantly, at tolerable [Ni(II)] the increase in cellular nickel content in all of the variant strains was much less than the change in set point of InrS. Thus, KNi(II) of InrS is not the point of reference that directly governs cellular [Ni(II)]. Rather, KNi(II) of InrS must be tuned near to the cellular [Ni(II)] in order to respond.
Histidine competed with InrS for Ni(II) at molar ratios (InrS:histidine) similar to those found in cyanobacteria (Fig. 5). Altering the ratio by ten-fold largely inverted the outcome of this finely poised competition. A lower histidine concentration which failed to compete with InrS still out competed H21L. Thus, an intracellular metal-buffer could define the range within which the metal-affinity of a metal-sensor must be tuned (Supplementary Fig. 17). Some components of the buffered pools for different metals are known but more await discovery, noting that macromolecules with labile metal-sites will also contribute to metal buffering in vivo33–35. Some sensors may have allosterically effective sites of differing KMetal generating more than one set point, as for Zn(II) and Zur36,37. Pathogens are exposed to metal fluctuations as a function of host nutritional immunity38–40, and these organisms may contain additional sensory-sites attuned to such transient metal pulses.
The Irving-Williams series describes a generic order of metal-binding affinities in the absence of steric selection (from weak to tight, Mg(II) < Mn(II) < Fe(II) < Co(II) < Ni(II) < Cu(II) [Cu(I)] > Zn(II))4,5,7, creating the challenge to correctly populate some proteins with tight-binding metals and others with weaker-binding ones. This challenge is partly met by intracellular metal-availabilities being held to the inverse of this affinity series1,5–7. Supplementary figure 17 shows how the metal-affinities of bacterial metal-sensors co-vary with these available metal concentrations over multiple orders of magnitude. Because the polydisperse buffer of the cytoplasm provides a multitude of sulfur, nitrogen and oxygen ligands that can be organised into diverse geometries with negligible steric selection, it represents a perfect medium for the Irving-Williams series. Metals that form more stable complexes will inevitably be buffered to lower concentrations than those forming weaker complexes. The activities of metal-transporters will be optimised to sustain these concentrations. Metal-sensors are tuned to these concentrations, to help to avoid saturation or depletion of the buffer, thereby creating the correlation in supplementary figure 17.
Online Methods
Site directed mutagenesis and purification of InrS variants
Site directed mutagenesis to create the H21E InrS variant was carried out as previously described22 using oligonucleotides 1 and 2 with plasmids sequenced to confirm their identity (Supplementary Fig. 18, Supplementary Table 4). The insert in all plasmids matched the sequence of inrS other than the introduced mutations in H21L (CAC → CTG), H21E (CAC → GAG) and H78L (CAC → CTG). All other InrS variants were generated previously22. InrS variants were expressed and purified as previously described11. InrS variants were transferred to an anaerobic glovebox applied to a 1 ml heparin column (GE Healthcare) equilibrated with 10 mM HEPES pH 7.8, 300 mM NaCl, 10 mM DTT, 10 mM EDTA. Inside the glovebox the column was washed with 20 column volumes of 10 mM HEPES pH 7, 50 mM NaCl, 200 mM KCl before elution with 10 mM HEPES pH 7, 200 mM NaCl, 800 mM KCl (both buffers chelex-treated and N2-purged). Protein concentration, reduced thiol content and metal content of anaerobic protein samples were assayed as described previously11. All proteins were routinely found to be >90% metal free and >95% reduced. All in vitro experiments involving UV-visible spectroscopy or fluorescence anisotropy were carried out under anaerobic conditions using chelex-treated and N2-purged buffers.
Determination of InrS crystal structure
InrS in 10 mM HEPES pH 7.8, 800 mM NaCl, 5 mM EDTA, 5 mM DTT was concentrated to approximately 7.3 mg ml–1 by centrifugation in a 10 kDa cut-off centrifugal concentrator. Sitting drop crystallization trays were prepared using a Screenmaker (Innovadyne) with drop ratios of protein to reservoir of 1:1 and 2:1, using a range of commercially available crystallization kits. Small crystals were obtained under a range of high-molecular weight polyethylene glycol (PEG) conditions. The best crystals were obtained with 0.2 M MgCl2, 0.1 M Tris pH 8.5, 20% w/v PEG 8000 from the JCSG+ screen (Molecular Dimensions)43. All crystals were mounted in loops and frozen in liquid nitrogen. All diffraction data were collected at beam line I02 (λ = 0.9795 Å) at the Diamond Light Source (T = 100 K, PILATUS pixel detector44). The data were processed with XDS45, and the structure was solved with PHASER46 using Mycobacterium tuberculosis CsoR (PDB entry 2HH7)12 and refined with REFMAC47, part of the CCP4 package48. The model contains two independent molecules in the asymmetric unit where N-terminal residues 1–21 (chain A) and 1–18 (chain B) are disordered, and hence not included. Surface residues that showed weak electron density for the side chains were refined as alanine. The protein model exhibits excellent geometry with all residues in the preferred region of the Ramanchandran plot. Coordinates and structure factors have been deposited with the PDB (www.rcsb.org) under accession code 5FMN.
Determination of Ni(II)-affinities
Increasing concentrations of NiCl2 were added to a solution of 10 mM HEPES pH 7, 100 mM NaCl, 400 mM KCl and variable [EGTA] before adding InrS or one of its variants to achieve the final concentrations stated in the figure legends. Exchange reactions were allowed to equilibrate overnight at room temperature before recording absorption intensity at 333 nm using a λ35 UV-vis spectrophotometer (Perkin Elmer). Data were fit to a model describing protein-competition for one molar equivalent of Ni(II) (four sites per tetramer) using Dynafit49 to determine Ni(II)-binding constants, with mean and standard deviation values determined from triplicate analyses. EGTA KNi(II) = 4.98 × 10–10 M at pH 7 (determined using Schwarzenbach’s α co-efficient method11,25).
Cu(II)- and Ni(II)-H78L co-migration by size exclusion chromatography
For Cu(II), H78L was pre-incubated with 1.5 molar equivalents of CuSO4 for 30 min then applied to Sephadex G75 (PD10 column, GE Healthcare) equilibrated with 10 mM HEPES pH 7, 100 mM NaCl, 400 mM KCl then eluted with the same buffer. For Ni(II), H78L was applied to Sephadex G75 equilibrated with 10 mM HEPES pH 7, 100 mM NaCl, 400 mM KCl, 20 μM NiCl2 then eluted with the same buffer. Fractions were analysed for metal by ICP-MS and for protein by Bradford assay.
Fluorescence anisotropy
InrS and its variants were prepared as described previously22 before titrating against hexachlorofluorescein labelled nrsDProFA or nikProFA (produced as for nrsDProFA11 using oligonucleotides 3 and 4, Supplementary Table 4) in 10 mM HEPES pH 7, 60 mM NaCl, 240 mM KCl with or without 5 mM EDTA depending on whether the InrS variant was apo or metal-loaded. Changes in anisotropy were measured using a modified Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies), with settings and data analysis performed as previously described22. Mean and standard deviation values determined from at least triplicate analyses with ‘n’ specified for each experiment.
Mathematical modelling
The model shown in Supplementary Figure 3 was produced as previously described15. An experimentally determined (Casy Cell Counter TT, Innovatis) cell volume of 6 fl was used in calculations along with determined KDNA and KNi(II) (Supplementary Table 2), and an assumption of 2 recognition sites and 100 tetramers per cell. The indicated set points were determined based on [Ni(II)] required to reduce DNA occupancy below 0.037 (95% reduction of InrS DNA occupancy on the nrsD promoter, equivalent to 50% occupancy of the nik promoter). The tuneable InrS KNi(II) range shown in supplementary figure 17 was determined by calculating InrS KNi(II) required to give a DNA occupancy of 0.037 on the nrsD promoter at [Ni(II)]buffered of 1.5 × 10–11 M and 3 × 10–10 M (the responsive set points of H21E on the nik promoter and InrS on the nrsD promoter).
Construction of strains expressing inrS variants
EcoRI excised, kanamycin (Km) resistance gene-disrupted nrs promoter region from pGEMPnrsBACD∷Km22 was used to transform ΔinrS cells11 to Km resistance, creating ΔinrS ΔPnrs. Deletion of the nrs promoter by insertion of the Km cassette and segregation to all chromosomal copies was confirmed by PCR with oligonucleotides 5 and 6.
Oligonucleotides 7 (containing a XhoI site) and 8 (containing XhoI and BamHI sites) were used to amplify a 717 bp fragment comprising the inrS gene and promoter from Synechocystis genomic DNA and the product was ligated to pGEM-T Easy vector (Promega). A streptomycin (Sm) and spectinomycin (Sp) cassette excised with BamHI from plasmid pCSE12050 was then inserted to create pGEMinrSregion∷SmSp cassette.
Oligonucleotides 9 and 10 were used to amplify a 1.6 kb internal fragment of slr0168 from Synechocystis genomic DNA51,52 and the product ligated to pGEM-T Easy vector. Oligonucleotides 11 and 12 were used to introduce a XhoI site into the slr0168 fragment, generating plasmid pGEMslr0168. A XhoI fragment from pGEMinrSregion∷SmSp (containing inrS, inrS-promoter and Sm/Sp cassette) was ligated to plasmid pGEMslr0168. The inrS gene in the resulting plasmid was modified by site directed mutagenesis to generate inrS variants using oligonucleotides 13 and 14 (H21L), 1 and 2 (H21E), and 15 and 16 (H78L). All plasmids were sequenced using oligonucleotides 17, 18 and 19. Plasmids containing disrupted slr0168 were incubated with the ΔinrS ΔPnrs deletion strain to transform the cells to Sm and Sp resistance. Transformants were selected on BG11 plates (5 μg ml-1 Sm, 5 μg ml-1 Sp). Insertion of the inrS variant plasmids into the slr0168 locus region, and segregation to all chromosomal copies, was confirmed by PCR using oligonucleotides 20, 21 and 22. Gels were imaged with a Gel Doc XR Gel Documentation System (Bio Rad).
Spotting assays and liquid growth experiments
For spotting assays exponentially growing cells were prepared at OD800 of 5, 1, 0.2, 0.04 and 0.008 and 5 µl of each cell suspension was spotted on BG11 (which contains 0.17 μM NiSO4) plates supplemented with the stated NiSO4 concentration. Plates were incubated for 6 days before imaging. Trends were repeated with biological replicates on 3 occasions. For liquid growth experiments exponentially growing cells were inoculated to OD800 of 0.075 with the stated NiSO4 concentrations. For all strains other than H21L this corresponds to ~1 × 107 cell ml-1, due to the slightly larger volume of H21L cells this corresponds to ~8.5 × 106 cell ml–1. Cells were cultured for 48 h before recording cell ml–1 (Casy Cell Counter TT). Mean and standard deviation values determined from triplicate biological analyses.
Isolation of RNA and reverse transcriptase PCR
Exponentially growing cells were inoculated to OD800 of 0.075 in BG11 (modified as noted in figure legends) and cultured for 48 h. RNA was extracted and cDNA produced as previously11. Reverse transcriptase was omitted from negative controls. Transcript abundance was assessed by PCR with oligonucleotides 23 and 24 (nrsD), 25 and 26 (nikM), and 27 and 28 (rps1, loading control), each pair designed to amplify ~ 300 bp. Trends were repeated with biological replicates on 3 occasions. Gels were imaged with a Gel Doc XR Gel Documentation System.
Hydrogenase activity assay
Hydrogenase activity was measured using an S1 Clark-type electrode (Hansatech) electro-plated for hydrogen detection53 according to manufacturer’s instructions. Exponentially growing cells were inoculated to OD800 of 0.075 in BG11 or BG11 lacking a basal Ni(II) supplement and cultured for 7 days. Cultures (8 ml) were pelleted by centrifugation. Cell pellets were suspended in 100 µl 50 mM Tris pH 7.5 and added to a solution containing 50 mM Tris pH 7.5, 20 mM sodium dithionite, 5 μM methyl viologen (total volume = 2 ml) in the Clark electrode reaction vessel. The vessel was sealed, covered to exclude light, and generated current measured (polarity = -0.7 V, 30 °C, with constant stirring). The current generated by a solution saturated with 10% v/v H2/N2 was used as a standard54. Total protein was determined using a cell pellet from a replicate aliquot of culture with the Biorad Protein Assay kit. The hox– strain55 lacking the bi-directional hydrogenase operon was included as a negative control. Mean and standard deviation values determined from triplicate biological analyses.
Nickel content of cells
Exponentially growing cells were inoculated to OD800 of 0.075 in standard BG11 or BG11 lacking basal Ni(II) supplement and cultured for 84 h (total), with addition of 0.5 μM NiSO4 (where stated) 12 h prior to harvest. Alternatively, exponentially growing cells were inoculated to OD800 of 0.075 in standard BG11 supplemented with 0.25 μM NiSO4 and cultured for 24 h prior to harvest. Cell numbers were recorded (Casy Cell Counter TT) before harvesting 40 ml of each culture by centrifugation. Cell pellets were washed with 50 mM Tris, pH 7.5, 0.5 M sorbitol, 100 μM EDTA then suspended in ultrapure 65% v/v HNO3 (1 ml) and left to digest prior to metal analysis by ICP-MS. Mean and standard deviation values determined from triplicate biological analyses.
Detection of cellular Ni(II) using Newport Green
Cells were cultured by the first method described above for determining the nickel content of cells. An aliquot (5 ml) of cell culture was pelleted by centrifugation then suspended in 5 ml of BG11 lacking a basal Ni(II) supplement. An aliquot (980 µl) of this suspended pellet was added to a LabTek Chambered #1 Borosilicate Cover Glass System microscope slide (Nunc) before addition of 20 µl 1 mM Newport Green DCH diacetate (cell permeant) (Molecular Probes), prepared in DMSO. Slides were incubated for 1 h at 30 °C before recording images using a SP5 II laser scanning confocal microscope (LSCM) (Leica) equipped with an optical resolution enhancing module, developed in house (PhMoNa)56. Newport Green emission was collected between 505–535 nm following excitation at 496 nm (×2 digital magnification to ensure maximum flatness of field, 1024 × 1024 frame-size, 2.44 ns pixel dwell time, 4 frame average).
In order to achieve excitation with maximal probe emission, the microscope was equipped with a triple channel imaging detector, comprising two conventional PMT systems and a HyD hybrid avalanche photodiode detector. The pinhole was always determined by the Airy disc size, calculated from the objective in use (HCX PL APO 63x/1.40 NA LambdaBlue), using the lowest excitation wavelength (496 nm, 3.25 mW). Spectral imaging on this Leica system is possible (with the xyλ-scan function) and was adapted in an unconventional way57 by using averaged contrast transfer function (CTF) calculations. This approach involves selecting well-separated virtual band-pass filter settings for the desired intensity bands by using an assigned detector and monitoring the relative standardised brightness (referenced to the lowest occurrence). After collecting images of the two bacterial strains exposed to different exogenous Ni(II) concentrations, the CTF was calculated and averaged from five selected 100 × 100 pixel sized areas that displayed an even brightness. This was repeated in triplicate for each observation point. Mean and standard deviation values were calculated from 10-15 observation points, trends were repeated with biological replicates on two occasions. These CTF values were considered as the measured intensity integral values, and their relative ratio was calculated. This allowed variation in brightness to be monitored as a function of cell and probe environment. Conditions were maintained at 25 °C and 5% v/v CO2, and humidity was maintained at 10% v/v with a bubble humidifier. The measured brightness values using standardised instrumental parameters can be interpreted using the following expanded relationship for B′58: B′(λ,c) = ηεφte–kt, where c is the dye or probe loading concentration, λ is the excitation wavelength, k is the radiative rate constant for emission, ε is the molar extinction coefficient and φ is the fluorescence quantum yield of the probe, η is an accumulation parameter, reflecting the amount of complex internalised, and t is the standardised pixel dwell time.
Time-resolved fluorescence microscopy used a home built system based on an Axiovert 135M Inverted UV epifluorescence microscope (Zeiss). The excitation source was a diode laser LDH-P-C-485 (PicoQuant) (485 nm, 70 ps pulses, full width at half maximum at 20 MHz). A 100× 1.40NA oil immersion lens (Zeiss) was used for the detection of the fluorescence selected by a 540 ± 25 nm band pass filter (Comar Instruments). Fluorescence was detected using a photon counting module (ID100-20, ID Quantique) in combination with a SPC-130 time correlated single photon counting module (Becker & Hickl). The data were fit to a sum of exponentials by deconvolution with the instrument response function (IRF) obtained from a light scattering solution of Ludox particles.
Determination of cellular amino acid and glutathione concentrations by LC-MS
Exponentially growing InrS cells were inoculated to OD800 of 0.075 in standard BG11 and cultured for 4 days before harvesting 40 ml of culture by centrifugation. Cell pellets were suspended in 1 ml H2O, pelleted by centrifugation, flash frozen in liquid nitrogen and stored at –80 °C. Pellets were suspended in 1 ml sodium phosphate pH 7.4 and lysed by vortexing with glass beads (≤106 μm, 1 min on vortex, 1 min on ice × 3). Solids were removed by centrifugation and the supernatant used to determine amino acid and glutathione concentrations.
Proteins were removed by the addition of HCl to 0.1 N, incubated on ice for 30 min and centrifuged (16,000 × g, 15 min, 4 °C). Fluorescent derivatives of thiols were detected plus/minus reduction with sodium borohydride by reaction with mono-bromobimane59. Derivatives of cysteine, plus reduced and oxidised glutathione were used to prepare standard curves and samples were spiked with these standards to estimate recoveries. Fluorescent derivatives were separated and quantified by chromatography, using an H-Class UPLC system (Waters) with an Acquity FLR detector (Waters) and HSS T3 column (Waters, 100 mm × 2 mm, 1.7 µm particle size) (mobile phase of 85:15 50 mM K2HPO4 pH 6.0 with HAc:MeOH, 200 µl min–1, 40 °C, with excitation at 390 nm and emission at 478 nm).
For detection and quantification of amino acids, extracts were sequentially derivatized with o-phthaldialdehyde (OPA) reagent and fluorenylmethyloxycarbonyl (FMOC) to detect primary amines and proline respectively (10 μl sample, 10 μl OPA reagent, 20 μl of 5 mM FMOC in acetonitrile, and 60 μl methanol). Separations were performed on a Cortecs C18 column (100 mm × 2.1 mm, 1.6 µm particle size, Waters) (40 °C, 400 µl min–1). Mobile phase A was 20 mM sodium acetate pH 6.0 and mobile phase B acetonitrile:methanol:water (45:45:10). Samples were analysed using 5% v/v B for 5 min and then an increasing linear gradient to 85% v/v B after 15 min (total run time = 20 min). For OPA detection the excitation and emission wavelengths were 340 nm and 455 nm, for FMOC, 266 nm and 305 nm. Quantification was made with reference to amino acid standards that were generated and run with each sample set. Mean and standard deviation values determined from triplicate biological analyses (Supplementary Table 3).
Titration of InrS variants with Ni(II) in reconstituted cytoplasmic buffer
NiCl2 was added to buffer containing 10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl, 283 μM L-aspartic acid, 2,390 μM L-glutamic acid, 607 μM L-serine, 634 μM L- glutamine, 45 μM L-histidine, 126 μM L-arginine, 205 μM L-threonine, 1,649 μM L-alanine, 43 μM L-tyrosine, 163 μM L-valine, 73 μM L-isoleucine, 77 μM L-leucine and 302 μM reduced glutathione, a control buffer containing 10 mM HEPES pH 7.0, 100 mM NaCl, 400 mM KCl (standard buffer) or variants of these buffers as stated. Amino acid and glutathione concentrations determined as above. InrS, or its H21L variant, was subsequently added to the stated concentration and Ni(II) exchange monitored by the increase in absorption feature at 333 nm using a λ35 UV-vis spectrophotometer. At equilibrium the absorption spectrum of the solution was recorded using the same spectrophotometer. Mean and standard deviation values determined from triplicate analyses.
Throughout, investigators were not blinded to sample identity.
Data availability
Protein expression plasmids, strains associated with this work and any additional primary data will be made available upon request.
Supplementary Material
Acknowledgements
This research was funded by BBSRC grants BB/K00817X/1, BB/H006052/2 and BB/L009226/1 to NJR. R. Pal is a Royal Society University Research Fellow. Thanks to the Diamond Light Source and staff of beam line I02. We thank M.K. Grøftehauge for support in crystal structure determination, J.E. Frías for plasmid pCSE120 and C. Eckert for hox- Synechocystis PCC 6803.
Footnotes
Author contributions A.W.F. was involved in all aspects of data interpretation, iterative experimental design and along with R. Pernil made equal contributions to the experiments, C.J.P generated crystals, P.T.C. and A.J.P.S. supported Ni(II)-binding studies in a synthetic cytoplasm. L-O.P. and R. Pal analysed Ni(II) with Newport Green, I.C. performed the metabolomics analyses while E.P. generated the X-ray crystal structure. All authors contributed to drafting the manuscript. N.J.R. planned, managed and had overall responsibility for the programme, for data interpretation and for writing the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Accession codes Atomic coordinates and structure factors for InrS are deposited in the Protein Data Bank (PDB) under the accession code 5FMN.
References
- 1.Tottey S, et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 2.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 3.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 4.Irving H, Williams RJP. Order of stability of metal complexes. Nature. 1948;162:746–747. [Google Scholar]
- 5.Fraústo da Silva JJR, Williams RJP. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Oxford University Press; 1991. [Google Scholar]
- 6.Reyes-Caballero H, Campanello GC, Giedroc DP. Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys Chem. 2011;156:103–114. doi: 10.1016/j.bpc.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster AW, Osman D, Robinson NJ. Metal preferences and metallation. J Biol Chem. 2014;289:28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmann JD, Soonsanga S, Gabriel S. In: Molecular Microbiology of Heavy Metals. Nies DH, Silver S, editors. Springer Berlin Heidelberg; 2007. pp. 37–71. [Google Scholar]
- 9.Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 10.Changela A, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 11.Foster AW, Patterson CJ, Pernil R, Hess CR, Robinson NJ. Cytosolic Ni(II) sensor in cyanobacterium: nickel detection follows nickel affinity across four families of metal sensors. J Biol Chem. 2012;287:12142–12151. doi: 10.1074/jbc.M111.338301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 13.Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli - the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol Microbiol. 2006;62:252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 14.Grossoehme N, et al. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem. 2011;286:13522–13531. doi: 10.1074/jbc.M111.220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osman D, et al. Generating a Metal-responsive Transcriptional Regulator to Test What Confers Metal Sensing in Cells. J Biol Chem. 2015;290:19806–19822. doi: 10.1074/jbc.M115.663427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smaldone GT, Helmann JD. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology. 2007;153:4123–4128. doi: 10.1099/mic.0.2007/011742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto K, Agari Y, Agari K, Kuramitsu S, Shinkai A. Structural and functional characterization of the transcriptional repressor CsoR from Thermus thermophilus HB8. Microbiology. 2010;156:1993–2005. doi: 10.1099/mic.0.037382-0. [DOI] [PubMed] [Google Scholar]
- 18.Festa RA, et al. A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol Microbiol. 2011;79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbett D, et al. The combined actions of the copper-responsive repressor CsoR and copper-metallochaperone CopZ modulate CopA-mediated copper efflux in the intracellular pathogen Listeria monocytogenes. Mol Microbiol. 2011;81:457–472. doi: 10.1111/j.1365-2958.2011.07705.x. [DOI] [PubMed] [Google Scholar]
- 20.Dwarakanath S, et al. Response to copper stress in Streptomyces lividans extends beyond genes under direct control of a copper-sensitive operon repressor protein (CsoR) J Biol Chem. 2012;287:17833–17847. doi: 10.1074/jbc.M112.352740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang FM, et al. Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR) J Biol Chem. 2014;289:19204–19217. doi: 10.1074/jbc.M114.556704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster AW, Pernil R, Patterson CJ, Robinson NJ. Metal specificity of cyanobacterial nickel-responsive repressor InrS: cells maintain zinc and copper below the detection threshold for InrS. Mol Microbiol. 2014;92:797–812. doi: 10.1111/mmi.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J Am Chem Soc. 2008;130:7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins KA, Chivers PT, Maroney MJ. Role of the N-terminus in determining metal-specific responses in the E. coli Ni- and Co-responsive metalloregulator, RcnR. J Am Chem Soc. 2012;134:7081–7093. doi: 10.1021/ja300834b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Z, Wedd AG. The challenges of determining metal-protein affinities. Nat Prod Rep. 2010;27:768–789. doi: 10.1039/b906690j. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Maury L, Garcia-Dominguez M, Florencio FJ, Reyes JC. A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol Microbiol. 2002;43:247–256. doi: 10.1046/j.1365-2958.2002.02741.x. [DOI] [PubMed] [Google Scholar]
- 27.Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J Bacteriol. 2006;188:317–327. doi: 10.1128/JB.188.1.317-327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandal A, et al. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe(2+)-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol Microbiol. 2010;75:637–657. doi: 10.1111/j.1365-2958.2009.06977.x. [DOI] [PubMed] [Google Scholar]
- 29.Guedon E, Helmann JD. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol Microbiol. 2003;48:495–506. doi: 10.1046/j.1365-2958.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- 30.Faulkner MJ, Ma Z, Fuangthong M, Helmann JD. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J Bacteriol. 2012;194:1226–1235. doi: 10.1128/JB.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamagnini P, et al. Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol Rev. 2007;31:692–720. doi: 10.1111/j.1574-6976.2007.00085.x. [DOI] [PubMed] [Google Scholar]
- 32.Kaczmarek P, Szczepanik W, Jezowska-Bojczuk M. Acid-base, coordination and oxidative properties of systems containing ATP, L-histidine and Ni(II) ions. Dalton Trans. 2005:3653–3657. doi: 10.1039/b508962j. [DOI] [PubMed] [Google Scholar]
- 33.Hider RC, Kong XL. Glutathione: a key component of the cytoplasmic labile iron pool. Biometals. 2011;24:1179–1187. doi: 10.1007/s10534-011-9476-8. [DOI] [PubMed] [Google Scholar]
- 34.Jiang LJ, Maret W, Vallee BL. The glutathione redox couple modulates zinc transfer from metallothionein to zinc-depleted sorbitol dehydrogenase. Proc Natl Acad Sci USA. 1998;95:3483–3488. doi: 10.1073/pnas.95.7.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Z, et al. Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol Microbiol. 2014;94:756–770. doi: 10.1111/mmi.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Z, Gabriel SE, Helmann JD. Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 2011;39:9130–9138. doi: 10.1093/nar/gkr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin JH, et al. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Natl Acad Sci U S A. 2011;108:5045–5050. doi: 10.1073/pnas.1017744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djoko KY, Ong CL, Walker MJ, McEwan AG. The Role of Copper and Zinc Toxicity in Innate Immune Defense against Bacterial Pathogens. J Biol Chem. 2015;290:18954–18961. doi: 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braymer JJ, Giedroc DP. Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr Opin Chem Biol. 2014;19:59–66. doi: 10.1016/j.cbpa.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker KW, Skaar EP. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol Rev. 2014;38:1235–1249. doi: 10.1111/1574-6976.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang FM, Lauber MA, Running WE, Reilly JP, Giedroc DP. Ratiometric pulse-chase amidination mass spectrometry as a probe of biomolecular complex formation. Anal Chem. 2011;83:9092–9099. doi: 10.1021/ac202154r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan BG, Vijgenboom E, Worrall JA. Conformational and thermodynamic hallmarks of DNA operator site specificity in the copper sensitive operon repressor from Streptomyces lividans. Nucleic Acids Res. 2014;42:1326–1340. doi: 10.1093/nar/gkt902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman J, et al. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG+ strategy. Acta Crystallogr D Biol Crystallogr. 2005;61:1426–1431. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- 44.Broennimann C, et al. The PILATUS 1M detector. J Synchrotron Radiat. 2006;13:120–130. doi: 10.1107/S0909049505038665. [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bunkoczi G, et al. Phaser.MRage: automated molecular replacement. Acta Crystallogr D Biol Crystallogr. 2013;69:2276–2286. doi: 10.1107/S0907444913022750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 50.Picossi S, Flores E, Herrero A. The LysR-type transcription factor PacR is a global regulator of photosynthetic carbon assimilation in Anabaena. Environ Microbiol. 2015;17:3341–3351. doi: 10.1111/1462-2920.12800. [DOI] [PubMed] [Google Scholar]
- 51.Williams J. In: Methods in Enzymology. Packer L, Glazer AN, editors. Vol. 167. Academic Press; 1988. pp. 766–778. [Google Scholar]
- 52.Nakasugi K, Alexova R, Svenson CJ, Neilan BA. Functional analysis of PilT from the toxic cyanobacterium Microcystis aeruginosa PCC 7806. J Bacteriol. 2007;189:1689–1697. doi: 10.1128/JB.01640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dutheil J, et al. The AbrB2 autorepressor, expressed from an atypical promoter, represses the hydrogenase operon to regulate hydrogen production in Synechocystis strain PCC6803. J Bacteriol. 2012;194:5423–5433. doi: 10.1128/JB.00543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiesenburg DA, Guinasso NL. Equilibrium solubilities of methane, carbon monoxide, and hydrogen in water and sea water. J Chem Eng Data. 1979;24:356–360. [Google Scholar]
- 55.Eckert C, et al. Genetic analysis of the Hox hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 reveals subunit roles in association, assembly, maturation, and function. J Biol Chem. 2012;287:43502–43515. doi: 10.1074/jbc.M112.392407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pal R. Phase modulation nanoscopy: a simple approach to enhanced optical resolution. Faraday Discuss. 2015;177:507–515. doi: 10.1039/c4fd00158c. [DOI] [PubMed] [Google Scholar]
- 57.Smith DG, Pal R, Parker D. Measuring equilibrium bicarbonate concentrations directly in cellular mitochondria and in human serum using europium/terbium emission intensity ratios. Chemistry. 2012;18:11604–11613. doi: 10.1002/chem.201201738. [DOI] [PubMed] [Google Scholar]
- 58.Butler SJ, et al. EuroTracker(R) dyes: design, synthesis, structure and photophysical properties of very bright europium complexes and their use in bioassays and cellular optical imaging. Dalton Trans. 2015;44:4791–4803. doi: 10.1039/c4dt02785j. [DOI] [PubMed] [Google Scholar]
- 59.Svardal AM, Mansoor MA, Ueland PM. Determination of reduced, oxidised, and protein-bound glutathione in human plasma with precolumn deivatization with monobromobimane and liquid chromatography. Anal Biochem. 1990;184:338–346. doi: 10.1016/0003-2697(90)90691-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein expression plasmids, strains associated with this work and any additional primary data will be made available upon request.