Abstract
The broad family of LEA proteins are intrinsically disordered proteins (IDPs) with several potential roles in desiccation tolerance, or anhydrobiosis, one of which is to limit desiccation-induced aggregation of cellular proteins. We show here that this activity, termed molecular shield function, is distinct from that of a classical molecular chaperone, such as HSP70 – while HSP70 reduces aggregation of citrate synthase (CS) on heating, two LEA proteins, a nematode group 3 protein, AavLEA1, and a plant group 1 protein, Em, do not; conversely, the LEA proteins reduce CS aggregation on desiccation, while HSP70 lacks this ability. There are also differences in interaction with client proteins – HSP70 can be co-immunoprecipitated with a polyglutamine-containing client, consistent with tight complex formation, whereas the LEA proteins can not, although a loose interaction is observed by Förster resonance energy transfer. In a further exploration of molecular shield function, we demonstrate that synthetic polysaccharides, like LEA proteins, are able to reduce desiccation-induced aggregation of a water-soluble proteome, consistent with a steric interference model of anti-aggregation activity. If molecular shields operate by reducing intermolecular cohesion rates, they should not protect against intramolecular protein damage. This was tested using the monomeric red fluorescent protein, mCherry, which does not undergo aggregation on drying, but the absorbance and emission spectra of its intrinsic fluorophore are dramatically reduced, indicative of intramolecular conformational changes. As expected, these changes are not prevented by AavLEA1, except for a slight protection at high molar ratios, and an AavLEA1-mCherry fusion protein is damaged to the same extent as mCherry alone. A recent hypothesis proposed that proteomes from desiccation-tolerant species contain a higher degree of disorder than intolerant examples, and that this might provide greater intrinsic stability, but a bioinformatics survey does not support this, since there are no significant differences in the degree of disorder between desiccation tolerant and intolerant species. It seems clear therefore that molecular shield function is largely an intermolecular activity implemented by specialist IDPs, distinct from molecular chaperones, but with a role in proteostasis.
Introduction
Extreme water loss through evaporation imposes severe challenges on biological systems: cell membranes lose integrity or undergo fusion; potentially damaging reactive oxygen species are generated; and protein structure is compromised due to the diminution of the hydrophobic effect at reduced water activity. Despite this, many organisms are able to survive drying, during which they enter a state of suspended animation known as anhydrobiosis.1–4 These organisms are widespread throughout nature and include yeasts such as Saccharomyces cerevisiae,5,6 resurrection plants like Craterostigma plantagineum,7,8 and certain invertebrates, exemplified by the nematode Aphelenchus avenae.9
Since desiccation is highly damaging to non-anhydrobiotic cells and organisms, those that are tolerant of this stress must have mechanisms for minimising injury. In principle, two different strategies might be adopted: either global differences in the genomes and proteomes of desiccation tolerant organisms evolve, such that all the molecular components of anhydrobiotes are individually resistant to water loss; or protection and repair systems exist in anhydrobiotes that prevent permanent damage to other molecules in the cell. A “belt and braces” combination of both approaches is also possible. The former strategy is seen in some extremophiles whose proteins are demonstrably different to mesophile homologues, allowing them to function in extreme environments.10,11 The latter strategy is typified by the mesophilic stress response to heat shock, for example, where molecular chaperones are upregulated to deal with protein denaturation and aggregation.12,13
Whole proteome remodelling seems unlikely as a general strategy for anhydrobiosis, however. In plants, desiccation tolerance has been lost and has re-emerged in single lineages;14 there is surely insufficient time, even over millions of years, for natural selection to modify a complete proteome in this way. Furthermore, the water soluble proteome of the anhydrobiotic nematode, A. avenae, is no less prone to desiccation-induced aggregation than that of human,15 suggesting there are no major differences in resistance to desiccation damage at the level of individual proteins. Nevertheless, some researchers16 have hypothesised that the degree of low complexity, which was assumed to correlate with disorder in prokaryotic proteomes, is linked with resistance to desiccation. In contrast, most work on anhydrobiosis has assumed that a discrete set of molecular adaptations are responsible, and that these act to minimise damage to molecular and cellular architecture. Among these adaptations, recent emphasis has been on intrinsically disordered proteins (IDPs), chiefly the LEA proteins, which are thought to have various roles in protein homeostasis, membrane stabilisation and bioglass formation, among others, during anhydrobiosis.17–20
LEA proteins are generally small (10–30 kDa), highly hydrophilic IDPs that, on the basis of protein sequence motifs and peptide composition, fall into three main groups.20,21 Evidence is accumulating that LEA proteins and other hydrophilic proteins have a protein stabilisation function since they preserve enzyme activity in vitro after desiccation or freezing.22–28 One mechanism for the protection observed is the prevention of water stress-induced aggregation of sensitive proteins.15,23,28,29 This anti-aggregation activity extends in vivo to spontaneously aggregating polyglutamine-containing (polyQ) and polyalanine-containing proteins.15,30 Although superficially this anti-aggregation function resembles that of classical molecular chaperones, several fundamental differences are apparent such that we have termed the former molecular shield activity.21,23 For example, molecular chaperones are largely well-structured proteins, unlike the LEA proteins, and in many cases they function through interaction with exposed hydrophobic regions on (partially) unfolded client proteins.12,31 Such interactions are sufficiently robust that co-immunoprecipitation experiments can be performed to recover chaperone-client complexes from cell extracts (e.g. the interaction of HSP60 with polyQ proteins32). A similar mode of action is unlikely for highly hydrophilic IDPs like the LEA proteins, at least via hydrophobic interfaces. Instead, we have suggested that the anti-aggregation activity of hydrophilic IDPs results from physical interference whereby the IDP reduces the encounter frequency of aggregating protein species. In this report, we explore the characteristics of molecular shield activity and examine this in the context of an intermolecular versus an intramolecular stabilisation function.
Results
The anti-aggregation activity of molecular shield proteins is distinct from that of a molecular chaperone
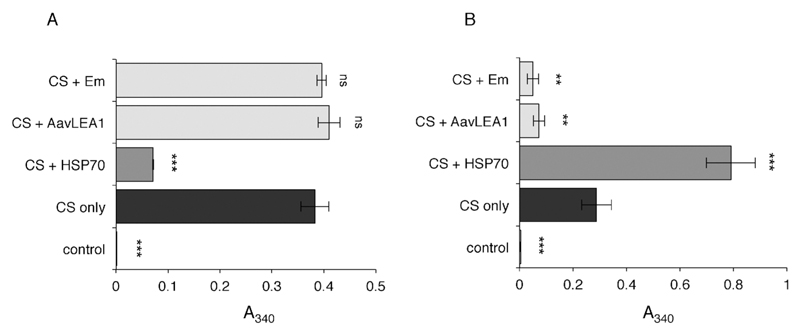
To compare the ability of molecular shields to prevent protein aggregation with that of a classical molecular chaperone, citrate synthase (CS) was induced to aggregate by either heating or vacuum drying. CS was first subjected to heat stress, either by itself, or in the presence of a molar excess of the chaperone HSP70, or one of two shield proteins, either the nematode group 3 LEA protein, AavLEA1, or the soyabean group 1 LEA protein, Em. While HSP70 significantly reduced CS aggregation on heating, neither of the LEA proteins was effective (Fig. 1a). These results are in accordance with the literature and show that group 1 and group 3 LEA proteins are ineffective against heat-induced CS aggregation,23 in contrast to molecular chaperones (for example, Zhai et al. 200833). Under conditions of desiccation stress, on the other hand, both LEA proteins were able to reduce CS aggregation markedly, while HSP70 did not (Fig. 1b). In fact, the mixture of HSP70 and CS resulted in increased levels of aggregation compared to CS alone, although HSP70 by itself did not aggregate (data not shown). One explanation for this observation might be that, although HSP70 is unable to prevent CS aggregation, it still binds to denatured CS and this might serve to increase the size of aggregates. These results demonstrate a clear distinction between the activities of a chaperone and molecular shields that nevertheless fits with their proposed physiological roles.
Fig. 1.
Light scattering measured as apparent absorbance (A340) of CS (black bars) after (A) heat stress, or (B) desiccation stress, in the presence of five-fold molar excess HSP70 (dark grey), AavLEA1, or Em (both light grey). Non-stressed CS is taken as control (white). *** denotes significance at p < 0.001 and ** denotes p < 0.01 using one-way ANOVA, plus Tukey post test; ns, not significant.
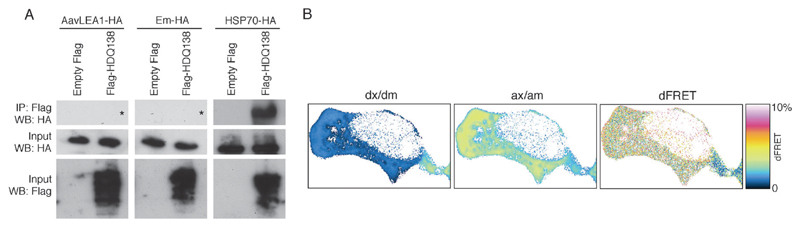
In its simplest form, the molecular shield hypothesis states that shield proteins use physical interference to reduce the frequency of cohesive interactions between aggregating species. Therefore, this does not necessitate the formation of complexes with client proteins as is proposed for chaperones. To attempt to contrast shield and chaperone interaction with aggregating proteins, we chose a target that both are known to act on under similar conditions, i.e. huntingtin-derived polyQ protein, expressed in mammalian cells.15,30,34 When HDQ138 and HSP70 are co-expressed in T-REx293 cells, the chaperone could be co-immunoprecipitated with the polyQ protein, indicating a strong interaction. However, neither AavLEA1 nor Em were co-immunoprecipitated with HDQ138, suggesting a much weaker association (Fig. 2a).
Fig. 2.
(A) Immunoprecipitation (IP) after expression of empty vector pFLAG-CMV5a or FlagHDQ138 and AavLEA1-HA (left panels), Em-HA (middle panels), or HSP70-HA (right panels). IP was performed with anti-Flag-M2 affinity gel followed by immunoblotting with anti-HA antibody (top row of panels). The inputs from the total cell lysates were probed with antibodies against HA (middle row) or Flag (bottom row) to detect the molecular shield or chaperone, and the polyQ protein, respectively. The asterisks in the top left and top middle panels show the expected position of any HA signal. (B) Example FRET analysis of EGFP-HDQ74 (donor) and AavLEA1-mCherry (acceptor) interactions in a live cell, showing signal in the donor channel upon excitation at donor wavelength (dx/dm), signal in the acceptor channel upon excitation at acceptor wavelength (ax/am), and donor normalized and unmixed FRET transfer efficiency dFRET.
To investigate the possibility of loose or transient interaction between a molecular shield protein and its target, we used mCherry-tagged AavLEA1 and EGFP-tagged HDQ74 to perform quantitative Förster resonance energy transfer (FRET) via sensitized acceptor (mCherry) fluorescence emission upon donor (EGFP) excitation. The method used yields FRET efficiencies normalized either by the prevailing acceptor concentration (aFRET), or the donor concentration (dFRET), the relative level of which depends on the stoichiometry of interaction.35,36 The positive control construct (EGFP tethered to mCherry by a 7-amino acid linker) was expressed in T-REx293 cells and gave FRET levels of 15.2% (here aFRET = dFRET, because the stoichiometry of interaction is 1). The negative control (co-expressed EGFP and mCherry proteins alone) did not yield significant FRET levels (0.6%). For the EGFP-HDQ74 and AavLEA1-mCherry pair, dFRET was measured at approximately 3%, indicating a weak interaction between the two proteins in vivo (Fig. 2b).
Neutral polysaccharides share some properties of molecular shields
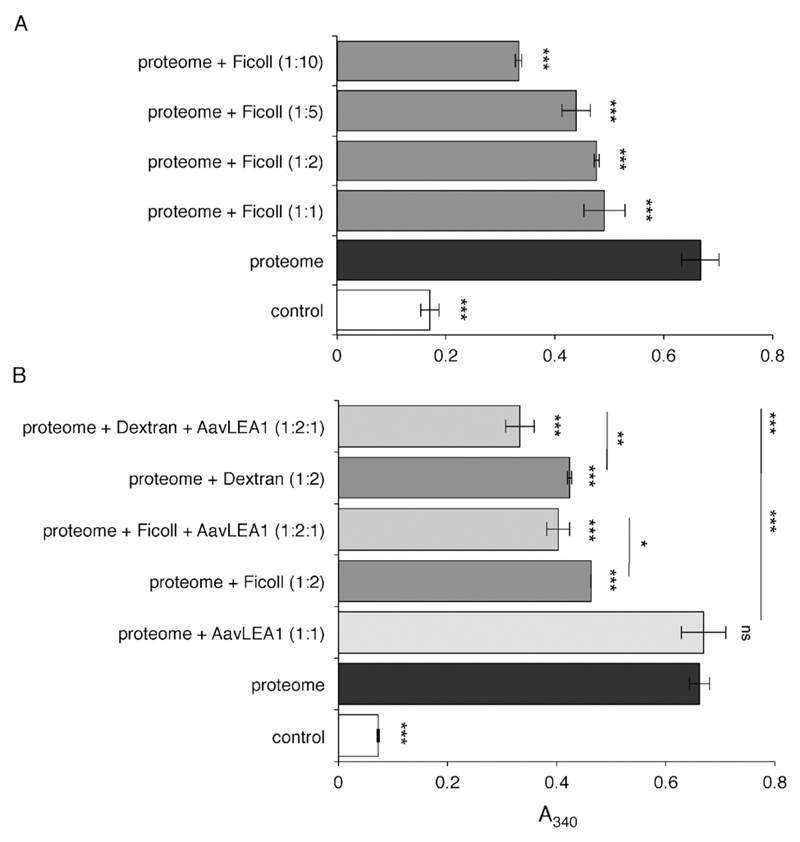
By analogy with the stabilisation of colloidal suspensions by hydrophilic polymers,37,38 molecular shields could behave sterically, electrostatically or both, i.e. electrosterically. We might therefore expect some synthetic hydrophilic polymers to behave as molecular shields, and to reduce protein aggregation. To test this, we used the polysaccharide ficoll 70, which is a cross-linked polymer of sucrose with average Mr 70 000. Vacuum drying of a complex mixture of proteins, the water-soluble proteome from a human cell line, was performed in the presence of varying concentrations of ficoll. In the absence of polysaccharide, the protein mixture aggregates markedly on drying and rehydration, but addition of ficoll led to reduction of aggregation in a concentration-dependent manner (Fig. 3a). Although ficoll is commonly used as a molecular crowding reagent to simulate conditions within a cell, the concentrations used here were more than an order of magnitude lower than those in crowding experiments. Therefore, the anti-aggregation effect we observed was unlikely to be an effect of crowding; indeed, when very high ficoll concentrations (e.g. 250 mg ml−1) were used in the proteome desiccation assay, aggregation was enhanced (data not shown), possibly because at these concentrations there is an increase in protein-protein association constants due to a depletion effect.39 We next asked whether there was an additive anti-aggregation effect of ficoll and LEA protein: when AavLEA1 was added to the proteome at a 1 : 1 molar ratio, it did not reduce aggregation significantly after desiccation, as reported previously.15 However, when the proteome was dried in the presence of both a sub-optimal concentration of ficoll (1 : 2 molar ratio) and the LEA protein at a 1 : 1 molar ratio, aggregation was reduced to a level below that observed with ficoll alone (Fig. 3b). A comparable effect was also obtained for AavLEA1 and another polysaccharide, Dextran 42 (Fig. 3b).
Fig. 3.
Light scattering measured as apparent absorbance (A340) of T-REx293 water-soluble proteome after in vitro desiccation stress (black bars), (A) in the presence of variable molar ratios of ficoll 70 (dark grey), and (B) in the presence of 1 : 1 or 1 : 2 molar ratio of AavLEA1 (light grey) or ficoll 70/Dextran 42 (dark grey), respectively, or with 1 : 2 : 1 ratio of proteome: ficoll 70/Dextran 42: AavLEA1 (mid grey). The non-dried water-soluble proteome is taken as control (white). *, ** and *** denote significance at p < 0.05, p < 0.01 and p < 0.001, respectively, using one-way ANOVA, plus Tukey post test; ns, not significant.
Molecular shields have a limited capacity to prevent intramolecular changes due to desiccation
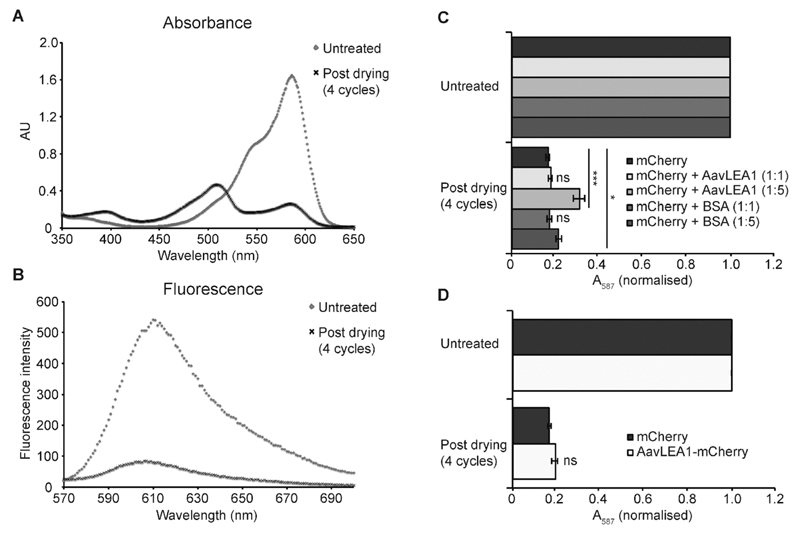
If molecular shields function chiefly to reduce the productive collision rate of potentially aggregating protein species, then we might expect they would be ineffective at protecting proteins from deleterious structural perturbations caused by desiccation, where these changes are distinct from the effects of aggregation. An ideal candidate protein to test this hypothesis is the monomeric fluorescent protein mCherry: minimal aggregation of mCherry was observed upon desiccation and rehydration (Fig. S1). However, both the absorbance and fluorescence spectra were substantially altered, indicating damage to the protein’s fluorophore, possibly due to intramolecular changes induced by desiccation (Fig. 4a and b). In the absorbance spectrum, the main peak at 587 nm consistently decreased with the number of drying cycles, and new peaks developed at 395 and 509 nm. The emission spectrum also decreased dramatically in intensity upon desiccation, consistent with the absorbance data. Notably, excitation at 395 and 509 nm did not give rise to appreciable levels of fluorescence, suggesting that the properties of the fluorophore have been lost entirely.
Fig. 4.
AavLEA1 provides limited protection of mCherry during desiccation. (A) Absorbance and (B) fluorescence emission spectra of mCherry, before and after four cycles of drying and rehydration. (C) Effect of drying on mCherry in the absence or presence of AvLEA1 or BSA (control) at molar ratios of 1 : 1 and 1 : 5. (D) Effect of desiccation on mCherry and an AavLEA1-mCherry fusion protein. Absorbance at 587 nm (the absorbance maximum of the intrinsic fluorophore) was measured before and after two and four cycles of drying and rehydration. Data were normalised, with the absorbance of the untreated sample represented as 1, for ease of comparison. All experiments were carried out in triplicate; error bars indicate ± 1 SD. *** denotes significance at p < 0.001 and * denotes p < 0.05 using one-way ANOVA, plus Tukey post test; ns, not significant.
To assess whether an LEA protein could protect against such an effect, mCherry was mixed with AavLEA1 at two different molar ratios prior to drying, and also with bovine serum albumin (BSA) as a control. Up to four drying cycles were performed and the effect on mCherry function was assessed by measuring A587, which gave the most consistent changes with stress (Fig. 4c). In all cases desiccation resulted in a dramatic decrease in absorbance at 587 nm, with ~70–80% reduction being observed after four cycles of drying and rehydration. This is in stark contrast to the citrate synthase aggregation assay23 (Fig. 1), indicating that AavLEA1 is much less effective at protecting mCherry, consistent with the proposed mode of action as a shield protein. Intriguingly, AavLEA1 afforded a moderate level of protection at a 5 : 1 molar ratio with the fluorescent protein; this was most apparent after four cycles of desiccation and rehydration, where A587 was almost double that of mCherry alone (Fig. 4c). The reason for this is not clear, as it is not predicted from the molecular shield hypothesis, but it is possible that the weak interactions between AavLEA1 and client proteins detected using FRET (Fig. 2b) are responsible. If so, we might expect that covalent linkage of the two proteins (increasing the effective local concentration of AavLEA1 with respect to mCherry), might afford better protection. Therefore, an AavLEA1-mCherry fusion protein was constructed and tested for protection of mCherry during desiccation. However, this proved ineffective, since drying of the fusion protein gave very similar results to mCherry alone (Fig. 4d), and to mCherry dried in the presence of AavLEA1 as a separate polypeptide at a molar ratio of 1 : 1 (Fig. 4c).
Molecular shield function is likely to derive from a limited set of IDPs
It has been hypothesised that intrinsically disordered regions (IDRs) are more abundant in the proteins of desiccation tolerant organisms compared to intolerant species and that these disordered regions, akin to an intramolecular shield, might improve protein stability where they occur.16 In ref. 16, disorder was identified with low sequence complexity regions within proteins, often located at the N- or C-terminus and therefore resembling the AavLEA1-mCherry fusion protein. Further work on a putative nudix hydrolase from Deinococcus radiodurans emphasised disordered “tails” that were proposed to lower the hydration free energy of the protein, thereby allowing it to remain hydrated for longer during desiccation.40 Unfortunately, the proteome is not currently defined for A. avenae, from which AavLEA1 derives, so that we were not able to look at the degree of disorder in this nematode. However, when we examined the proteomes of several well-characterised prokaryotes using the disorder prediction tool FoldIndex,41 we were unable to detect differences in the distribution of IDRs across the set, although this included D. radiodurans, a desiccation tolerant bacterium, as well as the desiccation sensitive Escherichia coli (Table 1). Specifically, genomes were obtained for a number of species understood to be durable in the face of abiotic stress, together with species that are understood to be more sensitive. Proteins greater than 100 amino acids derived from the genes were sent to FoldIndex, which returns a list of peptides predicted to be disordered. By dividing the total length of disordered segments by the length of the protein, a percent-disordered value was obtained for each protein, with values ranging from 0% (i.e. predicted to be totally folded) to 100% (predicted to be totally unfolded). Scanning each genome in turn, percent-disordered values were computed for each protein, with counts of percent-disordered values in decile bins being recorded. Because genome sizes vary across the set of genomes examined, the bins recorded the percentage of the total gene count found in the respective percent-disordered bins. The central observation is that the spectrum of percent-disordered values is similar across all the genomes, irrespective of whether they are from tolerant or sensitive species. For example, using the R statistical suite, the adjusted R2 value for the linear model of D. radiodurans versus E. coli was 0.9969 (p-value 4.3 e-14).
Table 1.
For each of a range of species, the table lists the percentage of proteins in each percent-disordered decile bin. For example, the 0..10 bin has the count of those proteins from Deinococcus radiodurans in which more than 0% and up to 10% of the amino acids are predicted by FoldIndex to be in an intrinsically disordered state. Zero percent and 100% are treated as special cases. The genomes being considered are: BURPS (Burkholderia pseudomallei) and DEIRA (Deinococcus radiodurans), both regarded as durable species, and BURMA (Burkholderia mallei), CAMJE (Campylobacter jejuni), ECOLI (Escherichia coli (strain K12)), PSEPK (Pseudomonas putida (strain KT2440)), SHEON (Shewanella oneidensis) and THET2 (Thermus thermophilus (strain HB27)), which are regarded as sensitive species
| Percentage of sequences with designated percentage of amino acids predicted as natively unfolded |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species code | N seqs | Ave. length | 0 | (0..10] | (10..20] | (20..30] | (30..40] | (40..50] | (50..60] | (60..70] | (70..80] | (80..90] | (90..100) | 100 |
| BURMA | 4238 | 347.09 | 40.44 | 23.12 | 15.22 | 7.57 | 3.63 | 2.45 | 1.56 | 1.6 | 1.16 | 0.71 | 0.73 | 1.79 |
| BURPS | 5293 | 367.86 | 41.62 | 25.07 | 16.59 | 7.82 | 3.72 | 2.38 | 1.15 | 0.77 | 0.42 | 0.19 | 0.13 | 0.13 |
| CAMJE | 1477 | 333.89 | 27.76 | 20.24 | 21.12 | 12.93 | 8.4 | 4.4 | 2.1 | 1.42 | 1.08 | 0.41 | 0 | 0.14 |
| DEIRA | 2889 | 322.35 | 35.48 | 21.18 | 18.86 | 11.21 | 5.61 | 2.77 | 2.04 | 1.25 | 0.9 | 0.31 | 0.1 | 0.28 |
| ECOLI | 3812 | 344.71 | 35.2 | 22.14 | 17.81 | 10.86 | 7.06 | 2.86 | 1.71 | 1.42 | 0.6 | 0.1 | 0.13 | 0.1 |
| PSEPK | 4853 | 357.14 | 36.91 | 23.49 | 18.5 | 9.83 | 5.09 | 2.7 | 1.55 | 1.24 | 0.37 | 0.14 | 0.06 | 0.12 |
| SHEON | 3740 | 350.59 | 36.23 | 21.26 | 18.9 | 11.28 | 5.53 | 2.73 | 2.01 | 1.07 | 0.59 | 0.29 | 0.05 | 0.05 |
| THET2 | 2017 | 323.28 | 41.5 | 23.75 | 17.65 | 8.43 | 3.77 | 2.43 | 1.34 | 0.55 | 0.2 | 0.15 | 0.1 | 0.15 |
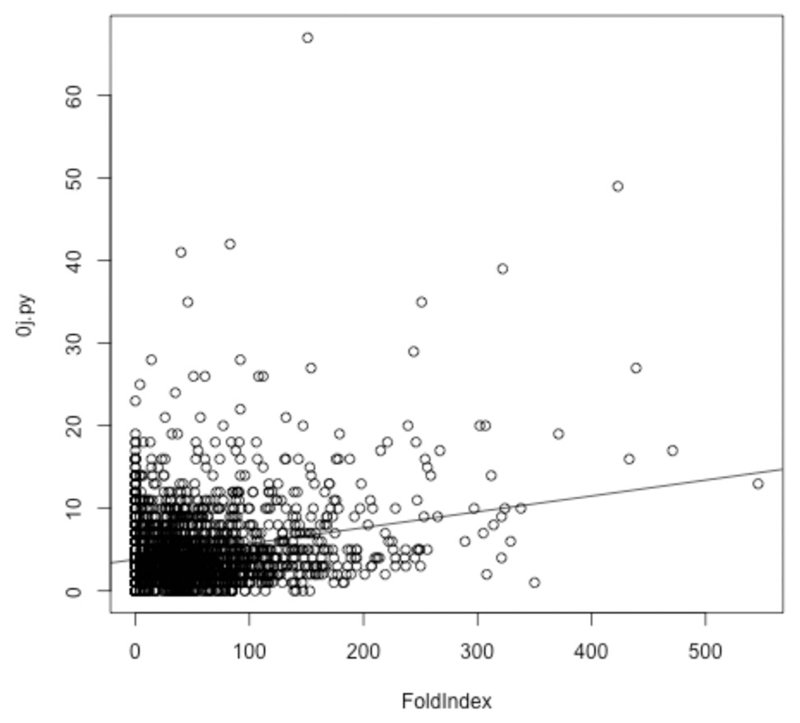
One reason for the discrepancy of this analysis with that of ref. 16 might be the methods used to define disordered protein regions. Krisko et al.16 equated disorder with low complexity (LC), whereas the data of Table 1 were generated using FoldIndex, an application that directly predicts disorder. To check the relationship between LC and disorder, LC software tools SEG42 (used by Krisko et al.16) and 0j.py43 were applied in turn to the proteome from D. radiodurans (2,833 sequences). For each sequence, the number of amino acids in LC regions was correlated (using the R statistical package) with number of amino acids predicted by FoldIndex to be in an IDR (Fig. 5). A linear model of SEG predictions versus FoldIndex predictions had an adjusted R2 value of 0.03644, while the equivalent linear model based on 0j.py had an adjusted R2 value of 0.06144 (both linear models have p-values less than 2e-16). While only the one IDR predictor was used, i.e. FoldIndex, it is unlikely anything other than a complete lack of correlation would have been observed using alternative predictors. It seems clear, therefore, that LC correlates very poorly with disorder and is unreliable as a predictor of IDRs. The corollary of this finding is that molecular shield function in desiccation tolerant organisms is likely to be derived from a relatively small set of IDPs that protect the whole proteome, rather than from the presence of IDRs within all or most proteins within the organism; i.e. it is an intermolecular activity of a few specialist IDPs rather than an intramolecular activity of all proteins.
Fig. 5.
For each protein in D. radiodurans, the score reported by 0j.py, reflecting low complexity, is plotted against the number of amino acids from that protein predicted by Foldindex to be in an intrinsically disordered domain. A linear model is shown, but is a poor fit given the lack of a correlation between low complexity (as measured by 0j.py) and intrinsic disorder. Use of SEG produces very similar results (data not shown).
Discussion
Maintaining the proper functioning of the proteome, a large and varied set of molecular operations broadly encapsulated by the term protein homeostasis or proteostasis,13 is crucial for the health and survival of the cell. This becomes particularly critical under environmental conditions that adversely impact protein structure and function, and one of the more extreme stresses that might be imposed is the loss of essentially all cellular water through evaporation, i.e. desiccation. For organisms to survive extreme water loss it is assumed they are able to put in place protection or repair mechanisms to minimise or eliminate permanent damage to cellular components including proteins.2 The alternative strategy, whereby the anhydrobiotic proteome is engineered through evolution with sufficient stability to withstand desiccation, seems unlikely, given the sporadic occurrence of desiccation tolerance in taxonomic groups.14
One protection mechanism studied increasingly in recent years is the ability of IDPs to stabilise other proteins during desiccation.2,17 IDPs such as the LEA proteins, whose expression is often associated with the acquisition of desiccation tolerance,44,45 can help maintain protein function under conditions of water stress, at least partly by reducing aggregation of denatured species.15,23,28 Superficially, the role of desiccation-relevant IDPs as anti-aggregants resembles that of classical molecular chaperones, but there are several points of difference such that we refer to IDPs in this context as molecular shields. For example, while molecular chaperones do contain disordered regions,31,46 they have three-dimensional structure essential for their activity. In contrast, molecular shield proteins tend to be fully or largely disordered and function as entropic chains. Furthermore, molecular chaperones form transient complexes with their client proteins through specific binding sites, often hydrophobic patches,12,32 whereas both in vivo and in vitro experiments suggest that LEA proteins act by slowing the cohesion rate, rather than by sequestration, of aggregating protein species.30 Any interaction of shield proteins with their clients is loose, although measurable in the case of the nuclear IDP anhydrin36 and AavLEA1 (Fig. 2b), and is unlikely to involve hydrophobic patches given their highly hydrophilic nature. Data presented in this paper illuminate further differences since two molecular shields show reciprocal activity to that of the chaperone HSP70 in heat stress and desiccation aggregation assays (Fig. 1), and the chaperone is able to co-immunoprecipitate with a polyQ-containing protein, while shield proteins do not, indicating differing strengths of interaction with clients (Fig. 2).
We have begun to investigate the mechanism of action of molecular shields using synthetic macromolecules: polysaccharides such as ficoll 70 and Dextran 42 display shield activity in desiccation-induced aggregation experiments, and at low concentrations polysaccharides and LEA proteins can act together to reduce aggregation further than they might alone (Fig. 3). One interpretation of these results is that both types of macromolecule function similarly in this assay. Polysaccharides are not considered to behave as molecular chaperones, but they are known to increase the stability of proteins (e.g. dextran/horse radish peroxidase mixtures are more stable at high temperatures and low pH than the enzyme alone47), most likely due to solution effects. ficoll and Dextran are neutral molecules, indicating that electrostatic interactions are not essential for the molecular shield function observed and that steric effects are at least partially responsible.
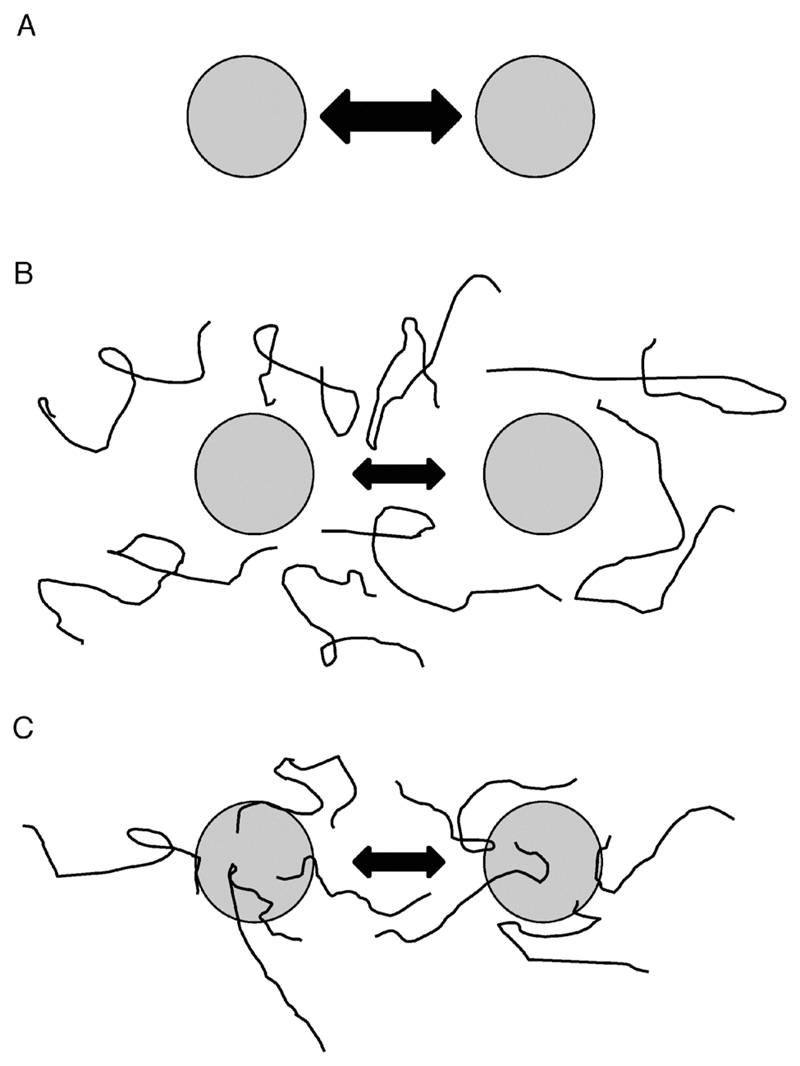
A model for molecular shield function based purely on steric interference (Fig. 6a and b), where shield proteins act as non-interacting space fillers that reduce collision rates between aggregating protein species, would address intermolecular effects of desiccation, i.e. aggregation, but not intramolecular effects, i.e. denaturation or structural modification of individual polypeptides. Experiments with the monomeric red fluorescent protein mCherry are relevant to this issue, since mCherry suffers relatively little aggregation on desiccation, while its spectral properties alter markedly, presumably due to conformational changes affecting its fluorescent centre (Fig. 4). To a large extent, the decreased absorbance and emission caused by desiccation occurs whether other proteins, i.e. the LEA protein AavLEA1 or the control protein BSA, are present or not, as expected based on the above model. Intriguingly, however, a very modest reduction in this effect is seen at a 5 : 1 molar ratio of AavLEA1 to mCherry, meaning that the LEA protein offers limited protection against denaturation of the fluorophore. This cannot be explained by a purely steric effect, suggesting the reality of molecular shield function is more complicated. Indeed, it is already clear that this must be the case because proteins such as BSA or RNaseA15 do not prevent desiccation-induced aggregation, as might be expected if molecular shields were merely non-interacting volume excluders. In fact, the observation that FRET can occur in cells between two different shield proteins, anhydrin36 and AavLEA1 (Fig. 2b), and their targets is consistent with a degree of interaction. In turn, this means that the stabilisation effect of the polysaccharides such as ficoll 70 in desiccation-induced aggregation assays (Fig. 3) is likely to involve some association between polysaccharide and target proteins. Accordingly, it is increasingly recognised that polysaccharides can interact with protein surfaces; one such polymer, polyethylene glycol, has been shown by NMR spectroscopy to interact with hydrophobic surfaces on cytochrome c, for example.48
Fig. 6.
Models for molecular shield function. (A) In the absence of molecular shields, partially denatured proteins (shaded circles) will interact and adhere at some rate, indicated by the double-headed arrow. (B) In the basic molecular shield model, shield proteins (represented by lines) are entropic chains that do not interact with other proteins but occupy space in solution and reduce the collision rate of aggregating species (indicated by a smaller double-headed arrow). (C) Evidence suggests a loose association of shield proteins with other polypeptides thereby forming a dynamic, three-dimensional protective barrier around aggregating species. Such interactions might also involve partial folding of the molecular shield on the surface of the misfolded client protein, potentially allowing a degree of entropy transfer that might facilitate refolding of the client, as proposed by Tompa and Csermely.46
Therefore, a modified model would involve a molecular shield associating loosely with the surface of a target protein to form a three-dimensional protective barrier around it that would limit the approach of other proteins (Fig. 6c). This arrangement would probably be dynamic, with each shield molecule having a limited residence time on the protein surface, but would lead to an averaged volume exclusion effect similar to that of the entropic bristles of MAP2, tau and neurofilament side arms, which act as spacers of cytoskeletal filaments.49 A further development of the model might incorporate the ideas of Tompa and Csermely,46 who postulate that disordered regions of some molecular chaperones can gain structure on interaction with misfolded client proteins, thus allowing the client to partially unfold through an entropy transfer effect, and then follow the correct folding pathway leading to its native conformation. There is evidence for LEA proteins gaining secondary structure on association with membrane surfaces50–52 and it is possible that similar folding could occur on the surface of client proteins. Such an entropy transfer model could explain the limited protection of mCherry fluorophore function by AavLEA1 (Fig. 4c).
Such a model of molecular shield function might suggest that the greatest protection would result from a covalent linkage of IDR to a labile protein, providing a shield in cis, rather than in trans. From an analysis of microbial proteomes, this would seem not to be a solution favoured by natural selection (Table 1). The data of Fig. 4d, where the fusion protein AavLEA1-mCherry was no less susceptible to desiccation damage than mCherry alone, indicates that the in cis strategy is not effective against intramolecular damage, but this needs further examination using target proteins more prone to aggregation under stress. Certainly, there is evidence to show that a covalently linked LEA protein can decrease the potential for aggregation under some circumstances: Singh et al.53 were able to reduce inclusion body formation and improve recovery of otherwise recalcitrant recombinant proteins by fusion with LEA proteins.
Molecular shield activity can be distinguished from that of molecular chaperones, although since it is increasingly recognised that many classical chaperones contain disordered regions apparently necessary for their function,31,46 it can be argued that both shields and chaperones populate a continuum of protein stabilisation activities, involving neutralisation of exposed hydrophobic surfaces by specific interaction, refolding of client proteins driven by ATP hydrolysis or entropy transfer (or both), steric or electrosteric stabilisation of crowded colloidal suspensions, and volume exclusion.17 In conclusion, we suggest there is a role in proteostasis for the stabilisation of the colloidal protein suspension of the cell interior by molecular shields, and that this is particularly significant under conditions of water stress.
Experimental
Constructs, recombinant proteins and antibodies
HSP70-pET28a+ and pcDNA3-HSP70-HA were gifts from C. Bertoncini (IRB Barcelona, Spain) and G. Kudla (University of Edinburgh, UK), respectively. The HSP70 used here encodes the human HSPA1A protein. pCI-FlagHDQ138 and pEGFP-HDQ74 constructs were gifts from D.C. Rubinsztein and S. Luo (CIMR, Cambridge, UK). pFLAG-CMV5a and pmCherry were obtained from Sigma-Aldrich (Poole, UK) and (Clontech, Saint-Germain-en-Laye, France), respectively. AavLEA1-mCherry-pcDNA3 and pET-15b-AavLEA1 have been described previously.30,54 AavLEA1 and Em cDNA sequences30,54 were PCR-amplified, the HA tag introduced at the C-terminus and then cloned into the HindIII and BamHI sites of pcDNA3.1 (Invitrogen, Paisley, UK). pET-28a-mCherry and pET-28-AavLEA1-mCherry were created as follows: the mCherry gene with engineered 5′ NdeI and 3′ SacI sites, and the AvLEA1 gene with engineered 5′ KpnI and 3′ BamHI sites, were PCR amplified from pmCherry and pET-15b-AavLEA1, respectively. Both products were inserted into pCR-2.1-TOPO (Invitrogen) by TOPO-TA cloning. The mCherry sequence was subcloned into pET-28a using the NdeI and SacI sites, while the AavLEA1 sequence was inserted upstream of the mCherry gene in pmCherry using the KpnI and EcoRI sites; the whole AavLEA1-mCherry sequence was then excised with NheI and EcoRI and inserted into pET-28a. All clones were verified by DNA sequencing. Expression and purification of recombinant His-tagged versions of HSP70, AavLEA1, and Em were performed as previously described.30,54,55 mCherry and AavLEA1-mCherry were produced largely as described for AavLEA1.54 Briefly, bacteria carrying expression constructs were grown to OD600 0.6 and expression induced by 1 mM IPTG with further growth for 12 h in sealed flasks at 37 °C or 30 °C for mCherry and AavLEA1-mCherry, respectively. Cells were harvested and lysed by sonication and His-tagged recombinant proteins purified using Ni-NTA sepharose (Qiagen, Crawley, UK). Eluted proteins were dialysed into phosphate buffered saline and the His-tag cleaved by overnight incubation at 4 °C with thrombin (GE Healthcare). The tag and any uncleaved protein were removed by Ni-NTA sepharose and thrombin was removed using benzamidine sepharose (GE Healthcare, Chalfont St Giles, UK). Primary antibodies used for immunoblotting were monoclonal anti-Flag (1 : 6000, Sigma-Aldrich) and monoclonal anti-HA (1 : 1000 for immunoprecipitation and 1 : 4000 for input; HA.11 clone 16B12, Covance, Princeton, NJ). Secondary antibodies used were horseradish peroxidase-linked ECL antimouse IgG (1 : 3333; GE Healthcare).
In vitro aggregation assay
CS aggregation assays under heat and desiccation stress were performed as described.23 Proteome extraction and subsequent partial proteome desiccation was performed as described15 (with a minor variation for CS: heat stress was for 2 h at 43 °C; vacuum drying was carried out at 1750 mTorr). ficoll 70 and Dextran 42 (analytical grade; Sigma-Aldrich) were of approximate Mr 70 000 and Mr 42 000, respectively. All assays were conducted in a total starting volume of 200 μl. In the case of desiccation, the reaction mix contained only water in addition to the indicated proteins/polysaccharides (which were dialysed or prepared in water prior to the experiments); in the case of heat stress, proteins were dialysed into PBS and the reaction mix also contained 50 mM HEPES pH7.5. For desiccation, CS was used at 5 μM, while protectants were used at 25 μM; for heat stress, CS was used at 1 μM, while protectants were used at 5 μM. The partial proteomes were used at 0.15 mg ml−1 (assuming an average Mr of 50 000) while the indicated molar excess of AavLEA1 protein or polysaccharide concentration was used.
mCherry drying experiments
Prior to drying experiments all proteins were dialysed extensively against water and the concentrations determined by absorbance at 280 nm using the following molar extinction coefficients: mCherry, 32 430 M−1 cm−1; AavLEA1-mCherry, 41 960 M−1 cm−1; AavLEA1, 8250 M−1 cm−1; BSA (Sigma-Aldrich, product number A7906), 43 824 M−1 cm−1. 100 μl samples in water were prepared containing 30 μM mCherry alone or in the presence of AavLEA1 or BSA (control) at molar ratios of 1 : 1 and 1 : 5, and 30 μM AavLEA1-mCherry fusion protein. Samples were dried in an Eppendorf 5301 vacuum concentrator, transferred to a Dura-Stop™ microprocessor-controlled vacuum tray drier (FTS Systems, Stone Ridge, NY) for 1 h at 500 mTorr with a tray temperature of 25 °C, and resuspended in 100 μl of water; four cycles of drying and rehydration were carried out. mCherry absorbance was measured at 23 °C in a Lambda 35 UV/visible spectrophotometer (Perkin Elmer, Cambridge, UK), using a 1 cm path-length UV-transparent cuvette. Wavelengths between 350 and 650 nm were measured with a scanning rate of 240 nm min−1 and a data interval of 1 nm. Fluorescence emission spectra were recorded using a Cary Eclipse fluorimeter (Agilent Technologies). Excitation was at 543 nm and emission spectra were recorded between 570 and 700 nm with a scan rate of 600 nm min−1 and a data interval of 1 nm. Emission and excitation slit width was set at ±10 nm and a photomultiplier tube voltage of 690 V was used.
Statistical analysis
Aggregation and drying experiments were performed in triplicate and the standard deviation is shown; statistical relevance was determined by one-way Anova and a Tukey post-test using InStat3 (GraphPad Software, La Jolla, CA).
Mammalian cells, transfections and immunoprecipitation
T-REx293 (Invitrogen) was grown at 37 °C in a 5% CO2, 100% relative humidity atmosphere in DMEM with 10% FBS, 5 mM glutamine, 500 units/ml penicillin, 0.5 mg ml−1 streptomycin (Sigma-Aldrich) and 5 μg ml−1 blasticidin (Invitrogen). Transient transfections were performed using GeneJammer (Agilent Technologies, Stockport, UK) according to the manufacturer’s instructions. For immunoprecipitation, 1 μg each of pFLAG-CMV5a/pCI-FlagHDQ13856 (mutant huntingtin, amino acids 1-588 with 138 glutamine residues) and HSP70-HA57/AavLEA1-HA/Em-HA were co-transfected for 24 h. For FRET analysis 0.75 μg each of pEGFP-mCherry linker,58 pEGFP-C1, pm-Cherry, pEGFP-HDQ7459 (mutant huntingtin exon 1 with 74 glutamine residues) and AavLEA1-mCherry30 were (co-)transfected as appropriate for 36 h. Immunoprecipitation and subsequent immunoblotting analysis were performed as described.36
FRET measurements
FRET between EGFP/EGFP-HDQ74 and pmCherry/pmCherry-tagged AavLEA1, or the positive control pEGFP-mCherry linker, was quantified via sensitized acceptor (mCherry) fluorescence upon donor (EGFP) excitation as previously described.35,36
Bioinformatics
Two applications were used to assess the extent of low-complexity amino acid patterns in input protein sequences, SEG42 and 0j.py,43 in case the results were just related to SEG. The web application FoldIndex was used for the IDR predictions via a purpose written Python script that allows programmatic interaction with the FoldIndex server. Purpose written Python scripts were also created to scan the FoldIndex predictions for each proteome and tally the percent-disordered predictions into decile bins. Statistical analysis (linear model creation and plotting) was done using the R package of statistical functions.
Supplementary Material
Acknowledgements
We thank C. Bertoncini, S. Luo, D.C. Rubinsztein and G. Kudla for gifts of plasmids. This work was funded by the European Research Council (Advanced Investigator Grant 233232; AT), the Broodbank Trust and Hughes Hall, Cambridge (SC), the Cambridge Trusts (RT), the Wellcome Trust/MRC Initiative for Neurodegenerative Disorders (CFK) and the EPSRC (CFK, GSK).
References
- 1.Crowe JH, Hoekstra FA, Crowe LM. Annu Rev Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- 2.Burnell AM, Tunnacliffe A. In: Molecular and Physiological Basis of Nematode Survival. Perry RN, Wharton D, editors. CABI Publishing; Wallingford, UK: 2010. [Google Scholar]
- 3.Clegg JS. Comp Biochem Physiol Part B: Biochem Mol Biol. 2001;128:613–624. doi: 10.1016/s1096-4959(01)00300-1. [DOI] [PubMed] [Google Scholar]
- 4.Alpert P. J Exp Biol. 2006;209:1575–1584. doi: 10.1242/jeb.02179. [DOI] [PubMed] [Google Scholar]
- 5.Gadd GM, Chalmers K, Reed RH. FEMS Microbiol Lett. 1987;48:249–254. [Google Scholar]
- 6.Ratnakumar S, Tunnacliffe A. FEMS Yeast Res. 2006;6:902–913. doi: 10.1111/j.1567-1364.2006.00066.x. [DOI] [PubMed] [Google Scholar]
- 7.Ingram J, Bartels D. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 8.Bartels D, Schneider K, Terstappen G, Piatkowski D, Salamini F. Planta. 1990;181:27–34. doi: 10.1007/BF00202321. [DOI] [PubMed] [Google Scholar]
- 9.Madin KAC, Crowe JH. Am Zool. 1975;15:802–802. [Google Scholar]
- 10.Siddiqui KS, Cavicchioli R. Annu Rev Biochem. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 11.Razvi A, Scholtz JM. Protein Sci. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saibil HR. Curr Opin Struct Biol. 2008;18:35–42. doi: 10.1016/j.sbi.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Balch WE, Morimoto RI, Dillin A, Kelly JW. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 14.Oliver MJ, Tuba Z, Mishler BD. Plant Ecology. 2000;151:85–100. [Google Scholar]
- 15.Chakrabortee S, Boschetti C, Walton LJ, Sarkar S, Rubinsztein DC, Tunnacliffe A. Proc Natl Acad Sci U S A. 2007;104:18073–18078. doi: 10.1073/pnas.0706964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krisko A, Smole Z, Debret G, Nikolic N, Redman M. J Mol Biol. 2010;402:775–782. doi: 10.1016/j.jmb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Tunnacliffe A, Hincha DK, Leprince O, Macherel D. In: Dormancy and Resistance in Harsh Environments. Lubzens E, Cerda J, Clark MS, editors. Springer; Berlin: 2010. pp. 91–108. [Google Scholar]
- 18.Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias A. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih M, Hoekstra F, Hsing Y. Adv Bot Res. 2008;48:211–255. [Google Scholar]
- 20.Tunnacliffe A, Wise MJ. Naturwissenschaften. 2007;94:791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- 21.Wise MJ, Tunnacliffe A. Trends Plant Sci. 2004;9:13–17. doi: 10.1016/j.tplants.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Ballesta MT, Rodrigo MJ, LaFuente MT, Granell A, Zacarias L. J Agric Food Chem. 2004;52:1950–1957. doi: 10.1021/jf035216+. [DOI] [PubMed] [Google Scholar]
- 23.Goyal K, Walton LJ, Tunnacliffe A. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grelet J, Benamar A, Teyssier E, Avelange-Macherel MH, Grunwald D, Macherel D. Plant Physiol. 2005;137:157–167. doi: 10.1104/pp.104.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reyes JL, Rodrigo MJ, Colmenero-Flores JM, Gil JV, Garay-Arroyo A, Campos F, Salamini F, Bartels D, Covarrubias AA. Plant Cell Environ. 2005;28:709–718. [Google Scholar]
- 26.Reyes JL, Campos F, Wei H, Arora R, Yang YI, Karlson DT, Covarrubias AA. Plant Cell Environ. 2008;31:1781–1790. doi: 10.1111/j.1365-3040.2008.01879.x. [DOI] [PubMed] [Google Scholar]
- 27.Haaning S, Radutoiu S, Hoffmann SV, Dittmer J, Giehm L, Otzen DE, Stougaard J. J Biol Chem. 2008;283:31142–31152. doi: 10.1074/jbc.M805024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama K, Okawa K, Kakizaki T, Inaba T. Biosci Biotechnol Biochem. 2008;72:1642–1645. doi: 10.1271/bbb.80214. [DOI] [PubMed] [Google Scholar]
- 29.Kovacs D, Kalmar E, Torok Z, Tompa P. Plant Physiol. 2008;147:381–390. doi: 10.1104/pp.108.118208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Chakrabortee S, Li RH, Zheng YZ, Tunnacliffe A. FEBS Lett. 2011;585:630–634. doi: 10.1016/j.febslet.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Uversky VN. Chem Rev. 2011;111:1134–1166. doi: 10.1021/cr100186d. [DOI] [PubMed] [Google Scholar]
- 32.Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, Frydman J. Nat Struct Mol Biol. 2009;16:1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhai RG, Zhang F, Hiesinger PR, Cao Y, Haueter CM, Bellen HJ. Nature. 2008;452:887–U888. doi: 10.1038/nature06721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sittler A, Lurz R, Lueder G, Priller J, Hayer-Hartl MK, Hartl FU, Lehrach H, Wanker EE. Hum Mol Genet. 2001;10:1307–1315. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 35.Elder AD, Domin A, Schierle GSK, Lindon C, Pines J, Esposito A, Kaminski CF. J R Soc Interface. 2009;6:S59–S81. [Google Scholar]
- 36.Chakrabortee S, Meersman F, Kaminski Schierle GS, Bertoncini CW, McGee B, Kaminski CF, Tunnacliffe A. Proc Natl Acad Sci U S A. 2010;107:16084–16089. doi: 10.1073/pnas.1006276107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bright JN, Woolf TB, Hoh JH. Prog Biophys Mol Biol. 2001;76:131–173. doi: 10.1016/s0079-6107(01)00012-8. [DOI] [PubMed] [Google Scholar]
- 38.Napper DH. Polymeric Stabilization of Colloidal Dispersions. Academic Press; London: 1983. [Google Scholar]
- 39.van den Berg B, Wain R, Dobson CM, Ellis RJ. EMBO J. 2000;19:3870–3875. doi: 10.1093/emboj/19.15.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Awile O, Krisko A, Sbalzarini IF, Zagrovic B. PLoS Comput Biol. 2010:6. doi: 10.1371/journal.pcbi.1000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 42.Wootton JC, Federhen S. Comput Chem. 1993;17:149–163. [Google Scholar]
- 43.Wise MJ. Bioinformatics. 2001;S17:288–295. [Google Scholar]
- 44.Hoekstra FA, Golovina EA, Buitink J. Trends Plant Sci. 2001;6:431–438. doi: 10.1016/s1360-1385(01)02052-0. [DOI] [PubMed] [Google Scholar]
- 45.Dure L. Protein Pept Lett. 2001;8:115–122. [Google Scholar]
- 46.Tompa P, Csermely P. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 47.Altikatoglu M, Basaran Y. Protein J. 2011;30:84–90. doi: 10.1007/s10930-011-9306-4. [DOI] [PubMed] [Google Scholar]
- 48.Crowley PB, Brett K, Muldoon J. ChemBioChem. 2008;9:685–688. doi: 10.1002/cbic.200700603. [DOI] [PubMed] [Google Scholar]
- 49.Mukhopadhyay R, Kumar S, Hoh JH. BioEssays. 2004;26:1017–1025. doi: 10.1002/bies.20088. [DOI] [PubMed] [Google Scholar]
- 50.Tolleter D, Hincha DK, Macherel D. Biochim Biophys Acta Biomembr. 2010;1798:1926–1933. doi: 10.1016/j.bbamem.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 51.Thalhammer A, Hundertmark M, Popova AV, Seckler R, Hincha DK. Biochim Biophys Acta Biomembr. 2010;1798:1812–1820. doi: 10.1016/j.bbamem.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 52.Popova AV, Hundertmark M, Seckler R, Hincha DK. Biochim Biophys Acta Biomembr. 2011;1808:1879–1887. doi: 10.1016/j.bbamem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 53.Singh J, Whitwill S, Lacroix G, Douglas J, Dubuc E, Allard G, Keller W, Schernthaner JP. Protein Expression Purif. 2009;67:15–22. doi: 10.1016/j.pep.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Goyal K, Tisi L, Basran A, Browne J, Burnell A, Zurdo J, Tunnacliffe A. J Biol Chem. 2003;278:12977–12984. doi: 10.1074/jbc.M212007200. [DOI] [PubMed] [Google Scholar]
- 55.Roodveldt C, Bertoncini CW, Andersson A, van der Goot AT, Hsu ST, Fernandez-Montesinos R, de Jong J, van Ham TJ, Nollen EA, Pozo D, Christodoulou J, Dobson CM. EMBO J. 2009;28:3758–3770. doi: 10.1038/emboj.2009.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo SQ, Vacher C, Davies JE, Rubinsztein DC. J Cell Biol. 2005;169:647–656. doi: 10.1083/jcb.200412071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. PLoS Biol. 2006;4:933–942. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lleres D, Swift S, Lamond AI. Curr Protoc Cytom. 2007;Chapter 12:Unit12 10. doi: 10.1002/0471142956.cy1210s42. [DOI] [PubMed] [Google Scholar]
- 59.Narain Y, Wyttenbach A, Rankin J, Furlong RA, Rubinsztein DC. J Med Genet. 1999;36:739–746. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.