Abstract
Limitations of animal infection models have engendered longstanding obstacles in basic science and translational research. Lack of suitable animal models, the need for better predictors of human immune responses, and pathogens that grow poorly or not at all outside the human host impact our ability to study infectious agents that cause human disease, generation of essential tools for genetic manipulation of microbial pathogens, and development of vaccines, therapeutics, and host-targeted immunotherapies. The advent of conceptual and methodological advances in tissue engineering along with collaborative efforts between the bioengineering and infectious diseases scientific communities holds great promise to overcome these significant barriers.
Keywords: tissue engineering, infectious diseases, microbial pathogenesis, 2D, 3D, stem cells
Teaser sentence
Bioengineered human tissue models demonstrate the need to balance ease of use for basic science research with recapitulation of human tissue architecture in organotypic models for infectious disease and preclinical product development studies.
The use of animal models in infectious diseases research enabled historic developments in vaccines and therapeutics, and contributed greatly to our understanding of microbial pathogenic mechanisms. However, limitations inherent to animal models of infection also hindered progress in these areas. Some pathogens have a uniquely human host range; their inability to infect any animal species confounds the development of products to combat these infections. Moreover, animal infections inadequately reproduce human disease pathophysiology [1–4]. Classical two dimensional (2D) in vitro culture systems typically used in research laboratory settings lack tissue-like architecture and mirror the inadequacies of animal models, i.e., the failure to mimic human disease or to reflect the immune response. Thus, preclinical studies in animal models and traditional in vitro cell culture models that use primarily unicellular, transformed cells cannot accurately predict clinical trial outcomes, a significant obstacle that underscores the need for better predictors of efficacy, host toxicity, and host-targeted therapies. Lastly, some pathogens cannot be cultured routinely, which precludes the development of genetic tools for these microbes and impedes insights into their pathogenesis.
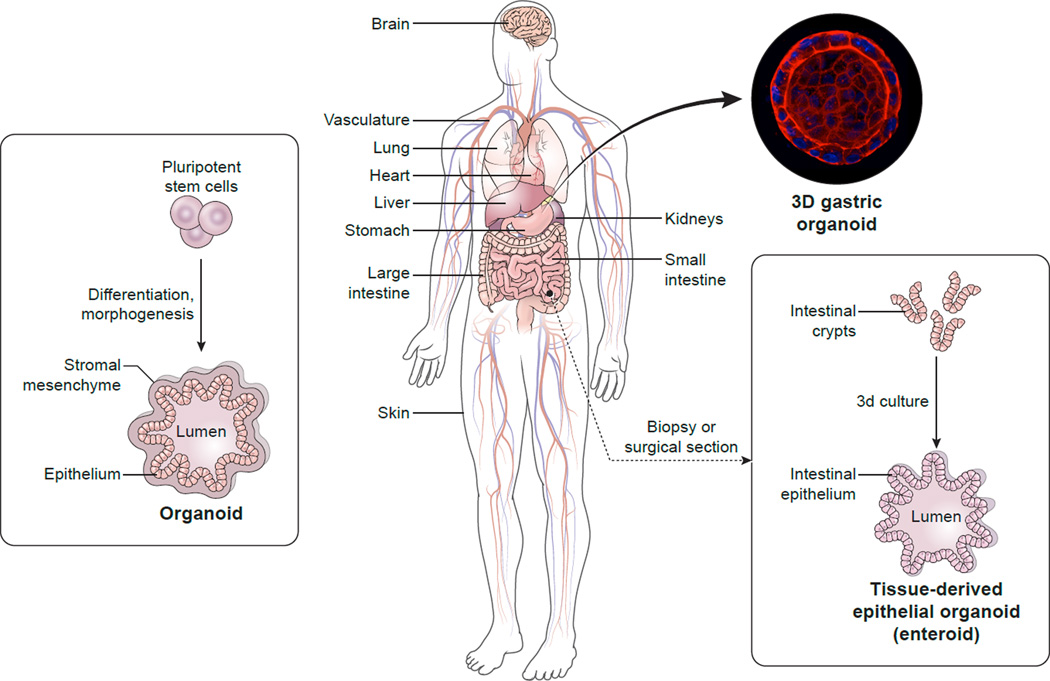
The potential to generate improved infection models evolved with breakthroughs in understanding embryogenesis, developmental biology and stem cells, in conjunction with technological advances in tissue engineering, a field that initially focused on regenerative medicine [5]. Biomimetic infection models applicable to basic and translational research represent critical advances for the infectious diseases field, and collaborations between bioengineers and infectious disease experts to generate new models represent a significant scientific frontier. Early studies of bacterial pathogenesis, some of which combined advances in tissue engineering with infectious diseases research, generated intriguing results [6–8]. Now, collaborations between tissue engineers and infectious diseases experts are poised to realize in-depth studies in human-pathogen interactions that address longstanding scientific obstacles. Here, we review selected, three dimensional (3D) and derived 2D nontransformed human experimental ex vivo tissue-like models developed to recapitulate major organ systems (Figures 1 and 2, Table 1) and their related pathogens. This exploding area of research explores many questions of normal human cell biology, physiology and pathophysiology, and these new tools are being applied to precision medicine, regenerative medicine and tissue engineering. Many types of human organs are being modeled, and varied nomenclature causes some confusion in the field. Pluripotent stem cell (PSC)-derived tissues and structures are most frequently called organoids. However, “organoids” may have different origins. For example, different models of “mini-intestine” cultures of the small intestinal tract can be distinguished; tissue-derived epithelial-only cultures, tissue-derived epithelial-mesenchymal cultures that are grown in air-liquid interface conditions, and PSC-derived cultures [9]. Although the Intestinal Stem Cell Consortium [10] proposed using the term organoid to refer to cultures derived from PSC and using the term enteroids to refer to cultures derived from crypts (containing intestinal stem cells) isolated from biopsy or surgical tissues (Figure 1), this has not been uniformly followed. We use these terminologies herein but also discuss specific models following the nomenclature in individual publications. A few methodology and review articles are included here [11–13] and in pertinent sections.
Figure 1. Generating human tissue biomimetics for studies of infectious diseases.
(Left) Stem cells can undergo directed differentiation, morphogenesis, and, potentially, organogenesis to develop human organ-specific organoids. The organoid shown contains both mesenchymal and epithelial tissue with an open interior to which may be added microbes, immune cells or other substances, e.g., therapeutics. An alternative method uses biopsy or surgical sections to generate organoids comprised of epithelial tissue; the tissue-derived epithelial organoid shown (Right) shows the use of a small intestinal tissue sample to obtain intestinal crypts that ultimately form an “inside-out” enteroid with an outer epithelial layer and an interior lumen. The photo inset shows an “inside-in” polarity gastric epithelial organoid derived from human stomach tissue and stained for nuclei and the actin cytoskeleton. Permission to use this confocal reconstruction image was generously provided by Drs. Calvin J. Kuo and Manuel R. Amieva (Huang, J.Y. et al. (2015) Cell Host Microbe 18 (2), 147–156. See [33] in main reference list.
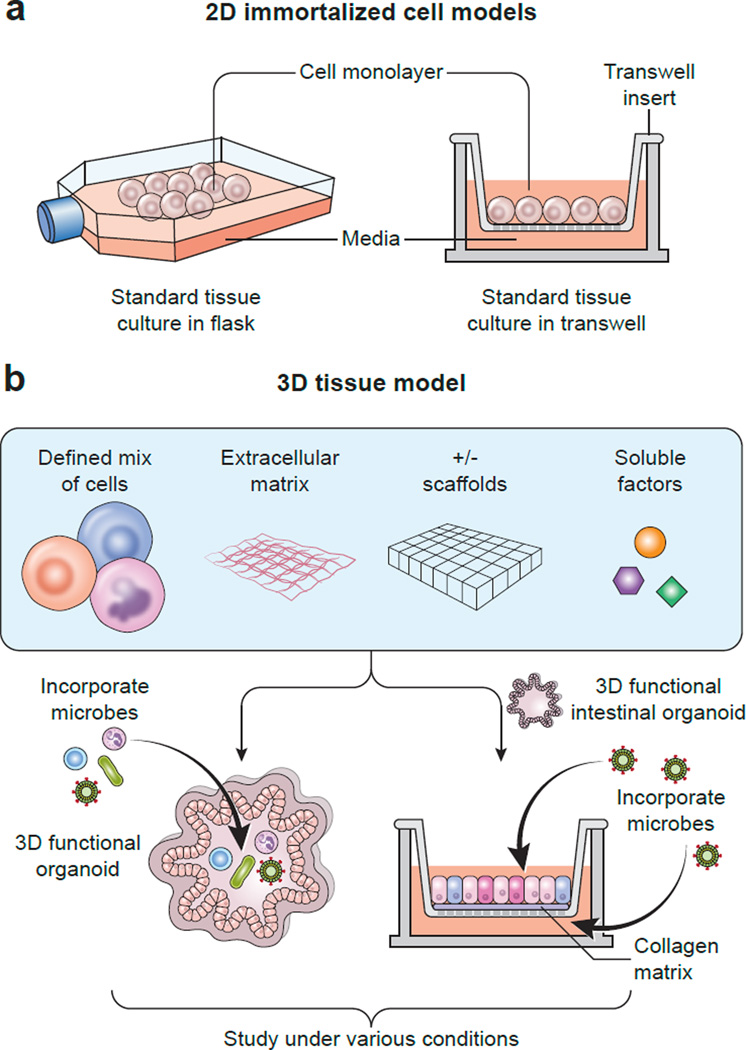
Figure 2. 2D and 3D tissue culture models differ in cell sources, additives, methods, and model complexity.
Panel A. This illustration shows a classical 2D cell culture model for studies of infectious diseases. An immortalized cell line was cultured in flasks, cells were harvested, recombined with media, and then plated and grown as monolayers comprised of a single cell type in various cell culture dishes, e.g, microtiter plates. The Transwell™ culture system represented a technological advance. Cell monolayers can be assessed for polarization; and microbes, immune cells, or other substances can be added either to the apical or basolateral side of the monolayer. Panel B. Increased complexity of 3D tissue culture models is depicted in this schematic. Cell mixtures contain different cell types; an extracellular matrix may consist of collagen or more complex materials, e.g., Matrigel; soluble factors, e.g., growth factors, are added to the growth medium; and these components may be cultured in the presence or absence of scaffolding materials, which can serve as structural guides for tissue development. (Left) Microbial organisms (commensals and/or pathogens) or immune cells, or other factors, can be added to functional 3D organoids, which then can be studied under various conditions, e.g., by differing the pH or oxygenation status, or by adding mechanical or electrical stimulation. (Right) 3D functional intestinal epithelial organoids, or enteroids, may be converted into 2D monolayers for culturing in Transwell™ systems; a matrix (matrigel or collagen) may be added to the porous membrane to serve as an extracellular matrix; pathogens or commensal microbes may be added (here shown as viruses added to medium in the apical and basolateral compartments); immune cells or other factors can be added, and then the model can be exposed to varying conditions, e.g., differing pH or oxygenation.
Table 1.
Pathogenic Models: Characteristics and Features
| Models | Cell Types | Matrix / Scaffold |
Stability | Responses | References |
|---|---|---|---|---|---|
| BRAIN | |||||
| Cortical brain tissue | Primary cortical neurons (rat) |
Collagen 1 and Silk-collagen protein |
Weeks to months | Physiological: Axon growth Neuronal clusters/networks Neuronal gene expression Electrophysiological function Pharmacophysiological response Pathophysiological: Traumatic brain injury: Neuronal damage Impact-induced hyperactivity Increased Glu levels post-injury |
Chwalek et al. 2015. Nat Protoc Tang-Schomer et al. 2014. Proc Natl Acad Sci U S A |
| Cerebral organoid | hPSCsa or iPSCsb or Patient-specific iPSCs (microcephaly) fibroblast feeder cells |
Matrigel or Growth factor- reduced Matrigel |
6–12 months | Physiological: Discrete, interdependent brain regions Fluid-filled ventricle-like structure Glutamatergic receptor activity Neural activity Pathophysiological: Microcephaly model: Smaller neural tissues Premature neural differentiation Neurogenic non-proliferative divisions Imprecise glial spindle orientation |
Lancaster et al. 2013. Nature Lancaster and Knoblich. 2014. Nature Protocols |
| Brain organoid | iPSCs | Matrigel | Up to 10 months | Physiological: Mimics 1st trimester development Pathophysiological: Zika virus infection: Growth rate reduction |
Garcez et al. 2016. Science Lancaster et al. 2013. Nature |
| Forebrain organoids | iPSCs | Matrigel | Months | Physiological: Embryonic cortical development Markers for 6 cortical layers Complex neuronal morphology Neuronal maturation features Pathophysiological: Zika virus infection: (differing fetal stage models) Cell type tropism Decreased organoid size Increased lumen/ventricle size Reduced neuronal layer thickness |
Qian et al. 2016. Cell |
| Cerebral organoid | hESCsc H9 hESCs (WA09) Irradiated mouse embryonic fibroblasts |
Matrigel | NPd | Physiological: Complex morphology Fluid-filled ventricle-like structure Regional specificity Neural activity Mimics early trimester fetal tissue Pathophysiological: Zika virus infection (MR766 strain): Organoid shrinkage Increase in viral copy number Neurogenesis dysregulation Activation of apoptosis TLR3 network perturbation |
Dang et al. 2016. Cell Stem Cell Lancaster et al. 2013. Nature |
| STOMACH | |||||
| Gastric epithelial organoid | hPSCs | Matrigel | > 1 month | Physiological: Recapitulate embryonic development Differentiated antral epithelial cells High degree of cellular complexity Pathophysiological: H. pylori infection: Epithelial responses CagA translocation CagA-dependent proliferation |
McCracken et al. 2014. Nature |
| Gastric epithelial organoid | Human adult tissues: (gastric glands from normal/cancerous stomach tissue) |
Matrigel | > 1 year - may be frozen and thawed indefinitely |
Physiological: Expressed markers of the 4 lineages Self assembled gland and pit domains Mucus producing pit or gland cells Pathophysiological: H. pylori infection: Inflammatory response (gland lineage) |
Bartfeld et al. 2015. Gastroenterology Bartfeld and Clevers. 2015. J Vis Exp |
| Gastric epithelial organoid | Human adult tissues: (gastric glands from normal stomach tissue) |
Matrigel | > 1 year - may be frozen and thawed indefinitely |
Physiological: Metabolite production (urea) Pathophysiological: H. pylori infection: Chemoreception Chemotaxis Host urea destruction |
Bartfeld and Clevers. 2015. J Vis Exp Huang et al. 2015. Cell Host Microbe |
| Gastric epithelial organoid | Human adult tissues: (gastric glands from normal stomach tissue) |
Matrigel | ~Indefinite | Physiological: Polarized epithelial cells Gland-like structures Mucus secretion Epithelial proliferation Pathophysiological: H. pylori infection: Morphological changes Inflammatory response |
Schlaermann et al. 2016. Gut |
| (GASTRO) INTESTINAL | |||||
| Gastrointestinal epithelial spheroids: Stomach Duodenum Ileum Colon |
Human adult tissues: (Patient-specific intestinal crypts/ stem cells) |
Matrigel | Spheroid cell lines amenable to freezing and storage |
Physiological: Barrier integrity Mucus layer production Pathophysiological: Pathogenic E. coli infections: (EAggEC, EHEC, EPEC): Adherence |
VanDussen et al. 2015. Gut |
| Intestinal epithelial tissue | Human intestinal cell lines: Caco-2 Ht29-MTX H-InMyoFib (Intestinal Myofibroblasts) |
Collagen 1 Silk fibroin protein |
≥ 2 months | Physiological: Mucus layer production Intraluminal oxygen gradients Pathophysiological: Y. pseudotuberculosis interaction: Colonization L. rhamnosus GG (probiotic): Colonization |
Chen et al. 2015. Sci Rep |
| Small intestinal enteroids: Duodenum Jejunum Ileum |
Human adult tissues: (Patient-specific intestinal crypts/stem cells) |
Matrigel | Amenable to freezing and storage |
Physiological: NP Pathophysiological: Human Rotavirus (HRV) and Animal Rotavirus (ARV) infection: Host range restriction HRV replication Fluid secretion mimicry Vaccine strain attenuation |
Saxena et al. 2015. J Virol |
| Duodenal enteroids | Human adult tissues: (intestinal crypts/stem cells) |
Matrigel | Up to 8 months | Physiological: Ion transport Absorptive processes Fluid secretion Pathophysiological: Cholera toxin responses |
Foulke-Abel et al. 2016. Gastroenterology |
| Ileal and colonic enteroids | Human adult tissues: (intestinal crypts/stem cells) |
Matrigel | NP | Physiological: Blood group antigen expression Pathophysiological: Cholera toxin treatment: cAMP activation |
Kuhlmann et al. 2016. Am J Trop Med Hyg |
| Intestinal Organoids | hPSC line: A1ATD-1 |
Matrigel | NP | Physiological: Intestinal cell type markers Crypt-like and microvilli structures Pathophysiological: Salmonella infection: Global transcriptional changes Bacterial invasion Salmonella-containing vacuoles |
Forbester et al. 2015. Infect Immun |
| Colonoids (HCMs) | Human adult tissues: (intestinal crypts/stem cells) |
Matrigel | NP | Physiological: Organized brush borders Mucus production Pathophysiological: EHEC infection: Colonization Brush border damage Mucus layer destruction Intermicrovillar bridge reduction |
In et al. 2016. Cell Mol Gastroenterol Hepatol Tyska, M.J. 2016. Cell Mol Gastroenterol Hepatol |
| Large intestine organoids | hPSCs | Matrigel | NP | Physiological: Secretory/absorptive functions Mucus production Pathophysiological: C. difficile infection: Inhibition of NHE3 Decreased MUC2 levels |
Engevik et al. 2015. Am J Physiol Gastrointest Liver Physiol 308:G497- G509 Engevik et al. 2015. Am J Physiol Gastrointest Liver Physiol 308:G510- G524 |
| KIDNEY | |||||
| Renal tissue | Human renal proximal tubule cell line: NKi-2 |
Matrigel- Collagen 1 |
NP | Physiological: NP Pathophysiological: Shiga toxin Type 2 treatment: Inhibition of protein synthesis Cytotoxicity Kim-1 secretion (injury marker) |
DesRoches et al. 2015. Infect Immun |
| LIVER | |||||
| Hepatic sinusoid tissue | Human hepatocyte cell line: Huh-7 Huh-NTCP Liver sinusoidal endothelial cell lines (unknown designation) |
Collagen 1 | NP | Physiological (select examples): LSEC barrier function Hepatic adhesive proteins Hepatic ECM release Pathophysiological: E. histolytica infection: Parasite adhesion Parasite migration Increased cytokine levels HBV infection: Viral replication |
Petropolis et al. 2014. PLoS Pathog Petropolis et al. 2016. PLoS One |
| MPCCs (Micropatterned Cocultures) |
Human primary hepatocytes Mouse embryonic fibroblast cell lines: 3T3-J2 or NIH-3T3 |
Collagen 1 on MP plates |
4–6 weeks | Physiological (select examples): CYP450 activity Albumin/urea secretion NTCP (HBV receptor) Pathophysiological: HBV/HCV entry and infection |
March et al. 2015. Nat Protoc Shlomai et al. 2014 Proc Natl Acad Sci U S A Ploss et al. 2010 Proc Natl Acad Sci U S A |
| Hepatocyte-like cells | iPSC lines: RC2 iPS.C2a LN4 Subject-specific iHLCse |
Matrigel | Months | Physiological: Polygonal morphology Hepatic markers Host entry factors (parasites) Pathophysiological: Plasmodium infection: Supports liver-stage malaria Antimalarial drug sensitivity |
Ng et al. 2015. Stem Cell Reports |
| LUNG | |||||
| Lung tissue | Human cell lines: Lung fibroblasts: MRC-5 Bronchial epithelial: 16HBE14o |
Collagen 1 | Weeks | Physiological: ECM protein synthesis Pathophysiological: M. tuberculosis infection: TB-infected macrophage migration Granuloma-like formation |
Braian et al. 2015. J Vis Exp |
| Lung tissue | Human cell lines: Lung fibroblasts: MRC-5 Bronchial epithelial: 16HBE14o- Alveolar epithelial: A549 |
Collagen 1 | NP | Physiological: Morphology Pathophysiological: S. aureus infection: Cytotoxicity Neutrophil-mediated damage Inflammatory response |
Shambat et al. 2015
Dis Model Mech Nguyen Hoang et al. 2014. J Leukoc Biol |
| Lung tissue | Human cell lines: Lung fibroblasts: MRC-5 Bronchial epithelial: 16HBE14o- |
Collagen 1 | NP | Physiological: NP Pathophysiological: Andes Hantavirus infection: Viral replication Immune responses |
Sundström et al. 2016. PLoS One |
| SKIN | |||||
| Skin tissue | Human primary fibroblasts Human cell line: Keratinocyte: HaCaT Pheochromocytoma cell line: PC12 |
Collagen | Weeks | Physiological: Epidermal stratification Epidermal-dermal interactions HSV-1 infected neuronal cells Pathophysiological: HSV-1 infection: Viral latency Reactivation of latent HSV-1 |
Hogk et al. 2013. Methods Mol Biol |
| Skin tissue | Human cell lines: Keratinocytes: N/TERT-1 HaCaT Human primary cells: Neutrophils PBMCsf |
Collagen | Weeks | Physiological: Epidermal stratification Epidermal-dermal interactions Pathophysiological: SDSEg infection: Colonization Bacterial replication Tissue damage |
Siemens et al. 2015.Sci Rep |
hPSCs (human pluripotent stem cells)
iPSCs (induced pluripotent stem cells)
hESCs (human embryonic stem cells)
NP Not provided
iHLCs (iPSC-derived hepatocyte-like cells)
PBMCs (Peripheral blood mononuclear cells)
SDSE (Streptococcus dysgalactiae subsp. equisimilis)
Pathogenesis Models
Brain
3D brain-like tissues were developed with primary rat cortical neuron [14,15] or human pluripotent stem cell (hPSC) cultures [16,17]. Models generated with primary cortical neuron cultures used biomaterials of differing mechanical properties, i.e., silk protein-based stiff, porous scaffolds for neuronal anchoring and softer extracellular matrix (ECM) gels for axon penetration and connectivity. This 3D model formed functional brain-like cortical tissue that was maintained for months in vitro. The modular format provides a flexible platform approach that can be used to fabricate other scaffold shapes and incorporate additional extracellular matrix (ECM) components, soluble factors, and cell types to achieve more complex brain-like organizations. This physiologically relevant biomimetic responded to traumatic injury induced via the weight-drop method and exhibited neuronal damage, injury-induced surge of the excitatory Glu (glutamate) neurotransmitter, and changes in electrophysiological activity consistent with in vivo responses to concussive brain injury [15]. Although these studies did not involve infectious agents, recently, organoid models have been used to study Zika virus and demonstrate viral tropism for human brain cells and disrupted neurogenesis [18–20]. Progress in recreating blood-brain barrier physiology in a neurovascular unit may also facilitate new therapeutics to overcome the challenge of delivery of drugs for neurotropic pathogens [21,22].
3D cerebral organoids generated from in vitro culture of hPSCs recapitulate fundamental mechanisms of mammalian neurodevelopment and display characteristics of human brain development. Within 1–2 months in culture, the brain-like tissue revealed essential features (developing cerebral cortex, ventral telencephalon, choroid plexus and retinal identities); these organoids could be maintained in long-term culture for > a year. Studies with cerebral organoids derived from a microcephalic patient modeled aspects of microcephaly; results showed smaller neuroepithelial tissues and a large degree of neuronal outgrowth compared to controls, findings that suggest premature neural differentiation at the expense of early progenitors, supporting a model wherein the failure of progenitor population expansion leads to smaller brain size and smaller skull formation [16].
Gastrointestinal Tract
Selected review articles relevant to novel gastrointestinal tract models and pathogenesis studies are provided [9,23–28].
Gastric Models
Human gastric organoid (hGO) systems were developed by several investigative groups and used to study gastric physiology, signaling pathways involved in proliferation of the epithelium, and the pathogenesis of Helicobacter pylori, the causative agent of peptic ulcer disease and gastric adenocarcinoma; the latter of which led to WHO designation of H. pylori as a Class 1 Carcinogen [29–34]. In one study, human pluripotent stem cells were used to develop de novo 3D human gastric tissue through directed differentiation. Organoid development was similar to in vivo gastric organogenesis; cellular complexity mimicked that of the human gastric antrum, and, importantly, via this first in vitro model of human stomach development, the investigators reported a new role for EGF receptor signaling during embryonic stomach formation. H. pylori microinjected into hGO lumens showed translocation of the CagA virulence factor, thought to be involved in malignant transformation, into epithelial cells; association of CagA with the host c-Met receptor; and a twofold increase in epithelial cell proliferation [29]. Another study showed that the stem cell marker, cluster-of-differentiation (CD44), plays a functional role in H. pylori-induced epithelial cell proliferation, and this is associated with the bacteria expressing a functional CagA [34]. Taken together, these data suggest that hGO models reflect physiological changes expected upon H. pylori infection.
Another group generated hGOs with multipotent stem cells isolated from adult human gastric tissue for studies of the epithelial response to H. pylori [30,31]. These hGOs were grown from single stem cells, and organoids could be directed into the four different lineages of the stomach (pit mucous cells, gland mucous cells, chief cells, and enteroendocrine cells), reflecting normal attributes of the human stomach. This system demonstrated long-term (>1 year) cultivation without loss of essential features, and organoids could be frozen and thawed similar to cell lines, characteristics that permit studies of long duration, and the ability to culture equivalent organoids for later experiments [30,31]. H. pylori-infected hGOs demonstrated robust NF-κB activation, revealing the capacity of this hGO model to mount an innate inflammatory response to H. pylori, as is seen in the human host. Interleukin (IL)-8 expression in hGOs was independent of bacterial viability, the bacterial cytotoxicity associated gene pathogenicity island (cagPAI), and of TLR4, 5, and 9 signalling [30], unlike earlier studies that showed cagA-dependent activation of NF-κB in transformed human epithelial cell lines [35–37]. The differential findings related to cagA and the inflammatory response from these two model systems highlights the changing landscape of information arising with the use of physiologically relevant tissue models.
Recently, hGOs developed with gastric glands isolated from normal human stomach tissue were used to identify the mechanism whereby H. pylori locates the gastric epithelia prior to infection (Fig. 1 photo inset shows an inside-in polarity hGO) [33]. Inside-out polarity hGOs allowed access to apical surfaces; bacteria were observed swimming towards organoids and adhering to tight junctions. These studies revealed that H. pylori detects minute quantities of urea metabolite via its chemoreceptor TlpB and rapidly responds in a chemotactic-dependent manner; TlpB is a high affinity chemoreceptor that binds urea directly. Importantly, the pathogen’s virulence factor urease plays a role in chemosensation. Urea concentration in human stomach ranges from 1–5mM, but TlpB detects urea in nanomolar quantities, suggesting that bacterial urease, which lowers urea concentrations, might be connected to urea sensing. Thus, the hGOs produced urea and the TlpB chemoreceptor was sufficiently sensitive to detect it in the minute quantities that diffused from the organoid epithelia, and the use of this novel model revealed a chemoreception system linked to a known virulence factor [31,33].
More recently, organoids were generated from gastric glands isolated from healthy human stomach tissue and shown to recapitulate aspects of normal gastric physiology [32]. Sheared gastric organoid cultures inoculated with a CagA+ H. pylori strain exhibited translocation and phosphorylation of CagA, as well as the hallmark ‘hummingbird phenotype’ of infection. A robust inflammatory response was observed via microarray analyses; here too, highly upregulated genes were associated with the NF-κB signaling pathway, highlighting another model that can potentially define novel mechanisms of gastric disease pathogenesis.
Intestinal Models
Human intestinal culture systems representing compartments throughout the intestine have been developed and used to study normal human gastrointestinal physiology as well as pathophysiology after infection with viral and bacterial pathogens [38–47]. A Transwell® system was used to establish cultures (called spheroids, but these would be called enteroids per the ISSC nomenclature proposal [10]) from a patient-specific panel of intestinal crypt units from biopsy specimens [38]. These cultures displayed a relatively rapid differentiation rate (~2–3 weeks) well suited to personalized medicine. Banked cultures include lines from healthy patients, patients with inflammatory diseases, and patients with prior intestinal surgery. Duodenal, ileal, and rectal cultures retained regional-specific cell markers, confirming other studies showing small intestinal adult stem cells are intrinsically programmed to recapitulate location-specific function [44]. For in vitro studies of physiological processes, the ileal and rectal enteroid cultures were dissociated and seeded onto coated Transwell® membranes to form polarized epithelial monolayers, which were shown to retain their complex multicellular phenotype and to possess a mucus layer and barrier function based on measuring transepithelial resistance. Adherence of diarrheagenic Escherichia coli (EC) strains [enteroaggregative (EaggEC), enteropathogenic (EPEC), and enterohemmorhagic (EHEC)] and a nonadherent EC control strain were compared in 2D cultures with HeLa cell monolayers. The control strain did not adhere to any cell line; EPEC and EaggEC adhered to ileal and rectal epithelial cells in patterns similar to those in HeLa cells; diffuse EHEC adherence was visualized only in ileal and rectal cell cultures. The latter result suggests that primary human epithelial enteroids are more suitable for adherence assays of diarrheagenic E. coli. An unexpected finding was that EPEC, a small bowel pathogen, adhered to rectal epithelial cells, suggesting there may be a role for this bacterial pathogen in the colonic pathology occasionally reported in patients with EPEC or EAggEc who have bloody diarrhea [38]. The ability to make monolayer enteroid cultures facilitates infections with many enteric pathogens and commensals [25,38,41]. Most importantly, the ability to bank host-specific tissue-derived enteroids supports patient-directed experimental approaches that not only are an important step toward the goal of personalized medicine, but also provide the potential to dissect the impact of host genotype on host-pathogen interactions and deleterious sequela.
Another novel 3D in vitro model of the human large intestine was bioengineered to mimic the structure and function of native intestine [39]. Modular intestinal constructs comprised functional intestinal epithelium cultured on silk-based protein scaffolds with hollow lumens; primary human intestinal myofibroblasts were seeded within scaffold bulk spaces, and human enterocyte-like and goblet-like cells from the cancer-derived Caco-2 and HT29-MTX cell lines were cultured on the surface of scaffold lumens. To explore whether cells in the hollow lumens would produce oxygen profiles different from the same cells grown on open surfaces of Transwells®, an oxygen probe was used to quantify oxygen levels. Unlike confluent 2D monolayers, which maintained an oxygen tension of 13%, cells lining interior surfaces of 3D scaffolds experienced decreased oxygen levels that mimicked in vivo conditions and exhibited depth-graded oxygen profiles in the luminal direction that ranged from microaerobic to nanaerobic to anaerobic conditions. The use of wild type pathogenic Yersina pseudotuberculosis strains engineered as oxygen-sensing reporters confirmed the presence of low oxygen levels in 3D models, and interactions with these enteric pathogens supported the feasibility of using this 3D system in pathogenesis studies. Further, co-culture of a probiotic Lactobacillus rhamnosus GG strain with luminal epithelial cells demonstrated that these novel models can be colonized by pathogens and may also support gut microbiota growth. This 3D system was viable for months, although tissue functions decreased a few weeks post-seeding, a limitation that might be overcome in a perfusion bioreactor system [39]. This model has not yet been reported with nontransformed human enteroid or organoid cultures.
Human intestinal enteroid (HIE) cultures represent a novel model that recapitulates properties of human rotavirus (HRV) infection [40], models small intestinal pathophysiology and ion transport physiology [43], and potentially aids in personalized medicine, as clinical samples from individuals retain genotypic and phenotypic characteristics [25,40,45]. HIEs were established with tissue samples from the duodenum, jejunum, and ileum of different individuals [40]. HIEs adopted either multilobular or cystic morphologies and comprised a single, continuous lumen surrounded by epithelial cells that could be differentiated into absorptive enterocytes, enteroendocrine cells and goblet cells. Undifferentiated and differentiated HIEs contain Paneth cells within the stem cell niche [25]. Functional relevance of these models was demonstrated first by robust replication of HRV strains, with fewer cells infected by animal rotavirus strains, which corresponds to the host range specificity of HRV. In addition, HRV was found to replicate in differentiated enteroids in both enterocytes and enteroendocrine cells, which is consistent with a proposed model in which rotavirus stimulates serotonin production/release from enteroendocrine cells to activate the enteric nervous system [48]. HRV-infected HIEs displayed structures involved in the viral replication cycle (viroplasms and lipid droplets), as well as virus-induced swelling (mimicking fluid secretion during diarrhea) after viral infection or exposure to the rotavirus enterotoxin. Importantly, viral replication of RV1 vaccine strain was attenuated in all HIEs from secretor-positive patients, providing the first in vitro demonstration of RV1 attenuation [40]. Differential replication in HIEs from different patients supports the utility of these cultures to understand genetically-determined host differences in susceptibility and resistance to infection. Clinical isolates of HRV can be cultured in both HIEs and iPSC-derived organoids, and antivirals (ribavirin, interferon-α) can be evaluated [49,50]. HIEs also were used to examine ion transport, and key ion transporters were expressed and localized similarly to human intestine. A duodenal enteroid model showed that cAMP stimulated duodenal HCO3− secretion; that CFTR, the cystic fibrosis transmembrane conductance regulator, and NBCe1, the electrogenic Na+/HCO3− cotransporter 1, are essential transporters for HCO3− transport; and that NBCe1 might be a target for treatment of secretory diarrheas [43]. A novel finding from these studies was the finding that Na absorption in villus and colonic surface and upper crypt cells and anion secretion in crypt cells may not be as restricted as previously thought.
Human enteroids derived from ileal or colonic biopsies from blood type O or blood type A patients were generated to model and elucidate clinical and epidemiological associations of blood group O with the severity of cholera [51]. Enteroid enterocytes expressed blood group antigens appropriately on their cell surface, and cholera toxin (CT) treatment of enteroids from type O patients elicited higher levels of cAMP, a known cellular response to CT, compared to enteroids from type A patients. Vibrio cholerae infects the small intestine, but both ileal- and colonic-derived enteroids responded to CT with cAMP activation. These data reinforce the observed correlation between severe cholera disease in individuals with blood type O, and again support the use of human enteroids to model bacterial infections with the potential to gain mechanistic insight into pathogenesis. [51]
Human Colonoid Monolayer cultures (HCMs) derived from proximal colon were used to model Shiga toxin-producing EHEC infection [41]. Cultures of 3D Matrigel-grown colonoids were converted into 2D monolayers for access to apical and basolateral surfaces during studies of EHEC-epithelial interactions. HCM differentiation was characterized by the appearance of mucus-covered brush borders (BB) with numerous intermicrovillar bridges and tightly packed microvilli. Differentiated HCM were colonized by higher numbers of EHEC than undifferentiated HCM, and EHEC seemed to target the mucus layer. Reduction of mucus layer thickness and attachment of EHEC to glycocalyx remnants suggests that the colonic mucus layer might promote initial colonization. Sites of attachment to the epithelium displayed apical and basolateral membrane perturbation with characteristic formation of actin pedestals and attaching and effacing lesions. Mucin 2, a main component of colonic mucus, and Protocadherin 24 (PCDH24), an essential building block of intermicrovillar bridges, were targeted at early stages of infection, and EHEC-secreted EspP serine protease initiated BB damage through reduction of PCDH24. These data reflected and confirmed earlier findings with in vitro and in vivo models. Converting and culturing 3D colonoids into 2D monolayers enabled mechanistic pathogenesis studies, suggesting this approach represents a “next-generation” gut model system [38,42,45].
Human Intestinal Organoids (HIOs) that resembled the human proximal colon and derived from hPSCs were used to delineate further results obtained from patients with Clostridium difficile infections (CDI) [23,52–54]. Studies with clinical isolates of C. difficile, fecal samples, colonic biopsies and surgical resections investigated multiple aspects of CDI pathogenesis. C. difficile toxin B production inhibited expression of the Na+/H+ exchanger isoform 3 (NHE3), which led to elevated stool [Na+] and more alkaline fecal pH; these alterations in the intestinal tissue milieu were associated with changes in the gut microbiota, i.e., decreased presence of Firmicutes and increases in both Bacteroidetes and Proteobacteria. Microinjection of HIOs with C. difficile or CDI stool supernatant (or fecal supernatant from healthy subjects) confirmed that NHE3 inhibition was specific to C. difficile [52]. Additional studies from the same group with the same specimen types found that intestinal mucus in CDI patients is more acidic, consists primarily of MUC1 with decreased MUC2 expression, and a presumable mucus barrier defect. The composition of oligosaccharides in intestinal mucus was also changed, with GlcNAc and galactose increased and decreased levels of GalNAc. HIOs injected with C. difficile alone showed decreased MUC2 production, but injection of CDI stool supernatant was necessary to change mucus oligosaccharide levels, suggesting that another factor, possibly related to the changes observed in the gut microbiota, was required. Thus, the HIO model not only replicated results obtained with human specimens, but also elucidated finer details of CDI pathogenesis [53]. Microinjection and colonization with C. difficile or its toxin TcdA, but not toxin TcdB, also resulted in disruption of the epithelial paracellular barrier, which contributes new insight into bacterial and toxin pathogenesis [54].
Human intestinal organoids (iHOs) cultured from human iPSCs were tested as a model for studies of Salmonella enterica Serovar Typhimurium pathogenesis. Microinjection of this microbial pathogen resulted in Salmonella invasion of the organoid, and formation of Salmonella-containing vacuoles. A Salmonella invasion-defective mutant was unable to invade, illustrating the ability to assess the pathogenesis of different bacterial mutants. RNA sequencing characterized significantly changed iHO gene expression; upregulation (1448 genes) and downregulation (577 genes) of host transcription provided the first information about the early innate human epithelial response to bacterial infection with altered cytokine expression predominating. Thus, this iHO infection model demonstrated established steps of Salmonella pathogenesis and provided clues about how epithelial cells may participate in defense against this infection [55].
The importance of building more complex models of the intestine is well recognized and a human gut-on-a-chip microdevice shows potential to analyze how gut microbiome, inflammatory cells and peristalsis-associated mechanical motion contribute to intestinal infection and inflammation [46,47]. A model of bacterial overgrowth established in a microengineered device showed that immune cells and endotoxin stimulate a proinflammatory response that induced villus injury and compromised intestinal barrier function. Lack of motion also contributed to bacterial overgrowth [47]. The application of this system to human intestinal organoid cultures is of high interest.
These gastrointestinal models are beginning to be utilized to test antimicrobial drugs and therapeutics; a recent publication illustrates the feasibility of testing drugs in tissue models. Multiple model systems were utilized, including small intestinal organoids generated from mouse intestinal crypts and human duodenal organoids generated from human biopsies; both mammalian intestinal organoid models exhibit increased growth when treated with recombinant mouse or human interleukin-22 (IL-22) after tissue damage. Recombinant IL-22 treatment targeted intestinal stem cells (ISCs) in the organoids, subsequently increasing proliferation and expanding ISCs. These and other findings suggest that components of the mammalian immune system play important roles in restoring intestinal epithelial barrier function, which is a critical host defense [56]. Other studies have shown that high throughput drug screening can be done using enteroids from colorectal carcinomas; these studies demonstrated the role of Wnt signaling in colorectal cancer (CRC) development and the potential for Wnt inhibition as a treatment option in a subset of CRC patients [45,57]. Importantly, enteroids exhibited a range of sensitivities to the drugs, and compounds with the same molecular targets showed similar activity profiles. These studies demonstrated that patient-derived enteroids could be used for high-throughput drug screening to test the efficacy of clinically available drugs and to identify new therapeutic targets in a “personalized or precision medicine” approach. This may be important for antimicrobial drugs where increasing genetic susceptibilities to infectious disease are being recognized.
Kidney
A 3D human renal model provided the first assessment of damage to kidneys caused by Shiga toxin and new insight into hemolytic uremic syndrome (HUS), characterized by renal failure, hemolytic anemia, and thrombocytopenia [58]. Shiga toxin and its variants are bacterial cytotoxins secreted by Shigella dysenteriae and the Shiga toxin-producing E. coli during bacterial intestinal infection. HUS is a serious sequela, especially in young children. No animal models of infection reproduce HUS or the various diarrheal diseases caused by the human-specific shigellae. Thus, the mechanism(s) whereby Shiga toxin, which gains access to the circulatory system after damaging intestinal epithelium, injures human kidneys are unknown. The 3D model encompasses immortalized renal proximal tubule cells combined with a collagen I - Matrigel mixture cultured in Transwells®. Upon exposure to purified, LPS-free Shiga toxin (Stx2), the 3D model and renal cells grown in 2D displayed similar dose-dependent protein synthesis inhibition. Stx2-induced cytotoxicity was demonstrated in both systems, and Stx2 treatment of 3D tissue resulted in elevated levels of the kidney injury marker 1 (Kim-1) compared to treated 2D cultures. Inflammatory cytokine secretion was seen in Stx2-treated 3D and 2D models with increasing levels of IL-6, IL-8, and TNF-α, despite toxin-induced protein synthesis inhibition in cells, which seemed consistent with Shiga toxin’s capacity to induce protein production via activating translational regulation pathways. This is the first use of a 3D bioengineered human kidney tissue model to study renal damage by Stx2, and results imply that Kim-1 might be an early biomarker of renal injury. The cellular complexity and functional diversity of the kidney are substantial challenges to achieving functional maturation of the organ [59]. However, progress is being made on individual kidney cell types such as the proximal tubule [60] that can be used for initial studies of the response of nontransformed human kidney cells to infection.
Liver
3D and 2D in vitro human liver models were established that are applicable to basic science studies of hepatotropic pathogens and translational research to facilitate product development [61–67]. Human 3D hepatic sinusoid biomimetics were derived from cultures of liver sinusoidal endothelial cells (LSEC) and Huh-7 hepatoma cells [or Huh7-NTCP hepatocytes that stably express the HBV receptor sodium taurocholate co-transporter polypeptide (NTCP)] within a collagen 1 matrix. These cultures were used as infection models for the protozoan parasite Entamoeba histolytica, a human-only pathogen that causes amoebiasis, and hepatitis B virus (HBV), which causes chronic infections that can lead to cirrhosis and liver cancer [61,62]. Four variations of layered models were built with the scaffold and established cell lines in order to study aspects of pathogen invasion, i.e. migration, hepatocyte barrier crossing, endothelial and hepatocyte barrier crossing, and human HBV replication quantification. These easily assembled models combined simplicity and reproducibility with enhanced physiological relevance compared to standard 2D cultures. LSECs were essential for matrix architecture, hepatocyte differentiation, and proinflammatory responses. E. histolytica infection of the construct comprised of single layers of LSECs and hepatocytes in a collagen matrix demonstrated, for the first time, dependence of amoebic Gal/GalNAc lectin for endothelial crossing and a crucial role for human galectins in parasite adhesion to endothelia. Additionally, galectins-1 and -3 stimulated pro-inflammatory cytokine release. Protozoan infection of a single hepatocyte layer construct showed directional migration; galactose, a known inhibitor of E. histolytica adhesion, reduced migration distance. Amoebae behavior in a collagen-only model showed that hepatocytes were needed to maintain protozoan viability, correlating with earlier findings that hepatic cells alter medium composition and scaffold structure and composition. Infection of a double hepatocyte/single LSEC layered construct, representing liver sinusoid endothelial and hepatocyte barriers, revealed amoebae squeezing between hepatic cells to cross that barrier. HBV infection of tissue comprised of single LSEC and hepatocyte (Huh7-NTCP cells) layers showed viral replication in the absence of PEG-8000, necessary in standard HBV infection protocols; replication rates were comparable to those seen in conventional 2D cultures. The 3D model demonstrated that HBVx protein is required for productive infection, as in 2D cultures and in vivo experiments [61,62].
Micropatterned cocultures (MPCCs) of primary human hepatocytes organized into microspheroid-like 2D islands surrounded by mouse stromal cells produced miniature in vitro models used to study infections caused by hepatitis C virus (HCV) and HBV [63,65,66]. MPCCs express all the required factors for HCV entry and support HCV infection for several weeks [66]. MPCCs enabled sustained expression of HBV NTCP receptor and supported productive HBV infection throughout the 3 week long culture. HBV infection was enhanced by inhibition of the innate immune pathway via treatment with the Janus kinase inhibitor (JAKi) that diminishes expression of Interferon (IFN)-stimulated genes or an inhibitor of the TANK-binding kinase 1 (TBK1), an upstream activator of the IFN response pathway. MPCCs also were used as anti-HBV drug-testing tools in experiments with the HBV reverse transcriptase inhibitor entecavir and IFN-β, an alternative antiviral [65]. Thus, the MPCC platform potentially can be used for drug and vaccine development. Newer models to generate tissue-derived epithelial organoids from adult human liver bile duct cells demonstrated long-term viability and improved hepatocyte function when compared to the standard/reference HepG2 cell line [68]. This model can produce cultures from diverse patients but comparisons of these cultures for their ability to support hepatitis virus replication remains to be reported.
Studies designed to generate metabolically active hepatocyte cells from hPSCs [HES2 human embryonic stem cell line (hESC)] identified key steps in hepatocyte maturation. Maturation of hESC-derived hepatocyte-like cells required an initial aggregation step, which increased the expression of genes associated with liver function, epithelial characteristics, such as E-Cadherin, and the hepatocyte cell surface marker asialo-glycoprotein receptor 1 (ASGR1). Cyclic adenosine monophosphate (cAMP) signaling constituted the second step and induced changes associated with increased metabolic activity, reduction in fetal gene expression and concomitant increase in adult gene expression, and global expression of changes that indicate hepatocyte maturation [67]. Development of a strategy for production of metabolically active mature hepatocytes from hPSCs holds great promise for numerous applications, including their use in human liver tissue mimics. Other groups have shown that human induced pluripotent stem cell (iPSC)-derived hepatocyte-like cells (iHLCs) can serve as in vitro models for liver-stage malaria infection [64]. iHLCs express genes encoding malaria parasite CD81 and SRB1 host entry factors for liver-stage disease, and their proteins localize appropriately on cell surfaces. Differentiated iHLCs were phenotypically stable and could be infected with human-specific Plasmodium falciparum parasite sporozoites up to 55 days after differentiation initiation. Human and rodent (P. berghei and P. yoelii) malaria parasite sporozoites infect iHLCs, and these in vitro-derived host cells supported growth and maturation of EEFs (HSP70-expressing exoerythrocytic forms) [64].
Lung
Human 3D lung tissue models were developed and used to study Mycobacterium tuberculosis, staphylococcal pneumonia, and Andes Hantavirus pathogenesis [69–71]. Indeed, the adaptability of these human lung tissue models to studies of multiple pathogens, both viral and bacterial, suggests their potential for broad application and impact on the field. One model consisted of type 1 collagen-embedded human lung fibroblasts cultured on porous membranes followed by the introduction of uninfected human primary monocytes and M. tuberculosis-infected human primary macrophages. Human bronchial epithelial cells were then seeded on top of the collagen-fibroblast matrix; this multi-cell tissue was subsequently exposed to air to initiate production of extracellular matrix proteins, mucus secretion, and stratification of the epithelial layer. This lung infection model displays clustering of macrophages and migration of monocytes into the tissue at the site of infection to yield a granuloma-like formation. Although the authors cite limitations such as the use of cell lines, the absence of vascularization, and the lack of neutrophils and lymphocytes, the model does exhibit the early stages of TB granulomas [69].
A similar 3D human lung tissue construct was used to study staphylococcal pneumonia pathogenesis; it was generated on Transwell® inserts with lung fibroblasts in a collagen mixture as a first layer, followed by seeding of bronchial epithelial cells, and subsequent air exposure [70,72]. Staphylococcal aureus strains from patients with severe pneumonia, including necrotizing pneumonia and lung empyema, were used here, as were bacterial supernatants and purified α-toxin; supernatants contained pore-forming α-toxin and PVL (Panton-Valentine leukocidin) cytotoxins. Supernatants from necrotizing pneumonia and lung empyema strains were highly cytotoxic to primary human neutrophils; only the former demonstrated severe toxicity to lung epithelial cells. Lung tissue models responded similarly to bacterial supernatants; the necrotizing pneumonia isolates secreted high levels of α-toxin and PVL and caused significant α-toxin-mediated lung epithelium damage, PVL-mediated neutrophil cytotoxicity, and inflammation via toxin-mediated release of cytokines and chemokines in lung epithelia. Toxin-mediated damage was inhibited by polyspecific intravenous immunoglobulin that contained antibodies to α-toxin, PVL, and other factors present in the clinical isolate supernatants [70].
To study Andes virus (ANDV), which causes hantavirus pulmonary syndrome (HPS), an analogous organotypic lung tissue model was developed that was comprised of a collagen-coated porous membrane with sequential cultivation of, first, a mixture of human lung fibroblast cells (MRC-5 cell line) in collagen and, second, a layer of human bronchial epithelial cells (16HBE14o- cell line), which was then air exposed. ANDV was applied to the apical, or air-exposed, surface, and viral replication was observed with a peak of progeny virus occurring ~ 1 week post-infection (p.i.). Moreover, the capacity of the tissue model to sustain a long-term viral infection (≥1 month p.i.) was demonstrated by the presence of infected cells and progeny virus production. Peak production of progeny virus was coincident with the induction of innate immune responses as detected by increased expression of IFN-λ1, IFN-λ2, IFN-β, and the IFN-stimulated gene ISG56 mRNAs with later production of the proinflammatory cytokines IL-6 and IL-8 ~ 3 weeks p.i. Thus, a transient antiviral response preceded a proinflammatory response that might reflect the clinical course of asymptomatic infection transitioning to an inflammatory state, which might parallel the development of HPS. Of crucial importance was the capacity of the 3D model to enable in vitro studies not permissible in cell monolayers as well as studies of infection over a long period of time [71].
Skin
3D skin equivalent models would represent another critical advance for infectious diseases research; recently, human skin tissue models have been developed and used to study both bacterial and viral infections. An in vitro herpes simplex virus type 1 (HSV-1) skin model was developed for studies of viral latency and reactivation that might provide insights not permitted via traditional cell culture monolayers. This skin mimic incorporated the human keratinocyte cell line HaCaT (epidermal layer) grown on a collagen substrate containing human primary fibroblasts (dermal layer); a critical improvement was addition of a component designed to mimic further the in vivo environment, i.e., a quiescent HSV-1 infected neuronal equivalent (latency component). Epidermal stratification and epidermal-dermal interactions were attained via cultivation at an air-liquid interface. The authors described successful reactivation of latent virus via UVB-light with a caveat about reproducibility issues that may be resolved by optimizing infection conditions. Importantly, this skin model shows potential for investigating mechanisms related to latency and possibly new therapeutics [73]. A related organotypic skin tissue model, also comprised of stratified epithelium and a fibroblast dermal layer, was used to investigate the pathogenesis of Streptococcus dysgalactiae subsp. equisimilis (SDSE) strains, three of which were invasive strains that cause necrotizing soft tissue infections and one non-invasive strain. All SDSE strains were able to infect this model with bacterial loads resembling those seen in tissue biopsies also utilized in this study, but the invasive strains disseminated throughout the epithelium and caused substantial tissue damage evidenced by disruption and detachment of the epithelium [74].
Conclusions and Future Directions
Microphysiological systems such as modular human organs-on-a-chip and integration of organs into a human-on-a-chip are under development [21,22,46,47,75]. These chips have great potential to facilitate product development; their use in pharmacokinetic and toxicology assays can provide more accurate predictors of clinical trial outcomes. The goal of integrating modules to understand physiologically relevant interactions is essential, although a common culture medium and optimized platforms to accommodate needs of each organ system for integrated modules are yet to be developed.
Expertise and methodologies afforded to infectious diseases researchers working with experienced bioengineers conversant in areas such as biomaterials, cell sources, microtissue platforms, and nondestructive imaging catalyze development of urgently needed models for host-pathogen interactions studies. There have been many advances as summarized above, but there is more to be accomplished (Key Advances and Challenges). Limitations are acknowledged by the authors for many of the models presented here; limitations include the lack of mechanical or force stimulation, such as peristalsis; vascularization, i.e., to simulate blood and/or lymph circulation; innervation in the non-neural models; and components of the immune system, such as the mucosal-associated lymphoid tissue and circulating or resident immune cells; to name a few crucial features of a whole body system. A practical limitation is the current cost of these systems. Research is progressing to address these limitations but the full complexity and maturity of tissues remains to be achieved. Nonetheless, the existing models provide sufficient features to enable discovery of new insights in microbial pathogenesis and host interactions. These models demonstrate the need to balance simplicity and ease of use for basic science research with the recapitulation of human tissue architecture in organotypic model systems for preclinical therapeutic and vaccine studies. Generation of specialized tissues with multiple cell types in precise geometric locations that engender a high level of spatial organization would provide optimal biomimetic models that could asymptotically approach organogenesis. Here, the development of an in vivo-like restricted O2 gut microenvironment [39]; the identification of pathways, such as the new role for EGF signaling [29]; the capacity for long-term cultivation without loss of architectural features, function, and cellular markers in some models; and the remarkable similarity to organogenesis observed in many constructs point to the feasibility of developing 3D models of infection that more closely mimic human physiology and human diseases pathology.
Supplementary Material
Acknowledgments
MM and MKE would like to thank their colleagues in the Novel, Alternative Model Systems for Enteric Diseases Cooperative Research Network for stimulating discussions. MKE extends thanks for the collegial interactions with Mark Donowitz and other colleagues in the NIH/NCATS Microphysiological Systems.
Contributor Information
Melody Mills, Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, (240) 627-3318.
Mary K. Estes, Molecular Virology and Microbiology and Medicine-GI, Baylor College of Medicine, Houston, TX, (713) 798-3585.
References
- 1.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110(9):3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren HS, et al. Mice are not men. Proc Natl Acad Sci U S A. 2015;112(4):E345. doi: 10.1073/pnas.1414857111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolker J. Model organisms: There's more to life than rats and flies. Nature. 2012;491(7422):31–33. doi: 10.1038/491031a. [DOI] [PubMed] [Google Scholar]
- 4.Martic-Kehl MI, et al. Can animal data predict human outcome? Problems and pitfalls of translational animal research. Eur J Nucl Med Mol Imaging. 2012;39(9):1492–1496. doi: 10.1007/s00259-012-2175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260(5110):920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 6.Birkness KA, et al. A tissue culture bilayer model to study the passage of Neisseria meningitidis. Infect Immun. 1995;63(2):402–409. doi: 10.1128/iai.63.2.402-409.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerneis S, et al. Conversion by Peyer's patch lymphocytes of human enterocytes into M cells that transport bacteria. Science. 1997;277(5328):949–952. doi: 10.1126/science.277.5328.949. [DOI] [PubMed] [Google Scholar]
- 8.Nickerson CA, et al. Three-dimensional tissue assemblies: novel models for the study of Salmonella enterica serovar Typhimurium pathogenesis. Infect Immun. 2001;69(11):7106–7120. doi: 10.1128/IAI.69.11.7106-7120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedhia PH, et al. Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology. 2016;150(5):1098–1112. doi: 10.1053/j.gastro.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stelzner M, et al. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302(12):G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wikswo JP. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med (Maywood) 2014;239(9):1061–1072. doi: 10.1177/1535370214542068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramanan V, et al. New Methods in Tissue Engineering: Improved Models for Viral Infection. Annu Rev Virol. 2014;1:475–499. doi: 10.1146/annurev-virology-031413-085437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 14.Chwalek K, et al. In vitro bioengineered model of cortical brain tissue. Nat Protoc. 2015;10(9):1362–1373. doi: 10.1038/nprot.2015.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang-Schomer MD, et al. Bioengineered functional brain-like cortical tissue. Proc Natl Acad Sci U S A. 2014;111(38):13811–13816. doi: 10.1073/pnas.1324214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang J, et al. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcez PP, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 20.Qian X, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown JA, et al. Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor. Biomicrofluidics. 2015;9(5):054124. doi: 10.1063/1.4934713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alcendor DJ, et al. Neurovascular unit on a chip: implications for translational applications. Stem Cell Res Ther. 2013;4(Suppl 1):S18. doi: 10.1186/scrt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leslie JL, Young VB. A whole new ball game: Stem cell-derived epithelia in the study of host-microbe interactions. Anaerobe. 2016;37:25–28. doi: 10.1016/j.anaerobe.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells JM. Regenerative medicine in 2015: Generating and regenerating the digestive system. Nat Rev Gastroenterol Hepatol. 2016;13(2):65–66. doi: 10.1038/nrgastro.2015.223. [DOI] [PubMed] [Google Scholar]
- 25.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340(6137):1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 26.Zachos NC, et al. Human Enteroids/Colonoids and Intestinal Organoids Functionally Recapitulate Normal Intestinal Physiology and Pathophysiology. J Biol Chem. 2016;291(8):3759–3766. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinagoga KL, Wells JM. Generating human intestinal tissues from pluripotent stem cells to study development and disease. Embo j. 2015;34(9):1149–1163. doi: 10.15252/embj.201490686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato T, Clevers H. SnapShot: Growing Organoids from Stem Cells. Cell. 2015;161(7) doi: 10.1016/j.cell.2015.06.028. 1700-1700.e1701. [DOI] [PubMed] [Google Scholar]
- 29.McCracken KW, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516(7531):400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartfeld S, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148(1):126–136. e126. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartfeld S, Clevers H. Organoids as Model for Infectious Diseases: Culture of Human and Murine Stomach Organoids and Microinjection of Helicobacter Pylori. J Vis Exp. 2015;(105) doi: 10.3791/53359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlaermann P, et al. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65(2):202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang JY, et al. Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium. Cell Host Microbe. 2015;18(2):147–156. doi: 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertaux-Skeirik N, et al. Co-culture of Gastric Organoids and Immortalized Stomach Mesenchymal Cells. Methods Mol Biol. 2016;1422:23–31. doi: 10.1007/978-1-4939-3603-8_3. [DOI] [PubMed] [Google Scholar]
- 35.Ferrero RL. Innate immune recognition of the extracellular mucosal pathogen, Helicobacter pylori. Mol Immunol. 2005;42(8):879–885. doi: 10.1016/j.molimm.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Bartfeld S, et al. High-throughput and single-cell imaging of NF-kappaB oscillations using monoclonal cell lines. BMC Cell Biol. 2010;11:21. doi: 10.1186/1471-2121-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer W, et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42(5):1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 38.VanDussen KL, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64(6):911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, et al. Robust bioengineered 3D functional human intestinal epithelium. Sci Rep. 2015;5:13708. doi: 10.1038/srep13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saxena K, et al. Human Intestinal Enteroids: a New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J Virol. 2015;90(1):43–56. doi: 10.1128/JVI.01930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.In J, et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol. 2016;2(1):48–62. e43. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyska MJ. Brush Border Destruction by Enterohemorrhagic Escherichia coli (EHEC): New Insights From Organoid Culture. Cellular and Molecular Gastroenterology and Hepatology. 2(1):7–8. doi: 10.1016/j.jcmgh.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foulke-Abel J, et al. Human Enteroids as a Model of Upper Small Intestinal Ion Transport Physiology and Pathophysiology. Gastroenterology. 2016;150(3):638–649. e638. doi: 10.1053/j.gastro.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Middendorp S, et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells. 2014;32(5):1083–1091. doi: 10.1002/stem.1655. [DOI] [PubMed] [Google Scholar]
- 45.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingber DE. Reverse Engineering Human Pathophysiology with Organs-on-Chips. Cell. 2016;164(6):1105–1109. doi: 10.1016/j.cell.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 47.Kim HJ, et al. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016;113(1):E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagbom M, et al. Towards a human rotavirus disease model. Curr Opin Virol. 2012;2(4):408–418. doi: 10.1016/j.coviro.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Finkbeiner SR, et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio. 2012;3(4) doi: 10.1128/mBio.00159-12. e00159-00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin Y, et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antiviral Res. 2015;123:120–131. doi: 10.1016/j.antiviral.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 51.Kuhlmann FM, et al. Blood Group O-Dependent Cellular Responses to Cholera Toxin: Parallel Clinical and Epidemiological Links to Severe Cholera. Am J Trop Med Hyg. 2016 doi: 10.4269/ajtmh.16-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engevik MA, et al. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol. 2015;308(6):G497–G509. doi: 10.1152/ajpgi.00090.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engevik MA, et al. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol. 2015;308(6):G510–G524. doi: 10.1152/ajpgi.00091.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leslie JL, et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun. 2015;83(1):138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forbester JL, et al. Interaction of Salmonella enterica Serovar Typhimurium with Intestinal Organoids Derived from Human Induced Pluripotent Stem Cells. Infect Immun. 2015;83(7):2926–2934. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindemans CA, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madan B, et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35(17):2197–2207. doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DesRochers TM, et al. Effects of Shiga toxin type 2 on a bioengineered three-dimensional model of human renal tissue. Infect Immun. 2015;83(1):28–38. doi: 10.1128/IAI.02143-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Little MH. Growing Kidney Tissue from Stem Cells: How Far from "Party Trick" to Medical Application? Cell Stem Cell. 2016;18(6):695–698. doi: 10.1016/j.stem.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelly EJ, et al. Innovations in preclinical biology: ex vivo engineering of a human kidney tissue microperfusion system. Stem Cell Res Ther. 2013;4(Suppl 1):S17. doi: 10.1186/scrt378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petropolis DB, et al. A new human 3D-liver model unravels the role of galectins in liver infection by the parasite Entamoeba histolytica. PLoS Pathog. 2014;10(9):e1004381. doi: 10.1371/journal.ppat.1004381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petropolis DB, et al. Human Liver Infection in a Dish: Easy-To-Build 3D Liver Models for Studying Microbial Infection. PLoS One. 2016;11(2):e0148667. doi: 10.1371/journal.pone.0148667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.March S, et al. Micropatterned coculture of primary human hepatocytes and supportive cells for the study of hepatotropic pathogens. Nat Protoc. 2015;10(12):2027–2053. doi: 10.1038/nprot.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng S, et al. Human iPSC-derived hepatocyte-like cells support Plasmodium liver-stage infection in vitro. Stem Cell Reports. 2015;4(3):348–359. doi: 10.1016/j.stemcr.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shlomai A, et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A. 2014;111(33):12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ploss A, et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A. 2010;107(7):3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogawa S, et al. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development. 2013;140(15):3285–3296. doi: 10.1242/dev.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160(1–2):299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braian C, et al. A 3D Human Lung Tissue Model for Functional Studies on Mycobacterium tuberculosis Infection. J Vis Exp. 2015;(104) doi: 10.3791/53084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mairpady Shambat S, et al. Modelling staphylococcal pneumonia in a human 3D lung tissue model system delineates toxin-mediated pathology. Dis Model Mech. 2015;8(11):1413–1425. doi: 10.1242/dmm.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sundstrom KB, et al. Andes Hantavirus-Infection of a 3D Human Lung Tissue Model Reveals a Late Peak in Progeny Virus Production Followed by Increased Levels of Proinflammatory Cytokines and VEGF-A. PLoS One. 2016;11(2):e0149354. doi: 10.1371/journal.pone.0149354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen Hoang AT, et al. Technical advance: live-imaging analysis of human dendritic cell migrating behavior under the influence of immune-stimulating reagents in an organotypic model of lung. J Leukoc Biol. 2014;96(3):481–489. doi: 10.1189/jlb.3TA0513-303R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hogk I, et al. 3D-tissue model for herpes simplex virus-1 infections. Methods Mol Biol. 2013;1064:239–251. doi: 10.1007/978-1-62703-601-6_17. [DOI] [PubMed] [Google Scholar]
- 74.Siemens N, et al. Increased cytotoxicity and streptolysin O activity in group G streptococcal strains causing invasive tissue infections. Sci Rep. 2015;5:16945. doi: 10.1038/srep16945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fabre KM, et al. Organs-on-chips (microphysiological systems): tools to expedite efficacy and toxicity testing in human tissue. Exp Biol Med (Maywood) 2014;239(9):1073–1077. doi: 10.1177/1535370214538916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.