Abstract
Gold nanoparticles (AuNPs) have been of recent interest due to their unique optical properties and their biocompatibility. Biomolecules spontaneously adsorb to their surface, a trait that could potentially be exploited for drug targeting. Currently, it is unclear whether protein–AuNP interactions at the nanoparticle surface are dependent on nanoparticle size. In this work, we investigate whether varying surface curvature can induce protein unfolding and multilayer binding in citrate-coated AuNPs of various sizes. A recently developed NMR-based approach was utilized to determine the adsorption capacity, and protein NMR spectra were compared to determine whether nanoparticle size influences protein interactions at the surface. In addition, transmission electron microscopy (TEM) and dynamic light scattering (DLS) were employed to corroborate the NMR studies. Over a broad range of AuNP sizes (14–86 nm), we show that adsorption capacity can be predicted by assuming that proteins are compact and globular on the nanoparticle surface. Additionally, roughly one layer of proteins is adsorbed regardless of AuNP size. Our results hold for two proteins of significantly different sizes, GB3 (6 kDa) and bovine carbonic anhydrase (BCA, 29 kDa). However, the unstable drkN SH3 domain (ΔḠ0 ≈ 0, 7 kDa) does not appear to follow the same trend seen for stable, globular proteins. This observation suggests that unstable proteins can deform significantly when bound to AuNP surfaces. Taken together, the results of this work can be used to improve our knowledge of the mechanism of protein–AuNP interactions to optimize their use in the biomedical field.
Graphical abstract
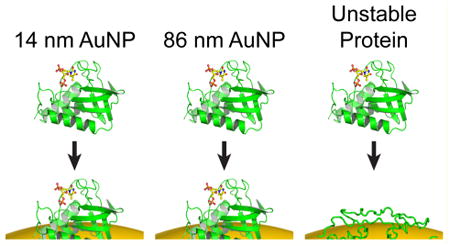
Introduction
Gold nanoparticles (AuNPs) have unique properties, which make them ideal for many fields of research such as chemistry, physics, and the health sciences.1–4 For instance, their small size and electronic properties result in unique spectroscopic qualities, causing them to be interesting research targets.5 Many methods have been developed for synthesizing gold colloids, enabling the study of these properties throughout the past 60 years.6–9 Throughout this time, much has been learned about gold nanoparticles, but many aspects of their applicability remain poorly understood. One notable success is the thermal targeting of AuNPs to tumors in cancer patients,10,11 but the therapeutic value of AuNP-based drugs is limited by our inadequate understanding of protein–nanoparticle interactions. Only when such an understanding is obtained will we be able to develop AuNP-based therapeutics with high-specificity and minimal cytotoxic effects.
Proteins are known to spontaneously adsorb to the surface of AuNPs.12–14 This can be both an advantage and a disadvantage. On one hand, this phenomenon can be exploited to adsorb a protein of interest to the nanoparticle, potentially introducing a therapeutic effect and functionalizing the nanoparticle. In addition, if nanoparticles come in contact with any biological fluids, they will become coated with the natural proteins found in the fluid (a biocorona), therefore making them effectively invisible to the organism and its immune system.15–17 However, this behavior can also produce unexpected results, severely complicating the design of protein-functionalized nanoconjugates. Not only can the biocorona mask the function of functionalized nanoparticles, the biocorona itself can change over time, making it impossible to predict the character of the nanoparticles after they have been exposed to a particular host.18,19 It is therefore important to determine the mechanism of interaction for protein–AuNP adsorption to optimize and characterize the activity of any drug molecules or enzymes adsorbed to the surface of the AuNPs and to prevent their activity from being blocked by the adsorption of a possible second layer, or soft corona, of native proteins of the organism.20
AuNPs exhibit low toxicity and can be taken up into cells.20,21 The optimal shape and size for AuNP uptake into cells has been determined to be spherical AuNPs with a diameter of 50 nm.22 However, AuNPs with a diameter of 15 nm or smaller are frequently used in studies due to their higher degree of monodispersity. Therefore, it is important to study a variety of AuNP sizes to understand which sizes are best suited for a particular application. There is a discrepancy as to whether varying the curvature, and therefore the size, of the AuNPs alters the mechanism of interaction between the proteins and the AuNPs. Using a variety of different analytical methods, several groups have suggested that AuNP surface curvature alters the structure and binding capacity of adsorbed proteins on the surface of the nanoparticles.23–25 However, others have suggested that the interactions between proteins and AuNPs are unchanged as the curvature is altered. For example, Boulous et al. found that for bovine serum albumin binding to AuNPs, the binding constant is roughly the same regardless of the nanoparticle shape and size.26 Together, these results suggest a complex relationship between protein structure, thermodynamic stability, and nanoparticle adsorption. To our knowledge, systematic investigations of how protein stability relates to structure on the nanoparticle surface have yet to be performed.
Here, we apply a recently developed NMR-based approach27 for quantifying protein adsorbed to AuNPs to determine how the mode of protein–AuNP interactions may change as the size of the protein or nanoparticle changes. The nanoparticles themselves are not detected by traditional solution NMR because of their slow rotational correlation time.28 As the protein is in slow exchange with the AuNP surface (on the time scale of minutes or longer),29 only the free protein is detected, which permits the rapid and accurate quantification of the bound protein in situ. We apply this method to four different diameters of spherical AuNPs: 14, 30, 43, and 86 nm, and three different proteins: GB3, a small immunoglobulin binding domain from Staphylococcus aureus; bovine carbonic anhydrase (BCA), an enzyme responsible for converting carbon dioxide to carbonic acid and bicarbonate; and the Drosophila drkN SH3 domain, a small, unstable (ΔḠ0 ≈ 0) domain involved in protein–protein interactions. Transmission electron microscopy (TEM) and dynamic light scattering (DLS) are also used to investigate the protein behavior on AuNP surfaces. We find that both GB3 and BCA form a single layer of compact protein on all AuNPs tested, consistent with a globular conformation on the surface. However, drkN SH3 deviates from this behavior, suggesting that protein deformation can occur on AuNP surfaces if the folding stability is sufficiently low.
Materials and Methods
Protein Preparation
Wild-type (WT) GB3 was prepared recombinantly as described previously.27 BCA was purchased from Sigma-Aldrich and used without further purification. The expression plasmid for drkN SH3 was generously provided by Dr. Nikolai Srkynnikov (Purdue University), and 15N-labeled protein was expressed and purified as described.30 Protein purity was established using SDS-PAGE electrophoresis, and the concentration of all proteins was determined using the UV absorbance at 280 nm.31 As DNA can interfere with protein–AuNP binding experiments, we also examined the ratio of UV absorbance at 280 nm to the absorbance at 260 nm. All proteins had a 280/260 nm absorbance ratio greater than one, suggesting no DNA contamination in any of the samples.
AuNP Preparation and Characterization
Gold(III) chloride trihydrate and sodium citrate dihydrate were used as received from Sigma-Aldrich. Approximately spherical AuNPs of different sizes (14, 43, and 86 nm) were synthesized using the protocol laid out by the Frens method.32 5 mg (0.01% by mass) of HAuCl4 was added to 50 mL of ultrapure 18.2 MΩ Milli-Q water. This solution was brought to a boil, and then varying amounts of 1% by mass of sodium citrate were added immediately after boiling began. This mixture was kept boiling for 20 min, and then allowed to cool to room temperature. Centrifugation at 7500g for 45 min was performed to concentrate the AuNPs, and then sonication was performed using a Branson bath sonicator. The 30 nm AuNPs used in this study were obtained commercially (nanoComposix, San Diego, CA).
All of the nanoparticle sizes were confirmed using a JEOL 2100 transmission electron microscope, operated at 200 kV (Figure S1).33,34 TEM samples were prepared on copper grids coated with a Formvar carbon-based film to provide support for the nanoparticles. A single droplet containing the concentrated AuNPs was placed on the copper grid and left on a clean surface at room temperature to dry overnight. ImageJ software was used to generate average size distributions of all synthesized AuNPs. Distances are reported as the average of at least 20 independent measurements, and the uncertainties are calculated using the sample standard deviation of observed distances.
The extinction coefficients of AuNPs were calculated using the method described by Liu et al.35 Briefly, the number of gold atoms per nanoparticle is calculated assuming a spherical AuNP. The total number of gold atoms then is calculated using simple stoichiometric relations. These two numbers were used to calculate the theoretical concentration of the prepared nanoparticles. Finally, the absorbance of the prepared sample was taken, and the Beer–Lambert law was used to estimate the extinction coefficient for each sample. This approach yielded values for the UV–visible extinction coefficient that were comparable to those found by Liu et al.,35 and absorbance maxima were in agreement with previously reported values.33,34 As GB3, BCA, and drkN SH3 were added to solutions containing AuNPs of varying sizes, the absorbance peaks exhibited a red shift, similar to what was observed previously for GB3 alone on 15 nM AuNPs (Figure S2).27 A complete listing of the nanoparticles and extinction coefficients used in this study is provided (Table S1).
Dynamic Light Scattering (DLS)
A Wyatt DynaPro NanoStar DLS instrument was used to measure solution distributions of nanoparticle size. AuNPs (at a final concentration of 1 nM), or AuNPs containing 20 μM protein, were filtered using a 0.1 μm Anotop syringe filter. After equilibration for 1 h at room temperature, the solution was diluted 5-fold before transfer to a disposable microcuvette for measurement. The nanoparticle hydration radii were measured using the regularization fit functionality of the DYNAMICS software. For each measurement (with or without protein), the average value of three independently prepared samples is reported, and the uncertainty is calculated as the standard error of the mean of these measurements.
NMR Measurements of Binding Capacity
Binding capacity measurements were performed at 25 °C as described previously.27 All NMR experiments were performed on a Bruker Avance III 600 MHz NMR with a cryogenically cooled (CP-QCI) probe. Briefly, 20 μM protein was premixed with a stock solution of 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), sodium phosphate at pH 6.6, and D2O. The final concentrations of DSS, sodium phosphate, and D2O were 55 μM, 40 mM, and 6% (v/v), respectively. To ensure consistency in the results, the same stock solution was added to all protein samples in equivalent volumes. Like trimethylsilylpropanoic acid (TMSP), DSS does not appear to interact with AuNPs and is less sensitive to small changes in pH. The AuNPs were added last, and to ensure that all concentrations were consistent, the same volume of either AuNPs or Milli-Q water was added to a final sample volume for NMR (550 μL). Thus, even small systematic errors in protein or AuNP stock solutions could be well controlled throughout our experiments. The sample was left undisturbed to equilibrate at room temperature for a minimum of 1 h before NMR experiments were performed to allow sufficient time for protein adsorption.36 Next, 1D 1H NMR spectra were recorded and processed as described previously, using manual integration of proton peaks to quantify the solution protein concentration relative to the DSS peak. The AuNP-bound protein ([Pbound]) is calculated as the difference between the total protein concentration (typically 20 μM) and the solution concentration. The apparent binding capacity (N), defined as the maximum number of proteins bound per nanoparticle, is calculated as the slope relating [Pbound] and the total nanoparticle concentration ([AuNP]):
| (1) |
Equation 1 holds when the association constant is tight (≤1 μM).27 In this work, at least three points were used to establish N for all measurements, and all experiments were performed in triplicate. Error bars are reported as the standard deviation of three independent experiments. Two-dimensional TROSY-HSQC spectra were recorded to confirm that no line broadening or chemical shift perturbations occurred when proteins were mixed with AuNPs.37
Geometric Predictions of Binding Capacity
The predicted binding capacity is calculated from geometric considerations using the following equation:
| (2) |
Here, RAuNP is the nanoparticle radius as determined by TEM measurements, and RG is the geometric radius of gyration calculated using the folded protein structure. This equation represents the surface area of the nanoparticle divided by the surface area occluded by an approximately spherical adsorbed protein.27,38,39 The following PDB structures were used in this study to calculate RG: GB3 (2OED, RG = 10.8 Å),40 BCA (1V9E, RG = 17.1 Å),41 and drkN SH3 (2A36, RG = 10.6 Å).42
Circular Dichroism-Based Protein Stability Measurements
The stability of WT GB3 was measured using an Olis DSM 20 circular dichroism (CD) spectropolarimeter. GB3 at a concentration of 50 μM in 20 mM sodium phosphate pH 7.0 and 50 mM NaCl was placed in a 0.2 cm cell. The CD signal at 220 nm was monitored as a function of guanidinium chloride (GdmCl). The final concentration of the GdmCl stock solution was measured using refractometry,43 and the stability of GB3 in H2O at 25 °C was determined using the linear extrapolation method.44
Results and Discussion
NMR-Based Measurements of Binding Capacity on 43 nM AuNPs
In this study, we sought to determine the behavior of two proteins, GB3 and BCA, during adsorption on to AuNPs of increasing size. Our previous work determined that, for a data set of six proteins, adsorption was consistent with a single layer of globular protein on the surface of a 15 nm AuNP, and that the number of proteins adsorbed could be accurately predicted by a simple surface area based approach.27 Here, when mixed with 43 nm AuNPs, we observe similar behavior for both GB3 and BCA (Figure 1A,B). When these proteins were mixed with varying concentrations of 43 nm AuNPs, the amide proton peaks in the 1D NMR spectrum decrease in intensity as the concentration of AuNPs increases. No line broadening or chemical shift perturbations are observed, suggesting that exchange between the bound and unbound protein is slower than the duration of the NMR acquisition time (2 s). These results are consistent with hydrogen–deuterium exchange (HDX) measurements, which indicate that the exchange rate is on the time scale of minutes or longer.29 Once adsorbed, proteins diffuse slowly with the AuNPs and do not contribute to the solution NMR spectrum. Thus, the behavior observed for GB3 and BCA results from both the slow exchange with the surface and the slow AuNP rotational diffusion time.
Figure 1.
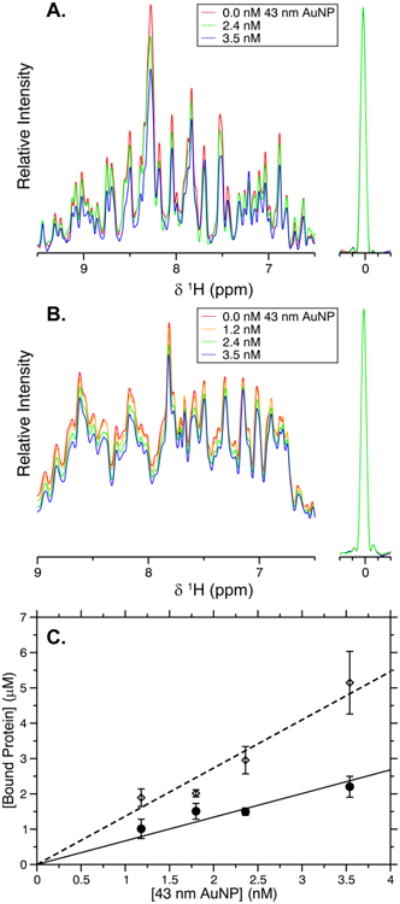
NMR-based adsorption measurements of GB3 and BCA. One-dimensional proton NMR spectra of GB3 (A) and BCA (B) as the concentration of 43 nm AuNPs is increased. The DSS peak used for concentration referencing is shown to the right. As protein adsorbs, the NMR signal decreases quantitatively. (C) The concentration of bound protein calculated from the data in (A) and (B). The bound concentration of GB3 (◇, dashed line) and BCA (●, solid line) increases linearly with AuNP concentration, and the slope of each line represents the adsorption capacity, as described in the text. Error bars here and elsewhere represent the standard error of the mean for three independently performed experiments.
Both GB3 and BCA show a linear decrease in intensity as 43 nm AuNPs are added (Figure 1A,B), which corresponds to an increase in the concentration of bound protein (Figure 1C). These data are obtained by manually integrating the NMR spectra to determine how much protein remains in solution after adding AuNPs. The linear behavior of these plots implies that the binding of the protein is saturated; that is, increasing the concentration of protein binding sites results in a proportional increase in the amount of protein bound.27 This behavior is expected when the apparent single-site dissociation constant (Kd) is stronger than 1 μM. Under these conditions, the apparent binding capacity, or the maximum number of proteins bound on a single AuNP, can be estimated as the slope of the curves shown. For GB3 on 43 nm AuNPs, an adsorption capacity of 1200 ± 500 proteins per AuNP is observed; for BCA, this number decreases to 440 ± 130 proteins per AuNP.
To confirm that the proteins were in slow exchange, and that all residues were experiencing similar behavior, we recorded two-dimensional TROSY-HSQC spectra for 15N-labeled GB3 (Figure 2A). As expected, no chemical shift perturbations were observed in the presence of AuNPs, and no line broadening was observed under the conditions measured. The relative decrease in intensity for each residue was observed to be roughly uniform throughout the protein with an average for all residues of 0.888 ± 0.010 (Figure 2B, red line). This is very close to the value of 0.90 that is expected from 1-D experiments for an AuNP concentration of 1.2 nM. Using the spectral noise to estimate the uncertainty in the intensity, we find that all but four peaks are within two standard deviations of the observed average (Figure 2B). This suggests that all residues are behaving in a statistically identical way. Thus, for GB3 (and presumably for BCA, which exhibits similar behavior in its 1D NMR spectrum), no residue-specific interactions are observed between the protein and 43 nm AuNPs. While this could be interpreted to mean that all orientations are equally likely on the AuNP surface, more likely it results from the fact that, once adsorbed, the protein becomes invisible to NMR, and no further information can be obtained using conventional spectral acquisition.45,46
Figure 2.
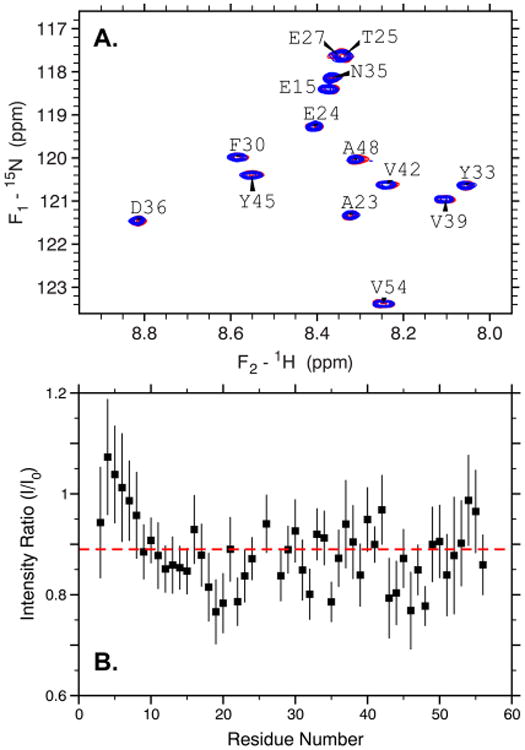
Residue-specific behavior of GB3 in the presence of 1.2 nM 43 nm AuNPs. (A) A 15N–1H TROSY HSQC spectrum showing selected residues of GB3 in the presence (blue) and absence (red) of AuNPs. No significant chemical shift changes or line broadening is observed. (B) The ratio of peak intensities in the presence versus absence of AuNPs for each residue in GB3. The red dashed line corresponds to the average of all residues, and nearly all residues fall close to this average, suggesting the absence of residue-specific effects.
Measuring Protein Adsorption to AuNPs of Varying Size
Both GB3 and BCA appear to exhibit similar behavior on 43 nm AuNPs as was observed previously with 15 nm AuNPs. To test whether the results were generalizable, we performed NMR adsorption measurements for both proteins with a range of nanoparticle sizes and curvatures, from 14 to 86 nm. All combinations of AuNP sizes exhibited the same slow exchange behavior with no apparent chemical shift perturbations as the AuNP concentration was increased (Figures S3 and S4). In addition, we also tested the behavior of a small, unstable SH3 domain from Drosophila melanogaster, drkN SH3, on 14 nm AuNPs. This SH3 domain was shown previously to be moderately unstable with .30 The folding of drkN SH3 is slow on the NMR time scale (kex = 1.4 s−1), but nevertheless appears to be largely two-state.47 The drkN SH3 domain also exhibits the same behavior seen for GB3: both unfolded and folded protein peaks decrease uniformly in the presence of AuNPs, with no preference for either the folded or the unfolded state peaks (Figure S5). It is possible to use both the folded and the unfolded peaks of drkN SH3 independently to calculate the concentration of bound protein, and these concentrations are statistically identical. For example, at a final concentration of 30 nM 14 nm AuNPs, and 20 μM total drkN SH3, the concentration of bound protein is calculated to be 7.1 ± 0.5 μM using the folded state peaks (spectrum shown in Figure S5A). Using the unfolded state peaks, the concentration is calculated to be 7.8 ± 0.5 μM. This is in good agreement with the value expected from 1D 1H spectra, 7 μM (Figure S5B). The correspondence between the two sets of resonances is expected because of the exchange between the folded and unfolded states. Even if one state were to bind to AuNPs preferentially, the unbound protein would rapidly re-equilibrate to show no difference in signal between the folded and unfolded states. Thus, while the behavior of the drkN SH3 domain is consistent with GB3 and BCA, one cannot easily determine whether the native or unfolded state binds preferentially to the AuNP surface from these measurements. A summary of all combinations of proteins and AuNPs measured in this work is shown in Table 1.
Table 1. Adsorption Data for Proteins Used in This Study.
| protein | RG (Å) |
|
AuNP size (nm) | observed adsorption capacity | surface density (nm−2) | predicted adsorption capacity | |
|---|---|---|---|---|---|---|---|
| GB3 | 10.8 | 4.6 | 14 | 179 ± 15 | 0.073 | 168 | |
| 30 | 720 ± 20 | 0.064 | 772 | ||||
| 43 | 1200 ± 500 | 0.052 | 1590 | ||||
| 86 | 8200 ± 1500 | 0.088 | 6340 | ||||
| BCA | 38.6 | 17a | 14 | 54 ± 8 | 0.022 | 67 | |
| 30 | 310 ± 11 | 0.027 | 308 | ||||
| 43 | 440 ± 130 | 0.019 | 632 | ||||
| 86 | 2900 ± 700b | 0.031 | 2530 | ||||
| drkN SH3 | 10.6 | 0.2c | 14 | 252 ± 7 | 0.102 | 174 | |
| 30 | 910 ± 15 | 0.080 | 801 |
Performing adsorption capacity measurements on 14–43 nm AuNPs was straightforward, but measurement was more difficult for 86 nm nanoparticles. Measurement for larger-sized nanoparticles is complicated by two related factors. First, the synthesis of these larger nanoparticles results in a comparatively low starting concentration. Second, the 86 nm AuNPs tend to aggregate during the concentration step, even at low centrifugal force. NMR-based measurements rely on a quantifiable difference between the concentration of protein in the presence and absence of AuNPs; therefore, when the AuNP concentration is low, the number of proteins bound is also low, and measurements become more challenging. This is revealed in the large error bars observed during measurements of GB3 binding to 86 nm AuNPs (Figure S3). Similarly, when the 86 nm AuNPs were mixed with BCA, there was only a very small change in peak intensity (4%), even when the total concentration of protein was lowered to 7 μM. On the basis of geometric considerations, fewer proteins are expected to bind as the protein size increases relative to the AuNP diameter, making measurements of large proteins (>30 kDa) on large AuNPs (>80 nM) even more difficult. More signal averaging is needed to obtain a statistically significant difference between protein spectra with and without AuNPs, and a complete measurement of binding capacity becomes impractical using five separate samples. Thus, the value reported for BCA represents a two-point measurement, and the uncertainty is estimated using the intensity loss of both amide and aliphatic protons within the spectrum. The value reported for BCA represents a concentration difference of ∼0.5 μM. Because it is difficult to measure smaller concentration differences accurately using NMR, this measurement likely represents the limit of reliability for measurements of this kind.
The adsorption capacities from our experiments can be converted to a surface density by dividing the adsorption capacity by the total AuNP surface area (Table 1). The density for each protein is consistent for all sizes of AuNPs observed. For GB3, the average value is (6.91 ± 0.07) × 10−2 proteins per nm2, and for BCA, the average is (2.49 ± 0.03) × 10−2. This consistency in density supports a hypothesis that the mechanism of adsorption for these proteins is similar regardless of nanoparticle size.
Predicting Adsorption Capacity from Protein Structure
Previously, we demonstrated that adsorption capacity and surface density could be predicted for a data set of six proteins using simple geometric considerations.27 This prior work focused on only 15 nm AuNPs. Here, we find that the same relationship holds for a variety of AuNP sizes and curvatures (Figure 3, Table 1). We assume that GB3 and BCA are compact on the AUNP surface and that their radii of gyration (RG) do not change upon adsorption. Further, we assume that the adsorbed protein forms a single layer of tightly packed protein. These assumptions can predict the adsorption capacity for both GB3 and BCA without any adjustable parameters. This leads us to hypothesize that both of these proteins are globular and most likely folded when adsorbed to AuNPs, forming a monolayer on the surface for all sizes tested. The drkN SH3 domain, however, exhibits a striking difference: This protein binds much more abundantly than would be predicted by the geometry of the folded state, and the predicted binding is severely underestimated for both 14 and 30 nm AuNPs (Table 1 and Figure 3, inset). The geometric model underestimates binding by more than 11 and 7 standard deviations, respectively, for 14 and 30 nm AuNPs. Given that no line broadening or chemical shift perturbations occur in the presence of AuNPs, we believe it is unlikely that additional layers of protein are forming on the surface. Instead, our observations are consistent with a uniform corona of bound, yet disordered, protein. The drkN SH3 domain exhibits a higher than expected degree of binding, and therefore each protein occludes a much smaller surface area than would be expected for a folded SH3 domain. Because of this, it is unlikely that this layer contains native protein structure, and if regular structure is retained, it is almost certainly highly distorted.
Figure 3.
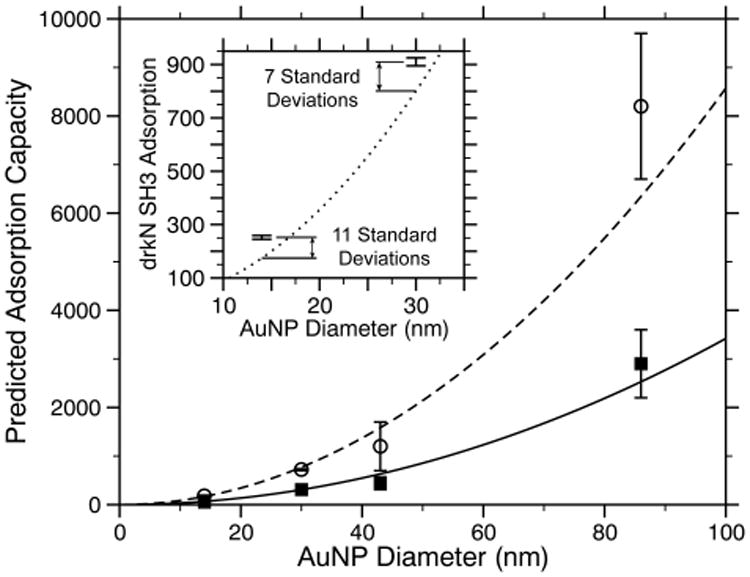
Comparison of observed versus predicted adsorption capacity for GB3 (○, dashed line) and BCA (■, solid lines). The lines represent a prediction that assumes a globular, densely packed monolayer of proteins on the surface and contains no adjustable parameters. The inset shows a similar prediction for the folded drkN SH3 domain (dotted lines) along with the observed adsorption capacity for drkN SH3 on 14 and 30 nm AuNPs.
While our results suggest that both GB3 and BCA are compact, it is possible that structural deformation occurs on the surface. Moreover, other groups have observed curvature-dependent behavior on AuNP surfaces. For example, Goy-López et al. observed a substantial change in the adsorption of human serum albumin from 2.7 to 4.5 layers of protein per AuNP as the nanoparticle diameter was increased from 10 to 100 nm,49 and Lacerda et al. have measured different adsorption association constants depending on AuNP size.50 In addition, it is well-known that enzymatic activity can decrease on AuNP surfaces, suggesting that the globular structure is not retained.25 These discrepancies may reveal that the behavior of protein–AuNP adsorption is strongly protein dependent; indeed, this would be supported by the extraordinary binding of the drkN SH3 domain. It is also possible that the NMR data presented here are only revealing information about the layer of proteins closest to the surface (hard corona), whereas other techniques may be more sensitive to the transient formation of additional layers (soft corona). A fast exchanging, weakly bound protein layer would not necessarily be visible using our results, because the NMR spectra would be dominated by the unbound protein at high protein to nanoparticle ratios. However, we believe the NMR-based approach employed here also offers several advantages over other previously published investigations. For example, the measurements of Goy-López et al. were performed using isothermal titration calorimetry (ITC).49 It has since been demonstrated that AuNP adsorption is kinetically controlled,51 suggesting that the ITC experiments may not be measuring a true equilibrium process. Similarly, fluorescence measurements used to measure adsorption isotherms may be adversely affected by the inner filter effect, which could lead to inaccurate values for the binding stoichiometry.52 NMR is not subject to either of these limitations, and our experiments therefore provide a complementary view of protein interactions at the AuNP surface.
Confirmation of Single-Layer Binding Using DLS and TEM
To supplement our NMR measurements, we employed two commonly used techniques to investigate the thickness of the BCA and GB3 layers on 30 nm AuNPs. By performing TEM on several of our protein conjugates, we observed the presence of several distinct halos surrounding the dark AuNP interior (Figure 4). Although we cannot confirm the specific identity of the species in the observed halo, no halos were observed in TEM samples prepared in the absence of proteins. Moreover, not every AuNP in protein samples contained a halo, but it is possible that the evaporation process used to prepare TEM samples disrupted the protein surface. Yang and Burkhard53 previously used TEM to visualize the protein coating on their self-assembled nanoparticles, suggesting that the halo we observe is indeed adsorbed protein. When present, the observed protein halo is approximately as thick as is expected for one layer of protein. For GB3, the observed thickness is 2.2 ± 0.2 nm, consistent with the predicted thickness of 2.2 nm for a single layer of compact protein (Figure 4A). The observed halo thickness for BCA is somewhat smaller than would be expected, 2.7 ± 0.2 nm as compared to the predicted value of 3.4 nm (Figure 4B). Nevertheless, this observation is consistent with a single layer of protein on the AuNP surface, as opposed to two or more layers of adsorbed protein.
Figure 4.
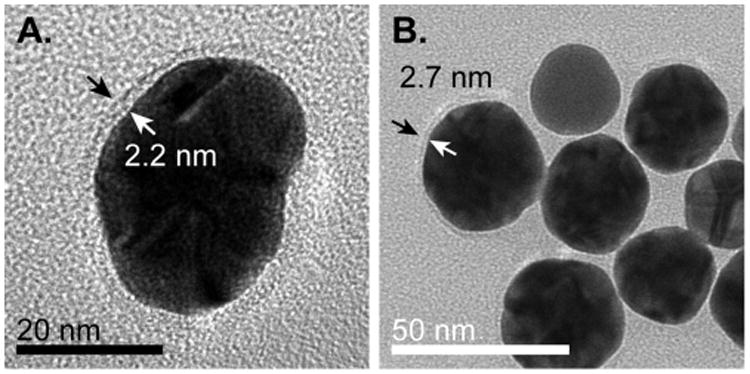
TEM images of protein coronas on AuNPs. Arrows indicate examples of halos surrounding AuNPs. (A) The thickness of the protein halo of GB3 adsorbed to 30 nm AuNPs is found to be 2.2 nm, consistent with the RG expected for a globular GB3 protein. (B) 15 nm AuNPs prepared with BCA. A somewhat thicker halo (2.7 nm) surrounds these nanoparticles, corresponding to the larger size of BCA as compared to GB3.
DLS was also used to investigate whether BCA and GB3 formed monolayers on 30 nm AuNP surfaces (Figure 5, Table 2). While DLS measures translational diffusion and therefore does not directly monitor size, it has nevertheless been useful for characterizing protein adsorption to AuNPs, and can measure size differences as small as 1–2%.54 Our results are consistent with the adsorption of one layer of BCA and GB3 to 30 nm AuNP surfaces. The larger apparent size of GB3-AuNPs as compared to BCA-AuNPs is counterintuitive considering that GB3 is the smaller protein, but the difference is within the uncertainty of the measurement. Stated differently, while the thickness of both protein layers is statistically significant as compared to the AuNPs themselves, the difference between the GB3 and BCA layers is smaller than 0.4 ± 0.5 nm and is therefore not statistically significant.
Figure 5.
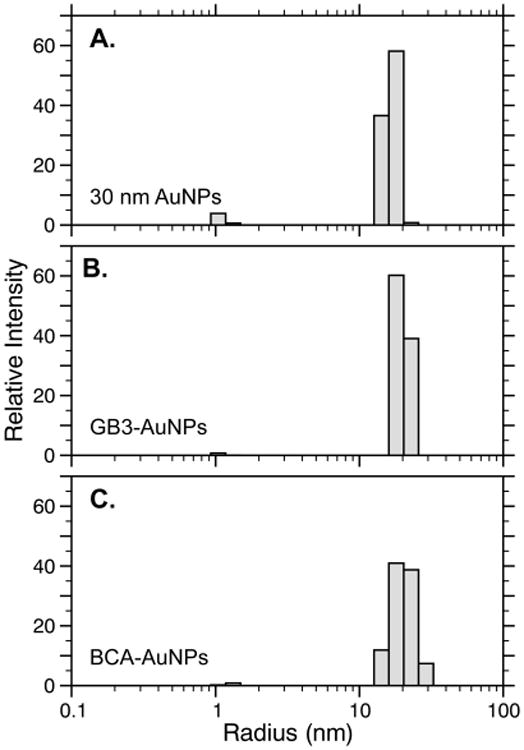
Typical DLS regularization fits for 30 nm AuNPs alone (A), 30 nm AuNPs in the presence of GB3 (B), and 30 nm AuNPs in the presence of BCA. The average values for three independently prepared samples support a small, but statistically significant, increase in the hydration radius when proteins are added to AuNP solutions. This increase is consistent with a single monolayer of compact protein on the AuNP surface (Table 2).
Table 2. Dynamic Light Scattering of Protein–AuNP Conjugates.
| sample | predicted single protein layer (nm) | RHa(nm) | RH increase (nm) | no. of protein layers |
|---|---|---|---|---|
| 30 nm AuNPs | 19.5 ± 0.3 | |||
| BCA-AuNPs | 3.4 | 22.1 ± 0.5 | 2.7 ± 0.6 | 0.8 ± 0.2 |
| GB3-AuNPs | 2.2 | 22.5 ± 0.2 | 3.1 ± 0.4 | 1.4 ± 0.2 |
Hydrodynamic radius, observed by DLS. Uncertainties are the standard error of the mean from three independently prepared samples.
While these data alone cannot confirm that all proteins bind in a monolayer to all sizes of AuNPs, the TEM and DLS data for BCA and GB3 support the hypothesis that these two proteins form a single layer of globular protein in the cases examined. In conjunction with the NMR studies presented above and in our previous work,27 this behavior appears to be a general feature for many proteins as they adsorb to citrate-coated AuNPs.
Relationship between Folding Free Energy and Adsorption
The striking difference in the behavior of drkN SH3 as compared to BCA and GB3 suggests that the stability of proteins will influence their adsorption behavior on AuNPs. Essentially, protein adsorption and unfolding represent a linked equilibrium where binding influences folding and vice versa. Previous studies have suggested that as the nanoparticle size grows and curvature decreases, protein–protein interactions on the surface can destabilize native structure.49,55,56 This is likely true; however, several aspects of protein–AuNP interactions make traditional measurements of protein stability challenging. For one, HDX measurements suggest that protein off-rates after AuNP adsorption are at least on the order of minutes,29 and they may be even slower.51 This severely complicates the measurement of binding equilibria because equilibrium is reached on a very slow time scale. Moreover, both urea and GdmCl tend to cause aggregation in AuNPs, which makes traditional protein stability measurements impractical if not impossible. Thus, modulating protein stability through mutagenesis or other means may be a potentially useful approach to understand the relationship between protein stability and surface adsorption (Figure 6). In particular, we hypothesize that unstable proteins or intrinsically disordered proteins will not follow the same trend observed for stable, globular proteins. This hypothesis is supported by the behavior of drkN SH3.
Figure 6.
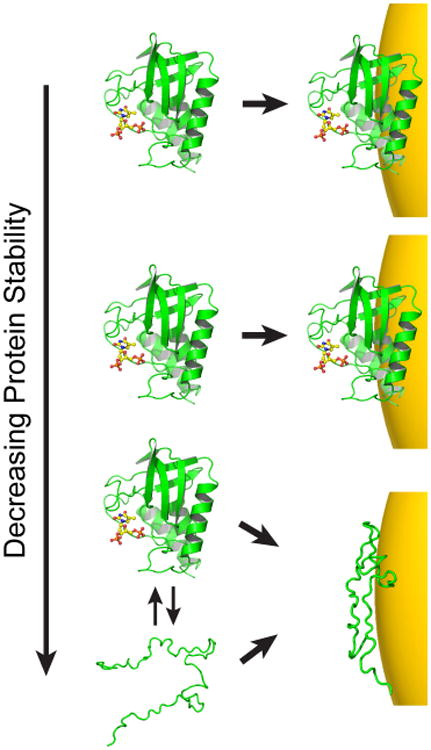
Hypothesized relationship between stability and AuNP adsorption. On the basis of the data presented here, stable proteins appear to remain globular when adsorbed, but the unstable drkN SH3 domain exhibits anomalous binding. Initially, as stability is lowered, protein adsorption may not be affected (top two panels). As folding stability continues to decrease, however, a protein may interact differently with the AuNP surface. This interaction may be modulated by direct binding of the unfolded state, or deformation of the folded state once bound. As a result, the apparent binding capacity can no longer be predicted by the native state structure.
In the work presented here, we find that GB3 and BCA behave as though they were compact and globular on the AuNP surface for all sizes measured. The stability of BCA is known to be 17 kcal mol−1,48 and we have determined the stability of GB3 to be 4.6 kcal mol−1 at 25 °C (Figure S6). The drkN SH3 domain, on the other hand, does not appear to remain compact and globular on either 14 or 30 nm AuNPs, and its stability is 0.2 kcal mol−1.30 This difference suggests that marginally stable proteins will not obey the same scaling law seen for stable, globular proteins. Mechanistically, there are several possible explanations for this: (1) First, it is possible that the unfolded state is favored over the folded state (kinetically or thermodynamically) to adsorb to the AuNP surface. In this case, the behavior of drkN SH3 would result from unstructured proteins binding directly to the AuNPs. (2) On the other hand, it is possible that the folded state is favored to bind the surface. In this case, adsorbed proteins would initially be globular, after which they would deform and lead to aberrant binding. (3) Finally, it is possible that some mixture of these two extremes is occurring. As discussed above, our experiments cannot distinguish whether the folded or unfolded state binds preferentially, as the unbound protein rapidly re-equilibrates between the folded and unfolded states.
Further studies are needed to determine the precise relationship between protein structure, stability, and adsorption. Other factors, such as protein–protein interactions, pH, temperature, and salt concentration, will also contribute to the behavior of protein structure on the AuNP surface.57 In addition, it is possible that structural deformation occurs to varying degrees once proteins reach the surface. For example, it may be that, while GB3 remains globular, its secondary structure is perturbed to the point where it is not strictly native once adsorbed. In this case, the energetics of adsorption to the AuNP surfaces would be poorly defined. Nevertheless, the dramatic difference observed between stable proteins and the unstable drkN SH3 domain suggests that a change in the nature of adsorption occurs as proteins become less stable.
Conclusions
In this work, we have investigated the nanoparticle size-dependence of the stoichiometry of protein–AuNP interactions. Because nanoparticle size directly affects surface curvature, this study addresses an open question whether decreased curvature can influence protein structure (and multilayer binding) at the nanoparticle surface.23–25 Several lines of evidence from this work suggest that AuNP size does not affect adsorbed protein structure, at least for GB3 and BCA. From our 1D NMR measurements, we were able to determine that there appears to be one layer of globular protein bound to the AuNPs at each size and for both proteins. While these measurements become substantially more challenging as protein and AuNP sizes increase, the data at each AuNP size strongly agree with our model that there is a three-step process for protein–AuNP adsorption that includes a hardening of the protein corona, but no unfolding or multilayer binding.27 These results are supported by direct visualization by TEM as well as by monitoring the marginal increase in AuNP radius using DLS. From our results, we hypothesize that most stable proteins remain globular on the AuNP surface, with only minor changes in their structure as they adsorb to the surface of the AuNPs. This phenomenon appears to be independent of both protein and AuNP size, and therefore can be assumed to be independent of curvature over the range we have studied.
At the same time, the aberrant behavior of the drkN SH3 domain provides useful insight into how protein stability may influence the typical behavior observed for stable proteins such as GB3 and BCA. The drkN SH3 domain is inherently unstable, and it represents the first case where we have observed a significant deviation from the adsorption that would be predicted for a well-packed monolayer of globular protein at the AuNP surface. Many more drkN SH3 molecules are adsorbed on the AuNP surface than would be predicted, suggesting that the unfolded state of this protein can accommodate a greater packing density than the folded state can. While other factors certainly affect the structure of adsorbed proteins, and while we cannot differentiate a truly native protein from a compact, surface-bound globule, the relatively low stability of GB3 (4.6 kcal mol−1) does not appear to affect its ability to bind as a compact globule, while the near-zero stability of drkN SH3 permits it to deform and bind to AuNPs at a much higher relative stoichiometry. Future work will be needed to identify precisely how protein stability influences adsorption. Given that protein deformation on nanoparticle surfaces can lead to adverse secondary affects, such as the formation of fibrils and aggregates,14,58 experiments such as these will be important for exploring the physiological consequences as protein–AuNP conjugates see increased use in clinical diagnostics and therapeutics.
Supplementary Material
Additional TEM images of AuNPs and NMR spectra, along with a data table of observed nanoparticle characteristics (PDF)
Acknowledgments
We thank Ailin Wang for assistance with nanoparticle preparation, and we thank Nikolai Skrynnikov for supplying the plasmid for drkN SH3. We also thank Jack Dunkle and Janet MacDonald for their suggestions on the TEM data presented here. Finally, we appreciate the assistance of an anonymous reviewer for their comments, which have greatly improved this manuscript. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R15GM113152. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- AuNPs
gold nanoparticles
- BCA
bovine carbonic anhydrase
- DLS
dynamic light scattering
- GdmCl
guanidinium chloride
- HDX
hydrogen-deuterium exchange
- RG
radius of gyration
- TEM
transmission electron microscopy
- WT
wild-type
Footnotes
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcc.6b08089.
Notes: The authors declare no competing financial interest.
References
- 1.Edelstein AS, Cammarata RC. Nanomaterials: Synthesis, Properties and Applications. Institute of Physics Publishing; Bristol, UK: 1998. [Google Scholar]
- 2.Ghosh SK, Pal T. Interparticle Coupling Effect on the Surface Plasmon Resonance of Gold Nanoparticles: From Theory toApplications. Chem Rev. 2007;107:4797–4862. doi: 10.1021/cr0680282. [DOI] [PubMed] [Google Scholar]
- 3.Daniel MC, Astruc D. Gold Nanoparticles: Assembly, Supramolecular Chemistry, Quantum-size-related Properties, and Applications Toward Biology, Catalysis, and Nanotechnology. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 4.Bethell D, Brust M, Schiffrin DJ, Kiely C. From Monolayers to Nanostructured Materials: An Organic Chemist's View of Self-Assembly. J Electroanal Chem. 1996;409:137–143. [Google Scholar]
- 5.Blakey I, Merican Z, Thurecht KJ. A Method for Controlling the Aggregation of Gold Nanoparticles: Tuning of Optical and Spectroscopic Properties. Langmuir. 2013;29:8266–8274. doi: 10.1021/la401361u. [DOI] [PubMed] [Google Scholar]
- 6.Turkevitch J, Stevenson PC, Hillier J. Nucleation and Growth Process in the Synthesis of Colloidal Gold. Discuss Faraday Soc. 1951;11:55–75. [Google Scholar]
- 7.Schmid G. Large Clusters and Colloids. Metals in the Embryonic State. Chem Rev. 1992;92:1709–1727. [Google Scholar]
- 8.Brown DH, Smith WE. The Chemistry of the Gold Drugs Used in the Treatment of Rheumatoid Arthritis. Chem Soc Rev. 1980;9:217–240. [Google Scholar]
- 9.Boyde A. Colloidal Gold. Principles, Methods and Application. J Anat. 1991;176:215–216. [Google Scholar]
- 10.Ghosh P, Han G, De M, Kim CK, Rotello VM. Gold Nanoparticles in Delivery Applications. Adv Drug Delivery Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Davis ME, Chen Z, Shin DM. Nanoparticle Therapeutics: An Emerging Treatment Modality for Cancer. Nat Rev Drug Discovery. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 12.Geoghegan WD, Ackerman GA. Adsorption of Horseradish Peroxidase, Ovomucoid and Anti-immunoglobulin to Colloidal Gold for the Indirect Detection of Concanavalin A, Wheat Germ Agglutinin and Goat Anti-human Immunoglobulin G on Cell Surfaces at the Electron Microscopic Level: A New Method, Theory and Application. J Histochem Cytochem. 1977;25:1187–1200. doi: 10.1177/25.11.21217. [DOI] [PubMed] [Google Scholar]
- 13.De Roe C, Courtoy PJ, Baudhuin P. A Model of Protein-Colloidal Gold Interactions. J Histochem Cytochem. 1987;35:1191–1198. doi: 10.1177/35.11.3655323. [DOI] [PubMed] [Google Scholar]
- 14.Zhang D, Neumann O, Wang H, Yuwono VM, Barhoumi A, Perham M, Hartgerink JD, Wittung-Stafshede P, Halas NJ. Gold Nanoparticles can Induce the Formation of Protein-Based Aggregates at Physiological pH. Nano Lett. 2009;9:666–671. doi: 10.1021/nl803054h. [DOI] [PubMed] [Google Scholar]
- 15.Lynch I, Salvati A, Dawson KA. Protein-Nanoparticle Interactions: What Does the Cell See? Nat Nanotechnol. 2009;4:546–547. doi: 10.1038/nnano.2009.248. [DOI] [PubMed] [Google Scholar]
- 16.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. What the Cell “Sees” in Bionanoscience. J Am Chem Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, Yu L, Monopoli MP, Sandin P, Mahon E, Salvati A, Dawson KA. The Biomolecular Corona is Retained During Nanoparticle Uptake and Protects the Cells from the Damage Induced by Cationic Nanoparticles Until Degraded in the Lysosomes. Nanomedicine. 2013;9:1159–1168. doi: 10.1016/j.nano.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 19.Milani S, Baldelli Bombelli F, Pitek AS, Dawson KA, Radler J. Reversible Versus Irreversible Binding of Transferrin to Polystyrene Nanoparticles: Soft and Hard Corona. ACS Nano. 2012;6:2532–2541. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- 20.Walkey CD, Chan WC. Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem Soc Rev. 2012;41:2780–99. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 21.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Gold Nanoparticles for Biology and Medicine. Angew Chem Int Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chithrani BD, Ghazani AA, Chan WCW. Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 23.Pramanik S, Banerjee P, Sarkar A, Bhattacharya SC. Size-Dependent Interaction of Gold Nanoparticles with Transport Protein: A Spectroscopic Study. J Lumin. 2008;128:1969–1974. [Google Scholar]
- 24.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc Natl Acad Sci USA. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagner JE, Lopez MD, Dordick JS, Siegel RW. Effect of Gold Nanoparticle Morphology on Adsorbed Protein Structure and Function. Biomaterials. 2011;32:7241–7252. doi: 10.1016/j.biomaterials.2011.05.091. [DOI] [PubMed] [Google Scholar]
- 26.Boulos SP, Davis TA, Yang JA, Lohse SE, Alkilany AM, Holland LA, Murphy CJ. Nanoparticle–Protein Interactions: A Thermodynamic and Kinetic Study of the Adsorption of Bovine Serum Albumin to Gold Nanoparticle Surfaces. Langmuir. 2013;29:14984–14996. doi: 10.1021/la402920f. [DOI] [PubMed] [Google Scholar]
- 27.Wang A, Vangala K, Vo T, Zhang D, Fitzkee NC. A Three-Step Model for Protein-Gold Nanoparticle Adsorption. J Phys Chem C. 2014;118:8134–8142. [Google Scholar]
- 28.Hostetler MJ, Wingate JE, Zhong CJ, Harris JE, Vachet RW, Clark MR, Londono JD, Green SJ, Stokes JJ, Wignall GD. Alkanethiolate Gold Cluster Molecules with Core Diameters from 1.5 to 5.2 nm: Core and Monolayer Properties as a Function of Core Size. Langmuir. 1998;14:17–30. [Google Scholar]
- 29.Wang A, Vo T, Le V, Fitzkee NC. Using Hydrogen–Deuterium Exchange to Monitor Protein Structure in the Presence of Gold Nanoparticles. J Phys Chem B. 2014;118:14148–14156. doi: 10.1021/jp506506p. [DOI] [PubMed] [Google Scholar]
- 30.Zhang O, Forman-Kay JD. Structural Characterization of Folded and Unfolded States of an SH3 Domain in Equilibrium in Aqueous Buffer. Biochemistry. 1995;34:6784–94. doi: 10.1021/bi00020a025. [DOI] [PubMed] [Google Scholar]
- 31.Grimsley GR, Pace CN. Spectrophotometric Determination of Protein Concentration. Curr Protoc Prot Sci. 2003:3.1.1–3.1.9. doi: 10.1002/0471140864.ps0301s33. [DOI] [PubMed] [Google Scholar]
- 32.Frens G. Controlled Nucleation for the Regulation of the Particle Size in Monodisperse Gold Suspensions. Nature Phys Sci. 1972;241:20–22. [Google Scholar]
- 33.Link S, El-Sayed MA. Size and Temperature Dependence of the Plasmon Absorption of Colloidal Gold Nanoparticles. J Phys Chem B. 1999;103:4212–4217. [Google Scholar]
- 34.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. Calculated Absorption and Scattering Properties of Gold Nanoparticles of Different Size, Shape, and Composition: Applications in Biological Imaging and Biomedicine. J Phys Chem B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Atwater M, Wang J, Huo Q. Extinction Coefficient of Gold Nanoparticles with Different Sizes and Different Capping Ligands. Colloids Surf B. 2007;58:3–7. doi: 10.1016/j.colsurfb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Vangala K, Ameer F, Salomon G, Le V, Lewis E, Yu L, Liu D, Zhang D. Studying Protein and Gold Nanoparticle Interaction Using Organothiols as Molecular Probes. J Phys Chem C. 2012;116:3645–3652. [Google Scholar]
- 37.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 Relaxation by Mutual Cancellation of Dipole–Dipole Coupling and Chemical Shift Anisotropy Indicates an Avenue to NMR Structures of Very Large Biological Macromolecules in Solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. Self-Assembly of CdSe-ZnS Quantum Dot Bioconjugates Using an Engineered Recombinant Protein. J Am Chem Soc. 2000;122:12142–12150. [Google Scholar]
- 39.Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S. Systematic Investigation of the Thermodynamics of HSA Adsorption to N-iso-propylacrylamide/N-tert-butylacrylamide Co-polymer Nanoparticles. Effects of Particle Size and Hydrophobicity. Nano Lett. 2007;7:914–920. doi: 10.1021/nl062743+. [DOI] [PubMed] [Google Scholar]
- 40.Ulmer TS, Ramirez BE, Delaglio F, Bax A. Evaluation of Backbone Proton Positions and Dynamics in a Small Protein by Liquid Crystal NMR Spectroscopy. J Am Chem Soc. 2003;125:9179–91. doi: 10.1021/ja0350684. [DOI] [PubMed] [Google Scholar]
- 41.Saito R, Sato T, Ikai A, Tanaka N. Structure of Bovine Carbonic Anhydrase II at 1.95 Å Resolution. Acta Crystallogr Sect D: Biol Crystallogr. 2004;60:792–5. doi: 10.1107/S0907444904003166. [DOI] [PubMed] [Google Scholar]
- 42.Bezsonova I, Singer A, Choy WY, Tollinger M, Forman-Kay JD. Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant. Biochemistry. 2005;44:15550–15560. doi: 10.1021/bi0512795. [DOI] [PubMed] [Google Scholar]
- 43.Grimsley GR, Huyghues-Despointes BM, Pace CN, Scholtz JM. Preparation of Urea and Guanidinium Chloride Stock Solutions for Measuring Denaturant-Induced Unfolding Curves. Cold Spring Harbor Protocols. 2006;2006 doi: 10.1101/pdb.prot4241. pdb.prot4241. [DOI] [PubMed] [Google Scholar]
- 44.Pace CN, Shaw KL. Linear Extrapolation Method of Analyzing Solvent Denaturation Curves. Proteins Struct FunctGenet. 2000;41(Suppl 4):1–7. doi: 10.1002/1097-0134(2000)41:4+<1::aid-prot10>3.3.co;2-u. [DOI] [PubMed] [Google Scholar]
- 45.Bodner CR, Dobson CM, Bax A. Multiple Tight Phospholipid-Binding Modes of Alpha-Synuclein Revealed by Solution NMR Spectroscopy. J Mol Biol. 2009;390:775–90. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Atomic-Resolution Dynamics on the Surface of Amyloid-Beta Protofibrils Probed by Solution NMR. Nature. 2011;480:268–72. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. Comparison of the Backbone Dynamics of a Folded and an Unfolded SH3 Domain Existing in Equilibrium in Aqueous Buffer. Biochemistry. 1995;34:868–78. doi: 10.1021/bi00003a021. [DOI] [PubMed] [Google Scholar]
- 48.Gitlin I, Gudiksen KL, Whitesides GM. Effects of Surface Charge on Denaturation of Bovine Carbonic Anhydrase. ChemBio-Chem. 2006;7:1241–1250. doi: 10.1002/cbic.200600191. [DOI] [PubMed] [Google Scholar]
- 49.Goy-Lopez S, Juarez J, Alatorre-Meda M, Casals E, Puntes VF, Taboada P, Mosquera V. Physicochemical Characteristics of Protein–NP Bioconjugates: The Role of Particle Curvature and Solution Conditions on Human Serum Albumin Conformation and Fibrillogenesis Inhibition. Langmuir. 2012;28:9113–9126. doi: 10.1021/la300402w. [DOI] [PubMed] [Google Scholar]
- 50.Lacerda SHDP, Park JJ, Meuse C, Pristinski D, Becker ML, Karim A, Douglas JF. Interaction of Gold Nanoparticles with Common Human Blood Proteins. ACS Nano. 2010;4:365–379. doi: 10.1021/nn9011187. [DOI] [PubMed] [Google Scholar]
- 51.Siriwardana K, LaCour A, Zhang D. Critical Sequence Dependence in Multicomponent Ligand Binding to Gold Nano-particles. J Phys Chem C. 2016;120:6900–6905. [Google Scholar]
- 52.Ameer FS, Ansar SM, Hu W, Zou S, Zhang D. Inner Filter Effect on Surface Enhanced Raman Spectroscopic Measurement. Anal Chem. 2012;84:8437–8441. doi: 10.1021/ac302073f. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y, Burkhard P. Encapsulation of Gold Nanoparticles into Self-Assembling Protein Nanoparticles. J Nanobiotechnol. 2012;10:42. doi: 10.1186/1477-3155-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jans H, Liu X, Austin L, Maes G, Huo Q. Dynamic Light Scattering as a Powerful Tool for Gold Nanoparticle Bioconjugation and Biomolecular Binding Studies. Anal Chem. 2009;81:9425–9432. doi: 10.1021/ac901822w. [DOI] [PubMed] [Google Scholar]
- 55.Lundqvist M, Sethson I, Jonsson BH. Protein Adsorption onto Silica Nanoparticles: Conformational Changes Depend on the Particles' Curvature and the Protein Stability. Langmuir. 2004;20:10639–10647. doi: 10.1021/la0484725. [DOI] [PubMed] [Google Scholar]
- 56.Shang W, Nuffer JH, Dordick JS, Siegel RW. Unfolding of Ribonuclease A on Silica Nanoparticle Surfaces. Nano Lett. 2007;7:1991–1995. doi: 10.1021/nl070777r. [DOI] [PubMed] [Google Scholar]
- 57.Wang A, Perera YR, Davidson MB, Fitzkee NC. Electrostatic Interactions and Protein Competition Reveal a Dynamic Surface in Gold Nanoparticle–Protein Adsorption. J Phys Chem C. 2016;120:24231. doi: 10.1021/acs.jpcc.6b08469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vacha R, Linse S, Lund M. Surface Effects on Aggregation Kinetics of Amyloidogenic Peptides. J Am Chem Soc. 2014;136:1776–82. doi: 10.1021/ja505502e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional TEM images of AuNPs and NMR spectra, along with a data table of observed nanoparticle characteristics (PDF)


