Abstract
Fibroblast growth factor-23 (FGF23) controls key responses to systemic phosphate increases through its phosphaturic actions on the kidney. In addition to stimulation by phosphate, FGF23 positively responds to iron deficiency anemia and hypoxia in rodent models and in humans. The disorder X-linked hypophosphatemia (XLH) is characterized by elevated FGF23 in concert with an intrinsic bone mineralization defect. Indeed, the Hyp mouse XLH model has disturbed osteoblast to osteocyte differentiation with altered expression of a wide variety of genes, including FGF23. The transcription factor Hypoxia inducible factor-1α (HIF1α) has been implicated in regulating FGF23 production and plays a key role in proper bone cell differentiation. Thus the goals of this study were to determine whether HIF1α activation could influence FGF23, and to test osteoblastic HIF1α production on the Hyp endocrine and skeletal phenotypes in vivo. Treatment of primary cultures of osteoblasts/osteocytes and UMR-106 cells with the HIF activator AG490 resulted in rapid HIF1α stabilization and increased Fgf23 mRNA (50–100 fold; p < 0.01–0.001) in a time- and dose-dependent manner. Next, the Phex gene deletion in the Hyp mouse was bred onto mice with a HIF1α/Osteocalcin (OCN)-Cre background. Although HIF1α effects on bone could be detected, FGF23-related phenotypes due to the Hyp mutation were independent of HIF1α in vivo. In summary, FGF23 can be driven by ectopic HIF1α activation under normal iron conditions in vitro, but factors independent of HIF1α activity after mature osteoblast formation are responsible for the disease phenotypes in Hyp mice in vivo.
Keywords: FGF23, Phosphate, Hypoxia inducible factor-1α, X-linked hypophosphatemia
Highlights
-
•
In vitro, a HIF activator stabilized HIF1α and increased Fgf23 mRNA expression.
-
•
A novel mouse model was generated by breeding the Hyp mouse onto the HIF1α/Osteocalcin (OCN)-Cre background.
-
•
Factors independent of HIF1α activity are responsible for the disease phenotypes in Hyp mice.
1. Introduction
The hormone FGF23 is produced in osteoblasts/osteocytes and plays a central role in renal phosphate handling. FGF23 acts in the kidney via its co-receptor αKlotho (αKL) and fibroblast growth factor receptors (FGFRs) to suppress renal phosphate reabsorption and reduce 1,25(OH)2 vitamin D (1,25D) production (Shimada et al., 2001, Larsson et al., 2004). Serum FGF23 is known to increase following dietary phosphate challenge (Antoniucci et al., 2006) as well as after 1,25D delivery (Shimada et al., 2005). Unique insight into the systemic control of FGF23 has been gained through the study of the disorder autosomal dominant hypophosphatemic rickets (ADHR) (ADHR-Consortium, 2000). Interestingly, patients with late-onset ADHR have elevated FGF23 and disease manifestations that originate during physiological situations of iron deficiency, including puberty and pregnancy (Econs and McEnery, 1997). We previously determined ADHR knock-in mice (carrying a corresponding human R176Q-Fgf23 mutant allele) markedly increased bone Fgf23 mRNA during iron deficiency anemia, leading to a hypophosphatemic rickets phenotype (Farrow et al., 2011). The relationships between iron metabolism and FGF23 are further reflected by highly significant negative correlations between total serum iron concentrations and FGF23 in ADHR kindreds (Imel et al., 2011), and in anemic patients treated with specific iron preparations (Shimizu et al., 2009, Wolf et al., 2013, Schouten et al., 2009a, Schouten et al., 2009b). In cultured osteoblastic cells, FGF23 can be induced by hypoxia and following iron chelation with deferoxamine (DFO), occurring in concert with marked protein accumulation of the transcription factor Hypoxia-induced factor-1α (HIF) (Farrow et al., 2011, Clinkenbeard et al., 2014). The HIF1α and 2α transcription factors play key roles in cellular responses to oxygen/iron sensing (Ivan et al., 2001). In addition to controlling genes such as erythropoietin (EPO) in kidney and vascular endothelial growth factor (VEGF) in many tissues (Yousaf and Spinowitz, 2016, Cubranic et al., 2015), HIFs plays essential roles in bone development. HIFs are required for proper osteogenesis and joint formation (Wang et al., 2007). Indeed, when deleted from mice, studies demonstrate that HIF1α is necessary for bone growth through control of a complex homeostatic response that allows chondrocyte to survival and differentiation under hypoxic conditions (Schipani et al., 2001).
The metabolic bone disorder X-linked hypophosphatemia (XLH) is caused by loss of function mutations in the Phosphate-regulating gene with homologies to endopeptidases on the X chromosome (PHEX gene) (Anon, 1995). Other members of this enzyme class include neutral endopeptidase (NEP) and endothelin-converting enzymes 1 and 2 (ECE-1 and ECE-2) (Turner and Tanzawa, 1997), which are known to cleave small peptides. XLH is characterized by elevated bioactive FGF23, inappropriately low or normal 1,25D, and a hypophosphatemic rickets/osteomalacia phenotype. The orthologous XLH mouse model, Hyp, has a large 3′ Phex deletion and phenocopies the XLH patient endocrine and skeletal disease phenotypes (Miao et al., 2001), including low serum phosphate, severe growth plate defects, and osteomalacia. In addition to ricketic bone disease, the Hyp mouse has a defect in osteoblast to osteocyte differentiation, with inappropriate expression of cell matrix genes (Bai et al., 2004). FGF23 is highly elevated in Hyp whole bone (Liu et al., 2006) as well as in Hyp-derived cultured osteoblasts/osteocytes (Liu et al., 2003). Primary cultures of osteoblastic cells from Hyp mice also fail to properly mineralize, thus underscoring an intrinsic cellular defect due to only partially resolved mechanisms (Bai et al., 2004). FGF23 is central to the Hyp metabolic bone disease phenotype, as demonstrated by the fact that global (Sitara et al., 2004) or osteoblast-specific (Clinkenbeard et al., 2016) knockout of Fgf23 in the Hyp genetic background reverses, or rescues, respectively, the low serum phosphate phenotype. However the molecular mechanisms controlling FGF23 in Hyp remain unknown.
Since FGF23 is elevated in the Hyp mouse, in concert with work linking HIF1α to increased FGF23 expression (Farrow et al., 2011, David et al., 2016) and this transcription factor's significant roles in bone cell differentiation, we sought to determine whether FGF23 could be induced under normal iron conditions following HIF1α activation, as well as whether bone-specific HIF1α deletion would correct FGF23 production and the Hyp disease phenotype. We found that in vitro, ectopic HIF1α activation strongly induced FGF23 production in primary cultures of differentiated osteoblasts/osteocytes, as well as in osteoblastic cell lines. In contrast, conditional deletion of HIF1α in vivo from mature osteoblasts demonstrated that FGF23 production in the Hyp mouse was independent of this transcription factor. Collectively, these findings further support the presence of unique mechanisms for abnormal FGF23 production among the disorders of phosphate handling involving this hormone.
2. Materials and methods
2.1. Animal studies
Animal studies were performed according to the Institutional Animal Care and Use Committee (IACUC) for Indiana University, and comply with the NIH guidelines for the use of animals. Hyp mice (Phex±) were mated to conditionally null flox(fl)-Hif1α/Osteocalcin (OCN)-Cre mice that lack Hif1α in the mature cells of the osteoblast-osteocyte lineage as reported (Wan et al., 2008). This cross generated Hyp/Hif1αfl/fl/OCN-Cre+, hereafter referred to as ‘Hyp/Hif1α(Cre+)’ mice. The other derived genotypes were: ‘WT,’ which were Phex+/+/Hif1αfl/fl/OCN-Cre−, as well as ‘WT/Hif1α(Cre+),’ which were: Phex+/+/Hif1αfl/fl/OCN-Cre+ genotype. Blood samples were collected at the time of sacrifice by cardiac puncture according to approved protocols. Mouse weights were taken every other week. Serum biochemistries were measured using an automated COBAS MIRA Plus Chemistry Analyzer (Roche Diagnostics; Indianapolis, IN). Serum intact bioactive FGF23 (iFGF23) concentrations were assessed using a commercial rodent-specific iFGF23 ELISA according to the manufacturer's protocol (Immutopics, Int'l/Quidel, Corp.).
2.2. Cell culture
UMR-106 cells (American Type Culture Collection; Manassas, VA) and primary cultures of differentiated osteoblasts/osteocytes were cultured in D-MEM/F-12 (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Hyclone; Thermo-Fisher, Inc.; Waltham, MA), and 25 mM penicillin-streptomycin in normoxia (21%) with 5% CO2/95% air at 37 °C. Primary cultures of osteoblasts/osteocytes were obtained from calvarial bones of neonatal C57BL/6 mice and were supplemented with 50 μg/ml ascorbic acid as previously reported (Bellido et al., 2005). Cells were treated with varying concentrations of AG490 in 12 well and 24 well plates, incubated 30 min to 24 h for time course trials and 8 and 24 h for dose-dependent studies. AG409 was diluted with the vehicle dimethyl sulfoxide (DMSO). UMR-106 cells were pre-treated with the MEK inhibitor (10 μM; U0126, R&D Systems).
2.3. RNA preparation and quantitative RT-PCR (qPCR)
The primary osteoblast/osteocyte cultures and UMR-106 cells were lysed in RLT buffer (Qiagen, Inc.) and homogenized in a Bullet Blender (MidSci Inc.). Total RNA from lysates was prepared using the RNeasy Kit (Qiagen, Inc.). RNA samples were tested with intron-spanning primers specific for mouse Fgf23 and normalized to β-actin as an internal control. All qPCR primers and probes were purchased as pre-optimized reagents (Applied Biosystems, Inc.). The TaqMan One-Step RT-PCR kit was used to perform qPCR. PCR conditions for all experiments were: 30 min 48 °C, 10 min 95 °C, followed by 40 cycles of 15 s 95 °C and 1 min 60 °C. The data was collected and analyzed by the 7500 Real Time PCR system and software (Applied Biosystems). RNA samples were analyzed in duplicate, and the 2−∆∆CT method described by Livak was used to analyze the data (Livak and Schmittgen, 2001).
2.4. Western blot analysis
UMR-106 cells were lysed with 100 μL 1 × Lysis buffer (Cell Signaling Technologies, Inc.) with 1 μg/mL AEBSF protease inhibitor (Sigma-Aldrich, Inc.). Cell lysate protein concentrations were determined with the Better Bradford Kit (Thermo-Fisher Scientific) according to the manufacturer's instructions. Western blot analysis was performed as previously described (Larsson et al., 2005) with 50 μg UMR-106 cellular lysates. The blots were incubated with 1:1000 primary anti-Hif1α; (Novus Biologicals) then incubated with the appropriate secondary antibody at 1:3000 (anti-rabbit IgG-HRP; Bio-Rad, Inc.); for normalization blots were stripped and incubated with 1:25,000 anti-β-actin-HRP (Sigma). Detection was performed using the ECL-Plus Western Blotting Detection Reagents (Amersham-GE Healthcare) and X-OMAT film (Eastman-Kodak Co.; Rochester, NY).
2.5. Micro-computed tomography
Morphological parameters of the distal femur were assessed using high resolution micro-computed tomography (μCT; Skyscan 1172). Bones were wrapped in parafilm to prevent drying during scanning. Scans were obtained using an X-ray source set at 60 kV and 167 mA over an angular range of 180° (rotational steps of 0.70°) with a 6 μm pixel size. Projection images were reconstructed using standard Skyscan software (NRecon). A 0.5 mm region of interest of the in the distal femur metaphysis was analyzed by segmenting the trabecular bone from the cortical shell and calculating trabecular bone volume per total volume (BV/TV) in accordance with recommended guidelines (13). On the most proximal slice of the ROI, the cortical shell was manually isolated from the trabecular bone by tracing the periosteal and endocortical edges for assessment of cortical bone geometry.
2.6. Statistical analysis
Where applicable ANOVA analysis with a Tukey HSD post-hoc test was conducted. Male and female Hyp, and Hyp/Hif1α(Cre+) were compared to their respective WT and WT/Hif1α(Cre+) genotype controls using Student's t-test. Significance for all tests was set at p < 0.05 and data are presented as means ± standard error of the mean (SEM).
3. Results
3.1. HIF activity is upstream of FGF23
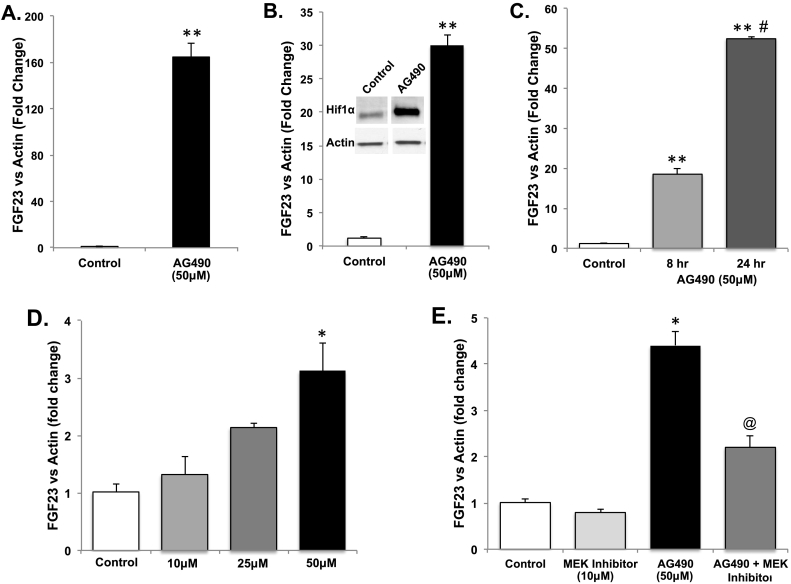
To test the role of HIF activation on the control of FGF23, the compound AG409 was used, as this analog strongly activates HIF1α in isolated cell systems (Fernandez-Sanchez et al., 2012). In this regard, primary cultures of differentiated osteoblasts/osteocytes and the osteoblastic UMR-106 cell line were treated with AG490, and Fgf23 mRNA expression was examined by qPCR. After 24 h, Fgf23 mRNA increased > 160-fold in the primary cultures (p < 0.001; Fig. 1A), and 30-fold in UMR-106 cells (p < 0.01, Fig. 1B). In addition, AG490 treatment also potently stabilized Hif1α protein as determined by immunoblot (Fig. 1B, inset). To determine the extent of the effects of AG490 on Fgf23 mRNA expression, time- and dose-dependence was examined in UMR-106 cells. Fgf23 mRNA increased from 4 to 24 h with a maximum response of 30-fold (p < 0.05–0.01; Fig. 1C), with doses of 10–50 μM (Fig. 1D). Pre-treatment with a MEK inhibitor (U0126) significantly blunted AG490-induced Fgf23 mRNA expression (p < 0.05; Fig. 1E).
Fig. 1.
AG490 activity on Fgf23 mRNA expression in vitro. A. AG490 (50 μM) stimulated FGF23 mRNA expression in primary cultures of differentiated osteoblasts/osteocytes (**p < 0.001) and B. UMR-106 cells (**p < 0.001). Inset: Hif1α protein was stabilized by both AG490 (50 μM) as assessed by immunoblot. (C.) Time- and (D.) dose-dependent effects on Fgf23 mRNA expression in UMR-106 cells versus vehicle (DMSO) after 2 h (*p < 0.05; **p < 0.01). E. AG490 (50 μM) activity on FGF23 mRNA expression is affected by pre-treatment with U0126 (*p < 0.05 vs untreated and U0126; @p < 0.05 vs AG490 treated).
3.2. Osteoblast lineage-specific Hif1α deletion on the Hyp background
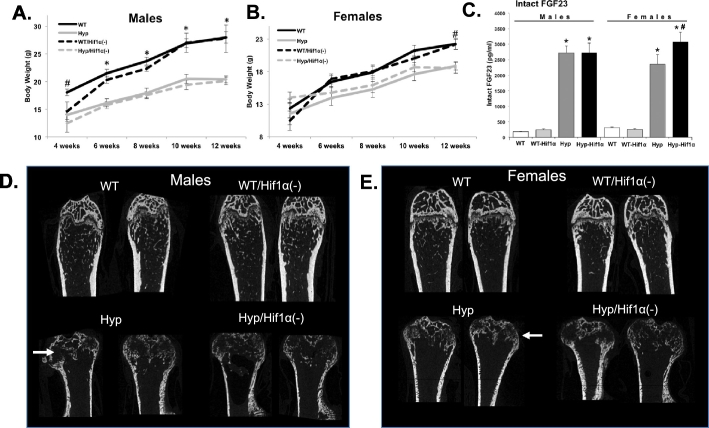
To test the role of Hif1α on the XLH-like phenotypes in vivo, the Phex gene deletion in the XLH model mouse Hyp was bred onto the HIF1α/Osteocalcin (OCN)-Cre background. Mice were serially assessed then sacrificed at 12 weeks of age. By 6 weeks of age, WT and WT/Hif1α(Cre+) males were significantly heavier than Hyp and Hyp/Hif1α(Cre+) (Fig. 2A; p < 0.01) and WT and WT/Hif1α(Cre+) females were significantly heavier than Hyp and Hyp/Hif1α(Cre+) female mice by 12 weeks (Fig. 2A; p < 0.05). There were no differences in the weights between either male or female WT and WT/Hif1α(Cre+) or between Hyp and Hyp/Hif1α(Cre+). Consistent with the known phenotype of Hyp, serum phosphate at sacrifice was reduced in both male and female Hyp and Hyp/Hif1α(Cre+) mice (Table 1; p < 0.01). Alkaline phosphatase was increased in Hyp and Hyp/Hif1α(Cre+) mice compared to their respective gender and genotype controls of WT and WT/Hif1α(Cre+) (Table 1; p < 0.01). No significant differences across genotypes occurred for calcium, creatinine or total iron (Table 1). Serum intact FGF23 was increased in male and female Hyp and Hyp/Hif1α(Cre+) mice compared to WT controls (Fig. 2B; p < 0.01).
Fig. 2.
Effects of Hif1α(Cre+) on Hyp phenotypes, A&B. Body weights (g) of male (left) and female (right) WT, WT/Hif1α(Cre+), Hyp, and Hyp/Hif1α(Cre+) mice were recorded every other week from weeks 4 through 12. (n ≥ 6 mice per group; *p < 0.01, #p < 0.05). C. Serum intact FGF23 was elevated in Hyp versus WT in the presence or absence of the Hif1α(Cre) allele. Female Hyp/Hif1α(Cre+) mice had slightly higher serum FGF23 compared with female Hyp. Bone structure in control and Hyp genetic crosses. Representative μCT images of distal femurs from 12 week old WT, WT/Hif1α(Cre+), Hyp, and Hyp/Hif1α(Cre+); D. male (left panel), E. female (right panel). Compared to genotype controls, Hyp mice had markedly widened distal femur (white arrows) consistent with their known ricketic phenotype, as well as far less trabecular bone.
Table 1.
Serum biochemistries for WT, WT/Hif1α(Cre+), Hyp, and Hyp/Hif1α(Cre+) mice (#p < 0.01; *p < 0.05; compared to genotype control).
| Alk. Phos. | Phosphate | Calcium | Creatinine | Total iron | ||
|---|---|---|---|---|---|---|
| Male | WT | 91.57 ± 5.35 |
11.74 ± 0.71 |
11.79 ± 0.54 |
0.42 ± 0.04 |
209.18 ± 12.08 |
| WT-Hif1α(Cre+) | 78.00 ± 12.00 |
14.63 ± 2.46 |
11.06 ± 0.20 |
0.43 ± 0.04 |
220.73 ± 16.95 |
|
| Hyp | 416.62# ± 17.57 |
8.92* ± 0.45 |
10.50 ± 0.37 |
0.40 ± 0.03 |
240.27 ± 10.60 |
|
| Hyp-Hif1α(Cre+) | 410.50# ± 28.52 |
9.55* ± 1.19 |
10.86 ± 0.26 |
0.35 ± 0.04 |
233.24 ± 12.13 |
|
| Female | WT | 114.90 ± 3.48 |
12.13 ± 1.03 |
10.95 ± 0.30 |
0.33 ± 0.04 |
206.89 ± 8.70 |
| WT-Hif1α(Cre+) | 114.00 ± 3.96 |
11.60 ± 0.47 |
10.65 ± 0.30 |
0.41 ± 0.04 |
214.77 ± 8.70 |
|
| Hyp | 275.50# ± 13.99 |
7.65* ± 0.46 |
10.43 ± 0.19 |
0.40 ± 0.01 |
200.77 ± 12.32 |
|
| Hyp-Hif1α(Cre+) | 276.43# ± 12.17 |
8.94* ± 0.85 |
9.95 ± 0.31 |
0.32 ± 0.05 |
231.68 ± 15.33 |
|
μCT analysis of the distal femur showed qualitative changes in Hyp and Hyp/Hif1α(Cre+) bone morphology including reduced metaphysis and growth plate versus gender- and genotype-matched WT controls (Fig. 2D & E). The Hyp and Hyp/Hif1α(Cre+) mice had lower trabecular bone volume (BV/TV), trabecular separation (Tb.Sp.), and trabecular number (Tb.N) (Table 2; Hyp male and females p < 0.01; Hyp/Hif1α(Cre+) male p < 0.01; Hyp/Hif1α(Cre+) female p < 0.05). Male Hyp and Hyp/Hif1α(Cre+) mice displayed larger bone area (B.Ar), compared to male WT and WT/Hif1α(Cre+) (p < 0.01) as well as lower cortical thickness (Ct.Th) (p < 0.01). Female WT/Hif1α(Cre+) mice exhibited decreased Ct.Th compared to WT females (p < 0.05), in line with previously reported bone effects of osteoblastic-specific Hif1α deletion (Wan et al., 2008). Collectively, these studies are consistent with the concept that in vitro Fgf23 mRNA is elevated during Hif1α activation, however FGF23 expression and the downstream metabolic bone disease phenotypes in the Hyp genetic syndrome are independent of the deletion of osteoblastic Hif1α.
Table 2.
μCT analysis in femur from WT, WT/Hif1α(Cre +), Hyp, and Hyp/Hif1α(Cre +) mice (#p < 0.01; *p < 0.05; compared to appropriate genotype control); (^p < 0.05; compared to female WT); bone volume/trabecular volume (BV/TV), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), trabecular number (Tb.N), cross-sectional area (CSA), bone area (B.Ar), marrow area (M.Ar), mean polar moment of inertia (MMI(polar)), cortical thickness (Ct.Th). Averages reported with ± the standard error of the mean.
| Trabecular bone |
Cortical bone |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| BV/TV | Tb.Th | Tb.Sp | Tb.N | CSA | B.Ar | M.Ar | MMI(polar) | Ct.Th | |
| Male | |||||||||
| WT | 12.33 ± 1.112 |
0.05 ± 0.002 |
0.20 ± 0.006 |
2.32 ± 0.137 |
2.07 ± 0.060 |
1.02 ± 0.043 |
1.05 ± 0.028 |
0.56 ± 0.046 |
0.15 ± 0.005 |
| WT/HIF1α (Cre+) |
17.43 ± 1.327 |
0.06 ± 0.001 |
0.19 ± 0.006 |
3.02 ± 0.199 |
2.27 ± 0.086 |
1.10 ± 0.031 |
1.18 ± 0.065 |
0.67 ± 0.049 |
0.16 ± 0.004 |
| Hyp | 3.62# ± 0.635 |
0.07# ± 0.003 |
0.40# ± 0.008 |
0.52# ± 0.075 |
2.02 ± 0.070 |
0.70# ± 0.028 |
1.32* ± 0.048 |
0.41* ± 0.035 |
0.10* ± 0.002 |
| Hyp/HIF1α (Cre+) |
2.70# ± 1.089 |
0.06 ± 0.002 |
0.41# ± 0.015 |
0.41# ± 0.142 |
1.91* ± 0.060 |
0.68* ± 0.018 |
1.23 ± 0.051 |
0.36* ± 0.021 |
0.10* ± 0.004 |
| Female | |||||||||
| WT | 4.88 ± 0.511 |
0.05 ± 0.002 |
0.26 ± 0.010 |
1.05 ± 0.096 |
1.90 ± 0.024 |
0.97 ± 0.016 |
0.93 ± 0.015 |
0.47 ± 0.016 |
0.20 ± 0.001 |
| WT/HIF1α (Cre+) |
5.77 ± 1.338 |
0.05 ± 0.002 |
0.27 ± 0.041 |
1.18 ± 0.227 |
1.84 ± 0.040 |
0.90 ± 0.064 |
0.95 ± 0.053 |
0.43 ± 0.030 |
0.16^ ± 0.017 |
| Hyp | 1.12# ± 0.184 |
0.07* ± 0.006 |
0.45# ± 0.008 |
0.16# ± 0.026 |
1.79 ± 0.049 |
0.78# ± 0.039 |
1.00 ± 0.083 |
0.38# ± 0.013 |
0.12# ± 0.008 |
| Hyp/HIF1α (Cre+) |
1.40* ± 0.253 |
0.08# ± 0.006 |
0.44* ± 0.007 |
0.18* ± 0.028 |
1.67 ± 0.061 |
0.77 ± 0.044 |
0.90 ± 0.037 |
0.35 ± 0.037 |
0.13 ± 0.009 |
4. Discussion
The heritable syndromes of mineral metabolism involving FGF23 are genetically heterogeneous, therefore defining the mechanisms underlying the distinct causes of elevated FGF23 and hypophosphatemia could lead to disease-specific therapies. HIF activity in the skeleton is critical for proper cell differentiation and mature bone development. In this regard, HIF1α ablation demonstrated that this factor is critical for chondrocyte survival and differentiation with the known hypoxia in bone tissue (Schipani et al., 2001), as well as for complete vascularization of the growing tissue (Kotch et al., 1999). The Hyp mouse model was shown to have defects in the maturation of osteoblasts to osteocytes, with over-expression of type I collagen, as well as inappropriate expression of matrix proteins including bone sialoprotein, and vitronectin at both the protein and mRNA levels (Bai et al., 2004). Our results showing that direct activation of HIF1α is upstream of Fgf23 mRNA production is consistent with studies demonstrating in vitro HIF1α activation and ChIP isolation of Fgf23 cis-promoter activity downstream of or potentially causing inflammatory responses (David et al., 2016, Singh et al., 2016), which can be associated with functional iron deficiency.
It remains unclear how the loss of function mutations in PHEX result in XLH, and cause both an intrinsic defect in bone mineralization and elevated FGF23. However reducing FGF23 bioactivity in patients (Carpenter et al., 2014) and mouse models of XLH (Bai et al., 2016, Erben et al., 2005) can reverse many of the severe manifestations of this disorder. Recent studies have shown that FGF23 can be elevated during situations of anemia in specific Fgf23-related disorders. This concept arose from treatment of anemic patients with specific iron preparations (Alleyne et al., 2008), prospective trials of iron formulations (Wolf et al., 2013), as well as in the heritable disorder ADHR (Farrow et al., 2011, Clinkenbeard et al., 2014). ADHR patients have FGF23 gain of function mutations and can develop late onset disease with variable penetrance, a unique finding among the FGF23 syndromes (Econs and McEnery, 1997, Econs et al., 1997). In some cases, patients clearly documented with elevated FGF23 had a natural reversal of their renal phosphate wasting or stark disease onset during physiological situations associated with anemia, such as puberty and pregnancy (Imel et al., 2007). From this observation, Imel and colleagues tested FGF23 and serum total iron levels in ADHR patients and healthy controls and found a significant association with bioactive FGF23 only in ADHR kindreds (Imel et al., 2011). These results were mimicked in mice containing ADHR R176Q-Fgf23 knock-in alleles. In this work, a low iron diet increased Fgf23 mRNA in bone and raised plasma FGF23 in parallel, leading to increased renal MEK signaling and to osteomalacia (Farrow et al., 2011). In the context of an ADHR FGF23 mutation, the enhanced transcription and more stable FGF23 protein led to elevated circulating hormone, providing the molecular mechanism for this disease.
The XLH syndrome, like anemia-induced ADHR, is associated with elevated Fgf23 mRNA, in concert with the inability to efficiently cleave and inactivate bioactive FGF23 (Harrell et al., 1985). In a clinical study, it was reported that although mean plasma FGF23 was elevated in patients with XLH compared to normals, FGF23 correlated negatively with serum iron in both patients and normal controls (Imel et al., 2014). This study suggested that the XLH disease phenotype was not dependent upon serum iron concentrations. In contrast, a female patient from a family with a documented FGF23 ADHR mutation received clinical benefit from iron (II) sulfate therapy (Kapelari et al., 2015). This treatment suppressed serum FGF23 and elevated 1,25D (Kapelari et al., 2015), supporting the idea that restoring serum iron concentrations to normal in anemic patients may help to reduce FGF23. Therefore, elevated bioactive FGF23 is the common denominator in multiple FGF23-related syndromes, however due to the lack of association between serum iron levels and the XLH phosphate wasting phenotype, XLH patients may have higher benefit from direct targeting of FGF23, such as with anti-FGF23 antibody therapy (Carpenter et al., 2014). Although we demonstrated HIF activity upstream of Fgf23 mRNA expression in vitro, our studies may not have observed an FGF23-related phenotype in Hyp mice for several reasons. First, HIF2α has been shown to potentially compensate for some phenotypes during osteoblastic HIF1α ablation in vivo (Wang et al., 2007). Therefore significant HIF2α expression could potentially influence FGF23 expression in whole bone. In prior studies, however, iron chelation with DFO in UMR-106 cells failed to activate HIF2α concomitant with elevated Fgf23 mRNA expression increases (Clinkenbeard et al., 2014), therefore whether HIF2α is activated in vivo in Hyp is unclear. Additionally, in the current study, the late osteoblast osteocalcin promoter was used to conditionally target HIF1α as reported (Wang et al., 2007). Since Fgf23 mRNA is expressed in a significant proportion of osteoblasts (Feng et al., 2006), due to the reported osteoblast to osteocyte transition defect in Hyp mice (Miao et al., 2001) Cre-mediated recombination of floxed-HIFs may have to be targeted earlier in the osteoblastic differentiation stages to more dramatically alter FGF23 production. Although we found in UMR-106 cells that an AG490-mediated increase in Fgf23 mRNA was potently induced, correlation between Fgf23 mRNA and FGF23 protein remains to be determined. Finally, there is emerging evidence that FGF23 is produced in cells that comprise bone outside of osteoblasts/osteocytes, such as chondrocytes (Kawai et al., 2013) and in marrow (Xiao et al., 2013). Therefore, whether these other sites compensate for osteoblast/osteocyte FGF23 production is currently unknown, and gives rise to the idea that in the Hyp genetic background, multiple cell types may have to be targeted to fully affect circulating FGF23 concentrations.
In summary, our studies support that HIF1α is functionally upstream of FGF23 production in osteoblast/osteocyte cells. The XLH phenotype of elevated FGF23 and hypophosphatemic rickets due to loss of function Phex mutations in the Hyp mouse however, is independent of mature osteoblastic HIF1α expression. Therefore, the genetic heterogeneity of diseases associated with elevated FGF23 must be considered during individual patient care, including targeting HIF-dependent pathways with iron therapy or HIF inhibitors, ADHR, but likely not XLH.
Disclosures
KEW receives royalties for licensing the FGF23 gene to Kyowa-Hakko-Kirin Pharmaceutics, Inc. The other authors have nothing to declare.
Acknowledgments
The authors acknowledge NIH grants R01-DK063934, R01-DK95784, R21-AR070329 (KEW); F32-AR065389 (ELC); T32-HL007910 and the American Heart Association postdoctoral fellowship 16POST27260108 (JMH); a Showalter Scholar award through the Ralph W. and Grace M. Showalter Research Trust (KEW). The authors thank Dr. Thomas L. Clemens (Johns Hopkins School of Medicine) for graciously providing the HIF1α/OCN-cre mice.
References
- ADHR-Consortium Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 2000;26(3):345–348. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- Alleyne M., Horne M.K., Miller J.L. Individualized treatment for iron-deficiency anemia in adults. Am. J. Med. 2008;121(11):943–948. doi: 10.1016/j.amjmed.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. The HYP Consortium. Nat. Genet. 1995;11(2):130–136. doi: 10.1038/ng1095-130. [DOI] [PubMed] [Google Scholar]
- Antoniucci D.M., Yamashita T., Portale A.A. Dietary phosphorus regulates serum fibroblast growth factor-23 concentrations in healthy men. J. Clin. Endocrinol. Metab. 2006;91(8):3144–3149. doi: 10.1210/jc.2006-0021. [DOI] [PubMed] [Google Scholar]
- Bai X. Transgenic mice overexpressing human fibroblast growth factor 23 (R176Q) delineate a putative role for parathyroid hormone in renal phosphate wasting disorders. Endocrinology. 2004;145(11):5269–5279. doi: 10.1210/en.2004-0233. [DOI] [PubMed] [Google Scholar]
- Bai X. CYP24 inhibition as a therapeutic target in FGF23-mediated renal phosphate wasting disorders. J. Clin. Invest. 2016;126(2):667–680. doi: 10.1172/JCI81928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellido T. Chronic elevation of parathyroid hormone in mice reduces expression of sclerostin by osteocytes: a novel mechanism for hormonal control of osteoblastogenesis. Endocrinology. 2005;146(11):4577–4583. doi: 10.1210/en.2005-0239. [DOI] [PubMed] [Google Scholar]
- Carpenter T.O. Randomized trial of the anti-FGF23 antibody KRN23 in X-linked hypophosphatemia. J. Clin. Invest. 2014;124(4):1587–1597. doi: 10.1172/JCI72829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinkenbeard E.L. Neonatal iron deficiency causes abnormal phosphate metabolism by elevating FGF23 in normal and ADHR mice. J. Bone Miner. Res. 2014;29(2):361–369. doi: 10.1002/jbmr.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinkenbeard E.L. Conditional deletion of murine Fgf23: interruption of the normal skeletal responses to phosphate challenge and rescue of genetic hypophosphatemia. J. Bone Miner. Res. 2016;31(6):1247–1257. doi: 10.1002/jbmr.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubranic A. Mystery story about erythropoietin (Epo) and erythropoietin receptor (EpoR) are disguised? Hepato-Gastroenterology. 2015;62(139):585–589. [PubMed] [Google Scholar]
- David V. Inflammation and functional iron deficiency regulate fibroblast growth factor 23 production. Kidney Int. 2016;89(1):135–146. doi: 10.1038/ki.2015.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Econs M.J., McEnery P.T. Autosomal dominant hypophosphatemic rickets/osteomalacia: clinical characterization of a novel renal phosphate-wasting disorder. J. Clin. Endocrinol. Metab. 1997;82(2):674–681. doi: 10.1210/jcem.82.2.3765. [DOI] [PubMed] [Google Scholar]
- Econs M.J. Autosomal dominant hypophosphatemic rickets is linked to chromosome 12p13. J. Clin. Invest. 1997;100(11):2653–2657. doi: 10.1172/JCI119809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erben R.G. Overexpression of human PHEX under the human beta-actin promoter does not fully rescue the Hyp mouse phenotype. J. Bone Miner. Res. 2005;20(7):1149–1160. doi: 10.1359/JBMR.050212. [DOI] [PubMed] [Google Scholar]
- Farrow E.G. Iron deficiency drives an autosomal dominant hypophosphatemic rickets (ADHR) phenotype in fibroblast growth factor-23 (Fgf23) knock-in mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108(46):E1146–E1155. doi: 10.1073/pnas.1110905108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J.Q. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat. Genet. 2006;38(11):1310–1315. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Sanchez R. AG490 promotes HIF-1alpha accumulation by inhibiting its hydroxylation. Curr. Med. Chem. 2012;19(23):4014–4023. doi: 10.2174/092986712802002554. [DOI] [PubMed] [Google Scholar]
- Harrell R.M. Healing of bone disease in X-linked hypophosphatemic rickets/osteomalacia. Induction and maintenance with phosphorus and calcitriol. J. Clin. Invest. 1985;75(6):1858–1868. doi: 10.1172/JCI111900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imel E.A., Hui S.L., Econs M.J. FGF23 concentrations vary with disease status in autosomal dominant hypophosphatemic rickets. J. Bone Miner. Res. 2007;22(4):520–526. doi: 10.1359/jbmr.070107. [DOI] [PubMed] [Google Scholar]
- Imel E.A. Iron modifies plasma FGF23 differently in autosomal dominant hypophosphatemic rickets and healthy humans. J. Clin. Endocrinol. Metab. 2011;96(11):3541–3549. doi: 10.1210/jc.2011-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imel E.A. Iron and fibroblast growth factor 23 in X-linked hypophosphatemia. Bone. 2014;60:87–92. doi: 10.1016/j.bone.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Kapelari K. Iron supplementation associated with loss of phenotype in autosomal dominant hypophosphatemic rickets. J. Clin. Endocrinol. Metab. 2015;100(9):3388–3392. doi: 10.1210/jc.2015-2391. [DOI] [PubMed] [Google Scholar]
- Kawai M. FGF23 suppresses chondrocyte proliferation in the presence of soluble alpha-Klotho both in vitro and in vivo. J. Biol. Chem. 2013;288(4):2414–2427. doi: 10.1074/jbc.M112.410043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotch L.E. Defective vascularization of HIF-1alpha-null embryos is not associated with VEGF deficiency but with mesenchymal cell death. Dev. Biol. 1999;209(2):254–267. doi: 10.1006/dbio.1999.9253. [DOI] [PubMed] [Google Scholar]
- Larsson T. Transgenic mice expressing fibroblast growth factor 23 under the control of the alpha1(I) collagen promoter exhibit growth retardation, osteomalacia, and disturbed phosphate homeostasis. Endocrinology. 2004;145(7):3087–3094. doi: 10.1210/en.2003-1768. [DOI] [PubMed] [Google Scholar]
- Larsson T. Fibroblast growth factor-23 mutants causing familial tumoral calcinosis are differentially processed. Endocrinology. 2005;146(9):3883–3891. doi: 10.1210/en.2005-0431. [DOI] [PubMed] [Google Scholar]
- Liu S. Regulation of fibroblastic growth factor 23 expression but not degradation by PHEX. J. Biol. Chem. 2003;278(39):37419–37426. doi: 10.1074/jbc.M304544200. [DOI] [PubMed] [Google Scholar]
- Liu S. Pathogenic role of Fgf23 in Hyp mice. Am. J. Physiol. Endocrinol. Metab. 2006;291(1):E38–E49. doi: 10.1152/ajpendo.00008.2006. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Miao D. Osteomalacia in hyp mice is associated with abnormal phex expression and with altered bone matrix protein expression and deposition. Endocrinology. 2001;142(2):926–939. doi: 10.1210/endo.142.2.7976. [DOI] [PubMed] [Google Scholar]
- Schipani E. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15(21):2865–2876. doi: 10.1101/gad.934301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten B.J. FGF23 elevation and hypophosphatemia after intravenous iron polymaltose: a prospective study. J. Clin. Endocrinol. Metab. 2009;94(7):2332–2337. doi: 10.1210/jc.2008-2396. [DOI] [PubMed] [Google Scholar]
- Schouten B.J. Iron polymaltose-induced FGF23 elevation complicated by hypophosphataemic osteomalacia. Ann. Clin. Biochem. 2009;46(Pt 2):167–169. doi: 10.1258/acb.2008.008151. [DOI] [PubMed] [Google Scholar]
- Shimada T. Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc. Natl. Acad. Sci. U. S. A. 2001;98(11):6500–6505. doi: 10.1073/pnas.101545198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. Vitamin D receptor-independent FGF23 actions in regulating phosphate and vitamin D metabolism. Am. J. Physiol. Ren. Physiol. 2005;289(5):F1088–F1095. doi: 10.1152/ajprenal.00474.2004. [DOI] [PubMed] [Google Scholar]
- Shimizu Y. Hypophosphatemia induced by intravenous administration of saccharated ferric oxide: another form of FGF23-related hypophosphatemia. Bone. 2009;45(4):814–816. doi: 10.1016/j.bone.2009.06.017. [DOI] [PubMed] [Google Scholar]
- Singh S. Fibroblast growth factor 23 directly targets hepatocytes to promote inflammation in chronic kidney disease. Kidney Int. 2016;90(5):985–996. doi: 10.1016/j.kint.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitara D. Homozygous ablation of fibroblast growth factor-23 results in hyperphosphatemia and impaired skeletogenesis, and reverses hypophosphatemia in Phex-deficient mice. Matrix Biol. 2004;23(7):421–432. doi: 10.1016/j.matbio.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A.J., Tanzawa K. Mammalian membrane metallopeptidases: NEP, ECE, KELL, and PEX. FASEB J. 1997;11(5):355–364. doi: 10.1096/fasebj.11.5.9141502. [DOI] [PubMed] [Google Scholar]
- Wan C. Activation of the hypoxia-inducible factor-1alpha pathway accelerates bone regeneration. Proc. Natl. Acad. Sci. U. S. A. 2008;105(2):686–691. doi: 10.1073/pnas.0708474105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. The hypoxia-inducible factor alpha pathway couples angiogenesis to osteogenesis during skeletal development. J. Clin. Invest. 2007;117(6):1616–1626. doi: 10.1172/JCI31581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M., Koch T.A., Bregman D.B. Effects of iron deficiency anemia and its treatment on fibroblast growth factor 23 and phosphate homeostasis in women. J. Bone Miner. Res. 2013;28(8):1793–1803. doi: 10.1002/jbmr.1923. [DOI] [PubMed] [Google Scholar]
- Xiao L., Esliger A., Hurley M.M. Nuclear fibroblast growth factor 2 (FGF2) isoforms inhibit bone marrow stromal cell mineralization through FGF23/FGFR/MAPK in vitro. J. Bone Miner. Res. 2013;28(1):35–45. doi: 10.1002/jbmr.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousaf F., Spinowitz B. Hypoxia-inducible factor stabilizers: a new avenue for reducing BP while helping hemoglobin? Curr. Hypertens. Rep. 2016;18(3):23. doi: 10.1007/s11906-016-0629-6. [DOI] [PubMed] [Google Scholar]