Abstract
The genetic underpinnings of recessively inherited moderate to severe sensorineural hearing loss are not well understood, despite its higher prevalence in comparison to profound deafness. We recruited 92 consanguineous families segregating stable or progressive, recessively inherited moderate or severe hearing loss. We utilized homozygosity mapping, Sanger sequencing, targeted capture of known deafness genes with massively parallel sequencing and whole exome sequencing to identify the molecular basis of hearing loss in these families. Variants of the known deafness genes were found in 69% of the participating families with the SLC26A4, GJB2, MYO15A, TMC1, TMPRSS3, OTOF, MYO7A and CLDN14 genes together accounting for hearing loss in 54% of the families. We identified 20 reported and 21 novel variants in 21 known deafness genes. Sixteen of the twenty reported variants, previously associated with stable, profound deafness were associated with moderate to severe or progressive hearing loss in our families. These data point to a prominent role for genetic background, environmental factors or both as modifiers of human hearing loss severity.
Keywords: Aetiology, Deafness, DFNB, Hearing loss, Modifiers, Mutation, Pakistan
Graphical abstract
INTRODUCTION
Hearing loss is a common sensory impairment that affects 278 million people worldwide (1). Dominant and recessive alleles account for approximately half of congenital or early onset childhood hearing loss that can have a broad range of severity (2). A majority of pathogenic recessive variants of genes associated with hearing loss cause congenital or prelingual, severe to profound, stable sensorineural deafness, which occurs in approximately 1 in 1,000 births (1). Autosomal dominant inheritance is typically associated with post-lingual, progressive hearing loss that can range from mild to profound at onset or over the course of many decades. In general, the hearing loss associated with dominant inheritance is less severe than that associated with recessive inheritance. A few mutant alleles of some profound deafness-associated genes such as MYO7A and CDH23, have been described to cause variable audiological phenotypes as well (3-5). The objective of this study was to determine if other genes previously associated with profound deafness also contribute to moderate or severe hearing loss. Our study implicates 21 different genes as well as modifiers in the etiology of recessively inherited moderate to severe hearing loss.
MATERIALS AND METHODS
Families
The study was approved by the Institutional Review Board at the School of Biological Sciences, University of the Punjab, Lahore and the Combined Neuroscience Institutional Review Board (protocol OH-93-N-016) at the National Institutes of Health, USA. Samples were collected with written informed consent from each participant.
We ascertained 92 consanguineous Pakistani families; 90 residing in the province of Punjab and two families from Khyber Pakhtunkhwa. Probands were identified through special schools in Punjab where children with different disabilities are educated, from schools for the deaf and through referrals from audiologists. In order to identify probands from schools, we first inquired about the extent of a child's hearing ability from teachers, which were verified by a distraction test that involves recording a subject's response to sound stimuli coming from a hidden source. Pure tone thresholds of the probands and other family participants, who expressed willingness for audiometric testing, were measured at 0.25, 0.5, 1, 2, 4 and 8 kHz in a quiet room due to the lack of a sound proof room. The pure-tone average (PTA) was calculated by averaging the hearing thresholds at 0.5, 1, 2 and 4 kHz (6). Hearing loss was classified on the basis of PTA as moderate (41 to 70 dB HL), severe (71 to 95 dB HL), and profound (above 95 dB HL) (6). Individuals with hearing thresholds between 31-40 dB HL were classified with mild hearing loss while those with thresholds up to 30 dB HL were considered to have normal hearing since performing audiometry in ambient noise conditions may result in slightly elevated thresholds for those with normal hearing (7). Subjects were questioned about the age of the onset and progression in severity of their hearing loss. Vestibular function was evaluated by tandem-gait and Romberg tests. Otoscopic examinations and medical histories were completed in order to exclude affected individuals due to active external or middle ear disease, and syndromic or environmental causes, though families with Usher syndrome were included. Funduscopy and electroretinography examinations were conducted for the oldest affected individuals in a subset of families.
Molecular characterization
Blood samples were collected from the participants and genomic DNA was extracted by a standard method (8). The sequence of GJB2 was analyzed for the affected participants from all families by Sanger sequencing (9). DNA samples from at least three affected individuals from 66 of 92 consanguineous families (Supplementary Table S1) were screened for linkage to genes associated with nonsyndromic recessively inherited deafness (DFNB) and Usher syndrome (USH), by homozygosity mapping as described (10, 11). In brief, samples from at least three affected individuals were chosen from each family, 2 to 5 tightly linked microsatellite markers for each deafness gene were PCR amplified using fluorescently tagged primers and the amplicons resolved by capillary electrophoresis. If affected individuals were homozygous for alleles of any of these markers, samples from all participants of the respective family were genotyped for the linked, as well as additional downstream or upstream markers to determine if there is co-segregation with the phenotype.
After homozygosity mapping, Sanger sequencing of the implicated genes was performed for 17 of the families (Supplementary Table S1). Samples from 53 families were screened with massively parallel sequencing (Supplementary Table S1). For targeted capture and massively parallel sequencing of 108 genes with a demonstrated role in hearing (Supplementary Table S2), custom probes were designed to capture the exons and approximately 50 bp of flanking intronic sequence. Fragment library preparation and target enrichment were performed according to the manufacturers’ instructions on one randomly selected sample from each family (Supplementary Table S3) and sequencing was performed on an ABI SOLiD 5500.
For whole-exome sequencing, one to three samples of affected individuals were randomly selected from the chosen families (Supplementary Table S3). Fragment libraries were prepared with a Nextera Rapid Capture library kit (REF#15034786) for paired end, dual index sequencing on an Illumina HiSeq 1000 platform at the NIDCD, USA, while some libraries were prepared and sequenced at Otogenetics, USA on an Illumina HiSeq 2000 platform. Variants were annotated using the web-based software wANNOVAR (http://wannovar.usc.edu/). Variants with overall population allele frequencies of >1% (0.01) in any of the public databases (1000 genome dataset, 6500 exome variant server, and Exome Aggregation Consortium, ExAC, database) were removed. Only variants in protein coding sequences or those affecting splice sites were considered further. For HGF (OMIM 142409), we also screened for the intronic variants that were shown to be associated with nonsyndromic deafness DFNB39 (OMIM 608265) (12).
All suspected pathogenic variants were evaluated for co-segregation with the hearing loss phenotype in families by PCR amplification and Sanger sequencing using Big Dye Terminator V3.1 (Applied Biosystems, Foster City, USA). Sequences of PCR primers are available on request. Allele frequencies of the novel variants were assessed by Sanger sequencing using genomic DNA from 100 to 200 ethnically matched unrelated individuals from Pakistan with threshold for assigning the variant as a polymorphism set to its detection in at least 1% control chromosomes. The pathogenicity of the missense variants was evaluated using in silico tools PROVEAN, http://provean.jcvi.org/, SIFT, http://sift.bii.a-star.edu.sg/, and Mutation Taster, http://www.mutationtaster.org/. Possible splicing effects introduced by variants identified near exon/intron junctions were determined by Human Splicing Finder (http://www.umd.be/HSF/) and Skippy (http://research.nhgri.nih.gov/skippy/). Alignments of multiple protein sequences were performed with Clustal Omega http://www.ebi.ac.uk/Tools/msa/clustalo/. The sequences of orthologs for alignment of proteins were obtained from UCSC genome server (https://genome.ucsc.edu/).
Calculation of percentage contributions and confidence intervals
Individuals from 92 families participated in the study. Percentage contribution of deafness genes to hearing loss in this cohort were calculated based on 91 families which included seven families for which linkage of hearing loss was detected to a reported deafness locus but a pathogenic variant was not identified. The 95% Confidence Intervals for contribution of genes to deafness were calculated as described (https://onlinecourses.science.psu.edu/stat200/node/48). A family with an USH1C variant was excluded from these calculations as Usher syndrome was manifested by the affected participants at the time of enrollment, and therefore for this family only the reported genes for Usher syndrome were analyzed.
RESULTS
Families
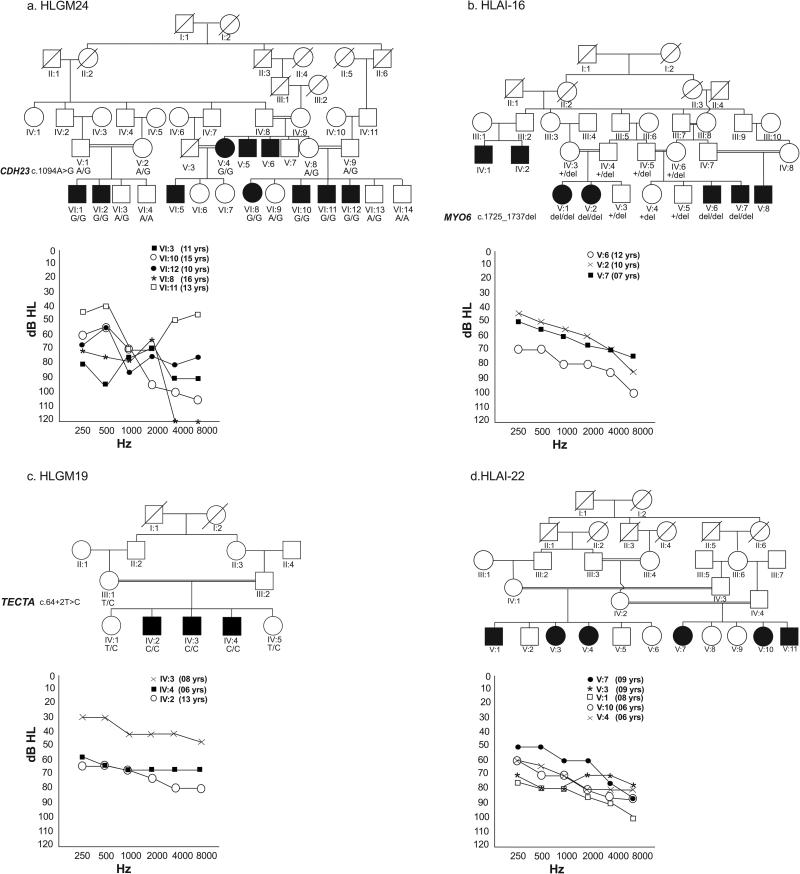
Most of the families we studied had multiple affected individuals who were born to unaffected parents with consanguineous marriages. Exceptions occurred in five families, in which one (HLGM10, HLAI-10, HLAI-19, HLAI-20) or both parents (HLM1) were also affected, with clear evidence for autosomal recessive inheritance of hearing loss in other branches of the respective pedigrees. Hearing loss in 71 families was moderate or severe (Fig. 1a-d and Table 1). Another twenty families had a majority of individuals with moderate or severe hearing loss while one to three affected individuals presented profound deafness (Table 1). The remaining one family (HLM1) had five members affected with profound deafness at the time of enrollment, which was said to have developed in all individuals gradually from a mild hearing loss in childhood. In most cases, previous audiograms were not available and the age of onset of hearing loss could not be reliably ascertained, though the hearing loss was reported to be progressive in nature for 87 individuals in 22% of the participating families (Table 1). None of the affected individuals exhibited an imbalance on tandem gait or Romberg tests.
Fig. 1. Selected Pedigrees and Pure tone audiograms.
Pedigree drawings are provided for four of the participating families with the Pure tone audiograms showing degree of hearing loss across the test frequency range. Hearing thresholds of unaffected age matched controls acquired in similar settings range between 20 dB HL and 30 dB HL. The ages of all affected individuals are noted on the right side of the audiogram. Results are plotted for the better hearing ear in each case. (a) Variable degree of mild-to-moderate, moderate-to-severe or moderate-to-profound hearing loss in family HLGM24 due to CDH23 missense mutation (b) Moderate, moderate-to-severe or moderate-to-profound hearing loss in family HLAI-16 due to MYO6 frameshift mutation (c) Mild-to-moderate, moderate, and moderate-to-severe hearing loss as observed in family HLGM19 which had a frameshift variant in TECTA. Note that one of the affected individuals has a significantly lesser degree of hearing loss (d) Moderate-to-severe or severe-to-profound hearing loss was present in family HLAI-22. The parents reported that the affected individuals had a progressive hearing loss.
Table 1.
Details on the participating families, their phenotypes and the identified mutations
| Gene | Family ID | aAffected individuals | Pure tone average (PTA500-4000 Hz) db HL | Variant | #Comment |
|---|---|---|---|---|---|
|
SLC26A4 NM_000441.1 |
*HLRB10 | 3 | 66, 78, 78 | c.170C>A:p.(Ser57X) | |
| *HLRB2 | 3 | 79, 83, 88 | c.716T>A:p.(Val239Asp) | ||
| *HLRB7 | 5 | 73, 89 | c.716T>A:p.(Val239Asp) | ||
| HLGM05 | 4 | 75, 83, 84, 98 | c.716T>A:p.(Val239Asp) | ||
| HLAI-20 | 6 [1] | 84, 85, 89, 104 (Progressive reported) | c.716T>A:p.(Val239Asp) | ||
| HLGM16 | 3 | 63, 69 | c.965dupA:p.(N322KfsX8) | ||
| HLAI-15 | 4 | 71, 73, 94, 94 | c.1337A>G:p.(Gln446Arg) | ||
| HLAI-21 | 5 | 79, 81, 94, 104 (Progressive reported) | c.1337A>G:p.(Gln446Arg) | ||
| HLAM01B | 4 | 78, 83, 89, 98 | c.1337A>G:p.(Gln446Arg) | ||
| HLAM06 | 5 | 64, 78, 79,79 | c.1337A>G:p.(Gln446Arg) | ||
| HLAM10 | 3 | 59, 70 | c.1337A>G:p.(Gln446Arg) | ||
| HLAM12A | 2 | 70 | c.1667A>G:p.(Tyr556Cys) | ||
| HLAM12B | 1 [7] | 70 | c.1667A>G:p.(Tyr556Cys) | ||
| HLMI01 | 7 | 73, 76, 80, 95 | - | ||
|
GJB2 NM_004004.5 |
*HLAI-02 | 8 | 81, 86, 88, 98, 103, 104 | c.71G>A:p.(Trp24X) | |
| *HLAI-12 | 3 | 59, 71, 90 | c.71G>A:p.(Trp24X) | ||
| *HLGM25 | 5 | 70, 70, 94 | c.71G>A:p.(Trp24X) | ||
| *HLMS16 | 2 | 74, 79 | c.71G>A:p.(Trp24X) | ||
| *HLMS34 | 2 | 75, 78 | c.71G>A:p.(Trp24X) | ||
| *HLRB1 | 4 | 54, 65, 71, 103 | c.231G>A:p.(Trp77X) | ||
| *HLRB8 | 6 | 70, 78, 78 | c.71G>A/c.231G>A p.(Trp24X)/p.(Trp77X) | ||
| *HLAM09 | 3 | 61 | c.71G>A/c.231G>A p.(Trp24X)/p.(Trp77X) | ||
| HLMR2 | 3 | 88, 91 | c.358_360 del: p.(Glu120del) | ||
|
MYO15A NM_016239.3 |
*HLRB3 | 3 | 78, 78, 86 | c.1185dupC:p.(Glu396ArgfsX36) | |
| HLAM05 | 3 [1] | 70, 83, 86 | c.1657delC:p.(Arg553GlyfsX76) | First report | |
| HLAI-06 | 3 | 68, 75 | c.2456C>A:p.(Ser819X) | First report | |
| HLAM07 | 6 | 65, 81, 98 | c.3866+1G>A | ||
| HLAI-10 | 4 | 83, 106 | c.6589C>T:p.(Gln2197X) | ||
| HLAI-25 | 2 | 68, 70 | c.8158G>A:p.(Asp2720Asn) | First report | |
| HLAI-08 | 10 | 68, 73, 79, 103, 104 | - | ||
|
TMC1 NM_138691.2 |
*HLAM02 | 3 | 71, 75, 89 | c.100C>T:p.(Arg34X) | |
| *HLAI-27 | 3 | 61, 80, 81(Progressive reported) | c.596A>T:p.(Asn199Ile) | ||
| *PHLAI-01 | 4 | 78, 86, 93, 93 (Progressive reported) | c.1166G>A:p.Arg389Gln) | ||
| *HLAI-04 | 4 | 65, 95, 101 | c.1404+1G>T | ||
| *HLAI-14 | 3 | 70, 75, 88 (Progressive reported) | c.1788C>A:p.(Ser596Arg) | ||
| *HLAI-17 | 6 | 80, 84, 95 | - | ||
|
TMPRSS3 NM_024022.2 |
HLAI-13 | 5 | 66, 78, 84, 99 (Progressive reported) | c.208delC:p.(His70ThrfsX19) | |
| HLAM08 | 3 | 80, 95 | c.1219T>C:p.(Cys407Arg) | ||
| HLM1 | 5 | 110 (Progressive reported) | c.323–6 G>A | ||
| HLMI02 | 4 | 70, 74, 85 | c.323–6 G>A | ||
|
OTOF NM_194248.2 |
HLAI-09 | 4 | 73, 76 | c.2965_2967del:p.(Phe989del) | First report |
| HLGM14 | 3 | 76, 86, 89 | c.3289-1G>T | First report | |
| HLAI-01 | 4 | 70, 84 | c.4805G>T:p.(Gly1602Val) | First report | |
| HLGM06 | 6 | 73, 86, 88 100 | - | ||
|
MYO7A NM_000260.3 |
HLAI-19 | 7 | 85, 85, 94 (Progressive reported) | c.1183C>T:p.(Arg395Cys) | First report |
| HLGM12 | 3 | 80, 89, 90 | c.6354G>C:p.(Lys2118Asn) | First report | |
| HLAI-07 | 3 | 73 | - | ||
|
CLDN14 NM_144492.2 |
*HLRB5 | 10 | 63, 74, 78, 79, 85, 88, 88, 88, 89, 91 | c.254T>A:p.(Val85Asp) | |
| HLAM04 | 4 | 76, 85, 94, 94 | c.254T>A: p.(Val85Asp) | ||
| HLAM11 | 9 | 73, 83, 83, 84, 88, 91 | c.254T>A: p.(Val85Asp) | ||
|
CDH23 NM_022124.5 |
HLGM24 | 7 | 58, 74, 78, 83, 84 | c.7814A>G:p(Asn2605Ser) | First report |
|
GIPC3 NM_133261.2 |
HLAI-11 | 6 | 64, 83, 86, 96 (Progressive reported) | c.662C>T:p.(Thr221Ile) | |
|
TECTA NM_005422.2 |
HLGM19 | 3 | 38, 69, 73 | c.64+2T>C | First report |
|
TRIOBP NM_001039141.2 |
HLAI-26 | 3 | 74, 86 | c.2968C>T:p.(Arg990X) | First report |
|
ESPN NM_031475.2 |
HLMR3 | 1 [2] | 68 | c.2019dupG:p.(Leu674AfsX72) | First report |
|
MYO6 NM_004999.3 |
HLAI-16 | 4 | 59, 63, 79 (Progressive reported) | c.1729_1741del:p.(Phe577IlefsX28) | First report |
|
HGF NM_000601.4 |
HLAI-18 | 2 [2] | 73 (Progressive reported) | c.482+1986_1988delTGA | |
|
DFNB59 NM_001042702.3 |
*HLGM15 | 6 | 79, 81,89, 90, 90, 109 (Progressive reported) | c.1028G>C:p.(Cys343Ser) | |
|
BSND NM_057176.2 |
HLAI-23 | 5 | 70 (Progressive reported) | c.35 T>C:p.(Ile12Thr) | |
|
TPRN NM_001128228.1 |
*HLRB6 | 3 | 79, 94 (Progressive reported) | c.42_52del11:p(Gly15AlafsX150) | |
|
PTPRQ NM_001145026.1 |
HLRB4 | 3 | 73 | c.189delC:p.(Glu65LysfsX95) | First report |
|
GPSM2/DFNB32 NM_013296.4 |
HLRB11 | 5 | 75, 76, 90, 94 | - | |
| HLAI-24 | 4 | 61, 69 (Progressive reported) | - | ||
|
MET NM_000245.2 |
*HLGM17 | 9 | 74, 83, 89, 89, 89, 108 | c.2521T>G:p.(Phe841Val) | |
|
GRXCR2 NM_001080516.1 |
*HLAI-05 | 3 [1] | 54, 63, 73 (Progressive reported) | c.714dupT: p.(Gly239TrpfsX74) | |
|
USH1C NM_005709.3 |
USH-1 | 3 | 64, 73 | c. 605dupC:p.(Gly203TrpfsX47) | First report |
|
USH1G NM_173477.4 |
*HLRB12 | 3 | 69, 76, 81 | c.163_164+13del | |
| Unknown | HLRB9 | 6 | 73, 73, 75, 80, 100 | ||
| HLRB13 | 3 | 69, 83 | |||
| HLGM02 | 3 | 78, 79, 88 | |||
| HLGM03 | 3 | 78 | |||
| HLGM04 | 3 | 85, 90, 94 | |||
| HLGM07 | 4 | 79, 79, 90, 94 | |||
| HLGM08 | 4 | 46, 49, 64, 84 | |||
| HLGM09 | 3 | 71, 89 | |||
| HLGM10 | 3 | 89, 93 | |||
| HLGM11 | 3 | 84, 85, 85 | |||
| HLGM13 | 3 | 83,88,111 (Progressive reported) | |||
| HLGM18 | 4 | 51, 56, 69 (Progressive reported) | |||
| HLGM20 | 4 | 60, 61, 63, 66 | |||
| HLGM21 | 3 | 58, 66, 90 | |||
| HLGM22 | 11 | 84, 85, 86, 90, 91, 95, 98, 101, 103 | |||
| HLGM23 | 4 | 75, 78, 80, 81 | |||
| HLGM26 | 7 | 45, 84, 99 | |||
| HLAI-03 | 4 | 69, 90, 94 (Progressive reported) | |||
| HLAI-22 | 6 | 61, 74, 75, 76, 83 (Progressive reported) | |||
| HLAM01A | 4 | 78, 81, 95 | |||
| HLAM03 | 5 | 76, 76, 78 | |||
| HLAM12B | 7 | 80, 84, 85, 89 | |||
| IHT01A | 3 | 43, 46, 74 (Progressive reported) | |||
| IHT02 | 3 | 79, 85, 94 | |||
| HLMR1 | 4 | 83, 88, 91, 99 | |||
| HLMR4 | 3 | 76, 89, 96 | |||
The number of affected participants or individuals with pathogenic mutations in deafness genes is denoted by an integer while in case of families exhibiting genetic heterogeneity number of those affected individuals without the causative variants are also shown by a bracketed integer. The PTA500-4000Hz of the latter group is not shown in the corresponding columns.
See Supporting Table S4 for references to the first reports of these mutations.
Data for these families were previously reported by us.
Genetic analyses
We identified 20 previously described and 21 novel pathogenic variants in reported deafness-associated genes for 57 of the 92 participating families (Table 1). For seven families, either a gene had not been identified for the implicated locus or no mutations in the reported exons of the corresponding genes were found although the phenotype was linked to markers of a deafness locus for which there are well-documented pathogenic alleles (Table 1). Of the twenty variants identified in this study that were previously reported to cause deafness (Supplementary Table S4), fifteen were present in the public nucleotide variant databases with overall allele frequencies ranging from 0.0000082 to 0.00024 (Supplementary Table S5). These variants were rare in the South Asian population as well, except for the p.W24X variant in GJB2 with an allele frequency of 0.004 and the p.V239D variant of SLC26A4 with an allele frequency of 0.0017. None of the 21 novel pathogenic mutations were detected in the DNA of ethnically matched controls (200 to 400 chromosomes) but five of these were identified in the ExAC database and had an overall allele frequency of 0.000008237 to 0.00005447 (Supplementary Table S5). The novel variants reported in our study have been deposited in the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/).
Novel variants and involvement with moderate or severe hearing loss
We identified novel variants in thirteen known deafness-associated genes in 21 of the 92 participating families (Table 1). Fifteen of the 21 variants are described in this manuscript (Table 1, Supplementary Information) while the other six variants were reported earlier by us (Table 1, Supplementary Table S4). These variants (Table 1) did not differ substantially from those previously reported in the type of mutation or predicted pathogenicity (Table 2, Supplementary Information). The variants included frameshift, stop-gain, in-frame deletions and missense mutations affecting evolutionary conserved amino acid residues in MYO7A (OMIM 276903), MYO15A (OMIM 602666), OTOF (OMIM 603681), CDH23 (OMIM 605516), TRIOBP (OMIM 609761), TECTA (OMIM 602574), ESPN (OMIM 606351), MYO6 (OMIM 600970), PTPRQ (OMIM 603317) and USH1C (OMIM 605242), (Table 1, Fig. 2, Supplementary Information).
Table 2.
Results of in silico predictions for novel missense variants
| Gene | Variant | MT Score | MT Pred | Prov Score | Prov Pred | SIFT Score | SIFT Pred |
|---|---|---|---|---|---|---|---|
|
MYO15A NM_016239.3 |
c.8158G>A:p.(Asp2720Asn) | 23 | D | −4.42 | D | 0.016 | D |
|
OTOF NM_194248.2 |
c.4805G>T:p.(Gly1602Val) | 109 | D | −8.87 | D | 0 | D |
|
MYO7A NM_000260.3 |
c.1183C>T:p.(Arg395Cys) | 180 | D | −7.75 | D | 0 | D |
| c.6354G>C:p.(Lys2118Asn) | 94 | D | −4.33 | D | 0.001 | D | |
|
CDH23 NM_022124.5 |
c.7814A>G:p.(Asn2605Ser) | 46 | D | −3.53 | D | 0 | D |
MT, Mutation Taster, Prov, Provean, SIFT, Sorting intolerant from tolerant, Pred, prediction, D, disease causing, deleterious and damaging predictions from Mutation Taster, Provean and SIFT, respectively.
Fig. 2. CLUSTALO alignments among diverse vertebrate species.
Alignments show absolute evolutionary conservation of novel amino acid changes reported in this study. The sequences for MYO15A, OTOF, MYO7A and CDH23 correspond to the proteins encoded from transcripts NM_016239.3, NM_194248.2, NM_000260.3 and NM_022124.5, respectively.
The variants of TRIOBP, ESPN and MYO6 described here implicate for the first time these genes in recessively inherited moderate or severe hearing loss. Though specific mutations of MYO15A and CDH23 are known to be involved with moderate or severe hearing loss (3, 13-15), some of the variants we identified do not correspond to the established genotype-phenotype correlations for these genes (Supplementary Information). For example, we found that homozygous mutations that affected both isoforms 1 and 2 of MYO15A (16) were associated with severe hearing loss (Table 1, Supplementary Information). This is in contrast to earlier findings in which it was hypothesized that mutations which affect both isoforms of MYO15A would result in profound deafness (13, 14).
We also found mutations in genes which are established causes of moderate to severe hearing loss (Supplementary Information) such as TECTA and PTPRQ (17, 18). It has been stated that PTPRQ mutations show weak evidence for association to deafness (19). However, different studies (18, 20-23) and the frameshift variant identified in three affected cousins in a participating family in our research (Table 1) together establishPTPRQ as a bona fide deafness gene.
Known variants as cause of moderate or severe hearing loss
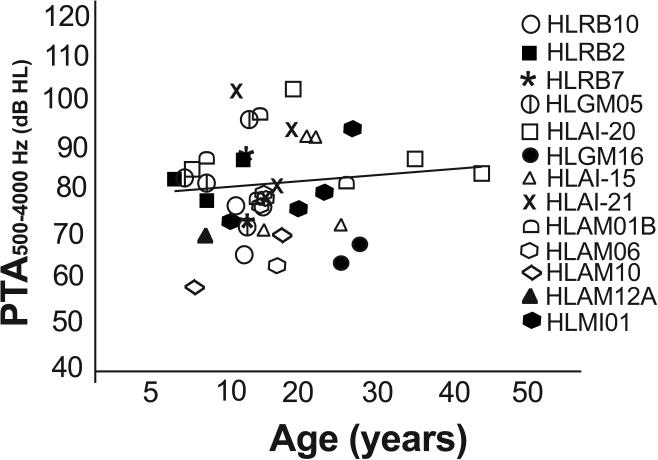
SLC26A4 (OMIM 605646) variants were the most numerous in our cohort, associated with hearing loss in twelve families and in one of the eight affected individuals in family HLAM12B. We found the same five nonsense, missense and frameshift mutations (24) (Table 1) as cause of the moderate or severe hearing loss, which were previously reported to result in profound deafness in other studies (Supplementary Table S5), (Fig. 3). Audiometric thresholds among individuals homozygous for these SLC26A4 variants were not significantly correlated with age (Ages, 6-45, Spearman's r = 0.226; p = 0.11) suggesting that other genetic or environmental factors can modify the phenotype associated with these mutations (Fig. 3). For example, an 18 year old individual in family HLAI-20 had profound deafness (PTA 104 dB HL) in contrast to the other older individuals in his family (ages 45 and 35 years; PTA 84 and 89 dB HL, respectively). Similarly, in families HLAI-15 and HLAM01B in which the affected individuals were homozygous for the p.Q446R variant of SLC26A4, the 25 and 26 years old individuals have better hearing (PTA 73 and 83 dB HL respectively) as compared to the younger affected individuals (ages 10 and 12 years, PTA 104 and 98 dB HL respectively). The type of mutation was also not correlated to the severity of the phenotype since three individuals homozygous for a nonsense (p.Ser57X) or a frameshift (c.965dupA) mutation in SLC26A4 had moderate hearing loss (PTA 66, 63 and 69 dB HL) at the ages of 12, 25 and 28 years, respectively (Table 1, Fig. 3).
Fig.3. PTA of individuals with SLC26A4 variants.
Pure tone averages (PTA) of hearing thresholds at 500, 1000, 2000 and 4000 Hz, for the better hearing ears in affected members with homozygous mutations in SLC26A4 are plotted as a function of age of the participants. The degree of hearing loss is different across and within families with mutations in SLC26A4 without correlation to age, an observation which is supported by the straight regression line in the graph.
In addition to SLC26A4, some known variants of other genes including GJB2 (OMIM 121011), MYO15A and TMPRSS3 (OMIM 605511), were found segregating with moderate or severe hearing loss in this study but were previously reported as a cause of profound deafness (Table 1, Supplementary Information). Comparison of audiograms of affected individuals with identical mutations in multiple families did not suggest that the observed hearing threshold differences were age related.
It has been hypothesized that homozygosity for TMPRSS3 frameshift variants results in profound deafness (25). We identified a homozygous frameshift mutation as a cause of moderate or severe hearing loss in one participating family (Table 1, Supplementary Information). In addition, the age of onset of hearing loss in one family with a cryptic acceptor splice site variant in intron 4 of TMPRSS3 (Table 1) was between 3-4 years and was moderate to severe at the age of 22 years in contrast to the deafness progressing to profound degree in two families with the identical mutation (26) (Supplementary Information).
Molecular heterogeneity
In families HLAI-05, HLAI-18, HLAI-20, HLAI-27, HLAM01A, HLAM12B and HLMR3, some of the affected individuals were not homozygous for the pathogenic mutation found in deaf siblings or close relatives (Supplementary Fig. S1), indicating that variants of other genes or environmental factors contribute to their hearing loss.
Overall contribution of deafness genes to the phenotype
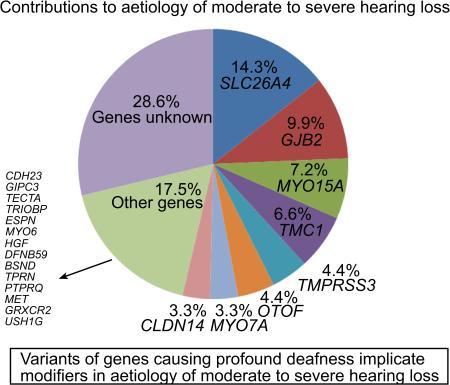
Mutations of eight genes SLC26A4, GJB2, MYO15A, TMC1, TMPRSS3, OTOF, MYO7A and CLDN14 together accounted for hearing loss in 54% of the families (Table 3). Variants of other genes were rare and collectively accounted for the phenotype in 15% of the families (Table 3).
Table 3.
Percentage contribution of genes to moderate or severe hearing loss in Pakistan
| Gene/Locus | Number of familiesa | Percentage | 95 % CI |
|---|---|---|---|
| SLC26A4 | 13 | 14.3 | 7.1-21.5 |
| GJB2 | 9 | 9.9 | 3.8-16 |
| MYO15A | 7 | 7.7 | 2.2-13.2 |
| TMC1 | 6 | 6.6 | 1.5-11.7 |
| TMPRSS3 | 4 | 4.4 | 0.1-8.7 |
| OTOF | 4 | 4.4 | 0.1-8.6 |
| MYO7A | 3 | 3.3 | 0-7 |
| CLDN14 | 3 | 3.3 | 0-7 |
| Other 13 genes or loci | 14 | - | - |
| “New genes”b | 2 | - | - |
| Unknown | 26 | 28.6 | - |
| Total | 91 | - | - |
CI, Confidence Interval.
Percentages are calculated on the basis of 91 pedigrees without the data of an USH1C family and includes data for families for which linkage was detected to a locus with a LOD score of ≥3 and a pathogenic mutation was not identified after sequencing the corresponding gene (MYO7A for HLAI-07, MYO15A for HLAI-08, SLC26A4 for HLMI01, TMC1 for HLAI-17, OTOF for HLGM06 and GPSM2 for HLRB11).
GRXCR2 and MET.
DISCUSSION
Persons with hearing loss of moderate or severe degree may derive satisfactory benefit from hearing aids, while cochlear implantation is a recommended treatment to restore access to sound for individuals with profound deafness. Since profound deafness is clinically distinct from moderate and severe hearing loss, we hypothesized that there would be a spectrum of variants of unique deafness genes as well as distinct mutant alleles of the reported deafness genes as causes of moderate to severe hearing loss. Although we did discover mutations of GRXCR2 (OMIM 615762) (27) and MET (OMIM 164860) (28) as novel causes of progressive or severe hearing loss, unexpectedly we found that exactly the same mutations previously reported to be associated with profound deafness can also cause moderate to severe hearing loss. In fact, the combined contribution of SLC26A4 and GJB2 to moderate or severe hearing loss in this cohort is as high as 24.2% in contrast to that of 13.35% due to the mutant alleles of these same two genes to profound deafness in Pakistan (29).
Phenotypic variability due to mutations in the same gene is usually attributed to a difference in severity of gene function disruption. Splice site variants not resulting in a complete loss of the protein and missense mutations can sometimes be less damaging than nonsense and frameshift mutations. However, of the 41 mutations that we identified in reported deafness genes, seven are nonsense (17%), eleven cause frameshifts in the canonical reading frames (27%), five are splice site mutations (12%), two are in-frame deletions (5%), one is a regulatory variant (2%) and the remaining fifteen are missense mutations (37%). The splice site mutations of MYO15A and USH1G are predicted to result in the loss of the encoded proteins. Phenotypic variation for the same mutant allele suggests the presence of genetic modifiers, environmental factors or both. We did not find variants in the mitochondrial genome that might account for phenotypic variability observed in families in which affected individuals had identical mutations. In addition, we found no rare variants of ATP2B2, a known modifier gene of CDH23 related deafness (15). The possibility remains to be explored that common variants of ATP2B2 are modifiers.
Few studies have reported population wide involvement of specific genes in moderate or severe hearing loss. For example, copy number variations and point mutations of STRC (OMIM 606440) contributed to approximately 6% of mild to moderate hearing loss in the USA and Germany in a cohort of 763 individuals (30, 31). In Qatar, GJB2 variants accounted for 15% of moderate to severe hearing loss in a cohort of 126 individuals (32). Our work has indicated the involvement of 23 genes in the genetics of moderate or severe hearing loss and implicates modifiers in reducing the severity of the phenotype. Future molecular genetic insight and epidemiological studies may be helpful in identifying specific modifier variants in the genetic background of our study subjects which, perhaps in combination with extrinsic factors, partially suppress profound deafness.
Supplementary Material
ACKNOWLEDGMENTS
We are highly grateful to the families for participating in the study. We thank Usman, Arif and Khalid for their untiring efforts throughout the project. We appreciate the Departments of Special Education, Punjab, Dr. Afzaal Alam, Dr. Rehan Saddiq Shaikh, Dr. Zeeshan Ahmed, Children's Hospital & the institute of Child Health, Lahore, and other hospitals and schools for their help in family ascertainment. We thank Davide Risso, Dr. Wade Chien and Dr. Andrew J. Griffith for reviewing the manuscript. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). The research was funded in part by the Intramural Research Program of the National Institute on Deafness and Other Communication Disorders, National Institutes of Health, DC000086 (2015) to R.J.M, DC000039 (2015) to T.B.F and R01TW007608 from the Fogarty International Center and National Institute of Deafness and other Communication Disorders, National Institutes of Health, USA, to S.N.
Footnotes
DISCLOSURE
The authors declare no conflict of interest
SUPPLEMENTARY MATERIAL
Supporting material is available for this article and includes a Supplementary file with Table S1, Table S2, Table S3, Table S4, Table S5, & Fig. S1 and Supplementary Information.
REFERENCES
- 1.Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Smith RJ, Bale JF, Jr., White KR. Sensorineural hearing loss in children. Lancet. 2005;365:879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 3.Astuto LM, Bork JM, Weston MD, et al. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am J Hum Genet. 2002;71:262–275. doi: 10.1086/341558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hildebrand MS, Thorne NP, Bromhead CJ, et al. Variable hearing impairment in a DFNB2 family with a novel MYO7A missense mutation. Clin Genet. 2010;77:563–571. doi: 10.1111/j.1399-0004.2009.01344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naz S. Naz S, editor. Genetics of nonsyndromic recessively inherited moderate to severe and progressive deafness in humans. Hearing Loss. Croatia: Intech. 2012:247–274. [Google Scholar]
- 6.Gendeaf Study Group. Mazzoli M, Van Camp G, et al. Recommendations for the description of genetic and audiological data for families with nonsyndromic hereditary hearing impairment. Audiological Medicine. 2003;1:148–150. [Google Scholar]
- 7.Riazuddin S, Castelein CM, Ahmed ZM, et al. Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat Genet. 2000;26:431–434. doi: 10.1038/82558. [DOI] [PubMed] [Google Scholar]
- 8.Grimberg J, Nawoschik S, Belluscio L, et al. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salman M, Bashir R, Imtiaz A, et al. Mutations of GJB2 encoding connexin 26 contribute to non-syndromic moderate and severe hearing loss in Pakistan. Eur Arch Otorhinolaryngol. 2015;272:2071–2075. doi: 10.1007/s00405-015-3523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman TB, Liang Y, Weber JL, et al. A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nat Genet. 1995;9:86–91. doi: 10.1038/ng0195-86. [DOI] [PubMed] [Google Scholar]
- 11.Imtiaz A, Naz S. A rapid and cost-effective protocol for screening known genes for autosomal recessive deafness. Pak J Zool. 2012;44:641–647. [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz JM, Khan SN, Ahmed ZM, et al. Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet. 2009;85:25–39. doi: 10.1016/j.ajhg.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashir R, Fatima A, Naz S. Prioritized sequencing of the second exon of MYO15A reveals a new mutation segregating in a Pakistani family with moderate to severe hearing loss. Eur J Med Genet. 2012;55:99–102. doi: 10.1016/j.ejmg.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nal N, Ahmed ZM, Erkal E, et al. Mutational spectrum of MYO15A: the large N-terminal extension of myosin XVA is required for hearing. Hum Mutat. 2007;28:1014–1019. doi: 10.1002/humu.20556. [DOI] [PubMed] [Google Scholar]
- 15.Schultz JM, Yang Y, Caride AJ, et al. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N Engl J Med. 2005;352:1557–1564. doi: 10.1056/NEJMoa043899. [DOI] [PubMed] [Google Scholar]
- 16.Liang Y, Wang A, Belyantseva IA, et al. Characterization of the human and mouse unconventional myosin XV genes responsible for hereditary deafness DFNB3 and shaker 2. Genomics. 1999;61:243–258. doi: 10.1006/geno.1999.5976. [DOI] [PubMed] [Google Scholar]
- 17.Naz S, Alasti F, Mowjoodi A, et al. Distinctive audiometric profile associated with DFNB21 alleles of TECTA. J Med Genet. 2003;40:360–363. doi: 10.1136/jmg.40.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schraders M, Oostrik J, Huygen PL, et al. Mutations in PTPRQ are a cause of autosomal-recessive nonsyndromic hearing impairment DFNB84 and associated with vestibular dysfunction. Am J Hum Genet. 2010;86:604–610. doi: 10.1016/j.ajhg.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abou Tayoun AN, Al Turki SH, Oza AM, et al. Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med. 2016;18:545–553. doi: 10.1038/gim.2015.141. [DOI] [PubMed] [Google Scholar]
- 20.Ammar-Khodja F, Bonnet C, Dahmani M, et al. Diversity of the causal genes in hearing impaired Algerian individuals identified by whole exome sequencing. Mol Genet Genomic Med. 2015;3:189–196. doi: 10.1002/mgg3.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, Su Y, Chen YL, et al. Identification of Two Novel Compound Heterozygous PTPRQ Mutations Associated with Autosomal Recessive Hearing Loss in a Chinese Family. PLoS One. 2015;10:e0124757. doi: 10.1371/journal.pone.0124757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakuma N, Moteki H, Azaiez H, et al. Novel PTPRQ mutations identified in three congenital hearing loss patients with various types of hearing loss. Ann Otol Rhinol Laryngol. 2015;124(Suppl 1):184S–1892S. doi: 10.1177/0003489415575041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sang Q, Mei H, Kuermanhan A, et al. Identification of a novel compound heterozygous mutation in PTPRQ in a DFNB84 family with prelingual sensorineural hearing impairment. Mol Genet Genomics. 2015;290:1135–1139. doi: 10.1007/s00438-014-0979-1. [DOI] [PubMed] [Google Scholar]
- 24.Khan MR, Bashir R, Naz S. SLC26A4 mutations in patients with moderate to severe hearing loss. Biochem Genet. 2013;51:514–523. doi: 10.1007/s10528-013-9582-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weegerink NJ, Schraders M, Oostrik J, et al. Genotype-phenotype correlation in DFNB8/10 families with TMPRSS3 mutations. J Assoc Res Otolaryngol. 2011;12:753–766. doi: 10.1007/s10162-011-0282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott HS, Kudoh J, Wattenhofer M, et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- 27.Imtiaz A, Kohrman DC, Naz S. A frameshift mutation in GRXCR2 causes recessively inherited hearing loss. Hum Mutat. 2014;35:618–624. doi: 10.1002/humu.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mujtaba G, Schultz JM, Imtiaz A, et al. A mutation of MET, encoding hepatocyte growth factor receptor, is associated with human DFNB97 hearing loss. J Med Genet. 2015;52:548–552. doi: 10.1136/jmedgenet-2015-103023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riazuddin S, Belyantseva IA, Giese AP, et al. Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat Genet. 2012;44:1265–1271. doi: 10.1038/ng.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francey LJ, Conlin LK, Kadesch HE, et al. Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am J Med Genet A. 2012;158A:298–308. doi: 10.1002/ajmg.a.34391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vona B, Hofrichter MA, Neuner C, et al. DFNB16 is a frequent cause of congenital hearing impairment: implementation of STRC mutation analysis in routine diagnostics. Clin Genet. 2015;87:49–55. doi: 10.1111/cge.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalifa Alkowari M, Girotto G, Abdulhadi K, et al. GJB2 and GJB6 genes and the A1555G mitochondrial mutation are only minor causes of nonsyndromic hearing loss in the Qatari population. Int J Audiol. 2012;51:181–185. doi: 10.3109/14992027.2011.625983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.