Abstract
Microbial species often exist in complex communities where they must avoid predation and compete for favorable niches. The type VI secretion system (T6SS) is a contact-dependent bacterial weapon that allows for direct killing of competitors through the translocation of proteinaceous toxins. Vibrio cholerae is a Gram-negative pathogen that can use its T6SS during antagonistic interactions with neighboring prokaryotic and eukaryotic competitors. The T6SS not only promotes V. cholerae's survival during its aquatic and host life cycles, but also influences its evolution by facilitating horizontal gene transfer. This review details the recent insights regarding the structure and function of the T6SS as well as the diverse signals and regulatory pathways that control its activation in V. cholerae.
Descriptive Key Terms: Type VI Secretion System (T6SS), Vibrio cholerae, structure, function, regulation
A Versatile Weapon for a Deadly Pathogen
The type VI secretion system (T6SS) is a contact-dependent contractile nanomachine used by bacteria to translocate a toxin-coated, membrane-puncturing device into neighboring cells [1–3]. Translocation of T6SS effector proteins is lethal unless target cells produce cognate immunity proteins that bind and sequester the incoming toxic effectors. Since its discovery, T6SS genes have been identified in over a quarter of sequenced Gram-negative bacteria [4,5]. This highly abundant system has been shown to mediate antagonistic interactions against a wide variety of prokaryotic and eukaryotic organisms[1,6–8]. One of the first bacterium shown to possess the T6SS was Vibrio cholerae and thus, much of our understanding of T6SS structure, function, and regulation has been developed from continued study of the T6SS in this pathogenic organism [1].
V. cholerae is a Gram-negative bacterium responsible for the diarrheal disease cholera. There have been seven recorded cholera pandemics in the past 200 years, with the seventh still ongoing [9]. Cholera infections continue to impact 1.4 to 4.3 million people globally and result in 21,000 to 143,000 deaths every year [9]. While over 200 serogroups of V. cholerae have been characterized, pandemics have only been attributed to the O1 serogroup [9]. V. cholerae exists primarily in the aquatic environment, where it can be transmitted to a human host through the ingestion of contaminated food or water. V. cholerae utilizes its T6SS to compete with the diverse prokaryotic and eukaryotic organisms that it encounters in both the aquatic environment and human host. Recent work on the activation of the T6SS suggests that it may contribute to the persistence and evolution of V. cholerae through direct antagonism of competing microbes [1,10,11]. Additionally, the V. cholerae T6SS is known to be active during infection, though its exact role in pathogenesis is not yet understood [12–16]. Indeed, there is still much to be uncovered about when and how the T6SS is deployed and the role it plays in environmental survival and infection. This review provides an update on the current knowledge of the structure, activity, and function of the T6SS, as well as the signals and regulatory networks important for its activation in V. cholerae.
The Structure of the T6SS
The T6SS is a multicomponent toxin delivery apparatus that has structural and functional homology to the T4 bacteriophage tail spike and tube [17,18]. Imaging studies suggest that translocation of T6SS effector proteins occurs through a contraction event that propels a membrane puncturing spike into neighboring cells (Figure 1) [19]. Assembly of the T6SS begins with the assembly of the membrane complex, followed by recruitment of baseplate proteins that anchor the outer sheath and the inner tube to the lipid membranes of the bacteria [20]. The membrane complex is comprised of the three proteins TssJLM (VasDFK in V. cholerae), which span the inner membrane and provide structural support to the system [20]. The cytoplasmic proteins that form the baseplate complex, TssEFGK (HsiF and VasABE in V. cholerae) and VgrG1-3, are then recruited to the membrane complex and are anchored to the inner membrane [20,21]. These baseplate subunits are required for proper formation of the tail complex, which is assembled onto the VgrG1-3 trimer that forms the tip of the T6SS apparatus [20]. Though not shown to be essential components of the baseplate, proteins containing repeating proline-alanine-alanine-arginine (PAAR) motifs cap the VgrG1-3 trimer and act to sharpen the T6SS spike complex [22]. The tail complex is composed of an inner tube formed by hemolysin-coregulated protein (Hcp) hexamers encased within an outer VipA/VipB tube complex [20,22,23]. The outer and inner tubes polymerize into the cytosolic space over the course of ∼30 seconds and can remain fully extended for several minutes until an unknown signal triggers rapid contraction of the outer sheath (∼5ms) and translocation of the inner tube into the extracellular space [19,24]. The signals and mechanisms that govern contraction remain elusive. After contraction and secretion occurs, the ClpV ATPase is recruited to disassemble and recycle the VipA/VipB tube components [25,26].
Figure 1. Contraction of the T6SS Results in the Translocation of Effector Proteins.
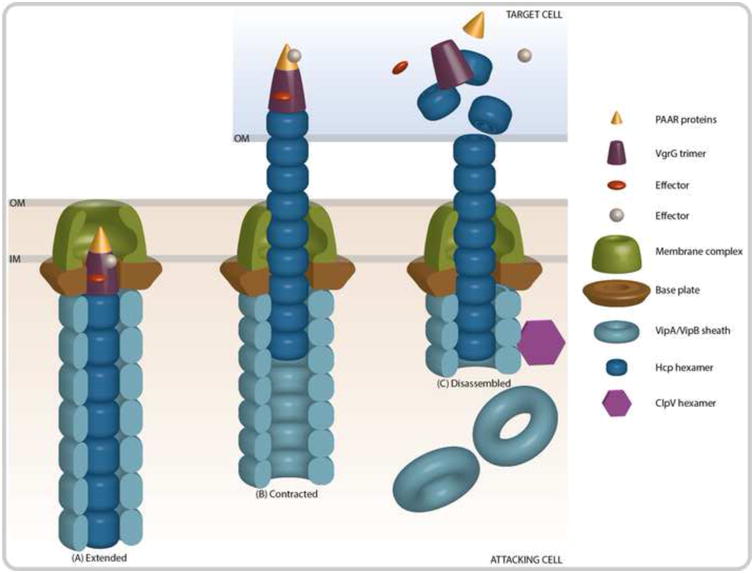
(A) The membrane complex is comprised of both a baseplate structure TssEFGK (HsiF and VasABE in V. cholerae), as well as membrane anchoring components TssJLM (VasDFK in V. cholerae). The outer sheath (VipA/VipB) and inner tube (Hcp) proteins polymerize to form an extended tube that is assembled onto VgrG and PAAR-motif proteins at the cytoplasmic side of the membrane complex. Effector proteins are recruited to the PAAR or VgrG proteins. (B) After assembly, the T6SS complex remains stable until an unknown signal results in the contraction of the outer sheath and the propulsion of the inner tube into a neighboring cell. (C) Upon translocation of the inner tube, tip, and effector complex, the effector proteins can exhibit their toxic activity. Concurrently, the ATPase ClpV disassembles the outer sheath so that the components can be recycled.
Analysis performed in a variety of species suggests that effectors can associate with Hcp, PAAR-motif proteins, and VgrG [27–31]. To date, all characterized V. cholera T6SS effectors are either loaded onto the VgrG tip or are part of the tip proteins themselves. ‘Cargo effectors’, are loaded directly onto the tip of the T6SS. It was recently determined that loading of the cargo effector TseL is facilitated by the chimeric protein Tap-1 (also called Tec-1), which contains a VgrG-binding N-terminal domain and a TseL-binding C-terminal domain [32,33]. Finally, the so-called PAAR-motif proteins, which assemble into a cone-like structure at the tip of the T6SS and form a sharp point that facilitates membrane puncture have also been shown to harbor C- or N-terminal effector domains or to bind and load additional effectors [22,34,35].
Activity and Function of the T6SS
V. cholerae encodes two T6SS effectors that target eukaryotic cells and can utilize these effectors as a means of escape from predatory amoeba in the environment. One of these effectors is the structural component VgrG-1, which contains a C-terminal effector extension that causes cytotoxic actin-crosslinking in the predatory amoeba Dictyostelium discoideum and J774 macrophages (Figure 2B) [1,3]. Additionally, VgrG-1 has been associated with intestinal inflammation and diarrheal symptoms in the infant rabbit, as well as efficient colonization of the infant mouse, suggesting a role for the T6SS during infection [13,16]. The cargo effector VasX targets both eukaryotic and prokaryotic cells by disrupting the cell membrane and has demonstrated activity against D. discoideum and Escherichia coli [36,37]. Efficient colonization of the infant mouse and rabbit intestinal tract is also significantly influenced by the presence of the peptidoglycan degrading effector VgrG-3 [15]. The host intestinal tract is colonized with commensal bacteria that act as a barrier between V. cholerae and its preferred niche within the lumen of the small intestine. It has been speculated that the antibacterial effector VgrG-3 may promote intestinal colonization through killing of the host microbiota (Figure 2C) [15,16].
Figure 2. Proposed Functions of the T6SS in the Life Cycle of V. cholerae.
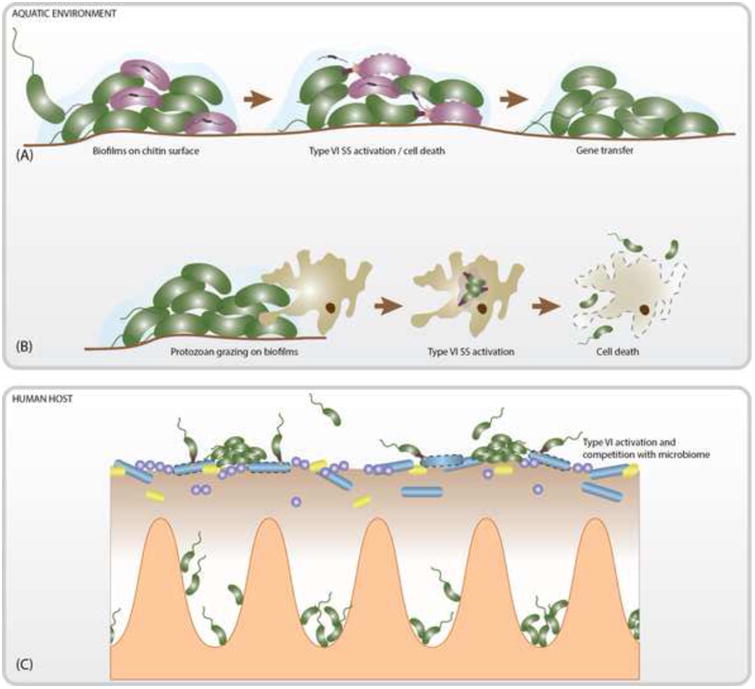
(A) In the aquatic environment V. cholerae often forms biofilms on chitin surfaces. Chitin oligomers serve as a signal for the co-activation of natural competency and the T6SS, which allows for the killing of competing microbes and the acquisition of released DNA (represented as linear fragments inside of intact cells and outside of compromised cells). (B) V. cholerae can also utilize the T6SS to escape from predatory protozoans. (C) During infection the T6SS is believed facilitate V. cholerae colonization and virulence through the killing of host microbes. It may additionally target host macrophages or cause intestinal inflammation that can facilitate infection.
The bactericidal activity of V. cholerae's T6SS plays a role in inter- and intra-species competition and clonal segregation [6,38]. When the V. cholerae T6SS targets another bacterial cell, two distinct outcomes can occur. If the neighboring bacterium encodes the same immunity genes as the predator cell, the delivered effectors are deactivated and the cell is protected [10]. These two bacteria are said to be compatible. For VgrG-3, this effector-immunity interaction appears to occur at 1:2 ratios, as dimerization of the immunity protein is critical to its function [39]. Alternatively, if the target bacterium does not encode the immunity genes, it will be subject to cell lysis. Though the T6SS gene clusters are widely conserved among V. cholerae strains, the effectors and corresponding immunity proteins encoded in these clusters have been reported to be highly variable [10]. This diversity in effector-immunity pairs is thought to contribute to intra-species competition in various environmental niches, though it is currently unknown which factors contribute to one strain outcompeting another. Some hypotheses include the arsenal of effectors, potential differences in growth rate, rate of firing of the T6SS, or how the T6SS is regulated under specific conditions [38,40].
Four effectors enact T6SS-mediated bacterial killing by pandemic V. cholerae. The putative lipase, TseL, and the pore forming colicin VasX, both target the cell membrane. Additionally, two effectors target peptidoglycan; the lysozyme VgrG-3 and the amidase TseH (Table 1) [39,41]. It is thought that the T6SS is only active against Gram-negative bacteria, likely because Gram-positive bacteria are protected from T6SS activity by their thick peptidoglycan [42]. Over the past decade, a plethora of effector classes have been identified in Gram-negative bacteria and it is likely more will be discovered. The study of T6SS effectors has illuminated the versatility and limitations of V. cholerae's T6SS during antagonistic interactions with competing microorganisms. The continued identification and characterization of T6SS effectors will remain an important means of enhancing our understanding of the role the T6SS plays in bacterial and host interactions.
Table 1.
T6SS Effector Proteins from Pandemic V. cholerae strains
| Effector | Target Kingdom | Adaptor | Immunity | Effector activity | Effector Class | Reference |
|---|---|---|---|---|---|---|
| VgrG1 | Eukaryotes | - | - | Actin Crosslinking | C-Terminal Extension | 3 |
| TseL | Prokaryotes | Tap-1 | TsiV1 | Putative Lipase | Cargo Effector | 29,38 |
| VasX | Prokaryotes Eukaryotes | VasW | TsiV3 | Pore Forming | Cargo Effector | 30,38 |
| VgrG3 | Prokaryotes | - | TsiV3 | Peptidoglycan | C-Terminal Extension | 19 |
| TseH | Prokaryotes | - | TsiH | Peptidoglycan | Cargo Effector | 36 |
T6SS Genetic Organization and Regulation
The V. cholerae T6SS genes are encoded in one large operon (VCA0105-VCA0124), known as the large or major cluster, and at least three smaller operons known as auxiliary clusters 1, 2, and 3 (VCA0017-VCA0021, VC1415-VC1419, and VCA0284-VCA0286) (Figure 3) [1,43,44]. The large cluster encodes the majority of the T6SS structural components, with the exception of the essential secreted inner tube component Hcp, two of the VgrG proteins, and one PAAR-motif protein (VCA0284) [1,44]. Auxiliary clusters 1 and 2 encode for the nearly identical hcp1 (VC1417) and hcp2 (VCA0017) genes, either of which is sufficient to produce Hcp and form the inner tube [44]. Auxiliary clusters 1 and 2 also encode for the VgrG-1 (VC1416) and VgrG-2 (VCA0018) tip components respectively, as well as unique effector sets and their cognate immunity proteins [41,44]. The recently identified third auxiliary cluster is predicted to encode for one PAAR motif protein (VCA0284) and a unique effector-immunity pair (VCA0285-86) [22,43]. In addition to the promoters found upstream of each operon, internal promoter activity has been identified within the large cluster and auxiliary clusters 1 and 2 that lie just upstream of the immunity genes [41,45]. The transcriptional regulators that drive expression from these internal promoters are not well understood; however it has been hypothesized that they may allow immunity proteins to be constitutively expressed, thus ensuring protection from neighboring kin cells even in the absence of an active T6SS [41].
Figure 3. Genetic Loci of the V. cholerae T6SS.
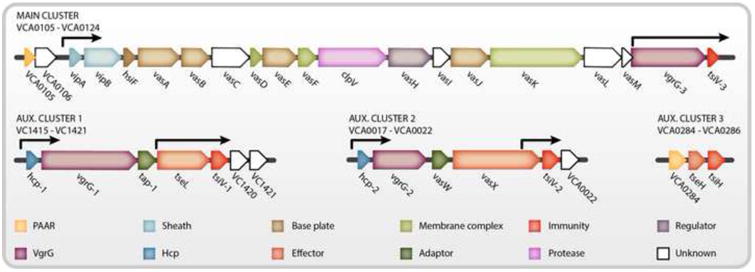
The genes of the T6SS are organized into one main cluster and at least 3 auxiliary clusters. Genes are color-coded based on predicted function and labeled with V. cholerae gene annotations. Black arrows denote known transcriptional start sites.
Significant advancements have been made in discerning the complex regulatory networks that govern the transcription and activation of the T6SS in V. cholerae. The transcriptional regulator VasH, which is encoded in the large T6SS cluster (VCA0117), was among the first T6SS regulators to be identified [1,46]. VasH is a bacterial enhancer-binding protein that complexes with the alternative sigma factor RpoN and coordinates transcription from the upstream promoters of auxiliary clusters 1 and 2 (Figure 4) [1,47-49]. The genetic organization of the T6SS clusters provides a mechanism by which activation of the large cluster prompts transcription of auxiliary clusters 1 and 2 and the production of an assembled T6SS. Since the discovery of VasH, a number of additional regulators have been identified and diverse signalling pathways that feed into T6SS activation in response to various environmental cues have been characterized. The T6SS is now known to be controlled via the quorum sensing, catabolite repression, and nucleoside scavenging pathways [13,49,50]. These signalling cascades are additionally integrated into the chitin-induced competency cascade, which coordinates co-expression of the T6SS and competency genes [11,51]. Finally, the T6SS is influenced by a number of additional environmental and host signals including, but not limited to, temperature, osmolarity, the secondary messenger cyclic dimeric (3′→5′) GMP (c-di-GMP), mucin, and bile [45,52-54]. Together, these regulatory mechanisms provide insights into how V. cholerae coordinates its T6SS activity in both the aquatic and host environments.
Figure 4. The Regulatory Network of the T6SS in V. cholerae.
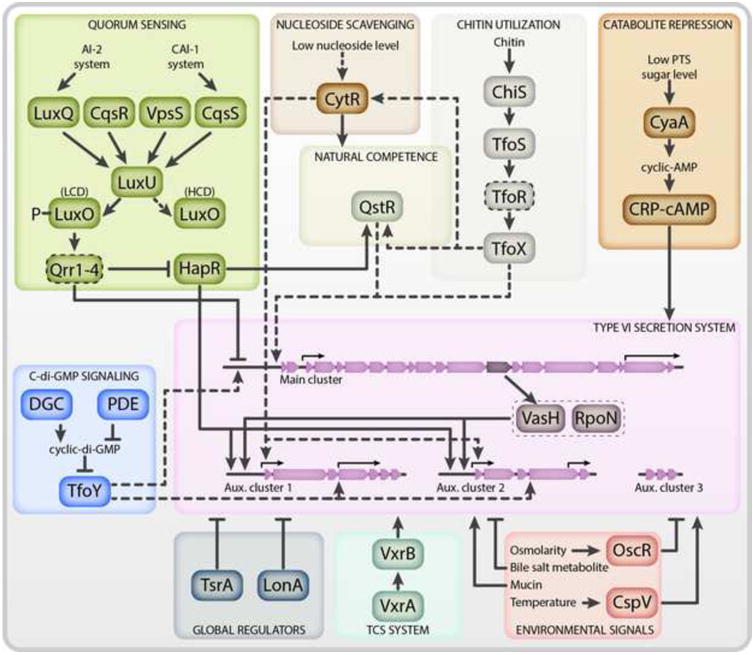
Activation is indicated by arrow-headed lines while inhibition is indicated by bar-headed lines. Lines that do not enter the Type VI Secretion System bubble represent regulation through an unknown mechanism. Regulators that directly bind to promoters are designated with solid lines while those that activate through as of yet unknown mechanisms have dashed lines. At low cell density, the quorum sensing small RNAs (sRNAs, denoted by a dashed outline) Qrr1-4 inhibit translation of hapR and the main T6SS cluster mRNAs. At high cell density, this inhibition is relieved. The chitin utilization cascade induces expression of TfoX, which acts in concert with HapR to activate QstR. TfoX and QstR facilitate transcription of the main cluster while HapR activates transcription of auxiliary clusters 1 and 2. When preferred carbohydrates are absent, CRP-cAMP accumulates and activates T6SS gene expression. At low nucleoside levels, CytR activates T6SS gene expression. Cyclic-di-GMP (c-di-GMP) is produced by diguanylate cyclases (DGCs) and degraded by phosphodiesterases (PDEs). When c-di-GMP levels are low, TfoY is translated and activates expression of the main cluster and the effector and immunity pairs located within auxiliary clusters 1 and 2. Activation of auxiliary clusters 1 and 2 requires the VasH-RpoN complex. The VxrAB two component system (TCS) senses an unknown signal and activates T6SS gene expression. OscR represses the T6SS under conditions of low osmolarity. CspV is required for activation of the T6SS at temperatures between 25°C and 37°C. Mucins activate the T6SS while bile salt metabolites inhibit T6SS tube formation. The H-NS-like protein TsrA and the LonA protease inhibit the T6SS. These regulatory cascades are described in detail within the main text.
Quorum Sensing
Quorum sensing (QS) is a form of bacterial communication that occurs through the production, secretion, and sensing of small molecules known as autoinducers [55,56]. This communication allows alterations in gene expression to occur across a population of bacteria in response to changing cell density, which is signaled by increasing levels of autoinducers. QS is known to regulate a variety of behaviors important for V. cholerae's aquatic and intestinal life cycles including biofilm formation, motility, natural competency, and virulence factor production [51,57–59]. It is now recognized that QS also coordinates T6SS activation by repressing the T6SS at low cell density (LCD) and upregulating the T6SS at high cell density (HCD) (Figure 4) [60]. In V. cholerae, QS-mediated gene regulation occurs through a phosphorelay cascade modulated by four sensor histidine kinases, CqsS, LuxPQ, CqsR, and VpsS. CqsS and LuxPQ sense the levels of cholerae autoinducer 1 (CAI-1) and autoinducer 2 (AI-2), respectively, while the ligands for CqsR and VpsS have not been identified [61–63]. At LCD, these four histidine kinases phosphorylate the phosphotransfer protein LuxU, which in turn phosphorylates LuxO [61,62]. Phosphorylated LuxO activates the expression of four small RNAs known as Qrr1-4, which bind to and destabilize the mRNA transcripts of the large cluster of the T6SS and HapR. At HCD, however, LuxO is unphosphorylated, and transcription of qrr1-4 is inactive, thus permitting the translation of the large T6SS cluster and HapR [60]. HapR positively regulates transcription of auxiliary clusters 1 and 2, likely via direct binding to HapR binding motifs [64].
Chitin-Induced Competency Pathway
V. cholerae spends a majority of its life cycle in the aquatic environment, where it is frequently found associated with chitinous surfaces, such as the exoskeletons of zooplankton. Mounting evidence suggests that growth on zooplankton facilitates V. cholerae's persistence, transmission, and virulence [65,66]. Thus, mechanisms and strategies that promote V. cholerae's successful colonization of chitinous surfaces are of great interest. V. cholerae has evolved several signaling cascades that are influenced by the presence of chitin, including the chitin utilization program, natural competency, and activation of the T6SS [11,67-69].
Chitin is an insoluble polymer consisting of repeating β-1,4-linked N-acetylglucosamine (GlcNAc) residues. Upon growth on chitin, the histidine kinase (HK) ChiS, senses GlcNac polymers and initiates a regulatory cascade that results in the expression of genes important for the transport, degradation, and utilization of chitin as a carbon source, as well as those required for natural competency, which allows bacteria to import extracellular DNA (eDNA) from the enviornment [68,69]. In V. cholerae, the signaling cascade initiated by ChiS couples natural competency to the induction of the T6SS via the transcriptional regulator TfoX (Figure 4) [70-73]. Briefly, ChiS is locked in an inactive state through interaction with the periplasmic chitin binding protein, CBP [74]. Upon growth on chitin, the extracellular chitinases ChiA-1 and ChiA-2 degrade chitin into short GlcNAc oligomers, which are then able to enter the cell through chitoporins [69]. GlcNAc oligomers bind to CBP resulting in disassociation of CBP from ChiS. In the absence of CBP, ChiS activates the transmembrane protein TfoS, which in turn promotes transcription of the sRNA tfoR [71-73]. The sRNA tfoR is necessary for the translation of TfoX and the activation of the TfoX regulon [71]. The ability of TfoX to activate competency and T6SS genes is dependent upon the presence of the QS and TfoX-dependent regulator, QstR, which is required for the production of T6SS structural components [11,51]. Thus, QS signals also appear to feed into the chitin-induced competency pathway to initiate activation of the T6SS; however, the molecular mechanisms underlying this activation have not been characterized. Additionally, the nucleoside scavenging regulator, CytR, is essential for natural transformation and contributes to T6SS gene activation; however, the means by which it regulates the T6SS remain unclear (Figure 4) [50,75]. During chitin-induced co-activation of the T6SS and natural competency, V. cholerae uses its T6SS to kill incompatible bacterial cells, freeing eDNA that is then taken up by its competency machinery [11]. Thus, the T6SS may facilitate the genetic diversity and evolution of V. cholerae strains in the environment via the acquisition of new genetic information through horizontal gene transfer, in addition to promoting V. cholerae's colonization and persistence on chitinous surfaces through targeted killing of competing microbes (Figure 2A).
Carbon Catabolite Repression
The T6SS is positively regulated by the small molecule cyclic adenosine monophosphate (cAMP) and global regulator cAMP receptor protein CRP (Figure 4) [49]. When preferred carbon sources are exhausted or unavailable, transcription of the adenylate cyclase gene, cyaA, is upregulated, which leads to increased levels of cAMP [76,77]. Free cAMP binds to CRP and the resulting complex acts as a transcriptional regulator, controlling the activation and repression of a number of essential V. cholerae pathways, including carbon uptake, QS, chitin utilization, chitin induced natural competency, and the T6SS [49,78–80]. Deletion of either cyaA or crp prevents production of Hcp, indicating that the cAMP-CRP complex is essential for T6SS production [49]. The mechanism by which cAMP-CRP regulates the T6SS is unclear; however, it is possible that cAMP-CRP influences T6SS production through its regulation of QS and chitin-induced competency [78,79]. Additional studies are needed to determine whether cAMP-CRP controls T6SS production through these pathways or through alternative regulatory mechanisms.
Influence of c-di-GMP
C-di-GMP is a secondary messenger that is capable of binding to a wide variety of targets and influencing transcriptional and enzymatic activities [81]. C-di-GMP was recently found to regulate the T6SS in V. cholerae through a protein homologous to TfoX, known as TfoY [45]. The 5'UTR of tfoY contains a c-di-GMP riboswitch that prevents translation of TfoY in the presence of high levels of c-di-GMP [45,82]. When c-di-GMP levels are decreased, T6SS-mediated killing is increased in a TfoY-dependent manner [45]. TfoY appears to upregulate both the large cluster and the effector immunity pairs in auxiliary clusters 1 and 2, though not auxiliary cluster 3 (Figure 4) [45]. Regulation of T6SS by TfoY is independent of TfoX's T6SS regulation through the natural competency regulon. Though further study is needed to fully elucidate the molecular mechanisms by which TfoY regulates the T6SS, it is speculated that TfoY may be involved in a danger sensing and defensive escape reaction based on the observation that effectors targeted to eukaryotic cells appear to be more highly expressed in a TfoY overexpression background [45,83].
Post-Translational Regulation
Little is known about the post-translational regulation of T6SS protein production, assembly, and activation in V. cholerae. However, the Lon protease was recently identified as a negative regulator of the T6SS (Figure 4) [84]. While a major role of the Lon protease is to degrade misfolded or otherwise aberrant proteins, it also has the ability to degrade specific protein targets [85]. This targeted degradation by Lon provides post-translational regulation of a wide array of processes in a variety of bacteria, including E. coli, P. aeruginosa, and B. subtilis, though V. cholerae is the only bacteria in which the Lon protease has been shown to regulate the T6SS. In the absence of Lon, transcription of hcp1 and 2 are upregulated ∼5-fold while ∼2-fold increases are observed in transcripts from the main cluster. Additionally, production and secretion of Hcp is increased, and killing of E. coli prey is increased by ∼2-fold in a standard killing assay [84]. Though this study demonstrated that Lon is a post-translational regulator of the T6SS, the mechanism by which Lon exerts this regulatory effect has not yet been determined. Given the global effect a Lon deletion has on T6SS transcription, translation, and activity, it is likely that Lon is responsible for directly degrading a key regulator or regulators of the T6SS, such as those discussed in this review [84].
Regulation of the T6SS in the Host
Infection by V. cholerae occurs through the ingestion of contaminated food or water. The invading bacteria navigate through the digestive system to the epithelial surface of the small intestine where V. cholerae produces virulence factors that promote colonization and disease onset. Recent studies suggests that the T6SS is active in the host and contributes to virulence and intestinal colonization [13,14,16]. The regulatory mechanisms that govern T6SS expression during pathogenesis remain poorly defined. It is likely that many of the regulators and signaling pathways mentioned earlier contribute to T6SS induction during pathogenesis; however, most in vivo assays have focused on the function of the T6SS within the host rather than its regulation. QS is known to play an essential role in coordinating V. cholerae's virulence cascade and current data suggests that the T6SS remains under the control of HapR and LuxO during infection [13,49,58,60]. Additionally, the H-NS-like protein TsrA was shown to repress virulence factors, including the T6SS; however, its mechanism of action remains unknown [13]. A recently identified two component system (TCS), VxrAB, was demonstrated to positively regulate intestinal colonization in a T6SS-dependent manner; however, the mechanism by which it regulates in vivo T6SS activity and the signal that feeds into this TCS is unclear [16].
The V. cholerae T6SS, however, is known to respond to host signals, such as mucin, bile, and indole (Figure 4). Mucins, the main component of the mucus layer in the intestine, are known to increase T6SS-mediated killing of bacterial prey, while the bile salt deoxycholic acid represses T6SS killing via inhibition of T6SS tube formation [54]. The production of deoxycholic acid is facilitated by the commensal bacterium Bifidobacterium bifidium, which is capable of metabolizing certain bile acids to deoxycholic acid [54]. Additionally, in vitro exposure to indole, a signaling molecule found in large concentrations in the mammalian intestinal tract, was shown to activate T6SS gene expression and may feed into in vivo control of the T6SS [86]. Given that efficient colonization of the intestinal tract is known to be significantly influenced by the peptidoglycan degrading effector VgrG-3, the V. cholerae's T6SS may target commensal bacteria to facilitate intestinal colonization (Figure 2C) [15,16]. Signals produced by the microbiota may serve to inhibit or activate V. cholerae's T6SS and may influence host susceptibility to disease. Indeed, host microbial communities are known to increase colonization resistance against many pathogens by preventing access to desirable niches, limiting nutrient availability, and producing inhibitory compounds [87– 90]. Thus, V. cholerae may overcome these obstacles through T6SS killing of the host's microbiota; however, additional work is required to characterize the influence of the microbial community on V. cholerae pathogenesis and T6SS activity. The important role the T6SS appears to play in pathogenesis prompts additional studies of T6SS regulation within the host, which may aid in our understanding of this system.
Influence of Environmental Signals on T6SS Regulation
The T6SS of V. cholerae is also influenced by a number of environmental signals encountered in the aquatic environment or the host, though many of the key regulators that mediate these signals have not yet been identified. Osmolarity is known to be an important signal that influences the T6SS through the osmoregulator OscR, which represses T6SS gene expression at low osmolarities (85mM NaCl). The repressive effect of OscR is relieved when V. cholerae is placed in high osmotic conditions (340mM NaCl) (Figure 4) [53,91]. Temperature additionally modulates the activity and expression of the T6SS, repressing T6SS gene expression at low temperatures (15°C) and activating T6SS gene expression at high temperatures (25°C – 37°C) (Figure 4) [52,53]. The activation of the T6SS in response to elevated temperature appears to be regulated in part by the cold shock protein CspV. Deletion of cspV, significantly decreases the transcription of hcp, resulting in less killing of bacterial prey at 25°C and 37°C [52]. While the mechanisms through which OscR and CspV regulate the T6SS remain unclear, some studies have shown that T6SS gene induction and activity is optimal at osmolarities of 340mM NaCl and temperatures of 25°C [52,53]. These conditions are similar to those found in the estuarine habitats where V. cholerae resides [92–95]. Thus, the regulatory mechanisms described here may have adapted to facilitate competition and survival within this niche.
Differential Regulation of the T6SS in V. cholerae
V. cholerae has over 200 serogroups based on LPS antigen classification [9]. While all sequenced V. cholerae isolates encode for T6SS genes, not all strains regulate their T6SS identically [1,13,96]. Many environmental isolates have a constitutively active T6SS, while most studied pandemic strains exercise more controlled regulation of the T6SS [1,13,49,96]. These differences are most apparent within the QS pathway [13,49,60]. In constitutively active strains, HapR has relatively little influence on T6SS gene expression and killing, while the influence of QS on strains that heavily regulate their T6SS differs, with some strains requiring the removal of LuxO for T6SS gene expression under standard laboratory conditions [13,60]. This diversity of regulatory strategies may be indicative of evolutionary adaptations that are advantageous to the specific isolate's niche.
Concluding Remarks
Over the past decade, significant advancements have been made on the structural and mechanistic properties of the T6SS, as well as the signals and regulatory pathways that govern its activation in V. cholerae. It is now understood that V. cholerae can use its T6SS on chitinous surfaces and co-regulates T6SS activation with natural competency pathways [11]. This provides a mechanism that likely increases the survival and persistence of V. cholerae in the aquatic environment through the direct killing of microbial competitors and may contribute to the evolution of V. cholerae through horizontal acquisition of new genetic information from killed cells (Figure 2A). In addition, V. cholerae may utilize its T6SS as a colonization and virulence determinant by targeting the host's epithelial and immune cells, as well as the commensal microbial population (Figure 2C). While numerous insights have been made into the structure, function, and regulation of this bacterial weapon, substantial gaps remain (see Outstanding Questions). For example, the extent to which the T6SS facilitates inter- and intra-species competition and the biologically relevant targets of the T6SS remain unclear. Furthermore, the regulatory mechanisms and pathways important for T6SS activation in the host are poorly characterized. Additional research is required to identify the activating signals and mechanisms through which most regulators function. Given the wealth of knowledge revealed about the T6SS in the past decade, it is exciting to consider what future study of the T6SS will yield as we continue to explore the complexities of the T6SS in V. cholerae and other important human pathogens.
Outstanding Questions.
To what extent does the T6SS facilitates inter- and intra-species competition in the aquatic and host environments and which targets are biologically relevant?
How does the T6SS contribute to fitness within the host? During infection, does V. cholerae use its T6SS for the killing of host commensal bacteria, escape from immune cells, attack of the epithelium or a combination of the three?
How does V. cholerae coordinate loading of multiple effectors? Can different effectors be loaded onto the same tip? Or are mechanisms in place to ensure only a single effector type is present on one T6SS complex?
What is the exact location of internal promoters responsible for constitutive production of the immunity genes? What are the regulatory mechanisms that govern activation from these promoter sites?
What are the regulatory networks and signals important for modulating T6SS activity during pathogenesis? Which of the known regulators take part in T6SS activation in the host? Are there host specific regulatory networks in place?
How do OscR, CspV, TfoY, TsrA, VxrB, Lon, and cAMP-CRP govern T6SS activation and functionality? Do any of these signaling pathways act in concert or with quorum sensing and natural competency pathways? If so, what regulators link these networks? Do the transcriptional regulators bind to and directly facilitate transcription from T6SS gene clusters? Or do they activate the T6SS indirectly through an as of yet uncharacterized pathway?
Trends Box.
The T6SS is a contact-dependent toxin delivery machine found in Gram-negative bacteria. Translocation of toxins occurs through a contraction event that propels a membrane puncturing spike, decorated with effector proteins, into neighboring cells.
V. cholerae can use its T6SS against a variety of organisms found in its aquatic and host environments including neighboring Gram-negative bacteria, phagocytic amoeba and immune cells.
In many V. cholerae isolates, T6SS induction is under the control of diverse regulatory mechanisms such as quorum sensing, carbon catabolite repression, and chitin induced competency pathways in addition to a variety of environmental signals found in the aquatic and host environments.
The T6SS enhances V. cholerae's survival and evolution in the aquatic environment and acts as a virulence factor to promote intestinal colonization in the host.
Acknowledgments
Work in the laboratory of FHY is supported by the US National Institute of Health (NIH) grants AI114261 and AI102584. Work in the laboratory of S.P. is supported by the Canadian Institutes of Health Research Operating Grants MOP-84473 and MOP-137106 and by Alberta Innovates Health Solutions. B.K. is a recipient of an NSERC CGS-D.
Work in the laboratory of S.P. is supported by the Canadian Institutes of Health Research Operating Grants MOP-84473 and MOP-137106 and by Alberta Innovates Health Solutions. B.K. is a recipient of an NSERC CGS-D.
The authors thank Ates Gurcan for figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pukatzki S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–33. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–30. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pukatzki S, et al. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–13. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingle LE, et al. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Das S, Chaudhuri K. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 2003;3:287–300. [PubMed] [Google Scholar]
- 6.MacIntyre DL, et al. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107:19520–4. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heckel BC, et al. Agrobacterium tumefaciens exoR controls acid response genes and impacts exopolysaccharide synthesis, horizontal gene transfer, and virulence gene expression. J Bacteriol. 2014;196:3221–33. doi: 10.1128/JB.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charles RC, Ryan ET. Cholera in the 21st century. Curr Opin Infect Dis. 2011;24:472–7. doi: 10.1097/QCO.0b013e32834a88af. [DOI] [PubMed] [Google Scholar]
- 10.Unterweger D, et al. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun. 2014;5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borgeaud S, et al. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015;347:63–7. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 12.Lombardo MJ, et al. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc Natl Acad Sci U S A. 2007;104:18229–34. doi: 10.1073/pnas.0705636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng J, et al. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A. 2010;107:21128–33. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A. 2010;107:4365–70. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, et al. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 2013;14:652–63. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng AT, et al. Vibrio cholerae Response Regulator VxrB Controls Colonization and Regulates the Type VI Secretion System. PLoS Pathog. 2015;11:e1004933. doi: 10.1371/journal.ppat.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pell LG, et al. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci U S A. 2009;106:4160–5. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–9. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunet YR, et al. The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization. PLoS Genet. 2015;11:1–21. doi: 10.1371/journal.pgen.1005545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoued A, et al. TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J Biol Chem. 2013;288:27031–41. doi: 10.1074/jbc.M113.499772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shneider MM, et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–3. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spínola-Amilibia M, et al. The structure of VgrG1 from Pseudomonas aeruginosa, the needle tip of the bacterial type VI secretion system. Acta Crystallogr Sect D, Struct Biol. 2016;72:22–33. doi: 10.1107/S2059798315021142. [DOI] [PubMed] [Google Scholar]
- 24.Basler M, et al. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–6. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bönemann G, et al. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–25. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietrosiuk A, et al. Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J Biol Chem. 2011;286:30010–21. doi: 10.1074/jbc.M111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho BT, et al. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverman JM, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51:584–93. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jobichen C, et al. Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS One. 2010;5:e12910. doi: 10.1371/journal.pone.0012910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hachani A, et al. The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J Biol Chem. 2014;289:17872–84. doi: 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unterweger D, et al. Adaptor Proteins of Type VI Secretion System Effectors. Trends Microbiol. 2016;0:51–62. doi: 10.1016/j.tim.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Unterweger D, et al. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 2015;34:2198–210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang X, et al. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci U S A. 2015;112:9106–11. doi: 10.1073/pnas.1505317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitney JC, et al. An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell. 2015;163:607–19. doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cianfanelli FR, et al. VgrG and PAAR Proteins Define Distinct Versions of a Functional Type VI Secretion System. PLoS Pathog. 2016;12:1–27. doi: 10.1371/journal.ppat.1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong TG, et al. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A. 2013;110:2623–8. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyata ST, et al. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun. 2011;79:2941–9. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong M, et al. Microbial herd protection mediated by antagonistic interaction in polymicrobial communities. Appl Environ Microbiol. 2016 doi: 10.1128/AEM.02210-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks TM, et al. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem. 2013;288:7618–25. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borenstein DB, et al. Established Microbial Colonies Can Survive Type VI Secretion Assault. PLoS Comput Biol. 2015;11:e1004520. doi: 10.1371/journal.pcbi.1004520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyata ST, et al. Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog. 2013;9:e1003752. doi: 10.1371/journal.ppat.1003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou S, et al. Structure of a peptidoglycan amidase effector targeted to Gram-negative bacteria by the type VI secretion system. Cell Rep. 2012;1:656–64. doi: 10.1016/j.celrep.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altindis E, et al. Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair. MBio. 2015;6:e00075. doi: 10.1128/mBio.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng J, et al. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metzger LC, et al. Independent Regulation of Type VI Secretion in Vibrio cholerae by TfoX and TfoY. Cell Rep. 2016;15:951–8. doi: 10.1016/j.celrep.2016.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernard CS, et al. Regulation of type VI secretion gene clusters by g54 and cognate enhancer binding proteins. J Bacteriol. 2011;193:2158–67. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitaoka M, et al. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol. 2011;193:6471–82. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012;40:7766–75. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa T, et al. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One. 2009;4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watve SS, et al. CytR Is a Global Positive Regulator of Competence, Type VI Secretion, and Chitinases in Vibrio cholerae. PLoS One. 2015;10:e0138834. doi: 10.1371/journal.pone.0138834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lo Scrudato M, Blokesch M. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 2013;41:3644–58. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Townsley L, et al. Response of Vibrio cholerae to Low-Temperature Shifts: CspV Regulation of Type VI Secretion, Biofilm Formation, and Association with Zooplankton. Appl Environ Microbiol. 2016;82:4441–52. doi: 10.1128/AEM.00807-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa T, et al. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun. 2012;80:575–84. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bachmann V, et al. Bile Salts Modulate the Mucin-Activated Type VI Secretion System of Pandemic Vibrio cholerae. PLoS Negl Trop Dis. 2015;9:e0004031. doi: 10.1371/journal.pntd.0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:1–25. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–114. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 58.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 2002;99:3129–34. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yildiz FH, et al. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 60.Shao Y, Bassler BL. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol Microbiol. 2014;92:921–30. doi: 10.1111/mmi.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung SA, et al. Quadruple Quorum-Sensing Inputs Control Vibrio cholerae Virulence and Maintain System Robustness. PLoS Pathog. 2015;11:1–19. doi: 10.1371/journal.ppat.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller MB, et al. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 63.Shikuma NJ, et al. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J Bacteriol. 2009;191:5147–5158. doi: 10.1128/JB.00401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsou AM, et al. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res. 2009;37:2747–56. doi: 10.1093/nar/gkp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huq A, et al. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl Environ Microbiol. 2005;71:4645–54. doi: 10.1128/AEM.71.8.4645-4654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vezzulli L, et al. Environmental reservoirs of Vibrio cholerae and their role in cholera. Environ Microbiol Rep. 2010;2:27–33. doi: 10.1111/j.1758-2229.2009.00128.x. [DOI] [PubMed] [Google Scholar]
- 67.Lo Scrudato M, et al. Regulatory elements involved in the expression of competence genes in naturally transformable Vibrio cholerae. BMC Microbiol. 2014;14:2. doi: 10.1186/s12866-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meibom KL. Chitin Induces Natural Competence in Vibrio cholerae. Science (80-) 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 69.Meibom KL, et al. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A. 2004;101:2524–9. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamamoto S, et al. Chitin disaccharide (GlcNAc)2 induces natural competence in Vibrio cholerae through transcriptional and translational activation of a positive regulatory gene tfoXVC. Gene. 2010;457:42–49. doi: 10.1016/j.gene.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto S, et al. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J Bacteriol. 2011;193:1953–1965. doi: 10.1128/JB.01340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamamoto S, et al. Regulation of natural competence by the orphan two-component system sensor kinase ChiS involves a non-canonical transmembrane regulator in Vibrio cholerae. Mol Microbiol. 2014;91:326–347. doi: 10.1111/mmi.12462. [DOI] [PubMed] [Google Scholar]
- 73.Dalia AB, et al. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. MBio. 2014;5:e01028–13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li X, Roseman S. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc Natl Acad Sci U S A. 2004;101:627–631. doi: 10.1073/pnas.0307645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antonova ES, et al. Natural competence in Vibrio cholerae is controlled by a nucleoside scavenging response that requires CytR-dependent anti-activation. Mol Microbiol. 2012;86:1215–1231. doi: 10.1111/mmi.12054. [DOI] [PubMed] [Google Scholar]
- 76.Houot L, et al. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J Bacteriol. 2010;192:3055–3067. doi: 10.1128/JB.00213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deutscher J, et al. How Phosphotransferase System-Related Protein Phosphorylation Regulates Carbohydrate Metabolism in Bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang W, et al. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology. 2007;153:2964–75. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 79.Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol. 2012;14:1898–1912. doi: 10.1111/j.1462-2920.2011.02689.x. [DOI] [PubMed] [Google Scholar]
- 80.Fong JCN, Yildiz FH. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J Bacteriol. 2008;190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sondermann H, et al. You've come a long way: C-di-GMP signaling. Curr Opin Microbiol. 2012;15:140–146. doi: 10.1016/j.mib.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.LeRoux M, et al. Bacterial danger sensing. J Mol Biol. 2015;427:3744–3753. doi: 10.1016/j.jmb.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rogers A, et al. The LonA Protease Regulates Biofilm Formation, Motility, Virulence, and the Type VI Secretion System in Vibrio cholerae. J Bacteriol. 2016;198:973–85. doi: 10.1128/JB.00741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Melderen L, Aertsen A. Regulation and quality control by Lon-dependent proteolysis. Res Microbiol. 2009;160:645–651. doi: 10.1016/j.resmic.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 86.Mueller RS, et al. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J Bacteriol. 2009;191:3504–16. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Talham GL, et al. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shikuma NJ, Yildiz FH. Identification and characterization of OscR, a transcriptional regulator involved in osmolarity adaptation in Vibrio cholerae. J Bacteriol. 2009;191:4082–96. doi: 10.1128/JB.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lipp EK, et al. Effects of global climate on infectious disease: the cholera model. Clin Microbiol Rev. 2002;15:757–70. doi: 10.1128/CMR.15.4.757-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miller CJ, et al. Response of toxigenic Vibrio cholerae 01 to physico-chemical stresses in aquatic environments. J Hyg (Lond) 1984;93:475–95. doi: 10.1017/s0022172400065074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Louis VR, et al. Predictability of Vibrio cholerae in Chesapeake Bay. Appl Environ Microbiol. 2003;69:2773–85. doi: 10.1128/AEM.69.5.2773-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Colwell RR, et al. Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science. 1977;198:394–6. [PubMed] [Google Scholar]
- 96.Bernardy EE, et al. Diversity of Clinical and Environmental Isolates of Vibrio cholerae in Natural Transformation and Contact-Dependent Bacterial Killing Indicative of Type VI Secretion System Activity. Appl Environ Microbiol. 2016;82:2833–42. doi: 10.1128/AEM.00351-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


