ABSTRACT
Pharmacodynamics of a polymyxin B, meropenem, and rifampin triple combination were examined against Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae (KPC-Kp) ST258. In time-kill experiments against three KPC-Kp isolates, triple combination generated 8.14, 8.19, and 8.29 log10 CFU/ml reductions within 24 h. In the hollow-fiber infection model, the triple combination caused maximal killing of 5.16 log10 CFU/ml at 78 h and the time required for regrowth was more than doubled versus the 2-drug combinations. Remarkably, combinations with a high single-dose polymyxin B burst plus rifampin preserved KPC-Kp polymyxin susceptibility (MIC240 h = 0.5 mg/liter) versus the same combination with traditionally dosed polymyxin B, where resistance was amplified (MIC240 h = 32 mg/liter).
KEYWORDS: polymyxin B, meropenem, rifampin, PK/PD, KPC-producing K. pneumoniae
TEXT
Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae (KPC-Kp) strains have spread worldwide and are associated with unacceptable mortality rates of up to 50% in the hospital setting (1, 2). The ST258 lineage of KPC-Kp is the most common clone globally and in the United States (3). The genes encoding KPC enzymes are commonly found on plasmids or transposons that contain additional antibiotic resistance determinants, resulting in multidrug-resistant (MDR) isolates susceptible to only a few preserved antimicrobial agents (4).
Although the polymyxin antibiotics (polymyxin E [colistin] and polymyxin B) fell out of favor after their introduction in the 1950s due to concerns over dose-limiting toxicities, their clinical use has seen a recent resurgence given the sharp rise in infections caused by MDR Gram-negative pathogens. In many clinical scenarios, polymyxin B is favored over colistin due its more favorable pharmacological properties that can be attributed to its administration as an active antibiotic. The polymyxins are among the only remaining classes of antibiotics that consistently maintain activity against KPC-Kp, with susceptibility rates of >85% (5). Despite sustained in vitro activity, treatment with polymyxin monotherapy frequently results in clinical failure, probably due to rapid proliferation of resistant subpopulations (6, 7). Polymyxin-carbapenem combination regimens have been associated with improved clinical outcomes and reduced mortality in KPC-Kp infections, providing a viable alternative to polymyxin monotherapy (8, 9). However, the clinical benefit of adding a carbapenem to the polymyxin is not apparent in infections due to KPC-Kp with elevated carbapenem MICs (≥16 mg/liter) (10). Interestingly, rifampin-containing combination regimens to combat KPC-Kp have shown promise in preclinical animal models and some in vitro studies (11–13). However, optimized combination regimens involving polymyxins, carbapenems, and rifampin have yet to be fully elucidated. Here, we describe the pharmacodynamic activity of single-drug regimens and double- and triple-drug combination regimens containing polymyxin B, meropenem, and rifampin against KPC-2-producing K. pneumoniae ST258 isolates with elevated meropenem MICs. To our knowledge, this study represents the first to have examined humanized polymyxin and polymyxin-based combination regimens over 10 days in the hollow-fiber infection model (HFIM) against KPC-Kp with high meropenem MICs.
Polymyxin B (Sigma-Aldrich, St. Louis, MO; lot WXBB4470V), meropenem (AK Scientific, Union City, CA; lot LC24337), and rifampin (Sigma-Aldrich, St. Louis, MO; lot 109K1417) were utilized in all experiments, and MICs were determined in duplicate using broth microdilution according to CLSI guidelines (14). The KPC-2-producing K. pneumoniae ST258 bloodstream isolates used in the present study (KPC-Kp 9A, KPC-Kp 24A, and KPC-Kp 27A) were obtained from cultures of blood from three different patients with hematologic malignancies, all of whom developed septic shock and died within 1 week of bacteremia onset (15). The KPC-Kp isolates were subjected to PCR with previously published primers for detection of blaSHV, blaTEM, blaCTX-M, blaNDM, blaVIM, blaIMP, and blaOXA-48 genes, followed by sequencing of positive results (16, 17). The presence and type of blaKPC gene were determined by a multiplex molecular-beacon real-time PCR assay (18), and PCR was performed for detection of mutations in K. pneumoniae outer membrane porin genes ompK35 and ompK36, as described previously (19). KPC-Kp 9A, KPC-Kp 24A, and KPC-Kp 27A were used in time-kill experiments at an ∼108 CFU/ml inoculum to initially assess the combinatorial pharmacodynamics over 24 h, as described previously (20). Time-kill experiments were carried out in duplicate with polymyxin B (2.41 and 6 mg/liter) (21), meropenem (49.6 mg/liter) (22), and rifampin (3.5 mg/liter) (23, 24) at concentrations representing the HFIM projected maximum free drug concentration (fCmax). The lower limit of quantification was 102 CFU/ml.
To build upon the time-kill data, HFIM experiments (cartridge C3008; FiberCell Systems Inc., Frederick, MD) were performed over 10 days, as previously described (7), to evaluate the effectiveness of dosing regimens of polymyxin B alone or combined with meropenem and rifampin. KPC-Kp 9A and KPC-Kp 27A were selected as representative isolates to use in the HFIM at an ∼108 CFU/ml inoculum for a proof-of-concept evaluation of the drug regimens described below. Population analysis profiles (PAPs) were also determined during the HFIM by plating a 50-μl aliquot of a bacterial sample onto an agar plate embedded with polymyxin B (0.5, 1, 4, or 10 mg/liter) or meropenem (4, 16, or 64 mg/liter) to simultaneously track the polymyxin- and meropenem-heteroresistant subpopulations that are often present in K. pneumoniae (25, 26). HFIM polymyxin B dosage regimens were based on the population pharmacokinetic model described by Sandri et al. from 24 critically ill patients who received intravenous polymyxin B doses ranging from 0.45 to 3.38 mg/kg of body weight/day (7, 21). The following polymyxin B (half-life [t1/2] = 8 h), meropenem (t1/2 = 2.5 h), and rifampin (t1/2 = 2.5 h) regimens were simulated using each drug alone or in double or triple combinations in the HFIM based on human pharmacokinetic data in critically ill patients (22–24):
-
i.
Polymyxin B loading dose: 2.22 mg/kg for 1 dose followed by 1.43 mg/kg q12h starting 12 h later, with an fCmax = 2.41 mg/liter and area under the concentration-time curve for the free, unbound fraction of a drug at 24 h (fAUC24) = 35.9 mg · h/liter across every day.
-
ii.
Polymyxin B burst: 5.53 mg/kg for 1 dose followed by no subsequent doses with an fCmax = 6.0 mg/liter and fAUC0–24 = 60.4 mg · h/liter.
-
iii.
Meropenem: 2 g q8h with an fCmax = 48.9 mg/liter and fAUC0–24 = 657.7 mg · h/liter across the first day and an fCmax = 49.0 mg/liter and fAUC24 = 707.3 mg · h/liter at the steady state.
-
iv.
Rifampin: 600 mg q24h with an fCmax = 3.5 mg/liter and fAUC24 = 14.0 mg · h/liter across every day.
During combination experiments where the HFIM system was set to a 2.5-h half-life, polymyxin B was supplemented into the central reservoir every 80 min to maintain a pharmacokinetic profile consistent with an 8-h half-life (27). Samples for validation of the antimicrobial pharmacokinetics were obtained from monotherapy HFIM runs over a 48-h period prior to the addition of bacteria. Polymyxin B concentrations were determined by a liquid chromatography single-quadrupole mass spectrometry (LC-MS) method (28). Meropenem concentrations were quantified using a liquid chromatography tandem mass spectrometry method (LC-MS/MS) that utilized an Agilent 1200 system and an Agilent 6430 system (Santa Clara, CA) (29). Rifampin concentrations were validated using a microbiological assay that utilized Staphylococcus aureus (ATCC 29213) as an indicator organism embedded in Mueller-Hinton agar (MHA) at 107 CFU/ml (30). Using 35 ml of agar, rifampin concentrations ranging from 0.0625 to 10 mg/liter were used to generate a logarithmic standard curve (R2 = >0.99). There was satisfactory agreement between the observed and targeted pharmacokinetic profiles for polymyxin B (R2 = 0.98, slope = 0.83, and intercept = −0.10), meropenem (R2 = 0.97, slope = 0.85, and intercept = 1.67), and rifampin (R2 = 0.99, slope = 1.15, and intercept = 0.08).
In time-kill experiments against KPC-Kp 9A, KPC-Kp 24A, and KPC-Kp 27A, monotherapies were unable to sustain bacterial killing through 24 h (Table 1). A double combination with polymyxin B at 6 mg/liter and rifampin was bactericidal (≥3 log10 CFU/ml reduction in viable colony count from the initial inoculum) and synergistic at 24 h against all three isolates, whereas polymyxin B at 6 mg/liter combined with meropenem was bactericidal and synergistic only against isolate KPC-Kp 27A at 24 h. Triple combination therapy with polymyxin B at 6 mg/liter yielded synergistic killing and undetectable bacterial counts at 24 h against all isolates.
TABLE 1.
KPC-Kp isolate characteristics, including β-lactamases and MIC values, followed by the antibiotic regimens used in the time-kill experiments and mean changes in bacterial log10 CFU/ml from baseline after 4, 8, and 24 ha
| KPC-Kp isolate | β-Lactamase(s) | MIC (mg/liter) |
Time-kill experiment antimicrobial(s)b,c | Time-kill experiment Δ bacterial log10 CFU/ml ± SDd,e |
||||
|---|---|---|---|---|---|---|---|---|
| PMB | MERO | RIF | 4 h | 8 h | 24 h | |||
| 9A | KPC-2, SHV-11, TEM-1 | 0.5 | 16 | 64 | PMB 2.41 | −2.93 ± 0.45 | −0.54 ± 0.09 | 0.86 ± 0.04 |
| PMB 6 | −4.19 ± 0.49 | −2.20 ± 0.15 | 0.82 ± 0.02 | |||||
| PMB 2.41 + MERO | −3.73 ± 0.05 | −0.67 ± 0.16 | 0.89 ± 0.05 | |||||
| PMB 2.41 + RIF | −3.16 ± 0.18 | −3.78 ± 1.73 | −0.98 ± 0.22* | |||||
| PMB 6 + MERO | −4.87 ± 0.04 | −3.51 ± 0.78 | 0.82 ± 0.12 | |||||
| PMB 6 + RIF | −5.87 ± 3.20 | −4.73 ± 1.55 | −5.06 ± 0.26* | |||||
| PMB 2.41 + MERO + RIF | −3.19 ± 0.19 | −5.81 ± 0.44* | −6.75 ± 1.91 | |||||
| PMB 6 + MERO + RIF | −8.13 ± 0.01 | −8.13 ± 0.01* | −8.13 ± 0.01* | |||||
| 24A | KPC-2 | 0.5 | 64 | >64 | PMB 2.41 | −0.55 ± 0.97 | 0.06 ± 0.75 | 0.90 ± 0.20 |
| PMB 6 | −5.85 ± 0.64 | −4.32 ± 0.78 | −0.28 ± 1.03 | |||||
| PMB 2.41 + MERO | −0.98 ± 1.25 | −0.15 ± 0.81 | 0.91 ± 0.14 | |||||
| PMB 2.41 + RIF | −3.27 ± 0.28 | −3.16 ± 0.44 | 0.70 ± 0.17 | |||||
| PMB 6 + MERO | −6.11 ± 0.69 | −4.28 ± 0.99 | −0.33 ± 1.22 | |||||
| PMB 6 + RIF | −8.15 ± 0.06 | −8.15 ± 0.06 | −7.05 ± 1.50* | |||||
| PMB 2.41 + MERO + RIF | −3.87 ± 0.40 | −3.59 ± 0.44* | 0.70 ± 0.12 | |||||
| PMB 6 + MERO + RIF | −8.23 ± 0.06 | −8.23 ± 0.06 | −8.23 ± 0.06 | |||||
| 27A | KPC-2, SHV-11, TEM-1 | 1 | 32 | >64 | PMB 2.41 | −3.34 ± 0.87 | −1.06 ± 0.57 | 0.74 ± 0.03 |
| PMB 6 | −5.14 ± 0.62 | −3.81 ± 0.96 | 0.20 ± 0.56 | |||||
| PMB 2.41 + MERO | −3.37 ± 0.47 | −1.42 ± 0.20 | 0.75 ± 0.05 | |||||
| PMB 2.41 + RIF | −0.93 ± 0.27 | −1.20 ± 0.50 | −0.46 ± 0.47 | |||||
| PMB 6 + MERO | −5.62 ± 0.21 | −5.40 ± 0.18 | −3.57 ± 2.74 | |||||
| PMB 6 + RIF | −2.78 ± 0.22 | −4.14 ± 0.60 | −3.92 ± 0.25* | |||||
| PMB 2.41 + MERO + RIF | −1.34 ± 0.09 | −1.43 ± 0.23 | −1.25 ± 0.21* | |||||
| PMB 6 + MERO + RIF | −2.92 ± 0.53 | −4.36 ± 0.75 | −8.24 ± 0.07* | |||||
Baseline, 0 h.
PMB 2.41, polymyxin B at 2.41 mg/liter; PMB 6, polymyxin B at 6 mg/liter; MERO, meropenem at 49.6 mg/liter; RIF, rifampin at 3.5 mg/liter.
The MERO, RIF, and MERO + RIF experimental arms are not displayed and did not reduce bacterial counts at 4, 8, or 24 h.
Change from 0 h bacterial concentration in time-kill experiments [log10(CFUxh) − log10(CFU0h)], where x = 4, 8, or 24 h; mean starting inoculum, 8.19 ± 0.07 log10 CFU/ml.
Data from synergistic combinations (defined as those resulting in a >2 log10 CFU/ml mean reduction in viable counts over the results seen with the most active agent administered alone) are bolded; bacterial reductions for combinations whose results were significantly different from those seen with the most active agent alone are denoted with an asterisk (P < 0.05).
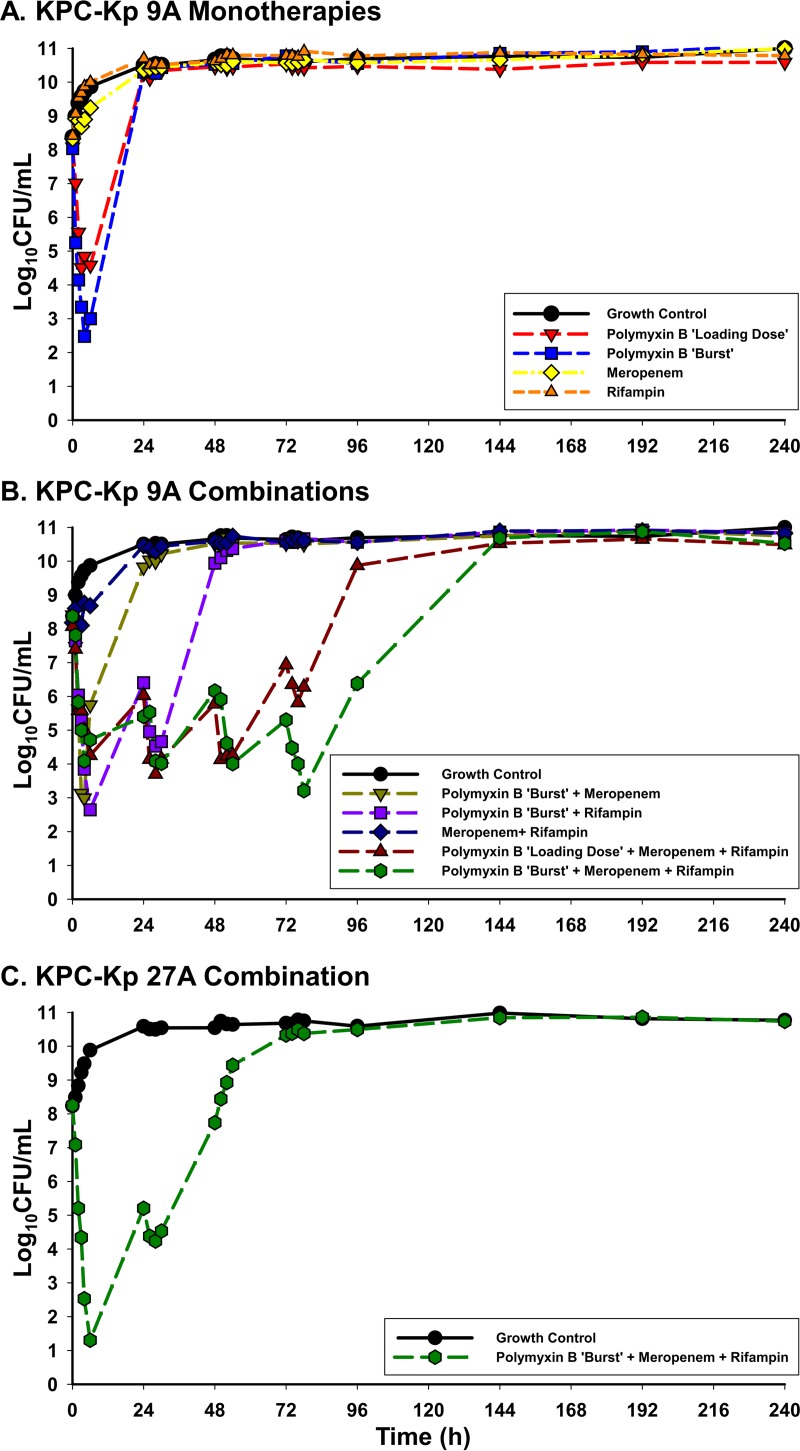
In the HFIM, neither meropenem nor rifampin as monotherapy was able to cause bacterial killing of KPC-Kp 9A at any time point (Fig. 1). Polymyxin B monotherapy administered as either a loading dose or a burst regimen caused >3 log10 CFU/ml bacterial reductions throughout the first 6 h. The polymyxin B loading dose achieved bactericidal activity and a maximal bacterial reduction of 3.57 log10 CFU/ml after 3 h, while the burst regimen provided bactericidal activity within 2 h and a maximal bacterial reduction of 5.56 log10 CFU/ml at 4 h. Bacterial killing with each polymyxin B monotherapy regimen was not maintained, and the total population regrew to growth control levels by 24 h. The polymyxin B burst regimen, which displayed greater killing than the loading dose, was essentially unaided by the addition of meropenem, where maximal log10 CFU/ml reductions of 5.41 were also achieved after 4 h. However, the addition of rifampin to the polymyxin B burst regimen enabled prolonged killing with bactericidal activity detected out to 30 h and synergy from 24 to 30 h before the total population began to regrow.
FIG 1.
HFIM total bacterial population counts in log10 CFU/ml over 240 h following exposure to polymyxin B loading dose (2.22 mg/kg for 1 dose followed by 1.43 mg/kg q12h starting 12 h later), polymyxin B burst (5.53 mg/kg × 1 dose followed by no subsequent doses), meropenem (2 g q8h), and rifampin (600 mg q24h). Experiments were conducted at a starting inoculum of ∼108 CFU/ml for KPC-Kp 9A versus monotherapies (A), KPC-Kp 9A versus combination therapies (B), and KPC-Kp 27A versus combination therapy (C).
Triple antimicrobial therapy combining polymyxin B, meropenem, and rifampin against KPC-Kp 9A in the HFIM substantially potentiated the activity of the individual agents. This triple combination using a polymyxin B loading dose maintained bactericidal activity until 54 h (3.81 log10 CFU/ml reduction) and doubled the time during which the combination could prevent bacterial regrowth compared to polymyxin B with rifampin. Despite administration of only a single polymyxin B dose, the burst in combination with meropenem and rifampin displayed the most bacterial killing of any regimen by tripling the time to bacterial regrowth over two-antibiotic combinations and causing maximal reductions of 5.16 log10 CFU/ml after 78 h. Against KPC-Kp 27A in the HFIM, the triple combination using a polymyxin B burst caused a 3.05 log10 CFU/ml reduction by 2 h and a maximal reduction of 6.95 log10 CFU/ml by 6 h. The triple combination maintained bactericidal activity through at least 30 h before the bacterial population completely regrew to the level seen with the growth control by 72 h.
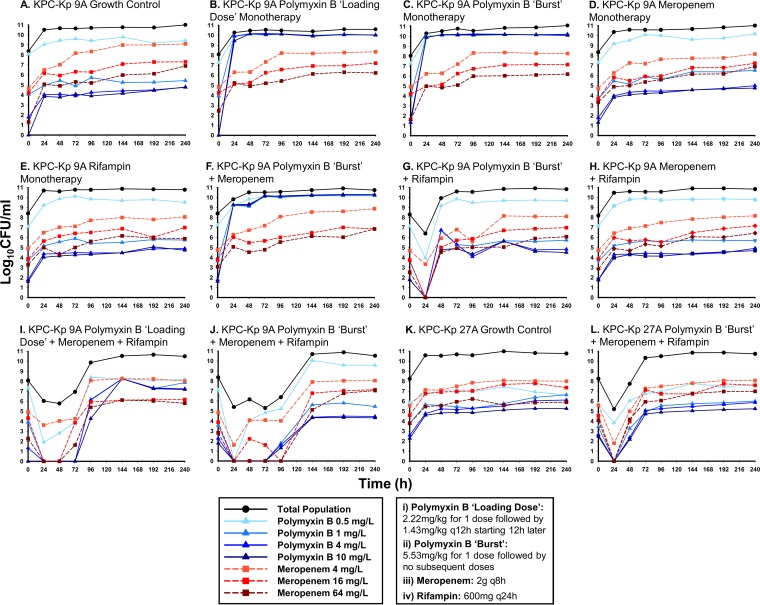
The levels of growth of KPC-Kp 9A and KPC-Kp 27A subpopulations resistant to polymyxin B and meropenem were tracked using PAPs over 240 h in the HFIM (Fig. 2). Irrespective of the administered antibiotic regimen, meropenem PAPs remained proportional to that seen with the growth control; there was not an appreciable expansion of any meropenem subpopulation, even after exposure to meropenem. Growth on plates containing polymyxin B at up to 10 mg/liter was noted for KPC-Kp 9A after exposure to polymyxin B without rifampin, including exposure to the burst and loading-dose monotherapies and the burst plus meropenem combination. In agreement with the PAPs, the KPC-Kp polymyxin B MICs following 240 h of exposure to these three antimicrobial regimens in the HFIM were >32 mg/liter. Conversely, when polymyxin B was administered as a burst in combination with rifampin against KPC-Kp 9A or KPC-Kp 27A, polymyxin B-resistant subpopulations displayed growth profiles similar to that of the growth control and, at 240 h postexposure, polymyxin B MICs remained 0.5 mg/liter. Further, the polymyxin B burst, meropenem, and rifampin triple combination suppressed the polymyxin B- and meropenem-resistant subpopulations to below baseline levels until at least 96 h for KPC-Kp 9A and 48 h for KPC-Kp 27A. In comparison to the polymyxin B burst triple combination, the three-drug regimen based on the polymyxin B loading dose caused ∼3 log10 CFU/ml more growth on plates treated with 4 or 10 mg/liter polymyxin B at 240 h and resulted in a postexposure polymyxin B MIC of 32 mg/liter.
FIG 2.
Population analysis profiles quantifying the growth of resistant KPC-Kp 9A (A to J) and KPC-Kp 27A (K to L) subpopulations on MHA plates containing the specified concentration of either polymyxin B (blue lines) or meropenem (red lines). Each panel represents a single antibiotic treatment regimen over 240 h in the HFIM and compares data from the antibiotic-resistant subpopulations, which are fractions of the respective total population (black lines). The black lines are displayed for comparison and represent the total population bacterial counts from Fig. 1 at time points when population analysis profiles were simultaneously performed (0, 24, 48, 72, 96, 144, 192, and 240 h). KPC-Kp 9A and KPC-Kp 27A polymyxin B MICs were initially 0.5 and 1 mg/liter, respectively. Polymyxin B MICs at the end of each experiment, following exposure for 240 h in the HFIM, were 0.5 mg/liter (A), >32 mg/liter (B), >32 mg/liter (C), 0.5 mg/liter (D), 0.5 mg/liter (E), >32 mg/liter (F), 0.5 mg/liter (G), 0.5 mg/liter (H), 32 mg/liter (I), 0.5 mg/liter (J), 0.5 mg/liter (K), and 0.5 mg/liter (L). KPC-Kp 9A and KPC-Kp 27A remained polymyxin susceptible following exposure to all combinations containing a polymyxin B burst plus rifampin.
Nontraditional combinations involving the polymyxin antibiotics are frequently employed in the fight against KPC-Kp because therapeutic options are extremely limited. However, it is unclear how the polymyxins should be dosed, especially when used in combination with other agents, such as carbapenems and rifampin. In the present investigation, against KPC-Kp isolates with meropenem MICs of ≥16 mg/liter (stop codons at amino acid [aa] 89 of the ompK35 porin gene), administration of polymyxin B with meropenem 2-drug combinations did not result in consistent bacterial killing. Unlike KPC-Kp 9A and KPC-Kp 24A, which harbored wild-type ompK36 genes, KPC-Kp 27A had an IS5 insertion in the promoter region of ompK36. Interestingly, synergy for the polymyxin B and meropenem combination was detected only against KPC-Kp 27A. The variations in meropenem MICs observed between KPC-Kp 9A, 24A, and 27A were likely due to these differing combinations of porin channel mutations in addition to fluctuations in expression of porin channels and the KPC-2 enzyme (31, 32). Similarly to polymyxin B with meropenem, polymyxin B and rifampin 2-drug combinations caused undetectable counts of only one KPC-Kp strain (isolate 24A) through 24 h; therefore, 3-drug regimens with rifampin were explored.
The addition of rifampin to polymyxin B and meropenem may enable prolonged bactericidal activity and may improve upon the potentially unreliable activity of 2-drug combinations against KPC-Kp with higher meropenem MICs. Administration of a polymyxin with a carbapenem and rifampin is a promising combination that was previously shown to be bactericidal (against 8/9 strains) in static time-kill experiments against carbapenemase-producing K. pneumoniae (33, 34) and was further validated in the present study. In the current investigation, rifampin added to a polymyxin B burst preserved polymyxin susceptibility through 10 days (polymyxin B MIC240 h = 0.5 mg/liter) for both KPC-Kp isolates. Contrastingly, polymyxin B monotherapy or polymyxin B plus meropenem or traditionally dosed polymyxin B plus meropenem and rifampin regimens resulted in amplified polymyxin resistance where KPC-Kp polymyxin B MICs were ≥32 mg/liter at 10 days postexposure. The benefit of coadministration of rifampin with the polymyxin B burst and meropenem may be multifactorial as a consequence of (i) directly reducing K. pneumoniae total population counts and (ii) suppressing polymyxin B resistance. Recurrent ∼1 to 2 log10 CFU/ml drops in bacterial density seen during administration of a polymyxin B-rifampin combination, but not rifampin monotherapy, in the HFIM support the concept that killing caused by rifampin is potentiated by polymyxin B, possibly via damage to the outer membrane of K. pneumoniae (35). Suppression of polymyxin B resistance by rifampin could be explained by an ability to inhibit the expression of lipopolysaccharide regulators such as phoP plus phoQ and pmrA plus pmrB, which can lower the negative charge of the K. pneumoniae outer membrane, thus decreasing polymyxin affinity (35). These two-component regulatory systems have also been shown to play an important clinical role in K. pneumoniae polymyxin heteroresistance, making them an enticing target for suppression (36).
Previous dynamic in vitro experiments (HFIM and one-compartment model) examining regimens against KPC-Kp have not described pharmacodynamic activity past 72 h (13, 37–40). Therefore, it is remarkable that in the present study, the triple combination that included a polymyxin B burst, meropenem, and rifampin was bactericidal until at least 78 h for KPC-Kp 9A and 30 h for KPC-Kp 27A despite combatting a starting inoculum that was 10 to 1,000× higher than those previously investigated. The use of KPC-Kp at an inoculum of ∼108 CFU/ml was employed to simulate the potentially high bacterial densities in deep-seated infections. In the context of infections with a lower bacterial burden, such as urinary tract infections and bacteremia, our results may underestimate the activity of the individual antibiotics (41). Our study should be examined in concert with the other studies that have explored combinations against KPC-Kp at lower inocula.
Polymyxin nephrotoxicity is the most clinically significant dose-limiting adverse event (42). Administration of a large one-time polymyxin B dose holds significant promise to circumvent cumulative drug exposure concerns which have been predictive of polymyxin-induced nephrotoxicity (43, 44). Remarkably, the triple combination incorporating the polymyxin B burst regimen displayed enhanced bacterial killing compared to the more traditional loading-dose-based combination, despite a cumulative polymyxin B exposure (AUC0–240) of <20% over 10 days. Furthermore, during exposure to polymyxin B burst-based combinations with rifampin, the K. pneumoniae isolates remained polymyxin B susceptible and there were fewer polymyxin B-resistant subpopulations present despite regrowth.
There are a number of important things to consider while interpreting the data contained in the present manuscript. One limitation of the time-kill experiments is that static fCmax concentrations were used, which may in part explain the greater killing that was observed compared to the HFIM. Polymyxin B, meropenem, and rifampin were simulated in the HFIM to approximate human pharmacokinetics in critically ill patients. However, there is likely significant interpatient variability that exists for the pharmacokinetics of these drugs in the clinical setting, which may influence the translation of these findings. Lastly, the polymyxin B burst, meropenem, and rifampin triple combination was tested only against 2 KPC-Kp isolates in the HFIM and its activity should be further validated in other clinically relevant strains.
In conclusion, the promising combination of polymyxin B, meropenem, and rifampin appears to provide a viable option to consider in the treatment of KPC-Kp, including against isolates with higher carbapenem MICs. Importantly, in combination with polymyxin B, rifampin seems to enable temporary suppression of polymyxin B resistance while acting to directly kill the K. pneumoniae strain, suggesting a multifactorial mode of attack. Further investigations in vitro and using animal models are warranted before translation to the clinic.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI111990. M.J.S. is the recipient of a career development award through the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number K23AI114994. The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, Prekates A, Themeli-Digalaki K, Tsakris A. 2011. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect 17:1798–1803. doi: 10.1111/j.1469-0691.2011.03514.x. [DOI] [PubMed] [Google Scholar]
- 2.Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y. 2012. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother 56:2108–2113. doi: 10.1128/AAC.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, Oethinger M, Paterson DL, Adams MD, Jacobs MR, Diekema DJ, Hall GS, Jenkins SG, Rice LB, Tenover FC, Bonomo RA. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother 63:427–437. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford PA, Kazmierczak KM, Biedenbach DJ, Wise MG, Hackel M, Sahm DF. 2015. Correlation of beta-lactamase production and colistin resistance among Enterobacteriaceae isolates from a global surveillance program. Antimicrob Agents Chemother 60:1385–1392. doi: 10.1128/AAC.01870-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Oliveira MS, de Assis DB, Freire MP, Boas do Prado GV, Machado AS, Abdala E, Pierrotti LC, Mangini C, Campos L, Caiaffa Filho HH, Levin AS. 2015. Treatment of KPC-producing Enterobacteriaceae: suboptimal efficacy of polymyxins. Clin Microbiol Infect 21:179.e171–179.e177. doi: 10.1016/j.cmi.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Tsuji BT, Landersdorfer CB, Lenhard JR, Cheah SE, Thamlikitkul V, Rao GG, Holden PN, Forrest A, Bulitta JB, Nation RL, Li J. 2016. Paradoxical effect of polymyxin B: high drug exposure amplifies resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 60:3913–3920. doi: 10.1128/AAC.02831-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tumbarello M, Trecarichi EM, De Rosa FG, Giannella M, Giacobbe DR, Bassetti M, Losito AR, Bartoletti M, Del Bono V, Corcione S, Maiuro G, Tedeschi S, Celani L, Cardellino CS, Spanu T, Marchese A, Ambretti S, Cauda R, Viscoli C, Viale P. 2015. Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J Antimicrob Chemother 70:2133–2143. [DOI] [PubMed] [Google Scholar]
- 9.Daikos GL, Tsaousi S, Tzouvelekis LS, Anyfantis I, Psichogiou M, Argyropoulou A, Stefanou I, Sypsa V, Miriagou V, Nepka M, Georgiadou S, Markogiannakis A, Goukos D, Skoutelis A. 2014. Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob Agents Chemother 58:2322–2328. doi: 10.1128/AAC.02166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Simmonds A, Nelson B, Eiras DP, Loo A, Jenkins SG, Whittier S, Calfee DP, Satlin MJ, Kubin CJ, Furuya EY. 4 April 2016. Combination regimens for the treatment of carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Antimicrob Agents Chemother doi: 10.1128/aac.03007-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michail G, Labrou M, Pitiriga V, Manousaka S, Sakellaridis N, Tsakris A, Pournaras S. 2013. Activity of tigecycline in combination with colistin, meropenem, rifampin, or gentamicin against KPC-producing Enterobacteriaceae in a murine thigh infection model. Antimicrob Agents Chemother 57:6028–6033. doi: 10.1128/AAC.00891-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaibani P, Lombardo D, Lewis RE, Mercuri M, Bonora S, Landini MP, Ambretti S. 2014. In vitro activity and post-antibiotic effects of colistin in combination with other antimicrobials against colistin-resistant KPC-producing Klebsiella pneumoniae bloodstream isolates. J Antimicrob Chemother 69:1856–1865. doi: 10.1093/jac/dku065. [DOI] [PubMed] [Google Scholar]
- 13.Wiskirchen DE, Koomanachai P, Nicasio AM, Nicolau DP, Kuti JL. 2011. In vitro pharmacodynamics of simulated pulmonary exposures of tigecycline alone and in combination against Klebsiella pneumoniae isolates producing a KPC carbapenemase. Antimicrob Agents Chemother 55:1420–1427. doi: 10.1128/AAC.01253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard-10th edition. CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Satlin MJ, Calfee DP, Chen L, Fauntleroy KA, Wilson SJ, Jenkins SG, Feldman EJ, Roboz GJ, Shore TB, Helfgott DC, Soave R, Kreiswirth BN, Walsh TJ. 2013. Emergence of carbapenem-resistant Enterobacteriaceae as causes of bloodstream infections in patients with hematologic malignancies. Leuk Lymphoma 54:799–806. doi: 10.3109/10428194.2012.723210. [DOI] [PubMed] [Google Scholar]
- 16.Eisner A, Fagan EJ, Feierl G, Kessler HH, Marth E, Livermore DM, Woodford N. 2006. Emergence of Enterobacteriaceae isolates producing CTX-M extended-spectrum beta-lactamase in Austria. Antimicrob Agents Chemother 50:785–787. doi: 10.1128/AAC.50.2.785-787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J Clin Microbiol 49:579–585. doi: 10.1128/JCM.01588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla(ACT-1) beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob Agents Chemother 50:3396–3406. doi: 10.1128/AAC.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ly NS, Bulman ZP, Bulitta JB, Baron C, Rao GG, Holden PN, Li J, Sutton MD, Tsuji BT. 2016. Optimization of polymyxin B in combination with doripenem to combat mutator Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:2870–2880. doi: 10.1128/AAC.02377-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhao RC, Wang J, Forrest A, Nation RL, Li J, Zavascki AP. 2013. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 22.Munz MR BJ, Höhl R, Jensen A, Junger A, Glaser R, Stelzer C, Kinzig M, Bertsch T, Holzgrabe U, Sörgel F. 2015. Individualizing meropenem prolonged infusions in intensive care unit patients via population modeling of renal function and infection biomarkers over time. Abstr Paracelsus Sci Get Together, Nuremberg, Germany, July 3, 2015. [Google Scholar]
- 23.Sanofi. 2013. Rifadin (R) rifampin (package insert). Sanofi, Bridegwater, NJ. [Google Scholar]
- 24.Acocella G. 1978. Clinical pharmacokinetics of rifampicin. Clin Pharmacokinet 3:108–127. doi: 10.2165/00003088-197803020-00002. [DOI] [PubMed] [Google Scholar]
- 25.Pournaras S, Kristo I, Vrioni G, Ikonomidis A, Poulou A, Petropoulou D, Tsakris A. 2010. Characteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical isolates of K. pneumoniae. J Clin Microbiol 48:2601–2604. doi: 10.1128/JCM.02134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poudyal A, Howden BP, Bell JM, Gao W, Owen RJ, Turnidge JD, Nation RL, Li J. 2008. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J Antimicrob Chemother 62:1311–1318. doi: 10.1093/jac/dkn425. [DOI] [PubMed] [Google Scholar]
- 27.Blaser J. 1985. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J Antimicrob Chemother 15(Suppl A):125–130. [DOI] [PubMed] [Google Scholar]
- 28.Cheah SE, Bulitta JB, Li J, Nation RL. 2014. Development and validation of a liquid chromatography-mass spectrometry assay for polymyxin B in bacterial growth media. J Pharm Biomed Anal 92:177–182. doi: 10.1016/j.jpba.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenhard JR, Bulitta JB, Connell TD, King-Lyons N, Landersdorfer CB, Cheah S-E, Thamlikitkul V, Shin BS, Rao G, Holden PN, Walsh TJ, Forrest A, Nation RL, Li J, Tsuji BT. 15 September 2016. High-intensity meropenem combinations with polymyxin B: new strategies to overcome carbapenem resistance in Acinetobacter baumannii. J Antimicrob Chemother doi: 10.1093/jac/dkw355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer SM, Rybak MJ. 1996. Pharmacodynamics of once- or twice-daily levofloxacin versus vancomycin, with or without rifampin, against Staphylococcus aureus in an in vitro model with infected platelet-fibrin clots. Antimicrob Agents Chemother 40:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landman D, Bratu S, Quale J. 2009. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol 58:1303–1308. doi: 10.1099/jmm.0.012575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tängdén T, Hickman RA, Forsberg P, Lagerbäck P, Giske CG, Cars O. 2014. Evaluation of double- and triple-antibiotic combinations for VIM- and NDM-producing Klebsiella pneumoniae by in vitro time-kill experiments. Antimicrob Agents Chemother 58:1757–1762. doi: 10.1128/AAC.00741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urban C, Mariano N, Rahal JJ. 2010. In vitro double and triple bactericidal activities of doripenem, polymyxin B, and rifampin against multidrug-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli. Antimicrob Agents Chemother 54:2732–2734. doi: 10.1128/AAC.01768-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velkov T, Deris ZZ, Huang JX, Azad MA, Butler M, Sivanesan S, Kaminskas LM, Dong YD, Boyd B, Baker MA, Cooper MA, Nation RL, Li J. 2014. Surface changes and polymyxin interactions with a resistant strain of Klebsiella pneumoniae. Innate Immun 20:350–363. doi: 10.1177/1753425913493337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik CC, Christensen H, Li P, Sutherland CA, Nicolau DP, Kuti JL. 2010. Comparison of the activity of a human simulated, high-dose, prolonged infusion of meropenem against Klebsiella pneumoniae producing the KPC carbapenemase versus that against Pseudomonas aeruginosa in an in vitro pharmacodynamic model. Antimicrob Agents Chemother 54:804–810. doi: 10.1128/AAC.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirsch EB, Ledesma KR, Chang KT, Schwartz MS, Motyl MR, Tam VH. 2012. In vitro activity of MK-7655, a novel beta-lactamase inhibitor, in combination with imipenem against carbapenem-resistant Gram-negative bacteria. Antimicrob Agents Chemother 56:3753–3757. doi: 10.1128/AAC.05927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman K, Levasseur P, Girard AM, Borgonovi M, Miossec C, Merdjan H, Drusano G, Shlaes D, Nichols WW. 2014. Activities of ceftazidime and avibactam against beta-lactamase-producing Enterobacteriaceae in a hollow-fiber pharmacodynamic model. Antimicrob Agents Chemother 58:3366–3372. doi: 10.1128/AAC.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulik CC, Nicolau DP. 2011. Double-carbapenem therapy for carbapenemase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 55:3002–3004. doi: 10.1128/AAC.01420-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adler A, Ben-Dalak M, Chmelnitsky I, Carmeli Y. 2015. Effect of resistance mechanisms on the inoculum effect of carbapenem in Klebsiella pneumoniae isolates with borderline carbapenem resistance. Antimicrob Agents Chemother 59:5014–5017. doi: 10.1128/AAC.00533-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akajagbor DS, Wilson SL, Shere-Wolfe KD, Dakum P, Charurat ME, Gilliam BL. 2013. Higher incidence of acute kidney injury with intravenous colistimethate sodium compared with polymyxin B in critically ill patients at a tertiary care medical center. Clin Infect Dis 57:1300–1303. doi: 10.1093/cid/cit453. [DOI] [PubMed] [Google Scholar]
- 43.Forrest A. 2013. Towards optimized dosing of intravenous colistin and polymyxin B: risk factors for nephrotoxicity. Proc 1st Int Conf Polymyxins, Prato, Italy. [Google Scholar]
- 44.Garonzik SM, Forrest A, Thamlikitkul V, Li J, Paterson DL, Silveira FP, Nation RL. 2011. Predictors of changes in renal function in critically-ill patients on colistin methanesulfonate, abstr A-1101, slide session 156. Abstr 51st Intersci Conf Antimicrob Agents Chemother, 17 to 20 September, Chicago, IL American Society for Microbiology, Washington, DC. [Google Scholar]