ABSTRACT
Pseudomonas aeruginosa is a highly virulent, multidrug-resistant pathogen that causes significant morbidity and mortality in hospitalized patients and is particularly devastating in patients with cystic fibrosis. Increasing antibiotic resistance coupled with decreasing numbers of antibiotics in the developmental pipeline demands novel antibacterial approaches. Here, we tested peptide-conjugated phosphorodiamidate morpholino oligomers (PPMOs), which inhibit translation of complementary mRNA from specific, essential genes in P. aeruginosa. PPMOs targeted to acpP, lpxC, and rpsJ, inhibited P. aeruginosa growth in many clinical strains and activity of PPMOs could be enhanced 2- to 8-fold by the addition of polymyxin B nonapeptide at subinhibitory concentrations. The PPMO targeting acpP was also effective at preventing P. aeruginosa PAO1 biofilm formation and at reducing existing biofilms. Importantly, treatment with various combinations of a PPMO and a traditional antibiotic demonstrated synergistic growth inhibition, the most effective of which was the PPMO targeting rpsJ with tobramycin. Furthermore, treatment of P. aeruginosa PA103-infected mice with PPMOs targeting acpP, lpxC, or rpsJ significantly reduced the bacterial burden in the lungs at 24 h by almost 3 logs. Altogether, this study demonstrates that PPMOs targeting the essential genes acpP, lpxC, or rpsJ in P. aeruginosa are highly effective at inhibiting growth in vitro and in vivo. These data suggest that PPMOs alone or in combination with antibiotics represent a novel approach to addressing the problems associated with rapidly increasing antibiotic resistance in P. aeruginosa.
KEYWORDS: PPMO, Pseudomonas aeruginosa, antibiotic resistance, antimicrobial agents, antisense, experimental therapeutics, phosphorodiamidate morpholino oligomer
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic pathogen that causes significant morbidity and mortality in patients with immunocompromised conditions, such as cystic fibrosis (CF). It is intrinsically resistant to a variety of antimicrobials due to the low permeability of its outer membrane and a high rate of biofilm production (1). In addition, P. aeruginosa has developed multidrug-resistant (MDR) phenotypes, which actively resist antibiotics through overexpression of defensive efflux pumps and enzymes such as β-lactamases (1, 2). Drugs with significant toxicities, such as tobramycin or colistin, are now often utilized to treat P. aeruginosa infections (3–7). Unfortunately, P. aeruginosa has been shown to develop resistance to tobramycin and colistin in clinical settings (8–10). This demonstrates the urgent need for new paradigms in the development of antimicrobials that can control this MDR pathogen (6, 11, 12).
Phosphorodiamidate morpholino oligomers (PMOs) are synthetic antisense oligomers retaining the natural nucleobase but containing a synthetic morpholino and phosphorodiamidate backbone, rendering the PMOs resistant to enzymatic degradation (13). The oligomer sequences are designed to be complementary to an mRNA of interest, specifically around the Shine-Dalgarno or translation start site (NTG) start site of the target gene, and are thought to exert their effect through inhibition of protein translation. To aid in intracellular delivery, these oligomers are conjugated to cell-penetrating peptides (peptide-conjugated PMOs [PPMOs]) that are hypothesized to help deliver the oligomer across Gram-negative membranes by incompletely understood mechanisms (14). Importantly, PPMOs have already shown in vitro and in vivo activity against a variety of pathogens, including Escherichia coli, Salmonella enterica, Acinetobacter baumannii, and members of the Burkholderia cepacia complex (15–18).
In this study, we designed and tested PPMOs targeted to essential genes in P. aeruginosa and present novel observations for peptide conjugates that enhances the entry of PPMOs into P. aeruginosa. We demonstrate that PPMOs can inhibit growth and reduce established biofilms, a critically important aspect of P. aeruginosa pathogenesis, and that PPMOs can inhibit P. aeruginosa growth synergistically with clinically relevant antibiotics in vitro. Crucially, PPMOs can also inhibit P. aeruginosa growth in a murine model of acute pneumonia. These data suggest that PPMOs can be used alone or as an adjunct to antibiotic therapy and may be of clinical utility against P. aeruginosa infections.
RESULTS
P. aeruginosa is inhibited by PPMOs in vitro.
We utilized a focused screening approach, designing PPMOs targeting genes that we and others have previously shown to be essential in other pathogens or whose protein products are currently the target of antibiotic therapy, such as ribosome components or cell membrane pathways. The initial screening in Mueller-Hinton II cation-adjusted medium (MHII) showed that PPMOs did not inhibit growth at concentrations of ≤16 μM in most P. aeruginosa strains (see Fig. S1a in the supplemental material). However, with further experimentation, we found that PPMOs targeting 3 genes (acpP, lpxC, and rpsJ) showed substantial potency (MIC50 values of 0.5 to 16 μM) in the presence of a subinhibitory concentration (1 μg/ml) of the antibiotic colistin (polymyxin E) or polymyxin B nonapeptide (PMBN; 2 μg/ml) (see Fig. S1b and c in the supplemental material). In addition, PPMOs had enhanced potency against P. aeruginosa grown in morpholinepropanesulfonic acid (MOPS) minimal medium (see Fig. S1d in the supplemental material). MOPS medium contains lower divalent cations compared to MHII, which is known to destabilize lipopolysaccharide. Increasing the bacterial outer membrane permeability with either polymyxins or cation-limited media allowed our PPMOs to inhibit P. aeruginosa growth, suggesting that our PPMO leads were effective but that the penetrating-peptide components were insufficient to allow entry into the bacterium.
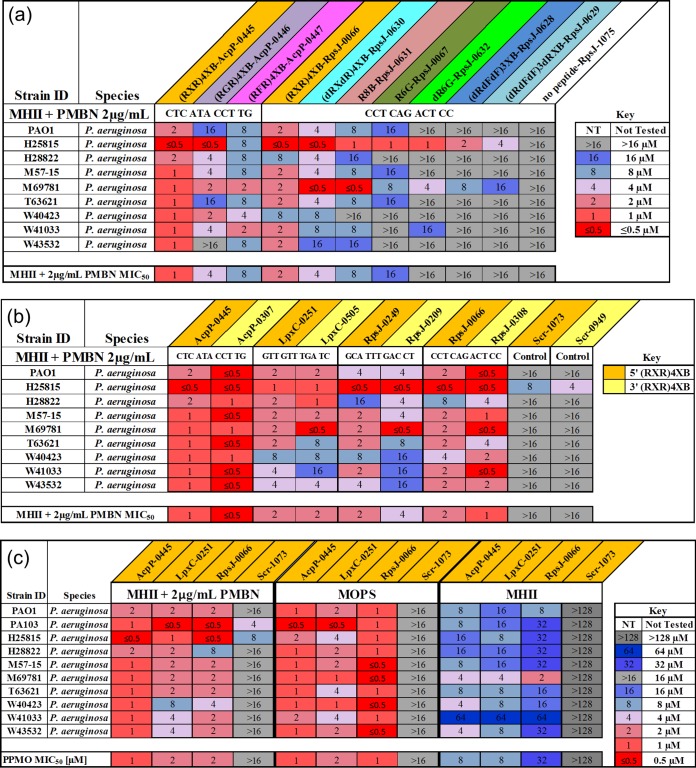
Therefore, we expanded our repertoire of cell-penetrating peptides and compared PPMOs with the same sequence but different peptides and sites of attachment. Of the nine different peptide conjugations examined, (RXR)4XB conjugates had the lowest MIC50s in MHII with PMBN at 2 μg/ml of (Fig. 1a, orange). In addition, various membrane-penetrating peptides were examined individually by substitution and attachment to either the 5′ or the 3′ end of the oligomer. (RXR)4XB conjugated PPMOs had similar MIC50 values across the nine P. aeruginosa strains tested, usually varying only 2-fold regardless of which side the peptide was conjugated (5′ versus 3′) (Fig. 1b).
FIG 1.
Peptide-conjugate (RXR)4XB was effective at delivering PMO into P. aeruginosa. (a) Heat map comparison of MICs for PPMOs with various peptide-conjugates against P. aeruginosa strains grown in MHII with PMBN at 2 μg/ml. (b) Heat map comparison of MICs for 5′ versus 3′ (RXR)4XB-conjugated PPMOs against P. aeruginosa strains grown in MHII with PMBN at 2 μg/ml. (c) Heat map comparison of MICs for lead sequence PPMOs against P. aeruginosa strains grown in MHII with PMBN at 2 μg/ml, MOPS, and MHII alone.
To confirm the optimal position of our AcpP PPMO, we designed 13 additional 11-mers spanning the −30 to +40 region of the acpP gene. These PPMOs were designed, utilizing our PPMO design tool, to avoid off-target hits spanning similar gene regions of other known essential genes. In vitro testing was performed and confirmed previous observations that sequences targeting the Shine-Dalgarno or ATG start site of the acpP gene were the most effective at bacterial inhibition (data not shown). Indeed, the MIC of AcpP-0445 (spanning the ATG) was 2 μM, with PPMOs losing activity the farther away they were placed from these two target sites. The RpsJ and LpxC PPMOs were designed to target these same regions, and PPMOs were selected based on the lowest number of off-target hits in other genes.
Having optimized sequence and peptide parameters, we next examined the gene target leads as 5′-(RXR)4XB conjugates in MHII without PMBN. AcpP-0445 and LpxC-0251 had MIC50s of 8 μM, whereas RpsJ-0066 had an MIC50 of 32 μM, revealing that the (RXR)4XB peptide was capable of P. aeruginosa outer membrane penetration, even in rich media, without additional membrane perturbation (compare panels in Fig. 1c). Both AcpP-0445 and LpxC-0251 were bactericidal at 2× their MICs by 24 h, and RpsJ-0066 was bactericidal at 4× the MIC by 24 h (see Fig. S2 in the supplemental material).
PPMOs can prevent and dissociate P. aeruginosa biofilm.
PPMOs were tested for their ability to prevent and break down biofilms. P. aeruginosa PAO1-GFP was grown in MBEC plates with PPMOs, and biofilm formation was measured by using crystal violet and confocal microscopy. PPMOs targeted to acpP, lpxC, and rpsJ prevented the formation of biofilm at 4 μM and above (see Fig. S3a in the supplemental material), and this effect was enhanced down to 1 or 2 μM in the presence of subinhibitory concentrations of PMBN (see Fig. S3b in the supplemental material). Since AcpP-0445 was the most effective at preventing biofilm, AcpP-0445-treated and control biofilms were also visualized using fluorescence microscopy. The images demonstrate that control cultures had thick biofilms (red) with large amounts of bacteria present (green), whereas cultures treated with 8 μM AcpP-0445 alone or 4 μM AcpP-0445 with PMBN (see Fig. S3c and d in the supplemental material) show little residual biofilm or bacteria.
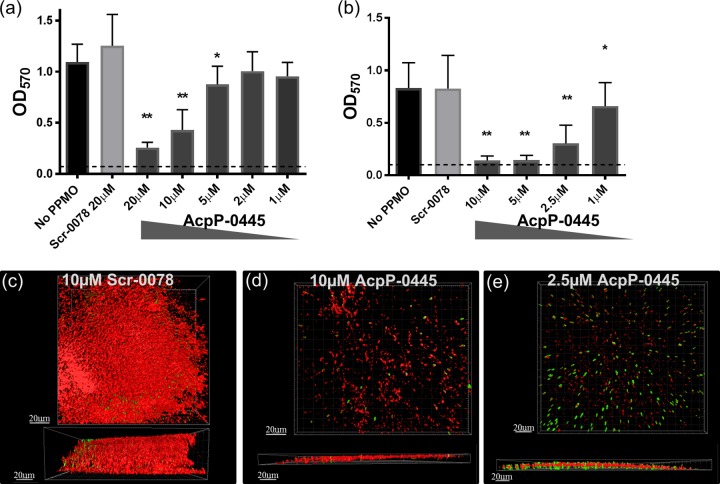
AcpP-0445 was further tested for its ability to decrease an existing biofilm. A thick biofilm was established by growing PAO1-GFP for 24 h, followed by exposure to PPMO at 24, 32, and 40 h, before analysis at 48 h. AcpP-0445 significantly reduced PAO1-GFP biofilm (77% reduction at 20 μM) in a dose-dependent manner, while Scrambled treatment had no effect (Fig. 2a). The addition of subinhibitory concentrations of PMBN enhanced this effect, (83% reduction at 10 μM) in a dose-dependent manner (Fig. 2b). Confocal microscopy of experiments revealed a thick biofilm (red, >50 μm), with numerous bacteria (green) in untreated cultures (data not shown) or in cultures treated with 10 μM a Scrambled PPMO (Fig. 2c). Established biofilm cultures treated with 10 or 2.5 μM AcpP-0445 were reduced to a thickness of <10 μm with reduced numbers of detectable bacteria (Fig. 2d and e).
FIG 2.
PPMO treatment reduces existing P. aeruginosa biofilm. Biofilm was grown for 24 h and then treated at 24, 32, and 40 h with the indicated PPMO in MHII (a) or in MHII with PMBN at 2 μg/ml (b) and analyzed at 48 h by crystal violet. The dashed line represents the lower limit of detection. These data are combined from three independent experiments, the error bars represent standard deviations, and statistics were determined with a one-way analysis of variance (ANOVA) and a Holm-Ŝidák's multiple-comparison test, where P < 0.0001 (**) and P < 0.05 (*) are indicated compared to no PPMO. Spinning disk confocal microscopy images of MBEC pegs are depicted from above and the side incubated as in panel b with 10 μM Scr-0078 (c), 10 μM AcpP-0445 (d), or 2.5 μM AcpP-0445 (e). Green indicates PAO1-GFP, and red indicates biofilm stained with concanavalin A-Alexa Fluor 647.
PPMOs synergize with existing antibiotics.
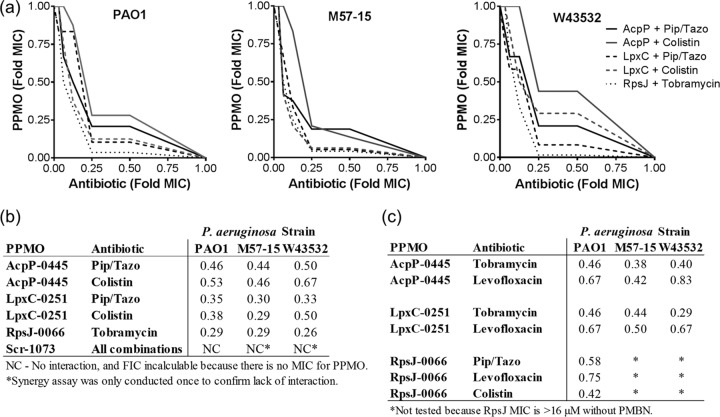
Based on the original finding that PPMOs worked better with the antibiotic colistin (see Fig. S1b in the supplemental material) and based on synergy findings with similar antisense modalities (19), we sought to determine whether PPMOs would have enhanced and perhaps synergistic activity with other approved antibiotics. We hypothesized that there would be enhanced interactions when the antibiotic and the PPMO targeted similar cellular processes (i.e., both targeting components in the bacterial membrane or both targeting translation). Synergy was consistently identified when AcpP-0445 or LpxC-0251 PPMO was combined with piperacillin-tazobactam (fractional inhibitory concentration [FIC] indices = 0.30 to 0.50), and when LpxC-0251 was combined with colistin (FIC indices = 0.38 to 0.5) (Fig. 3a and b). However, the most synergistic combination identified was RpsJ-0066 with tobramycin (FIC indices = 0.26 to 0.29) in PAO1 and two clinical P. aeruginosa strains (Fig. 3a and b). AcpP-0445 combined with colistin had both synergistic and additive interactions (FIC indices = 0.46 to 0.67) depending on the P. aeruginosa strain utilized (Fig. 3a and b). Tobramycin was also synergistic in all strains when combined with PPMOs targeting dissimilar cellular processes (AcpP and LpxC; Fig. 3c), whereas the other dissimilar cellular process pairs ranged from FIC indices of 0.42 to 0.83 (Fig. 3c).
FIG 3.
PPMOs display synergy with traditional antibiotics. Three representative strains of P. aeruginosa (PAO1, M57-15, and W43532) were utilized to examine if PPMOs enhanced activity when combined with current traditional antibiotics. (a) Synergy isobolograms of PPMOs and antibiotics that target similar cellular processes. (b) Fractional inhibitory concentration (FIC) indices of PPMOs and antibiotics that target similar cellular processes. (c) FIC indices of PPMOs and antibiotics that target dissimilar cellular processes. FIC indices were calculated as described in Materials and Methods. An FIC index is considered synergistic when ≤0.5. Data represent the average of three to four independent experiments, and FIC indices were calculated from the best interaction point of these averages. Pip/Tazo, piperacillin-tazobactam; Scr, Scrambled.
PPMOs reduce P. aeruginosa burden in infected mice.
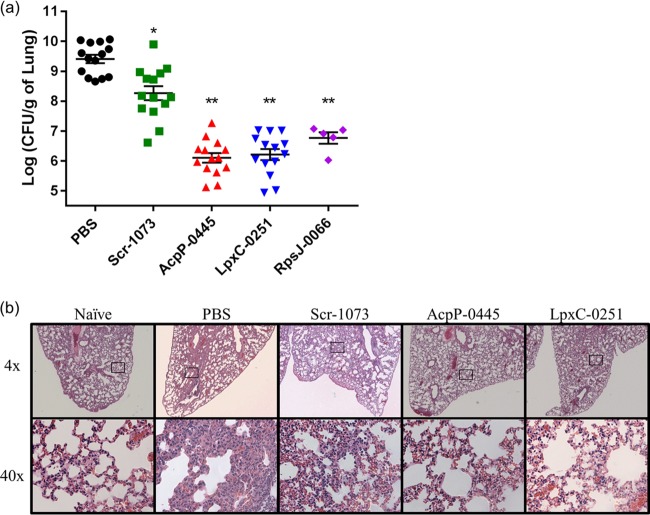
The therapeutic potential of PPMOs was further assessed by an in vivo model of acute pneumonia. BALB/c mice were infected by intratracheal instillation with P. aeruginosa PA103 and treated 6 h postinfection with PPMOs. Intranasal treatment of infected mice with AcpP-0445, LpxC-0251, or RpsJ-0066 significantly reduced the CFU in the lungs of infected mice by 24 h compared to phosphate-buffered saline (PBS) or Scramble PPMO controls (Fig. 4a). Interestingly, bacterial burden in Scramble PPMO-treated mice was intermediate to PBS and targeted PPMOs. In addition, histological examinations of lungs treated with targeted PPMOs showed a reduction in inflammation and cellular infiltrate (Fig. 4b). These data suggest that PPMOs have therapeutic potential in vivo.
FIG 4.
PPMOs decreased P. aeruginosa burden in the lungs of infected mice. BALB/c mice were infected intratracheal with P. aeruginosaPA103 and treated with PPMOs or PBS control at 6 h postinfection. (a) The CFU/g of whole lung were analyzed. Statistics were determined with a one-way ANOVA and a Holm-Ŝidák's multiple-comparison test where P < 0.001 (**) compared to PBS and Scr-1073 controls and P < 0.001 (*) compared to PBS control. (b) H&E stains of the left distal lungs of infected mice treated as indicated compared to a naive control. The magnification in the top row is at ×4, with black boxes to indicate the regions pictured at ×40 magnification below. These data are combined from three independent experiments, except for RpsJ-0066, which was one experiment.
DISCUSSION
Antibiotic resistance is on the rise; therefore, new antibiotic development is desperately needed. Targeting P. aeruginosa is particularly challenging given that it is intrinsically more resistant to numerous antibiotics compared to other Gram-negative bacteria due to a lower abundance of general diffusion porins and a high abundance of efflux pumps (20). To our knowledge, this is the first study to demonstrate the effectiveness of PPMOs in P. aeruginosa. We demonstrate that at 2× or 4× the MIC, PPMOs were bactericidal with a 3-log reduction (see Fig. S2 in the supplemental material). Critically, within these proof-of-concept studies, we demonstrate additional properties of PPMOs that have not been previously described, such as activity in biofilms and synergy with traditional antibiotics (Fig. 2 and 3).
(RXR)4XB-conjugated PMOs have now been identified as highly effective against P. aeruginosa, E. coli (21), A. baumannii (17), K. pneumoniae (22), and even the eukaryotic parasite T. gondii (23) but are ineffective against Burkholderia cepacia complex (15). In comparing inhibitory PPMO concentrations needed for other pathogens, the concentrations of (RXR)4XB-conjugated PMOs utilized against P. aeruginosa were slightly higher. However, the concentrations needed for inhibition could be enhanced by subinhibitory concentrations of polymyxins, including PMBN and colistin, or culture in cation-limiting media, which increases membrane permeability (Fig. 1; see also Fig. S1b, c, and d in the supplemental material). These data are consistent with the idea that most antimicrobial peptides, including the amphipathic peptide conjugates used here, directly penetrate through the lipid membrane (21).
Our initial screen (data not shown) identified three candidate essential genes (acpP, lpxC, and rpsJ) that were effectively inhibited by (RXR)4XB PPMOs. Acyl carrier protein (acpP) is involved in fatty acid biosynthesis, specifically as a small protein that carries all the needed intermediates in the pathway for membrane synthesis. AcpP-0445 targets a region of acpP that includes the ATG translation start site and, within the NCBI taxon ID 287 for Pseudomonas aeruginosa, is 100% conserved, so a single oligomer can target any strain that might be clinically encountered. We and others have previously identified acpP as a good therapeutic target for PPMOs and peptide nucleic acids (PNAs) in a variety of Gram-negative opportunistic pathogens (15–18, 24, 25).
LpxC-0251 targets the metalloamidase that catalyzes the first committed step in lipid A biosynthesis, which is also involved in membrane formation. Previous work has demonstrated that lpxC is essential in P. aeruginosa (26) and is well conserved in Gram-negative bacteria. Several small-molecule inhibitors of LpxC are in development, as reviewed previously (27), and are effective in vitro against E. coli, K. pneumoniae, and P. aeruginosa but not A. baumannii (A. W. Serio, presented at the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Denver, CO, 10 to 13 September 2013).
Finally, rpsJ encodes the 30S ribosomal protein S10. The ribosomal 30S subunit is a common target of aminoglycoside antibiotics, several of which are commonly used to treat CF, such as tobramycin and amikacin (28). However, the incidence of resistance to aminoglycosides is increasing due to a variety of mechanisms, including enzymatic inactivation of the antibiotic, efflux, and decreased outer membrane permeability (29). Therefore, PPMOs targeting the mRNA of the 30S ribosome subunit components have the potential to circumvent these antibiotic resistance mechanisms since they are enzymatically stable and (RXR)4XB conjugation assists in penetration of the membrane.
These lead PPMOs were highly effective in vivo, decreasing P. aeruginosa lung burden in a mouse model of acute infection by nearly 3 logs at 24 h. Simultaneously, there was an apparent reduction in cellular infiltrate and airway collapse in infected lungs in PPMO-treated mice compared to controls. The lead PPMO, which will ultimately move forward to advanced preclinical development, will be largely based on more extensive in vivo studies.
The ability of P. aeruginosa to form biofilms is clinically important both in patient tissues and on abiotic surfaces of medical devices. P. aeruginosa embedded in biofilms is inherently more resistant to antibiotics due to a variety of intrinsic properties, including the composition of the biofilm matrix, which may bind certain antibiotics and limit initial diffusion (30), and the reduced metabolic activity of bacteria deep within the matrix (31). A single treatment of PPMO prevented the formation of biofilm, most likely by inhibiting planktonic growth, which seeds the biofilm. Importantly, repeated treatment with PPMO was successful at decreasing the amount of established biofilm, and this effect was enhanced with PMBN. Our data show that PPMOs are effective at penetrating biofilms and suggest that PPMOs could have clinical utility in environments where biofilm formation is a prominent feature of disease. Given that PPMOs are high-molecular-weight molecules, it is intriguing that they would work in this setting. We hypothesize that this effect is related to the cationic nature of the peptide conjugate, and ongoing studies are focused on dissecting the mechanism behind this effect.
Due to the increasing levels of antibiotic resistance in clinical P. aeruginosa infections, it was encouraging to find that PPMOs demonstrated synergy with traditional antibiotics, of which the most effective combination was RpsJ-0066 with tobramycin (FIC indices = 0.26 to 0.29) (Fig. 3). Interestingly, PPMOs demonstrated synergy with antibiotics targeting both similar and dissimilar cellular processes (i.e., both targeting components in the bacterial membrane or one targeting the membrane and the other targeting translation) (Fig. 3). However, the synergy observed between tobramycin and AcpP-0445 or LpxC-0251, which target seemingly dissimilar cellular processes, may be explained by previous observations that aminoglycosides are also able to permeabilize the outer membrane of P. aeruginosa (32). The range and multitude of synergistic interactions suggests PPMO and antibiotic combination therapy could be clinically useful. However, although CF patients are often prescribed multiple antibiotics, there is currently no evidence for improved patient outcomes when using combination in vitro susceptibility testing results (compared to standard single compound susceptibility testing results) to inform clinical therapy for acute pulmonary exacerbation of CF (33, 34).
There are several limitations of this study that are worthy of mention. First, we do not show target specificity of our three lead PPMOs. Studies have demonstrated that PMOs do not bind double-stranded DNA and in cell-free protein synthesis reactions the mechanism of action for PMOs is inhibition of translation (35, 36). It is unclear whether this is due to steric blockade of the actual binding of the ribosome complex or to an inability to continue with translation once the complex binds. We have recently shown that for the nonessential efflux pump AcrA, there is a dose-dependent decrease in AcrA protein when the targeted PPMO is used against E. coli (37). Methods to show decreases in protein levels of our essential gene leads are being developed. Second, we do not address the development of resistance in Pseudomonas to our lead PPMOs. To date, we have not detected mutants with resistance to the (RXR)4XB-linked PMOs. The only example of resistance to PPMOs thus far was shown to be due to the transporter SbmA in E. coli. Interestingly, SbmA mutants were resistant to RFF-linked PMOs but retained activity to the same PMO linked to an RXR-based peptide (38). In addition, PNAs have been shown to be transported by SbmA in E. coli; however, certain peptide-conjugated PNAs, including (RXR)4XB, enter through an SbmA-independent mechanism (39). Other potential mechanisms of resistance for PPMOs could be related to sequence-related changes of the target and efflux of the compound. Studies are ongoing to generate (RXR)4XB-resistance-related mutants and determine the mechanism and frequency of resistance. Third, our in vivo experiments, as well as other prior studies, show a mild but measurable effect of the control (Scrambled) PPMO despite a lack of in vitro activity. It is unclear whether this is related to nonspecific immune activation of the peptide itself; however, the recently reported scavenger receptor binding of PPMOs in eukaryotic cells (40) could make this a possible explanation. Recent studies by our group show that, at least in a sepsis infection model, the control PPMO acts similarly to a PBS control in terms of serum proinflammatory cytokine stimulation, and only the active PPMO significantly reduced serum proinflammatory cytokines (interleukin-6 and tumor necrosis factor alpha), presumably due to pathogen inhibition. Again, more extensive in vivo studies will better assess the cellular response to PPMO administration in a pulmonary setting.
The recent approval of the PMO-based Eteplirsen and the previously approved antisense therapeutics mipomersen, fomivirsen, and pegaptanib suggest that antisense therapeutics utilizing multiple chemistries are viable for development. In our experiments, PPMOs were delivered by intranasal administration to avoid any systemic PK/PD issues and to demonstrate inherent efficacy at the site of infection prior to studies of antibiotic synergy with currently aerosolized antibiotics. Further, direct administration of a PPMO to the lung is a clinically viable delivery approach, as has been done with tobramycin (6). Prior in vivo PK/PD analyses with (RXR)4XB-conjugated 20- to 24-mers (11-mers are used in this study) suggests that PPMOs were well tolerated in mice and rats at doses of ≤30 mg/kg and that (RXR)4XB conjugation increased the tissue uptake of PPMO compared to PMO, while prolonging the elimination half-life (41, 42). In addition, a single dose (300 μg; ∼15 mg/kg) of PPMOs used in our mouse experiments was well within the dose (30 to 50 mg/kg/week) of PMOs (Eteplirsen) used in phase II clinical trials, which was well tolerated (43).
MDR strains of P. aeruginosa routinely complicate the clinical treatment of infections in immunocompromised patients. Consequently, the current paradigm in the antimicrobial treatment of MDR strains often must rely on antibiotics that present additional toxicity to patients and increases the risk of developing extensively resistant or panresistant strains. Our data represent a novel approach for the treatment of P. aeruginosa infection. PPMOs (i) inhibited a large panel of clinical strains, (ii) are bactericidal, (iii) prevent and reduce biofilm, (iv) act synergistically with currently available antibiotics, and (v) reduce bacterial burden in vivo. Notably, similar 5′(RXR)4XB-conjugated PPMOs had no toxicity in cell culture (37) and demonstrated low toxicity at therapeutic doses in rodents (41, 42, 44, 45). PPMOs are a promising choice for future clinical investigations since they have now demonstrated efficacy in many Gram-negative genera.
MATERIALS AND METHODS
All MIC determinations were performed in 96-well tissue culture plates (Thermo Fisher Scientific, Waltham, MA). All reagents were of analytical grade and were obtained from Sigma-Aldrich (St. Louis, MO), Thermo Fisher Scientific (Waltham, MA), or Acros Organics (Geel, Belgium). MHII was obtained from Becton Dickinson & Company (Franklin Lakes, NJ). Morpholinopropanesulfonic acid (MOPS) minimal medium was made according to the method of Neidhardt et al. with 1 g/liter glucose and 100 μg/liter thiamine (46). Colistin (colistimethate) and polymyxin B nonapeptide (PMBN; Sigma-Aldrich) were used to enhance cellular uptake, where indicated, at 1 and 2 μg/ml, respectively.
Bacterial strains.
The P. aeruginosa strains used in this study were obtained from the American Type Culture Collection and are clinical isolates (the strains and sources are listed in Table S1 in the supplemental material). Green fluorescent protein (GFP)-expressing PAO1 (pSMC2 encoding GFP) was previously described (47).
PPMOs.
PMO sequences were designed using a custom webtool that has inputs for a taxon ID, gene, alignment region, and oligomer length. We designed PMOs to target either the Shine-Dalgarno or translation start sites (NTG), while having the lowest off-target hits in the start sites of other genes. Peptide-conjugated phosphorodiamidate morpholino oligomers (PPMOs), all of which are 11-mers, were synthesized by Sarepta Therapeutics (Cambridge, MA). PPMO gene targets, sequences, peptides, and peptide conjugation sites (5′ versus 3′) are indicated in Table S2 in the supplemental material. Scrambled (Scr) PPMO controls are 11-mer sequences conjugated to the same peptides and in the same orientation as active PPMOs; however, they are not complementary to the essential gene target (16, 18).
Bacterial susceptibility testing.
The MICs of the PPMOs were determined according to the Clinical and Laboratory Standards Institute (CLSI) broth microdilution method, with minor modifications. Briefly, each bacterial strain was diluted to a final concentration of approximately 5 × 105 CFU/ml in MHII or MOPS medium, and the PPMO was serially diluted 2-fold from 16 to 0.5 μM in a 96-well tissue culture plate. The plates were then covered with gas permeable membrane strips (MIDSCI, St. Louis, MO) and incubated at 37°C and 225 rpm in a shaking incubator for 18 to 20 h. The optical densities at 600 nm (OD600s) were measured using a microtiter plate reader and the lowest dose of PPMO at which the average OD600 that measured ≤0.06 was recorded as the MIC. Three independent experiments were performed for each PPMO. PPMOs were tested across multiple strains of P. aeruginosa to determine the MIC50 of each PPMO. The MIC50 was defined as the minimum concentration at which 50% of the strains were inhibited. The minimum bactericidal concentration (MBC), the concentration that results in a 3-log reduction in CFU/ml, was determined by plating MIC assays for enumeration. After the MIC assay OD600 measurement, wells containing 0.5×, 1×, 2×, and sometimes 4× the MIC were serially diluted and plated for CFU enumeration. If no detectable colonies were observed, the limit of detection was (1.3 log10 CFU/ml).
Biofilm assays.
Biofilm prevention assays were performed in 96-well minimum biofilm eradication concentration (MBEC) plates (Innovotech, Inc., Edmonton, Alberta, Canada). These plates have special lids with 96 pegs that dip into the growth media in the wells, allowing biofilm to form on the peg surface. P. aeruginosa PAO1-GFP was inoculated at 5 × 105 CFU/ml in 150 μl of filtered MHII per well with or without PMBN at 2 μg/ml, followed by incubation at 37°C and 110 rpm. In prevention assays, PPMO was added immediately after inoculation. For breakdown assays, plate lids containing pegs with formed biofilms were moved to a new 96-well plate with fresh media treated with control or active PPMOs at 24, 32, and 40 h postinoculation. This method was chosen to avoid medium exhaustion and to model the repeated dosing schedules often utilized in clinical settings. Biofilm growth on the peg was quantified utilizing a crystal violet assay. For analysis at 24 h (prevention) or 48 h (breakdown), the lids containing the pegs were removed from the 96-well plate and processed by placing the lid into new 96-well plates in the following sequence: 1 min in 150 mM NaCl to remove any nonadherent planktonic bacteria, 15 min in 100% methanol for fixation, air dried for a minimum of 3 h, stained for 20 min in an 80 μM crystal violet solution with gentle rocking, and then placed in 100% acetic acid for 10 min to elute the crystal violet. The OD of the eluted stain was then measured at 570 nm for quantitation of the biofilm.
Spinning disk confocal microscopy.
For both biofilm assays described above, pegs from separate, unstained MBEC plates were imaged by spinning disk confocal microscopy. After incubation, the lids were washed in 150 mM NaCl as described above and then fixed in 5% glutaraldehyde at 37°C for 30 min. After fixation, the pegs were cleaved from the plate with a hot scalpel and stained with concanavalin A-Alexa Fluor 647 conjugate (Life Technologies, Grand Island, NY) at 200 μg/ml. The pegs were imaged using a 40× objective lens (Nikon; oil immersion; numerical aperture, 1.3) on an Axiovert 200M inverted microscope (Carl Zeiss, Thornwood, NJ) equipped with an UltraVIEWERS spinning-disk confocal head (Perkin-Elmer, Waltham, MA) and obtained with an EM-CCD C9100 (Hamamatsu, Bridgewater, NJ). The acquired images were deconvoluted using AutoQuant X3 (Media Cybernetics, Inc., Rockville, MD) and rendered with Imaris 7.2 (Bitplane USA, Concord, MA).
Synergy assays.
Checkerboard synergy assays, originally described by Bourque et al. (48), were performed according to CLSI guidelines similarly to the MIC determinations in MHII media. Antibiotics were diluted 2-fold vertically in a 96-well plate and then PPMOs were diluted 2-fold horizontally. Each plate contained antibiotic- and PPMO-only MIC controls, as well as positive- and negative-growth controls. The plates were inoculated with bacteria at a final concentration of 5 × 105 CFU/ml, followed by incubation for 18 h. Inhibition of growth in synergy and MIC wells was determined visually with comparison to medium control wells. Each synergy combination was assayed in triplicate and represented as an isobologram and as the FIC index, where FIC = [(MICantibiotic in combination/MICantibiotic alone) + (MICPPMO in combination/MICPPMO alone)]. The FIC index was considered synergistic when ≤ 0.5 (49, 50).
Mouse experiments.
Six-week-old female BALB/c mice (Jackson Laboratories, Bar Harbor, ME) were anesthetized with a cocktail of ketamine (25 mg/ml) and xylazine (12 mg/ml) and infected by noninvasive intratracheal instillation as previously described (51) with 3 × 104 CFU (25 μl) of P. aeruginosa PA103. At 6 h postinfection, the mice were anesthetized by isoflurane and treated intranasally with 300 μg of the indicated PPMOs or PBS in a 25-μl volume. Mice were euthanized at 24 h postinfection, and whole lungs were taken for CFU enumeration or histology. Lungs used for CFU determination were weighed and homogenized for 10 s in 1 ml of PBS, followed by serial dilution and plating for CFU enumeration. Lungs used for histology were fixed in 4% formalin overnight before transfer to PBS, embedded in paraffin, and sectioned for hematoxylin and eosin staining (H&E). Histology images were taken on a Leica DM2000 upright compound research photomicroscope using a bright field at ×4 and ×40 magnifications. All experimental procedures were carried out with the approval from the Institutional Animal Care and Use Committee (IACUC) at Emory University (Atlanta, GA).
Supplementary Material
ACKNOWLEDGMENTS
We thank our collaborators at Sarepta Therapeutics for their generous contribution of the PPMOs used in these experiments. B.L.G. is a consultant to Sarepta Therapeutics and an inventor on numerous patents and patent applications involving PPMOs. M.W. and S.M.B. are employees of Sarepta Therapeutics that hold numerous patents on the methods of synthesis and use of PPMOs. D.E.G. receives research support from Sarepta Therapeutics and is an inventor on numerous patent applications involving PPMOs. B.L.G. and D.E.G. receive royalties related to their patents. For all other authors, there no conflicts to declare.
This study was funded by the National Institutes of Health (AI105980 [D.E.G.] and AI111753 [D.E.G. and B.L.G.]). We acknowledge the assistance of the UT Southwestern Live Cell Imaging Facility, a Shared Resource of the Harold C. Simmons Cancer Center, supported in part by the National Cancer Institute at the National Institutes of Health (1P30 CA142543-01).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01938-16.
REFERENCES
- 1.Lambert PA. 2002. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med 95:22–26. [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson DH, Krikstopaityte V, Coote PJ. 2015. Enhanced efficacy of putative efflux pump inhibitor/antibiotic combination treatments versus MDR strains of Pseudomonas aeruginosa in a Galleria mellonella in vivo infection model. J Antimicrob Chemother 70:2271–2278. doi: 10.1093/jac/dkv111. [DOI] [PubMed] [Google Scholar]
- 3.Ahya VN, Doyle AM, Mendez JD, Lipson DA, Christie JD, Blumberg EA, Pochettino A, Nelson L, Bloom RD, Kotloff RM. 2005. Renal and vestibular toxicity due to inhaled tobramycin in a lung transplant recipient. J Heart Lung Transplant 24:932–935. doi: 10.1016/j.healun.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Edson RS, Brey RH, McDonald TJ, Terrell CL, McCarthy JT, Thibert JM. 2004. Vestibular toxicity due to inhaled tobramycin in a patient with renal insufficiency. Mayo Clin Proc 79:1185–1191. doi: 10.4065/79.9.1185. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann IM, Rubin BK, Iskandar SS, Schechter MS, Nagaraj SK, Bitzan MM. 2002. Acute renal failure in cystic fibrosis: association with inhaled tobramycin therapy. Pediatr Pulmonol 34:375–377. doi: 10.1002/ppul.10185. [DOI] [PubMed] [Google Scholar]
- 6.Canton R, Cobos N, de Gracia J, Baquero F, Honorato J, Gartner S, Alvarez A, Salcedo A, Oliver A, Garcia-Quetglas E; Spanish Consensus Group for Antimicrobial Therapy in the Cystic Fibrosis. 2005. Antimicrobial therapy for pulmonary pathogenic colonization and infection by Pseudomonas aeruginosa in cystic fibrosis patients. Clin Microbiol Infect 11:690–703. doi: 10.1111/j.1469-0691.2005.01217.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Zhang Q. 2015. Clinical efficacy and safety of colistin treatment in patients with pulmonary infection caused by Pseudomonas aeruginosa or Acinetobacter baumannii: a meta-analysis. Arch Med Sci 11:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saderi H, Owlia P. 2015. Detection of multidrug-resistant (MDR) and extremely drug resistant (XDR) P. aeruginosa isolated from patients in Tehran, Iran. Iran J Pathol 10:265–241. [PMC free article] [PubMed] [Google Scholar]
- 9.Johansen HK, Moskowitz SM, Ciofu O, Pressler T, Hoiby N. 2008. Spread of colistin resistant non-mucoid Pseudomonas aeruginosa among chronically infected Danish cystic fibrosis patients. J Cyst Fibros 7:391–397. doi: 10.1016/j.jcf.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Murray L, Barclay EJB, Stephen T, Chambers Peter E, Thornley Philip K, Pattemore Keith Grimwood. 1996. Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J Antimicrob Chemother 37:1155–1164. doi: 10.1093/jac/37.6.1155. [DOI] [PubMed] [Google Scholar]
- 11.Parkins MD, Floto RA. 2015. Emerging bacterial pathogens and changing concepts of bacterial pathogenesis in cystic fibrosis. J Cystic Fibrosis 14:293–304. doi: 10.1016/j.jcf.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Coburn B, Wang PW, Diaz-Caballero J, Clark ST, Brahma V, Donaldson S, Zhang Y, Surendra A, Gong Y, Tullis ED, Yau YCW, Waters VJ, Hwang DM, Guttman DS. 2015. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 5:10241. doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudziak RM, Barofsky E, Barofsky DF, Weller DL, Huang SB, Weller DD. 1996. Resistance of morpholino oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev 6:267–272. doi: 10.1089/oli.1.1996.6.267. [DOI] [PubMed] [Google Scholar]
- 14.Wu RP, Youngblood DS, Hassinger JN, Lovejoy CE, Nelson MH, Iversen PL, Moulton HM. 2007. Cell-penetrating peptides as transporters for morpholino oligomers: effects of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res 35:5182–5191. doi: 10.1093/nar/gkm478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg DE, Marshall-Batty KR, Brinster LR, Zarember KA, Shaw PA, Mellbye BL, Iversen PL, Holland SM, Geller BL. 2010. Antisense phosphorodiamidate morpholino oligomers targeted to an essential gene inhibit Burkholderia cepacia complex. J Infect Dis 201:1822–1830. doi: 10.1086/652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geller BL, Deere JD, Stein DA, Kroeker AD, Moulton HM, Iversen PL. 2003. Inhibition of gene expression in Escherichia coli by antisense phosphorodiamidate morpholino oligomers. Antimicrob Agents Chemother 47:3233–3239. doi: 10.1128/AAC.47.10.3233-3239.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geller BL, Marshall-Batty K, Schnell FJ, McKnight MM, Iversen PL, Greenberg DE. 2013. Gene-silencing antisense oligomers inhibit acinetobacter growth in vitro and in vivo. J Infect Dis 208:1553–1560. doi: 10.1093/infdis/jit460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tilley LD, Hine OS, Kellogg JA, Hassinger JN, Weller DD, Iversen PL, Geller BL. 2006. Gene-specific effects of antisense phosphorodiamidate morpholino oligomer-peptide conjugates on Escherichia coli and Salmonella enterica serovar Typhimurium in pure culture and in tissue culture. Antimicrob Agents Chemother 50:2789–2796. doi: 10.1128/AAC.01286-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dryselius R, Nekhotiaeva N, Good L. 2005. Antimicrobial synergy between mRNA- and protein-level inhibitors. J Antimicrob Chemother 56:97–103. doi: 10.1093/jac/dki173. [DOI] [PubMed] [Google Scholar]
- 20.Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellbye BL, Puckett SE, Tilley LD, Iversen PL, Geller BL. 2009. Variations in amino acid composition of antisense peptide-phosphorodiamidate morpholino oligomer affect potency against Escherichia coli in vitro and in vivo. Antimicrob Agents Chemother 53:525–530. doi: 10.1128/AAC.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sully EK, Geller BL, Li L, Moody CM, Bailey SM, Moore AL, Wong M, Nordmann P, Daly SM, Sturge CR, Greenberg DE. 2016. Peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO) restores carbapenem susceptibility to NDM-1-positive pathogens in vitro and in vivo. J Antimicrob Chemother 2016 Dec 20 pii:dkw476. doi: 10.1093/jac/dkw476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai BS, Witola WH, El Bissati K, Zhou Y, Mui E, Fomovska A, McLeod R. 2012. Molecular target validation, antimicrobial delivery, and potential treatment of Toxoplasma gondii infections. Proc Natl Acad Sci U S A 109:14182–14187. doi: 10.1073/pnas.1208775109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maekawa K, Azuma M, Okuno Y, Tsukamoto T, Nishiguchi K, Setsukinai K, Maki H, Numata Y, Takemoto H, Rokushima M. 2015. Antisense peptide nucleic acid-peptide conjugates for functional analyses of genes in Pseudomonas aeruginosa. Bioorg Med Chem 23:7234–7239. doi: 10.1016/j.bmc.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 25.Ghosal A, Nielsen PE. 2012. Potent antibacterial antisense peptide-peptide nucleic acid conjugates against Pseudomonas aeruginosa. Nucleic Acid Ther 22:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mdluli KE, Witte PR, Kline T, Barb AW, Erwin AL, Mansfield BE, McClerren AL, Pirrung MC, Tumey LN, Warrener P, Raetz CR, Stover CK. 2006. Molecular validation of LpxC as an antibacterial drug target in Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:2178–2184. doi: 10.1128/AAC.00140-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalinin DV, Holl R. 2016. Insights into the zinc-dependent deacetylase LpxC: biochemical properties and inhibitor design. Curr Top Med Chem 16:2379–2430. doi: 10.2174/1568026616666160413135835. [DOI] [PubMed] [Google Scholar]
- 28.Ratjen F, Brockhaus F, Angyalosi G. 2009. Aminoglycoside therapy against Pseudomonas aeruginosa in cystic fibrosis: a review. J Cyst Fibros 8:361–369. doi: 10.1016/j.jcf.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Sobel ML, McKay GA, Poole K. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 47:3202–3207. doi: 10.1128/AAC.47.10.3202-3207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bordi C, de Bentzmann S. 2011. Hacking into bacterial biofilms: a new therapeutic challenge. Ann Intensive Care 1:19. doi: 10.1186/2110-5820-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor PK, Yeung AT, Hancock RE. 2014. Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol 191:121–130. doi: 10.1016/j.jbiotec.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Loh B, Grant C, Hancock RE. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother 26:546–551. doi: 10.1128/AAC.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elphick HE, Jahnke N. 2014. Single versus combination intravenous antibiotic therapy for people with cystic fibrosis. Cochrane Database Syst Rev 2014:CD002007. doi: 10.1002/14651858.CD002007.pub3. [DOI] [PubMed] [Google Scholar]
- 34.Waters V, Ratjen F. 2015. Combination antimicrobial susceptibility testing for acute exacerbations in chronic infection of Pseudomonas aeruginosa in cystic fibrosis. Cochrane Database Syst Rev 2015:CD006961. doi: 10.1002/14651858.CD006961.pub3. [DOI] [PubMed] [Google Scholar]
- 35.Summerton J, Stein D, Huang SB, Matthews P, Weller D, Partridge M. 1997. Morpholino and phosphorothioate antisense oligomers compared in cell-free and in-cell systems. Antisense Nucleic Acid Drug Dev 7:63–70. doi: 10.1089/oli.1.1997.7.63. [DOI] [PubMed] [Google Scholar]
- 36.Deere J, Iversen P, Geller BL. 2005. Antisense phosphorodiamidate morpholino oligomer length and target position effects on gene-specific inhibition in Escherichia coli. Antimicrob Agents Chemother 49:249–255. doi: 10.1128/AAC.49.1.249-255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayhan DH, Tamer YT, Akbar M, Bailey SM, Wong M, Daly SM, Greenberg DE, Toprak E. 2016. Sequence-specific targeting of bacterial resistance genes increases antibiotic efficacy. PLoS Biol 14:e1002552. doi: 10.1371/journal.pbio.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puckett SE, Reese KA, Mitev GM, Mullen V, Johnson RC, Pomraning KR, Mellbye BL, Tilley LD, Iversen PL, Freitag M, Geller BL. 2012. Bacterial resistance to antisense peptide phosphorodiamidate morpholino oligomers. Antimicrob Agents Chemother 56:6147–6153. doi: 10.1128/AAC.00850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosal A, Vitali A, Stach JE, Nielsen PE. 2013. Role of SbmA in the uptake of peptide nucleic acid (PNA)-peptide conjugates in Escherichia coli. ACS Chem Biol 8:360–367. doi: 10.1021/cb300434e. [DOI] [PubMed] [Google Scholar]
- 40.Ezzat K, Aoki Y, Koo T, McClorey G, Benner L, Coenen-Stass A, O'Donovan L, Lehto T, Garcia-Guerra A, Nordin J, Saleh AF, Behlke M, Morris J, Goyenvalle A, Dugovic B, Leumann C, Gordon S, Gait MJ, El-Andaloussi S, Wood MJ. 2015. Self-assembly into nanoparticles is essential for receptor mediated uptake of therapeutic antisense oligonucleotides. Nano Lett 15:4364–4373. doi: 10.1021/acs.nanolett.5b00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein DA, Huang CY, Silengo S, Amantana A, Crumley S, Blouch RE, Iversen PL, Kinney RM. 2008. Treatment of AG129 mice with antisense morpholino oligomers increases survival time following challenge with dengue 2 virus. J Antimicrob Chemother 62:555–565. doi: 10.1093/jac/dkn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amantana A, Moulton HM, Cate ML, Reddy MT, Whitehead T, Hassinger JN, Youngblood DS, Iversen PL. 2007. Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide-morpholino oligomer conjugate. Bioconjug Chem 18:1325–1331. doi: 10.1021/bc070060v. [DOI] [PubMed] [Google Scholar]
- 43.Mendell JR, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, Lowes LP, Alfano L, Gomez AM, Lewis S, Kota J, Malik V, Shontz K, Walker CM, Flanigan KM, Corridore M, Kean JR, Allen HD, Shilling C, Melia KR, Sazani P, Saoud JB, Kaye EM; Eteplirsen Study Group. 2013. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol 74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 44.Yin H, Moulton HM, Seow Y, Boyd C, Boutilier J, Iverson P, Wood MJ. 2008. Cell-penetrating peptide-conjugated antisense oligonucleotides restore systemic muscle and cardiac dystrophin expression and function. Hum Mol Genet 17:3909–3918. doi: 10.1093/hmg/ddn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jearawiriyapaisarn N, Moulton HM, Buckley B, Roberts J, Sazani P, Fucharoen S, Iversen PL, Kole R. 2008. Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice. Mol Ther 16:1624–1629. doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bloemberg GV, O'Toole GA, Lugtenberg BJ, Kolter R. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol 63:4543–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourque M, Quintiliani R, Tilton RC. 1976. Synergism of cefazolin-gentamicin against enterococci. Antimicrob Agents Chemother 10:157–163. doi: 10.1128/AAC.10.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elion GB, Singer S, Hitchings GH. 1954. Antagonists of nucleic acid derivatives. VIII. Synergism in combinations of biochemically related antimetabolites. J Biol Chem 208:477–488. [PubMed] [Google Scholar]
- 50.Parsley TL, Provonchee RB, Glicksman C, Zinner SH. 1977. Synergistic activity of trimethoprim and amikacin against gram-negative bacilli. Antimicrob Agents Chemother 12:349–352. doi: 10.1128/AAC.12.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Revelli DA, Boylan JA, Gherardini FC. 2012. A noninvasive intratracheal inoculation method for the study of pulmonary melioidosis. Front Cell Infect Microbiol 2:164. doi: 10.3389/fcimb.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.