ABSTRACT
We report on the coexistence of mcr-1 and blaCTX-M in multidrug-resistant, extended-spectrum β-lactamase-producing Escherichia coli belonging to the sequence type 10 complex isolated from well water in rural China. Raoultella ornithinolytica with blaKPC-2 was also detected in well water from the same area. This study shows that genes coding for resistance to last-resort antibiotics are present in wells in rural China, indicating a potential source of antibiotic resistance.
KEYWORDS: ESBL, antibiotic resistance, mcr-1
TEXT
Extended-spectrum β-lactamase (ESBL)- and carbapenemase-producing members of the family Enterobacteriaceae such as Escherichia coli have spread worldwide both as commensal bacteria and as causative agents of infection in humans and animals (1–3). Carbapenems and polymyxins are regarded as last-resort antibiotics for the treatment of severe infections in humans caused by multidrug-resistant members of the family Enterobacteriaceae (4, 5). However, this paradigm was challenged when the plasmid-mediated colistin resistance gene mcr-1 was reported in members of the family Enterobacteriaceae isolated from humans and food animals (6). E. coli with both mcr-1 and the EBSL-encoding gene blaCTX-M (7–9) and the carbapenemase-encoding gene blaNDM (10, 11) have been isolated from both humans and animals, indicating that resistance to these last-resort antibiotics is emerging. The coexistence of mcr-1 and blaCTX-M in E. coli has also been reported in surface waters such as rivers and ponds in Switzerland and Malaysia (9, 12).
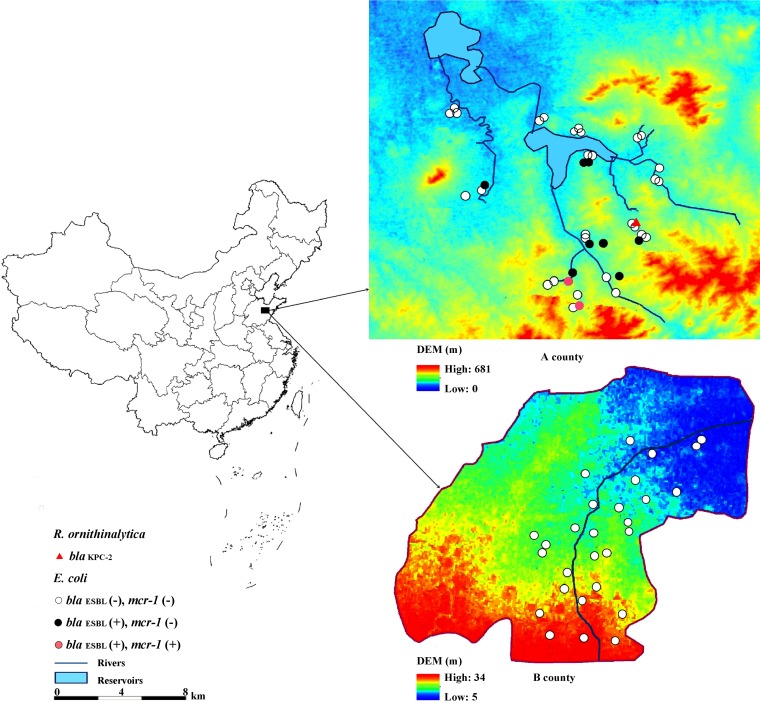
In rural areas of China, well water is the primary source of irrigation water and drinking water for humans and farm animals. In this study, 71 wells were sampled in 12 rural villages of county A (n = 43) and 12 villages in county B (n = 28) in the Shandong province of China during July and September 2015 (Fig. 1). There was a long history of pig breeding in county A and a long history of chicken manure application for intensive vegetable cultivation in county B. About 1,000 ml of well water was collected in sterile bottles and filtered through sterile membrane filters with a pore size of 0.45 μm (Millipore, Billerica, MA). The filters were aseptically put into ESwab tubes (Copan, Brescia, Italy). Each sample was subsequently cultured on chromID ESBL, chromID CARBA, and chromID OXA-48 agar plates (bioMérieux, Marcy l'Etoile, France) for 18 to 24 h at 37°C to identify presumptive ESBL- and/or carbapenemase-producing members of the family Enterobacteriaceae. Identification to the species level was obtained by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Antimicrobial susceptibility profiles were determined by the agar dilution method recommended by the Clinical and Laboratory Standards Institute (CLSI, 2016) and the EUCAST criteria (http://www.eucast.org/clinical_breakpoints/). The mcr-1, blaCTX-M, blaNDM, blaKPC, blaVIM, and blaOXA-48 genes were screened for by PCR (6, 13, 14). PCR products were verified by sequencing (BGI Company, Beijing, China), and amplicons with at least 99% similarity to the mcr-1 amplicon were considered mcr-1. Multilocus sequence typing (MLST) was performed according to the scheme at http://mlst.warwick.ac.uk/mlst/mlst/dbs/Ecoli.
FIG 1.
The genotypes of E. coli and R. ornithinolytica isolates from agricultural counties A and B. DEM, digital elevation model.
Ten ESBL-producing E. coli isolates were detected in a total of 71 samples, all of which came from county A (10/43, 23.3%) (Table 1). The mcr-1 gene was identified in two isolates from two different samples (ECcz1 and ECdw2) that also carried the blaCTX-M-14 and blaCTX-M-65 genes, respectively. Antibiotic susceptibility testing revealed that both strains exhibited multidrug-resistant phenotypes (resistance to more than two antibiotic classes), but both were susceptible to meropenem, amikacin, and tigecycline (Table 1). No carbapenemase genes were detected in the E. coli isolates. MLST showed that isolate ECdw2 belonged to sequence type 10 (ST10) and isolate ECcz1 belonged to ST48 (Table 1). ST48 is part of the ST10 clonal complex and differs from ST10 by only a single nucleotide.
TABLE 1.
Characteristics of ESBL-producing E. coli with mcr-1 and carbapenemase-producing R. ornithinolytica isolated from well water
| Strain | Deptha (m) | MIC (mg/liter)b |
ESBL/carbapenemase type | mcr-1d | MLST | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CL | AMC | TZP | CTX | CAZ | MEMc | GEN | AMK | TE | TGC | CIP | SXT | FOF | F | FFC | |||||
| ECaz1 | 5.0 | 1 | 16 | 32 | >8 | 8 | 0.06 | 4 | 4 | 64 | 0.5 | 0.25 | >16 | >256 | 32 | 4 | CTX-M-55 | − | ST2973 |
| ECaj4 | 4.5 | 2 | 16 | 32 | >8 | 4 | 0.06 | 4 | 4 | 64 | 0.25 | 0.25 | >16 | >256 | 32 | 4 | CTX-M-65 | − | ST2526 |
| ECaw1 | 4.5 | 1 | 16 | 256 | >8 | 1 | 0.06 | 2 | 2 | 64 | 0.25 | 2 | >16 | 128 | >256 | >32 | CTX-M-65 | − | ST2526 |
| ECbw1 | 20.0 | 1 | 16 | 32 | >8 | 32 | 0.06 | 2 | 4 | 64 | 0.5 | 0.25 | >16 | 128 | 128 | 4 | CTX-M-55 | − | ST2973 |
| ECbj3 | 5.0 | 1 | 16 | 32 | >8 | 8 | 0.03 | >32 | >128 | >128 | 2 | >16 | >16 | 256 | 32 | >32 | CTX-M-65 | − | ST1642 |
| ECcz1 | 7.0 | 16 | 16 | 8 | >8 | 8 | 0.06 | >32 | 4 | 64 | 0.5 | 16 | >16 | >256 | 128 | >32 | CTX-M-14 | + | ST48 |
| ECdw2 | 8.0 | 16 | 16 | 32 | >8 | 16 | 0.06 | >32 | 8 | 64 | 0.5 | >16 | >16 | 64 | 256 | >32 | CTX-M-65 | + | ST10 |
| ECdw3 | 30.0 | 1 | 16 | 8 | >8 | 1 | 0.03 | 2 | 2 | 128 | 0.25 | 2 | >16 | 8 | 256 | >32 | CTX-M-27 | − | ST162 |
| ECej3 | 5.0 | 2 | 8 | ≤2 | >8 | 2 | 0.06 | 2 | 4 | 64 | 0.5 | 8 | >16 | >256 | 32 | 4 | CTX-M-65 | − | ST2309 |
| ECiw1 | 20.0 | 2 | 4 | 64 | >8 | 2 | 0.5 | 2 | 4 | 128 | 0.5 | 1 | 0.5 | >256 | >256 | >32 | CTX-M-55 | − | ST4762 |
| ROfj3 | 6.0 | 1 | 8 | 32 | 1 | 16 | 16 | 2 | 4 | 128 | 0.5 | 0.5 | 16 | >256 | 32 | 8 | KPC-2 | − | NAe |
Depth of the well from which the water sample was taken.
MICs were determined by agar dilution methods. Resistance is indicated by boldface type. Abbreviations: CL, colistin; AMC, amoxicillin-clavulanate; TZP, piperacillin-tazobactam; CTX, cefotaxime; CAZ, ceftazidime; MEM, meropenem; GEN, gentamicin; AMK, amikacin; TE, tetracycline; TGC, tigecycline; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; FOF, fosfomycin; F, nitrofurantoin; FFC, florfenicol. Antimicrobial susceptibility profiles were determined by the agar dilution method recommended by the EUCAST (http://www.eucast.org/clinical_breakpoints/), except for tetracycline and florfenicol, for which the Clinical and Laboratory Standards Institute criteria (CLSI, 2016) were applied.
The meropenem epidemiological cutoff for E. coli is ≤0.125 mg/liter. According to EUCAST, a MIC of >0.125 mg/liter is used to detect suspected carbapenemase-producing isolates (http://www.eucast.org/mic_distributions_and_ecoffs/).
Symbols: +, present; −, absent.
NA, not available.
No ESBL-producing E. coli strains were detected in well water samples from county B. However, there are some essential differences between counties A and B that could have influenced the occurrence of resistant bacteria and ESBL-producing E. coli. The well depth in county B (40 to 80 m) was greater than that in county A (4.5 to 30 m), suggesting that depth and temperature can influence the growth or dissemination of resistant bacteria. Shallower wells are also more likely to be influenced by anthropogenic activity. There is also a higher intensity of livestock farming in county A, particularly pig farms, which could have increased the risk of contaminating the shallower wells. Furthermore, in county B, a larger part of the well water is used for irrigation; thus, the wells have a shorter hydraulic retention time, which might influence the dissemination of resistant bacteria.
The two E. coli strains that harbored mcr-1 and blaCTX-M genes in our study belonged to the ST10 complex, which is the most common ST of E. coli detected in human feces in southeastern China (15). The CTX-M ESBL has been reported as the dominant ESBL family in China, with blaCTX-M-14 being the major variant detected in community onset infections in county hospitals across China (16). blaCTX-M-14 is also frequently detected in farm animals, including pigs, and in pets in China. Although blaCTX-M-65 does occur in both humans and pets, it appears to be more common in pigs and broiler chickens (17–20). The other blaCTX-M genes identified in this study (Table 1) are also frequently detected in humans and animals, especially blaCTX-M-55 among clinical isolates. These findings demonstrate the wide spread of blaCTX-M among E. coli isolates from humans, animals, and the environment in China.
Multidrug-resistant E. coli with carbapenemase genes and mcr-1 has previously been reported in clinical samples and constitutes a health risk (10, 21). While none of the E. coli isolates carrying mcr-1 in this study were carbapenemase producers, an isolate of Raoultella ornithinolytica carrying the carbapenemase gene blaKPC-2 was also detected in a well water sample. R. ornithinolytica is a member of the family Enterobacteriaceae, and blaKPC-2-carrying strains of this species have previously been reported in clinical samples in China (22). Although the isolate was not detected in the same wells as the E. coli isolates carrying mcr-1, its presence in the same area and its close relatedness to E. coli indicate a potential risk of co-occurrence and transfer of resistance genes. In rural areas, well water is essential in daily life for activities such as tooth brushing and bathing and as drinking water for humans and pigs. Such activities could potentially increase the risk of dissemination of antibiotic-resistant bacteria from the environment to humans and animals.
As only two mcr-1-positive isolates were detected and well water was the only environmental matrix investigated in this study, generalization of our findings should be done with caution. A limitation of this study is that phenotypic screening for ESBL- and/or carbapenemase-producing members of the family Enterobacteriaceae was performed and thus mcr-1-carrying isolates without either phenotype would have remained undetected. From a One Health perspective, further studies involving more samples from humans, animals, and different environmental matrices are urgently needed to study the dissemination dynamics of mcr-1 and other resistance genes.
ACKNOWLEDGMENTS
Thanks to the IMPACT Consortium for its efforts in this project (https://www.folkhalsomyndigheten.se/impact/). We thank Sun Qiang, Shandong University, for his contributions.
This project was supported by the National Natural Science Foundation of China (grants 81361138021 and 41541013), the Fundamental Research Funds of Shandong University (2015JC011), and the Swedish Research Council, Public Health Agency of Sweden (grant D0879801).
We have no conflicts of interest to declare.
REFERENCES
- 1.Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford PA. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaye KS, Pogue JM, Tran TB, Nation RL, Li J. 2016. Agents of last resort: polymyxin resistance. Infect Dis Clin North Am 30:391–414. doi: 10.1016/j.idc.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Paterson DL, Harris PNA. 2016. Colistin resistance: a major breach in our last line of defence. Lancet Infect Dis 16:132–133. doi: 10.1016/S1473-3099(15)00463-6. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Haenni M, Poirel L, Kieffer N, Châtre P, Saras E, Métayer V, Dumoulin R, Nordmann P, Madec J-Y. 2016. Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis 16:281–282. doi: 10.1016/S1473-3099(16)00007-4. [DOI] [PubMed] [Google Scholar]
- 8.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zurfuh K, Poirel L, Nordmann P, Nuesch-Inderbinen M, Hachler H, Stephan R. 2016. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-beta-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother 60:2594–2595. doi: 10.1128/AAC.00066-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng B, Dong H, Xu H, Lv J, Zhang J, Jiang X, Du Y, Xiao Y, Li L. 2016. Coexistence of MCR-1 and NDM-1 in clinical Escherichia coli isolates. Clin Infect Dis 63:1393–1395. doi: 10.1093/cid/ciw553. [DOI] [PubMed] [Google Scholar]
- 11.Delgado-Blas JF, Ovejero CM, Abadia-Patino L, Gonzalez-Zorn B. 2016. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother 60:6356–6358. doi: 10.1128/AAC.01319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu CY, Ang GY, Chin PS, Ngeow YF, Yin WF, Chan KG. 2016. Emergence of mcr-1-mediated colistin resistance in Escherichia coli in Malaysia. Int J Antimicrob Agents 47:504–505. doi: 10.1016/j.ijantimicag.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Batchelor M, Hopkins K, Threlfall EJ, Cliftonhadley FA, Stallwood AD, Davies RH, Liebana E. 2005. bla(CTX-M) genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob Agents Chemother 49:1319–1322. doi: 10.1128/AAC.49.4.1319-1322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Chen X, Zheng S, Yu F, Kong H, Yang Q, Cui D, Chen N, Lou B, Li X, Tian L, Yang X, Xie G, Dong Y, Qin Z, Han D, Wang Y, Zhang W, Tang YW, Li L. 2014. Serotypes, genotypes and antimicrobial resistance patterns of human diarrhoeagenic Escherichia coli isolates circulating in southeastern China. Clin Microbiol Infect 20:52–58. doi: 10.1111/1469-0691.12188. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Zheng B, Zhao L, Wei Z, Ji J, Li L, Xiao Y. 2014. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis 14:659. doi: 10.1186/s12879-014-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Zeng Z, Chen S, Ma J, He L, Liu Y, Deng Y, Lei T, Zhao J, Liu JH. 2010. High prevalence of bla CTX-M extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin Microbiol Infect 16:1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Zhai Z, Li Q, Liu L, Guo S, Li Q, Yang L, Ye C, Chang W, Zhai J. 2016. Characterization of extended-spectrum β-lactamase–producing Escherichia coli isolates from pigs and farm workers. J Food Prot 79:1630–1634. doi: 10.4315/0362-028X.JFP-16-093. [DOI] [PubMed] [Google Scholar]
- 19.Gao L, Tan Y, Zhang X, Hu J, Miao Z, Wei L, Chai T. 2015. Emissions of Escherichia coli carrying extended-spectrum β-lactamase resistance from pig farms to the surrounding environment. Int J Environ Res Public Health 12:4203–4213. doi: 10.3390/ijerph120404203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng H, Zeng Z, Chen S, Liu Y, Yao Q, Deng Y, Chen X, Lv L, Zhuo C, Chen Z. 2012. Prevalence and characterisation of CTX-M β-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int J Antimicrob Agents 39:305–310. doi: 10.1016/j.ijantimicag.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Kasbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T. 2016. Colistin resistance gene mcr-1 in extended-spectrum beta-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 22.Zheng B, Zhang J, Ji J, Fang Y, Shen P, Ying C, Lv J, Xiao Y, Li L. 2015. Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob Agents Chemother 59:7086–7089. doi: 10.1128/AAC.01363-15. [DOI] [PMC free article] [PubMed] [Google Scholar]