ABSTRACT
The antimicrobial resistance (AMR) rates and levels recorded for Clostridium difficile are on the rise. This study reports the nature, levels, diversity, and genomic context of the antimicrobial resistance of human C. difficile isolates of the NAPCR1/RT012/ST54 genotype, which caused an outbreak in 2009 and is endemic in Costa Rican hospitals. To this end, we determined the susceptibilities of 38 NAPCR1 isolates to 10 antibiotics from seven classes using Etests or macrodilution tests and examined 31 NAPCR1 whole-genome sequences to identify single nucleotide polymorphisms (SNPs) and genes that could explain the resistance phenotypes observed. The NAPCR1 isolates were multidrug resistant (MDR) and commonly exhibited very high resistance levels. By sequencing their genomes, we showed that they possessed resistance-associated SNPs in gyrA and rpoB and carried eight to nine acquired antimicrobial resistance (AMR) genes. Most of these genes were located on known or novel mobile genetic elements shared by isolates recovered at different hospitals and at different time points. Metronidazole and vancomycin remain the first-line treatment options for these isolates. Overall, the NAPCR1 lineage showed an enhanced ability to acquire AMR genes through lateral gene transfer. On the basis of this finding, we recommend further vigilance and the adoption of improved control measures to limit the dissemination of this lineage and the emergence of more C. difficile MDR strains.
KEYWORDS: Clostridium difficile, NAPCR1, multidrug resistance, comparative genomics
INTRODUCTION
Clostridium difficile has attracted attention as an emerging pathogen on account of the worldwide spread of outbreak-causing strains (1), its changing epidemiology (2), and the growing incidence, severity, and health care costs associated with C. difficile infections (CDI) (3). Indeed, some consider that CDI have surpassed methicillin-resistant Staphylococcus aureus infections as the most common hospital-onset, health care facility-associated infections (4).
Partly because of the rapid spread of successful clones, such as the fluoroquinolone-resistant NAP1 strains with mutations in gyrA (5), but also because of the rich array of conjugative transposons (CTn), transposons (Tn), and mobilizable transposons (mobTn) that characterize some strains of this species (6), the antimicrobial resistance (AMR) rates and levels recorded for C. difficile are increasing around the globe (7–9).
This worrisome issue, along with the recognized role of several antibiotics in the etiology of CDI (10) and the mobility of the gut resistome (11), justifies continuous monitoring of antimicrobial resistance in C. difficile, its etiology, and the mechanisms through which it spreads., To date, these issues have not been explored using whole-genome sequencing in Latin American hospitals.
Along with the epidemic strain NAP1/RT027/ST01, a novel genotype of C. difficile termed NAPCR1/RT012/ST-54 (NAPCR1 variant with ribotype [RT] 012 and sequence type [ST] 54) caused an outbreak of CDI in a major Costa Rican hospital in 2009 (12). NAPCR1 isolates induced a severe clinical presentation, were associated with mortality and recurrence rates comparable to those of NAP1/RT027 strains, and produced a strong inflammatory reaction in animal models (12). These strains continue to circulate in several hospitals of this Central American country (13), and their closest known relative is the multidrug-resistant (MDR) strain CD630, which is also classified as RT012/ST-54 and has been claimed to carry bona fide determinants for tetracycline, erythromycin, daunorubicin, bacitracin, nogalamycin, beta-lactam, tellurite, streptogramin, and lantibiotic resistance (14).
We compared the susceptibilities to 10 antimicrobials of seven pulsed-field gel electrophoresis (PFGE) subtypes of NAPCR1 isolates recovered from patients with diarrhea over the past decade. This phenotypic analysis was followed by a detailed examination of whole-genome sequences (WGS) in order to determine the etiology and genomic context of the resistance phenotypes observed.
RESULTS
Antimicrobial susceptibility profiles.
In Costa Rica, epidemiological surveillance of C. difficile is performed using pulsed-field gel electrophoresis (PFGE). We analyzed 38 NAPCR1 isolates with 10 different PFGE SmaI patterns (Table 1) on account of their epidemic potential, high virulence, and close relationship to CD630 (Fig. 1). Although different resistance levels and patterns were seen across and within PFGE SmaI patterns (Table 2), all these bacteria were resistant to 4 to 7 classes of antimicrobials and were therefore categorized as MDR (Table 2). With a single exception, all isolates were categorized as resistant to clindamycin, ciprofloxacin, levofloxacin, moxifloxacin, and chloramphenicol. Moreover, resistance rates above 85% were recorded for tetracycline (33 of 38 isolates [87%]) and rifampin (31 of 32 isolates [97%]), and 90% of the isolates studied showed diminished susceptibility to linezolid (34/38 [92%]). In contrast, roughly two-thirds of the isolates were susceptible to tigecycline (24/38 [63%]), 4 isolates had diminished susceptibility to vancomycin (11%), and none of the isolates showed resistance to metronidazole (Table 2). Across all isolates and their antimicrobial resistance (AMR) profiles, no obvious patterns of increased resistance over time could be seen in the data set.
TABLE 1.
Clostridium difficile NAPCR1 isolates included in the study
| Isolate | PFGE SmaI pattern (no. of isolates) | Hospitala | Yr(s) of isolation |
|---|---|---|---|
| LIBA-3147 | 442 (1) | HSJD | 2003 |
| LIBA-2994 | 447 (6) | HCG | 2009 |
| LIBA-5701 | HSJD | 2009 | |
| LIBA-5711 | HSJD | 2009 | |
| LIBA-5767 | HCG | 2009 | |
| LIBA-5771 | CENARE | 2009 | |
| LIBA-6281 | HMX | 2011–2012 | |
| LIBA-2784 | 448 (12) | HBC | 2003 |
| LIBA-2954 | HMX | 2009 | |
| LIBA-2991 | HCG | 2010 | |
| LIBA-2993 | HCG | 2010 | |
| LIBA-3125 | HSJD | 2003 | |
| LIBA-3137 | HSJD | 2003 | |
| LIBA-5434 | HBC | 2003 | |
| LIBA-5704 | HSJD | 2009 | |
| LIBA-5707 | HSJD | 2009 | |
| LIBA-5751 | HMX | 2009 | |
| LIBA-5774 | HMX | 2009 | |
| LIBA-6275 | HMX | 2011–2012 | |
| LIBA-3129 | 449 (6) | HSJD | 2003 |
| LIBA-5719 | HSJD | 2009 | |
| LIBA-5750 | HMX | 2009 | |
| LIBA-5755 | HMX | 2009 | |
| LIBA-5772 | HSVP | 2009 | |
| LIBA-6276 | HMX | 2011–2012 | |
| LIBA-5734 | 452 (1) | HSJD | 2009 |
| LIBA-2945 | 487 (4) | CENARE | 2009 |
| LIBA-5763 | HCG | 2009 | |
| LIBA-5769 | HCG | 2009 | |
| LIBA-5770 | HCG | 2009 | |
| LIBA-2992 | 488 (1) | HCG | 2009 |
| LIBA-5761 | 489 (2) | HEB | 2009 |
| LIBA-5762 | HEB | 2009 | |
| LIBA-3145 | 558 (2) | HSJD | 2003 |
| LIBA-6285 | HMX | 2011–2012 | |
| LIBA-3144 | 578 (3) | HSJD | 2003 |
| LIBA-3150 | HSJD | 2003 | |
| LIBA-5436 | HBC | 2003 |
HSJD, San Juan de Dios Hospital; HCG, Calderón Guardia Hospital; CENARE, Centro Nacional de Rehabilitación; HMX, México Hospital; HBC, Blanco Cervantes Hospital; HSVP, San Vicente de Paúl Hospital; HEB, Enrique Baltodano Hospital.
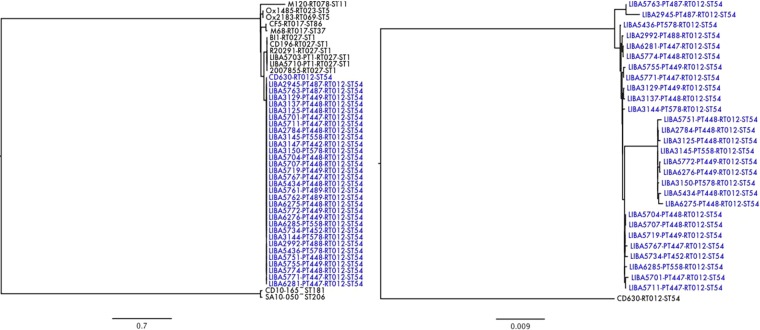
FIG 1.
Despite clustering with CD630 in a core SNP tree that includes representatives of all six C. difficile MLST clades (left), the NAPCR1 isolates (highlighted in blue) can be clearly distinguished from this MDR reference strain (right). SmaI patterns (PT), ribotypes (RT), and sequence types (ST) are given after isolate designations when available. Bars represent the average numbers of substitutions per site.
TABLE 2.
Antibiotic susceptibility profiles of Clostridium difficile NAPCR1 isolates
| Isolate | SmaI pattern | MIC (μg/ml)a |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tetracyclines |
Lincosamides (CLI) | Quinolones |
Nitroimidazoles (MET) | Ansamycins (RIF) | Glycopeptides (VAN) | Oxazolidinones (LIN) | Phenicols (CHL) | |||||
| TET | TIG | CIP | LEV | MOX | ||||||||
| LIBA-3147 | 442 | 0.38 | 0.064 | ≥256 | 256 | 256 | NAb | ≤0.25 | 128 | 1 | 1 | 64 |
| LIBA-2994 | 447 | 64 | 0.5 | ≥256 | 128 | ≥256 | 16 | ≤0.25 | ≥32 | 1 | 8 | ≥256 |
| LIBA-5701 | 64 | 0.064 | ≥256 | ≥32 | ≥32 | ≥32 | 0.19 | NAb | 1 | 1 | 128 | |
| LIBA-5711 | 64 | 0.094 | ≥256 | ≥32 | ≥32 | ≥32 | 0.5 | NA | 2 | 1 | 128 | |
| LIBA-5767 | 64 | 0.125 | ≥256 | 128 | ≥256 | 16 | ≤0.25 | 256 | 1 | 6 | ≥256 | |
| LIBA-5771 | 64 | 0.125 | ≥256 | 128 | ≥256 | 16 | 1 | 128 | 2 | 8 | ≥256 | |
| LIBA-6281 | 64 | 0.125 | ≥256 | ≥32 | ≥32 | ≥32 | ≤0.25 | ≥32 | 0.75 | 8 | ≥256 | |
| LIBA-2784 | 448 | 64 | 0.125 | 8 | ≥32 | ≥32 | ≥32 | 2 | NA | 1 | 8 | ≥256 |
| LIBA-2954 | 32 | 0.25 | ≥256 | 128 | ≥256 | ≥32 | 0.5 | 128 | 4 | 8 | ≥256 | |
| LIBA-2991 | 64 | 0.5 | ≥256 | 128 | ≥256 | 16 | 0.5 | ≥32 | 1 | 8 | ≥256 | |
| LIBA-2993 | 64 | 0.5 | ≥256 | 128 | ≥256 | 16 | ≤0.25 | ≥32 | 0.125 | 8 | ≥256 | |
| LIBA-3125 | 64 | 0.25 | ≥256 | 256 | 256 | ≥32 | ≤0.25 | 128 | 1 | 6 | ≥256 | |
| LIBA-3137 | 64 | 0.125 | 8 | 256 | 256 | ≥32 | ≤0.25 | ≤0.125 | 1 | 8 | >256 | |
| LIBA-5434 | 32 | 0.19 | ≥256 | 256 | 256 | ≥32 | ≤0.25 | 128 | 2 | 6 | 64 | |
| LIBA-5704 | 32 | 0.064 | ≥256 | ≥32 | ≥32 | ≥32 | 0.125 | NA | 1 | 6 | 64 | |
| LIBA-5707 | 64 | 0.094 | ≥256 | ≥32 | ≥32 | ≥32 | 0.75 | NA | 2 | 6 | 128 | |
| LIBA-5751 | 64 | 0.25 | ≥256 | 128 | ≥256 | 16 | ≤0.25 | 128 | 1 | 8 | ≥256 | |
| LIBA-5774 | 96 | 0.25 | ≥256 | 128 | ≥256 | ≥32 | 0.5 | 128 | 4 | 8 | ≥256 | |
| LIBA-6275 | 64 | 0.064 | ≥256 | ≥32 | ≥32 | ≥32 | ≤0.25 | ≥32 | 1.5 | 6 | 128 | |
| LIBA-3129 | 449 | 32 | 0.125 | ≥256 | ≥256 | ≥256 | ≥32 | ≤0.25 | 128 | 2 | 8 | ≥256 |
| LIBA-5719 | 64 | 0.094 | ≥256 | ≥32 | ≥32 | ≥32 | ≤0.25 | NA | 4 | 6 | 128 | |
| LIBA-5750 | 64 | 0.5 | ≥256 | 128 | ≥256 | 16 | 0.5 | 128 | 1 | 8 | ≥256 | |
| LIBA-5755 | 64 | 0.25 | ≥256 | 128 | ≥256 | 16 | 0.5 | 128 | 2 | 8 | ≥256 | |
| LIBA-5772 | 64 | 0.125 | ≥256 | 128 | ≥256 | 16 | 0.5 | 128 | 1 | 8 | ≥256 | |
| LIBA-6276 | 64 | 0.064 | ≥256 | ≥32 | ≥32 | ≥32 | ≤0.25 | ≥32 | 0.75 | 8 | 64 | |
| LIBA-5734 | 452 | 0.25 | 0.064 | ≥256 | ≥32 | ≥32 | ≥32 | ≤0.25 | NA | 4 | 12 | 64 |
| LIBA-2945 | 487 | 64 | 0.094 | ≥256 | ≥32 | ≥32 | ≥32 | 0.5 | >32 | 3 | 8 | 128 |
| LIBA-5763 | 64 | 0.5 | ≥256 | ≥256 | ≥256 | 16 | ≤0.25 | 128 | 0.5 | 8 | ≥256 | |
| LIBA-5769 | 128 | 0.5 | ≥256 | ≥256 | ≥256 | 32 | ≤0.25 | ≥32 | 2 | 2 | ≥256 | |
| LIBA-5770 | 128 | 0.5 | ≥256 | ≥256 | ≥256 | 16 | ≤0.25 | ≥32 | 1 | 4 | ≥256 | |
| LIBA-2992 | 488 | 64 | 0.5 | ≥256 | 128 | ≥256 | 16 | 0.5 | ≥32 | 1 | 8 | ≥256 |
| LIBA-5761 | 489 | 0.5 | 0.094 | ≥256 | 128 | ≥256 | 16 | ≤0.25 | 128 | 1 | 6 | 128 |
| LIBA-5762 | 0.25 | 0.125 | ≥256 | 128 | ≥256 | 16 | ≤0.25 | 128 | 1 | 6 | ≥256 | |
| LIBA-3145 | 558 | 64 | 0.25 | ≥256 | ≥256 | ≥256 | ≥32 | ≤0.25 | 128 | 1 | 6 | ≥256 |
| LIBA-6285 | 1 | 0.064 | ≥256 | ≥32 | ≥32 | ≥32 | ≤0.25 | ≥32 | 0.75 | 8 | 64 | |
| LIBA-3144 | 578 | 32 | 0.125 | ≥256 | ≥256 | ≥256 | ≥32 | ≤0.25 | 128 | 2 | 8 | ≥256 |
| LIBA-3150 | 32 | 0.047 | ≥256 | ≥256 | ≥256 | ≥32 | ≤0.25 | 128 | 1 | 8 | 128 | |
| LIBA-5436 | 32 | 0.19 | ≥256 | ≥256 | ≥256 | ≥32 | ≤0.25 | 16 | 2 | 12 | ≥256 | |
TET, tetracycline; TIG, tigecycline; CLI, clindamycin; CIP, ciprofloxacin; LEV, levofloxacin; MOX, moxifloxacin; MET, metronidazole; RIF, rifampin; VAN, vancomycin; LIN, linezolid; CHL, chloramphenicol. MIC values that indicate resistance or diminished susceptibility are shown in boldface. MICs were obtained with Etests or macrodilution tests.
NA, not assayed.
Genotypic mapping of the observed phenotypic resistance profiles.
Twenty-nine sequenced NAPCR1 isolates with PFGE SmaI pattern 442, 447, 448, 449, 452, 487, 488, 558, or 578 had the gene encoding the ribosomal protection protein TetM carried on Tn5397 and were, accordingly, tetracycline resistant (Table 3). On the other hand, both NAPCR1 isolates with the SmaI macrorestriction pattern 489 studied lacked Tn5397 and were susceptible to tetracycline.
TABLE 3.
Mechanisms and molecular context of the antibiotic resistance of Clostridium difficile NAPCR1 isolates from Costa Rican hospitals
| SmaI pattern(s) (isolate) | Resistance-associated SNPa | Acquired resistance gene(s) | Molecular context of acquired resistance |
|---|---|---|---|
| 442, 448, 449, 452, 447,b 488, 558, 578 | gyrA (Thr82Ile) rpoB (His502Asn Arg505Lys) | tetM | Tn5397 |
| ermB | Tn5398 variant | ||
| aacA-aphD | Tn4001 inserted into putative mobTn | ||
| catD | Tn4453a inserted into CD630-CTn2 and CD630-CTn5 | ||
| ant6–sat4–aphA-3–cfr-like gene | Putative Tn916-like CTn-Ac | ||
| 489 | gyrA (Thr82Ile) rpoB (His502Asn Arg505Lys) | ermB | Tn5398 variant |
| aacA-aphD | Tn4001 inserted into putative mobTn | ||
| catD | Tn4453a inserted into CD630-CTn2 and CD630-CTn5 | ||
| ant6–sat4–aphA-3–cfr-like gene | Putative Tn916-like CTn-Ac | ||
| 487 | gyrA (Thr82Ile) rpoB (His502Asn Arg505Lys) | tetM | Tn5397 |
| ermB | Tn5398 variant | ||
| catD | Tn4453a inserted into CD630-CTn2 and CD630-CTn5 | ||
| Putative bifunctional aac-aph | Phage-like element | ||
| ant6–sat4–aphA-3–cfr-like gene | Putative Tn916-like CTn-Bc | ||
| 447 (LIBA-5701) | gyrA (Thr82Ile) rpoB (His502Asn Arg505Lys) | tetM | Tn5397 |
| ermB | Tn5398 variant | ||
| aacA-aphD | Tn4001 inserted into putative mobTn | ||
| catD | Tn4453a inserted into CD630-CTn2 and CD630-CTn5 | ||
| ant6–sat4–aphA-3 | Putative Tn916-like CTn-Cc |
The MDR strain C. difficile 630 was used as a reference.
Data shown for SmaI pattern 447 here apply to all isolates with this pattern except isolate LIBA-5701.
Three variants of a putative Tn916-like CTn carrying the ant6–sat4–aphA-3 cluster (designated CTn-A, CTn-B, and CTn-C) were seen among the NAPCR1 strains.
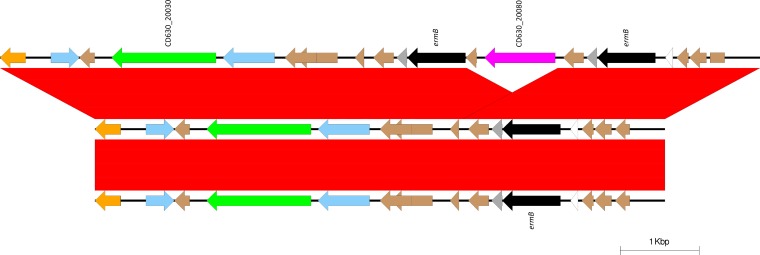
All NAPCR1 isolates carried a single copy of ermB, the product of which confers resistance to clindamycin, inserted into the Tn5398 variant element that is characteristic of C. difficile 630Δerm (Table 3; Fig. 2).
FIG 2.
Relative to strain CD630 (top), the NAPCR1 isolates lost one copy of ermB and a gene coding for a hydrolase (purple) in Tn5398 (center). This variant is characteristic of strain CD630Δerm (bottom). Arrows and arrowheads represent open reading frames. Black arrows represent ermB copies.
All isolates sequenced showed a Thr82Ile substitution in the gyrA gene, encoding DNA gyrase, as well as two rpoB mutations leading to the His502Asn and Arg505Lys substitutions (Table 3). The Thr82Ile mutation correlated with fluoroquinolone resistance and the other two mutations did so with rifampicin resistance.
In addition, all NAPCR1 WGS include two copies of the chloramphenicol acetyltransferase-carrying transposon Tn4453a inserted into homologues of the CD630_04230 and CD630_18620 helicases of CTn2 and CTn5, respectively (Table 3; Fig. 3 and 4). Accordingly, all NAPCR1 isolates are resistant to chloramphenicol.
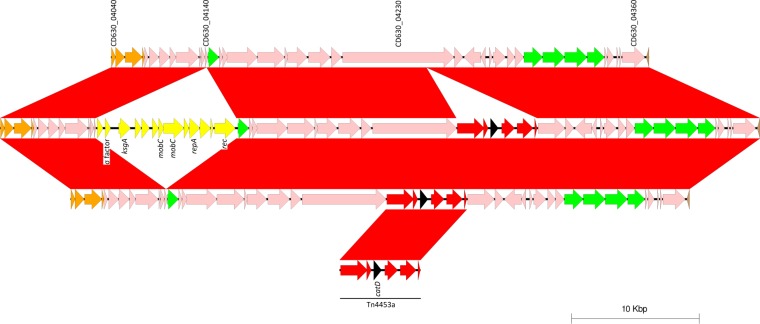
FIG 3.
(Top) Two different variants of the CTn2 of strain CD630 were seen among the NAPCR1 isolates. (Center) In isolates with PFGE SmaI pattern 487, a mobilizable transposon with a putative 16S rRNA methyltransferase gene (ksgA) is inserted into CD630_04140 (yellow arrows and arrowheads). (Bottom) All of the NAPCR1 isolates carry a second copy of Tn4453a (red arrows), and therefore of catD (black arrow), inserted into homologues of the CD630_04230 helicase. mobC, DNA mobilization gene; repA, replication gene; ksgA, gene encoding rRNA methyltransferase; rec, recombinase gene.
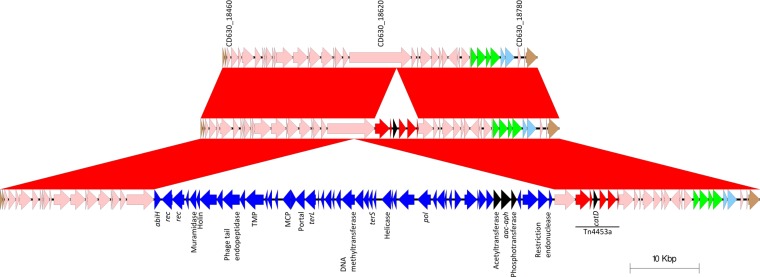
FIG 4.
(Top) Two variants of the CTn5 of strain CD630 were seen among the NAPCR1 isolates. (Center and bottom) All isolates have the catD gene (black arrowhead) of Tn4453a (red arrows and arrowheads) inserted into homologues of the CD630_18620 helicase. In addition, isolates with PFGE SmaI pattern 487 have a phage-like sequence (blue arrows and arrowheads) disrupting the same helicase at another position (bottom). This phage-like element contains a cluster of three putative aminoglycoside resistance genes (black arrowheads), one of which seems to encode a bifunctional Aac-Aph enzyme. abiH, phage abortive infection gene; rec, recombinase gene; TMP, tail tape measure gene; MCP, major capsid; terL, terminase large-subunit gene; terS, terminase small-subunit gene; pol, DNA polymerase gene; aac-aph, genes encoding a bifunctional aminoglycoside acetyltransferase/phosphotransferase.
Other resistance genes and resistance-related mobile genetic elements (MGE).
Although all sequenced NAPCR1 isolates included a vanG-like cluster (data not shown), only a few isolates showed decreased vancomycin susceptibility. This incongruence has been reported previously (15).
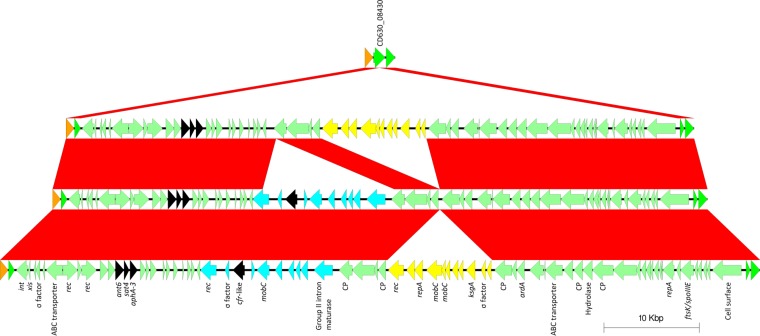
A gene coding for a putative radical SAM protein marginally resembling the rRNA dimethyltransferase Cfr was found, along with the aminoglycoside-streptothricin resistance gene cluster ant6–sat4–aphA-3, in three variants of a Tn916-like element in both linezolid-resistant and linezolid-susceptible NAPCR1 isolates (Table 3; Fig. 5). The largest and most common variant of this MGE was shared by isolates with PFGE SmaI patterns 442, 447, 448, 449, 452, 488, 489, 558, and 578. It included two putative mobTn: one with the cfr-like gene mentioned above and one with a gene showing partial matches to the gene encoding the 16S rRNA methyltransferase KsgA and an adjacent sigma factor (Fig. 5). The second variant was restricted to isolates with PFGE SmaI pattern 487 and lacked the mobTn with the ksgA-like gene (Fig. 5), which, instead, was found inserted into a putative conjugative gene of CTn2 (CD630_04140) (Fig. 3). The third variant did not have the mobTn with the cfr-like gene and was unique to the NAPCR1 isolate with PFGE SmaI pattern 447, LIBA-5701 (Fig. 5).
FIG 5.
The NAPCR1 isolates have three variants of a composite Tn916-like element inserted into the putative group 1 glycosyltransferase CD630_08430 of strain CD630 (top). All three variants include an ant6–sat4–aphA-3 aminoglycoside-streptothricin cluster (array of three black arrowheads). Two variants have a gene coding for a radical SAM enzyme showing partial matches to cfr in a putative mobilizable transposon (single black arrow within blue arrows and arrowheads). One of these also has the mobilizable transposon positive for the putative ksgA shown in Fig. 3 (yellow arrows and arrowheads), as does the variant without the cfr-like gene. The mobilizable transposon containing the ksgA-like gene is inserted into CTn2 in isolates with the PFGE SmaI 487 pattern. int, gene encoding integrase; xis, gene encoding excisionase; rec, gene encoding recombinase; mobC, DNA mobilization gene; CP, conjugative gene; repA, replication gene; ksgA, gene encoding rRNA methyltransferase; ardA, antirestriction gene; ftsK-spoIIIE, DNA transporter gene.
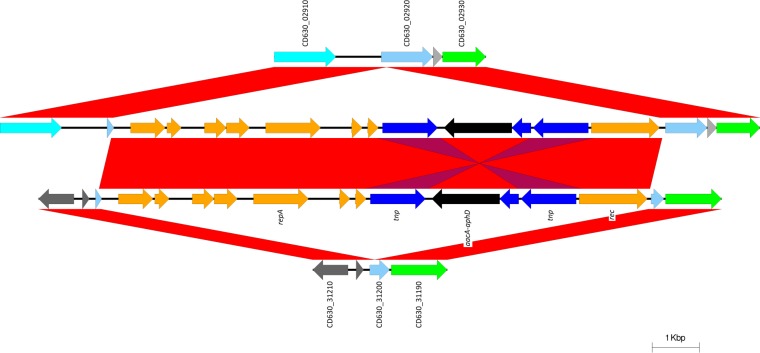
Except for isolates with PFGE SmaI pattern 487, the NAPCR1 WGS possessed two copies of a putative mobTn element inserted into genes homologous to those encoding transcriptional regulators CD630_02920 and CD630_31200 (Table 3; Fig. 6). This novel mobTn includes in its sequence a copy of the composite transposon Tn4001, which encodes a bifunctional aminoglycoside-modifying enzyme (AME) with both acetyltransferase and phosphotransferase functions (aacA-aphD) (Table 3; Fig. 6). The NAPCR1-487 isolates carried another putative bifunctional aminoglycoside-modifying enzyme, but in a phage-like sequence inserted into the CD630_18620 helicase of CTn5 (Fig. 4). The latter bifunctional aminoglycoside-modifying enzyme shows 56% identity to orthologous proteins from Campylobacter jejuni and C. difficile (NCBI protein accession numbers AGV79342.1 and WP_004452859.1) and is flanked by a gene encoding a protein with a GNAT acetyltransferase domain and a phosphotransferase from C. difficile (61% identity to NCBI protein accession number WP_021424056.1).
FIG 6.
Except for isolates with PFGE SmaI pattern 487, the NAPCR1 isolates have two copies of an uncharacterized MGE, each of which is inserted into a homologous gene of the putative transcriptional regulator CD630_02920 (top) or CD630_31200 (bottom). Along with other functions (orange arrows), this putative MGE includes a bifunctional aacA-aphD gene (black arrows) within Tn4001 (blue arrows). repA, replication gene; tnp, transposase-encoding gene; rec, recombinase-encoding gene.
DISCUSSION
We have completed the first genomic study of C. difficile in Costa Rica to determine the underlying diversity of AMR-associated single nucleotide polymorphisms (SNPs) and genes of isolates causing disease. We showed that NAPCR1 isolates are noteworthy not only because of the number of classes of antibiotics to which they are resistant but also because of their high MICs, the numbers of resistance genes detected in their genomes, and the association of these AMR-related genes with multiple transmissible or potentially transmissible MGE, some of which are novel.
Tetracycline resistance in human-associated strains of C. difficile is commonly explained by carriage of Tn5397 or Tn916-like elements that, in addition to tetM variants (16), include ermB (17). In contrast C. difficile strains of swine origin have been shown to be strongly associated with tetW (18). In agreement with their human intestinal origin, those sequenced isolates with phenotypically confirmed tetracycline resistance carried Tn5397.
The widespread clindamycin resistance of C. difficile is often linked to the rRNA adenine N-6-methyltransferase encoded by ermB. The best-known erm+ MGE in this species is the nonconjugative mobTn Tn5398, which contains two ermB copies (19). Laboratory strains positive for Tn5398 may become clindamycin susceptible through the loss of one ermB copy, although reversion of the phenotype may also occur (20). This might be the case for our sequenced isolates, since they naturally lost one ermB copy but retained their phenotypic resistance to clindamycin.
Mutations in the DNA gyrase GyrA or GyrB and in RNA polymerase subunit B (RpoB) confer resistance to fluoroquinolones and rifampin, respectively. Our NAPCR1 isolates share the fluoroquinolone resistance mutation Thr82Ile in GyrA, in addition to the RpoB substitutions Arg505Lys and His502Asn, known to confer high levels of resistance to rifampin on epidemic strains of C. difficile (21). In this regard, it is tempting to speculate that the Thr82Ile mutation in GyrA contributed to the epidemic potential shown by the NAPCR1 strains when they caused an outbreak in 2009.
Although all strains possessed VanG-like sequences, most remained susceptible to vancomycin. The genomic and experimental results at hand indicate that all NAPCR1 isolates share the same slpA allele, but in the future, we aim to address the cell wall structure and antibiotic binding characteristics of these isolates in order to understand the lack of congruence between genotypic and phenotypic data.
With regard to chloramphenicol resistance, this antibiotic is not widely used in human medicine, although different phenicols are used in animal farming (F. García, personal communication). It is not known at present whether animal- and farm-associated isolates of C. difficile from Costa Rica carry Tn4453 or cat genes in other molecular contexts.
The WGS of the isolates sequenced in this study, in addition to carrying genes known to confer resistance to antimicrobials used in clinical therapy, also carried genes conferring resistance to antimicrobials that are not used to treat C. difficile but are implicated in the development of CDI. This finding suggests either that lateral DNA transfer in this lineage is very active and is independent of strong selective pressure or that such resistance is maintained because it is coselected or offers other selective advantages. This is not unexpected, considering that C. difficile may be part of the human gut microbiota. For instance, the rRNA methyltransferase encoded by cfr modifies the 23S rRNA and thereby provides protection against phenicols, lincosamides, pleuromutilins, streptogramin A antibiotics, and selected 16-membered macrolides (22). This gene has been found in Tn6218 in C. difficile (23). However, our NAPCR1 isolates, irrespective of their sensitivity to linezolid, carry a gene showing partial matches to cfr as part of a new Tn916-like element.
Notwithstanding the fact that aminoglycosides are not active against anaerobic bacteria, many NAPCR1 isolates have at least two copies of a bifunctional aminoglycoside-modifying enzyme (AME) with Aac(6′) and Aph(2″) activities. This gene has already been seen in C. difficile as part of a novel family of transposons termed Tn6218 (24). However, we found two copies, each of which was inserted into an element that resembles a mobTn and contains Tn4001. Tn4001 was originally reported in Staphylococcus aureus but was later found in Enterococcus sp., Enterococcus faecalis, Enterococcus faecium, and Staphylococcus epidermidis strains from human guts (25). We found indirect evidence of its mobility in C. difficile. In a few NAPCR1 isolates, another allele for a bifunctional AME was seen in a putative phage as part of a gene array including genes with matches to other types of potential AMEs. This phage shows multiple features of the family Siphoviridae and is more closely related to phage from Enterococcus spp. than to cognates from Clostridium spp. (data not shown), suggesting that the NAPCR1 isolates actively exchange DNA with other intestinal Firmicutes. Although it has been shown that C. difficile phage can mediate the transduction of MGE containing AMR genes (26), if the NAPCR1 phage and its AME are indeed functional, this is the first report of a C. difficile phage with AMR genes.
The NAPCR1 isolates also have the aminoglycoside-streptothricin resistance gene cluster ant6–sat4–aphA-3, which characterizes staphylococci, Campylobacter coli, and E. faecium strains of different origins (27). In most staphylococci, the ant6–sat4–aphA-3 array is integrated into Tn5405 (27). However, our isolates have this array inserted into a putative Tn916-like CTn related to CD630-CTn1. The chimeric nature of this composite element strongly suggests that its mobTn are functional.
In addition to these AMR genes, all of the NAPCR1 isolates carry three genes previously annotated in strain CD630 (14) as encoding a beta-lactamase (CD0458), a beta-lactamase regulatory protein (CD0470), and a beta-lactamase repressor (CD0471) (data not shown). Although these antibiotics are not regularly used to treat CDI, some of them clearly seem to lead to CDI (1).
In parallel to the NAPCR1 isolates, we performed this survey on one NAP2 strain, one NAP4 strain, one NAP6 strain, and five NAP1 strains that cocirculated in time and space. A principal-component analysis on this extended susceptibility data set clearly distinguished the profiles of NAPCR1 strains from those of the other genotypes (data not shown), highlighting the fact that the NAPCR1 strains are different. In agreement with this result, these non-NAPCR1 isolates lacked the MGE that characterized the NAPCR1 strains.
In summary, a phenotypic and genomic survey revealed that during the past decade, MDR was widespread among NAPCR1 isolates from various Costa Rican hospitals. This trait was due to a variety of resistance mechanisms, some of which are absent in the closely related MDR strain CD630 and also in cocirculating strains. Our results highlight the usefulness of comparative genomics in the epidemiological surveillance of antibiotic resistance, point to the need for further monitoring of circulating strains for early detection of metronidazole and vancomycin resistance, and justify further development of antibiotic-independent treatments for CDI.
MATERIALS AND METHODS
Strains and C. difficile isolation.
A total of 38 NAPCR1 C. difficile isolates recovered from seven Costa Rican hospitals between 2003 and 2010 were studied (Table 1). These bacteria were taken from a collection maintained at the Anaerobic Bacteriology Research Laboratory of the University of Costa Rica. Without exception, they were derived from stool samples positive for C. difficile toxins as revealed by rapid immunochromatographic assays. For isolation, ethanol-treated samples were inoculated onto cefoxitin-cycloserine-fructose agar (CCFA; Oxoid) plates that were incubated under anaerobic conditions (90% N2, 5% H2, and 5% CO2) in a Bactron II chamber. To confirm their identification, we used the Rapid ID 32A system (bioMérieux) based on preformed enzymes, a PCR targeting tpi (28), and, in some cases, determination of fatty acid methyl ester profiles through gas chromatography (MIDI, microbial identification system).
PFGE typing.
The PFGE procedure followed was derived from a published protocol (12). Briefly, agarose plugs were prepared by mixing equal volumes of bacteria from 6- to 8-h cultures and SeaKem Gold agarose (Lonza) in 1× Tris-EDTA buffer containing SDS (Sigma). These plugs were then incubated in a buffer composed of lysozyme, RNase A, and mutanolysin (Sigma). After overnight digestion with SmaI (Roche), DNA fragments were separated on 1% agarose gels (Bio-Rad) prepared with 0.5× Tris borate-EDTA buffer and 50 μM thiourea (Sigma) using a CHEF-DR II system (Bio-Rad). Digitized images were analyzed with BioNumerics software (version 6.0; Applied Maths), and the resulting macrorestriction patterns were compared with those deposited in the databases of the National Microbiology Laboratory (Public Health Agency of Canada).
Antimicrobial susceptibility testing.
The MICs of tetracycline (resistance breakpoint, ≥16 mg/liter), tigecycline (≥0.25 mg/liter), clindamycin (≥8 mg/liter), cefotaxime (≥64 mg/liter), ciprofloxacin (≥8 mg/liter), levofloxacin (≥8 mg/liter), moxifloxacin (≥8 mg/liter), metronidazole (≥32 mg/liter), rifampin (≥0.004 mg/liter), chloramphenicol (≥32 mg/liter), vancomycin, and linezolid were determined using Etests (bioMérieux) or macrodilution tests. These assays were repeated 2 to 3 times using independent cultures. Resistance breakpoints were set in compliance with Clinical and Laboratory Standards Institute guidelines (29) or EUCAST epidemiological cutoff values (30), when available. For vancomycin and linezolid, we categorized MICs between 4 and 8 mg/liter as indicating reduced susceptibility (31). Isolates for which linezolid MICs were >8 mg/liter were categorized as resistant. For ciprofloxacin, we used the breakpoint defined for other fluoroquinolones. According to standardized international terminology (32), MDR was defined as acquired nonsusceptibility to at least one agent in each of three or more antimicrobial classes. The panel of antibiotics tested included drugs used in the treatment of CDI (tigecycline), the development of CDI (expanded-spectrum fluoroquinolones), front-line treatments (metronidazole, vancomycin), and alternative treatments for CDI (linezolid) and CDI relapses (rifampin).
Whole-genome sequencing and genomic analyses.
WGS for 31 isolates were obtained by sequencing by synthesis using multiplexed paired-end libraries (see Table S1 in the supplemental material). Reads were first assembled into contigs using Velvet (33) or Edena (34) and were then mapped back to assembly contigs to check for misassemblies. Single nucleotide polymorphisms (SNPs) were identified using Burrows-Wheeler alignment (BWA) (35), SAMtools (36), BCFtools (37), and the genome of the MDR reference strain CD630 (NCBI nucleotide accession number AM180355.1). For automated annotation, we used Prokka, version 1.11 (38), and proprietary databases containing publicly available C. difficile genomes from reference strains. If required, annotations were refined manually using BLAST, BLASTP, PSI-BLAST, BYPASS, PRODOM, SMART, and UniProt searches. Contig files were scanned against C. difficile PubMLST typing schemes using multilocus sequence typing (MLST) (39). AMR genes were identified manually, with ABRICATE and SRST2 (40), or through BLAST/BLAT searches against the CARD (41), ARDB (42), and ARG-ANNOT (43) databases. Putative lateral gene transfer events were predicted using Alien Hunter (44), PhiSpy (45), or ICEberg (46) and were verified though visual inspection of contigs ordered with Mauve (47). Core genome SNP phylogenies were done with Parsnp (48) using the genomes of the following strains: CD630 (NCBI nucleotide accession number AM180355.1), 2007855 (FN665654.1), LIBA-5703 (EBI-ENA accession number ERR467551), LIBA-5710 (ERR467558), R20291 (NCBI nucleotide accession number FN545816.1), CD196 (FN538970.1), BI1 (FN668941.1), OX1485 (EBI-ENA accession number ERR232375), Ox2183 (ERR126286), M68 (NCBI nucleotide accession number FN668375.1), CF5 (FN665652.1), M120 (FN665653.1), CD10-165 (JRHN00000000.1), and SA10-050 (JRHM00000000.1). All genomes and genome comparisons were visualized in Artemis (49) or ACT (50). Linear comparison figures of multiple genomic loci were prepared with Easyfig (51).
Accession number(s).
Sequencing data may be downloaded from the European Nucleotide Archive (study PRJEB5034). The corresponding EBI-ENA accession numbers are ERR467530, ERR467534, ERR467536, ERR467538, ERR467539, ERR467541 to ERR467547, ERR467550, ERR467552, ERR467555, ERR467559, ERR467565, ERR467569, ERR467578, ERR467581, ERR467586 to ERR467588, ERR467592, ERR467594 to ERR467596, ERR467601, ERR467602, ERR467607, and ERR467610 (Table S1).
Supplementary Material
ACKNOWLEDGMENTS
Pablo Vargas and Robin Cárdenas are acknowledged for technical assistance. Michael R. Mulvey, George Golding, and Tim Du provided us with access to the NAP database of the National Microbiology Laboratory (Public Health Agency of Canada) and helped us to interpret PFGE patterns.
This study was funded by grants from the Ministry of Science, Technology, and Telecommunications of the Republic of Costa Rica (FORINVES grant 803-B4-510), the National Rector's Council (grant 803-B4-652), and the Vice-Rectorate for Research of the University of Costa Rica (grants 803-B5-600 and 803-B5-770). Wellcome Trust grant 098051 funded N.T. and the sequencing.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02054-16.
REFERENCES
- 1.Goudarzi M, Seyedjavadi SS, Goudarzi H, Mehdizadeh Aghdam E, Nazeri S. 2014. Clostridium difficile infection: epidemiology, pathogenesis, risk factors, and therapeutic options. Scientifica 2014:916826. doi: 10.1155/2014/916826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubberke ER, Olsen MA. 2012. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis 55:S88–S92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller BA, Chen LF, Sexton DJ, Anderson DJ. 2011. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol 32:387–390. doi: 10.1086/659156. [DOI] [PubMed] [Google Scholar]
- 5.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D'Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullany P, Allan E, Roberts AP. 2015. Mobile genetic elements in Clostridium difficile and their role in genome function. Res Microbiol 166:361–367. doi: 10.1016/j.resmic.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, Wilcox MH. 2015. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect 21:248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Tickler IA, Goering RV, Whitmore JD, Lynn ANW, Persing DH, Tenover FC. 2014. Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011 to 2013. Antimicrob Agents Chemother 58:4214–4218. doi: 10.1128/AAC.02775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah D, Dang M-D, Hasbun R, Koo HL, Jiang Z-D, DuPont HL, Garey KW. 2010. Clostridium difficile infection: update on emerging antibiotic treatment options and antibiotic resistance. Expert Rev Anti Infect Ther 8:555–564. doi: 10.1586/eri.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owens RC, Donskey CJ, Gaynes RP, Loo VG, Muto CA. 2008. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin Infect Dis 46:S19–S31. doi: 10.1086/521859. [DOI] [PubMed] [Google Scholar]
- 11.Perry JA, Wright GD. 2013. The antibiotic resistance “mobilome”: searching for the link between environment and clinic. Front Microbiol 4:138. doi: 10.3389/fmicb.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quesada-Gómez C, López-Ureña D, Acuña-Amador L, Villalobos-Zúñiga M, Du T, Freire R, Guzmán-Verri C, del Mar Gamboa-Coronado M, Lawley TD, Moreno E, Mulvey MR, de Castro Brito GA, Rodríguez-Cavallini E, Rodríguez C, Chaves-Olarte E. 2015. Emergence of an outbreak-associated Clostridium difficile variant with increased virulence. J Clin Microbiol 53:1216–1226. doi: 10.1128/JCM.03058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López-Ureña D, Quesada-Gómez C, Montoya-Ramírez M, del Mar Gamboa-Coronado M, Somogyi T, Rodríguez C, Rodríguez-Cavallini E. 2016. Predominance and high antibiotic resistance of the emerging Clostridium difficile genotypes NAPCR1 and NAP9 in a Costa Rican hospital over a 2-year period without outbreaks. Emerg Microbes Infect 5:e42. doi: 10.1038/emi.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MTG, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 15.Ammam F, Marvaud J-C, Lambert T. 2012. Distribution of the vanG-like gene cluster in Clostridium difficile clinical isolates. Can J Microbiol 58:547–551. doi: 10.1139/w2012-002. [DOI] [PubMed] [Google Scholar]
- 16.Spigaglia P, Barbanti F, Mastrantonio P. 2006. New variants of the tet(M) gene in Clostridium difficile clinical isolates harbouring Tn916-like elements. J Antimicrob Chemother 57:1205–1209. doi: 10.1093/jac/dkl105. [DOI] [PubMed] [Google Scholar]
- 17.Spigaglia P, Barbanti F, Mastrantonio P. 2007. Detection of a genetic linkage between genes coding for resistance to tetracycline and erythromycin in Clostridium difficile. Microb Drug Resist 13:90–95. doi: 10.1089/mdr.2007.723. [DOI] [PubMed] [Google Scholar]
- 18.Fry PR, Thakur S, Abley M, Gebreyes WA. 2012. Antimicrobial resistance, toxinotype, and genotypic profiling of Clostridium difficile isolates of swine origin. J Clin Microbiol 50:2366–2372. doi: 10.1128/JCM.06581-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrow KA, Lyras D, Rood JI. 2000. The macrolide-lincosamide-streptogramin B resistance determinant from Clostridium difficile 630 contains two erm(B) genes. Antimicrob Agents Chemother 44:411–413. doi: 10.1128/AAC.44.2.411-413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J Med Microbiol 54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- 21.O'Connor JR, Galang MA, Sambol SP, Hecht DW, Vedantam G, Gerding DN, Johnson S. 2008. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother 52:2813–2817. doi: 10.1128/AAC.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706. doi: 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]
- 23.Hansen LH, Vester B. 2015. A cfr-like gene from Clostridium difficile confers multiple antibiotic resistance by the same mechanism as the cfr gene. Antimicrob Agents Chemother 59:5841–5843. doi: 10.1128/AAC.01274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH, Peto TE, Walker AS, Riley TV, Crook DW, Didelot X. 2014. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol 6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fouhy F, Ogilvie LA, Jones BV, Ross RP, Ryan AC, Dempsey EM, Fitzgerald GF, Stanton C, Cotter PD. 2014. Identification of aminoglycoside and β-lactam resistance genes from within an infant gut functional metagenomic library. PLoS One 9:e108016. doi: 10.1371/journal.pone.0108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. 2013. Phage ϕC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio 4:e00840–13. doi: 10.1128/mBio.00840-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner G, Hildebrandt B, Witte W. 2001. Aminoglycoside-streptothricin resistance gene cluster aadE-sat4-aphA-3 disseminated among multiresistant isolates of Enterococcus faecium. Antimicrob Agents Chemother 45:3267–3269. doi: 10.1128/AAC.45.11.3267-3269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemee L, Dhalluin A, Testelin S, Mattrat A, Maillard K, Lemeland J-F, Pons J-L. 2004. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol 42:5710–5714. doi: 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard. CLSI document M11-A7, 7th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 30.Erikstrup LT, Danielsen TKL, Hall V, Olsen KEP, Kristensen B, Kahlmeter G, Fuursted K, Justesen US. 2012. Antimicrobial susceptibility testing of Clostridium difficile using EUCAST epidemiological cut-off values and disk diffusion correlates. Clin Microbiol Infect 18:E266–E272. doi: 10.1111/j.1469-0691.2012.03907.x. [DOI] [PubMed] [Google Scholar]
- 31.Peláez T, Alcalá L, Alonso R, Martín-López A, García-Arias V, Marín M, Bouza E. 2005. In vitro activity of ramoplanin against Clostridium difficile, including strains with reduced susceptibility to vancomycin or with resistance to metronidazole. Antimicrob Agents Chemother 49:1157–1159. doi: 10.1128/AAC.49.3.1157-1159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 33.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez D, Tewhey R, Veyrieras J-B, Farinelli L, Østerås M, François P, Schrenzel J. 2014. De novo finished 2.8 Mbp Staphylococcus aureus genome assembly from 100 bp short and long range paired-end reads. Bioinformatics 30:40–49. doi: 10.1093/bioinformatics/btt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJM, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE. 2010. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B, Pop M. 2009. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res 37(Database issue):D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain J-M. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vernikos GS, Parkhill J. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- 45.Akhter S, Aziz RK, Edwards RA. 2012. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou H-Y. 2012. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res 40(Database issue):D621–D626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream M-A. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.