ABSTRACT
Pythiosis is a life-threatening infectious disease caused by the oomycete Pythium insidiosum. Direct exposure to Py. insidiosum zoospores can initiate infections of the eye, limb, gastrointestinal tract, or skin/subcutaneous tissue. Treatments for pythiosis have mostly relied on surgery. Antifungal drugs are generally ineffective against Py. insidiosum. However, one patient with an invasive Py. insidiosum infection recovered completely following treatment with terbinafine and itraconazole. Additionally, the drug target sterol biosynthetic enzymes have been identified in the oomycete Aphanomyces euteiches. It remains an open question whether Py. insidiosum is susceptible to the antifungal drugs and harbors any of the known drug target enzymes. Here, we determined the in vitro susceptibilities of terbinafine and itraconazole against 30 isolates of Py. insidiosum. We also analyzed endogenous sterols and searched for genes encoding the sterol biosynthetic enzymes in the genomes of Py. insidiosum and related oomycetes. The susceptibility assay showed that the growth of each of the Py. insidiosum isolates was inhibited by the antifungal agents, but only at difficult-to-achieve concentrations, which explains the clinical resistance of the drugs in the treatment of pythiosis patients. Genome searches of Py. insidiosum and related oomycetes demonstrated that these organisms contained an incomplete set of sterol biosynthetic enzymes. Gas chromatographic mass spectrometry did not detect any sterol end products in Py. insidiosum. In conclusion, Py. insidiosum possesses an incomplete sterol biosynthetic pathway. Resistance to antifungal drugs targeting enzymes in the ergosterol biosynthetic pathway in Py. insidiosum was due to modifications or losses of some of the genes encoding the drug target enzymes.
KEYWORDS: Pythium insidiosum, pythiosis, sterol biosynthetic enzyme, in vitro susceptibility, evolution, antifungal drug
INTRODUCTION
Pythiosis is a life-threatening infectious disease and has been increasingly reported from tropical, subtropical, and temperate countries (1–4). The etiologic agent is the fungus-like organism Pythium insidiosum (1). The microscopic morphology of Py. insidiosum is similar to that of filamentous fungi (1, 3). However, based on phylogenetic analyses, Py. insidiosum belongs to the oomycetes, a unique group of microorganisms in several genera, such as Pythium, Phytophthora, Hyaloperonospora, Albugo, Saprolegnia, and Aphanomyces (5, 6). Unlike most other pathogenic oomycetes, which infect plants, Py. insidiosum infects humans and animals (i.e., horses and dogs) (1–3, 6). In the environment, Py. insidiosum presents as mycelia and motile zoospores (7). Py. insidiosum infection occurs when a patient contacts or ingests pathogen-contaminated water. Common sites of the Py. insidiosum infection include the skin, eye, artery, and gastrointestinal tract (1–3).
Due to the aggressive nature of the infections, patients or animals with pythiosis often die if proper management is not provided. The treatment of choice for pythiosis has mostly relied on extensive surgery to remove the infected organ (2, 3). Use of an immunotherapeutic vaccine (which is a crude protein extract of Py. insidiosum) exhibits various efficacies, depending on the affected host, with 97% favorable responses in cattle, 60 to 85% in horses, 56% in humans, and only 33% in dogs (8, 9). Conventional antifungal drugs, inhibiting the production of fungal ergosterol, have been used for treatment of pythiosis, but drug resistance is usually observed (2, 4, 10). The oomycetes, including Phytophthora and Pythium species, are sterol auxotrophs and thus require exogenous sterols for basic metabolism (11–13). Lack of the drug target sterol biosynthetic enzymes in these organisms explains treatment failure of the antifungal drugs (12, 14).
Contrary to the above findings, Shenep et al. reported on successful medical treatment of a patient with invasive Py. insidiosum infection using a combination of two antifungal drugs: terbinafine and itraconazole (15). In addition, Madoui et al. analyzed the transcriptomic data of Aphanomyces euteiches (16, 17) and showed that this oomycete microorganism contains several sterol biosynthetic enzymes responsible for production of endogenous sterols (12). Such discordant results raise questions about Py. insidiosum's susceptibility to the antifungal drugs and whether the pathogen produces the drug target enzymes in the sterol biosynthetic pathway.
A better understanding of the drug resistance mechanism(s) of Py. insidiosum could lead to the development of more efficient agents for the control of this and related pathogens. Recently, genome sequences and transcriptomic data of many oomycetes (i.e., Py. insidiosum [18–21], non-insidiosum Pythium species [22, 23], Phytophthora species [14, 24, 25], Hyaloperonospora species [26], Albugo species [27], Saprolegnia species [28], and Aphanomyces species [17]) have been made publicly available. These genetic resources were used to explore the sterol biosynthetic pathway in Py. insidiosum and related oomycetes. In this study, we aimed to demonstrate the in vitro growth inhibitory effects of two key antifungal drugs (terbinafine and itraconazole) against clinical isolates of Py. insidiosum. We also sought to identify endogenous sterols and search for genes encoding sterol biosynthetic enzymes in this pathogen.
RESULTS
Antifungal drug susceptibility of 30 Py. insidiosum isolates.
Two key antifungal drugs, terbinafine and itraconazole (concentrations of 0 [no drug], 0.5, 2, 8, 32, 64, and 128 mg/liter), were tested against 30 isolates of Py. insidiosum. In relation to the no-drug control, terbinafine at 2 mg/liter modestly, but significantly, increased the average growth of Py. insidiosum to 116% (Fig. 1A). In contrast, at the concentrations 32, 64, and 128 mg/liter, terbinafine significantly reduced the average growth of Py. insidiosum to 39%, 7%, and 0.7%, respectively (Fig. 1A). Terbinafine at concentrations of 32, 64, and 128 mg/liter completely inhibited the growth of 1, 8, and 13 of the Py. insidiosum isolates, respectively.
FIG 1.
In vitro drug susceptibility assay of 30 Py. insidiosum isolates against antifungal drugs. The drug concentrations of terbinafine (A) and itraconazole (B) used in the in vitro susceptibility assay are 0.5, 2, 8, 32, 64, and 128 mg/liter. A dot represents the percentage of radial growth of an individual isolate in relation to the growth of the same isolate exposed to no drug (control). *, statistical significance compared to the no-drug control.
When Py. insidiosum was tested against 0.5 mg/liter itraconazole, the average growth was modestly, but not significantly, increased to 110% of that of the no-drug control (Fig. 1B). However, when the organism was tested against the higher drug concentrations, the average growth of Py. insidiosum was significantly decreased to 78% (at 8 mg/liter itraconazole), 65% (32 mg/liter), 61% (64 mg/liter), or 59% (128 mg/liter) of that of the no-drug control (Fig. 1B). None of the Py. insidiosum isolates was completely inhibited by itraconazole.
Identification of Py. insidiosum sterols.
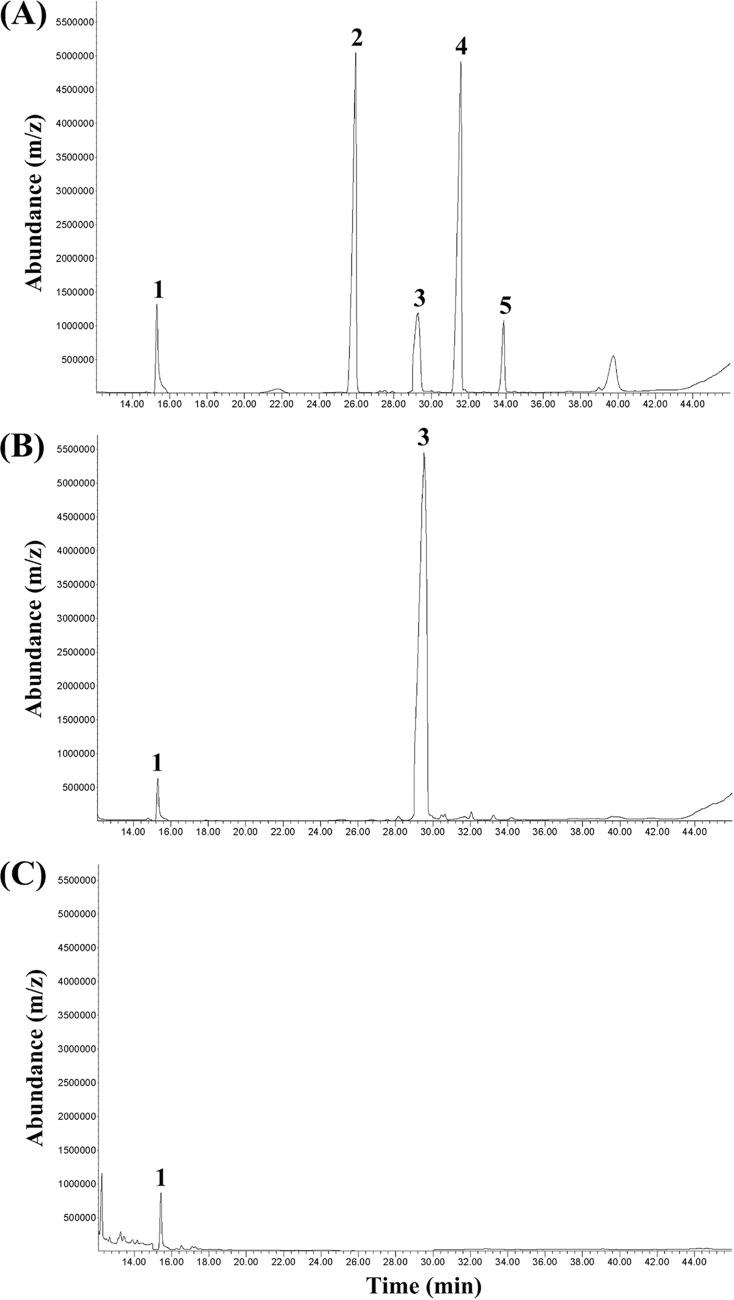
The sterol extract of Py. insidiosum (strain Pi-S) was analyzed for the presence of the sterol end products, such as cholesterol, stigmasterol, ergosterol, and fucosterol. Commercially available sterol standards (cholesterol, stigmasterol, ergosterol, and fucosterol) were used to generate the reference gas chromatography-mass spectrometry (GC-MS) peaks. The sterol extract from Candida albicans strain ATCC 90028 was also prepared and analyzed in parallel with the extract from Py. insidiosum. To serve as the internal control, 5α-cholestane was added to the sterol extracts of Py. insidiosum and Ca. albicans. Two hundred milligrams of lyophilized cells from either Py. insidiosum or Ca. albicans was derivatized for GC-MS analysis. The retention times of the reference sterols were 15.34 min for 5α-cholestane (peak 1), 25.80 min for cholesterol (peak 2), 29.26 min for ergosterol (peak 3), 31.56 min for stigmasterol (peak 4), and 33.88 min for fucosterol (peak 5) (Fig. 2A). Two GC-MS peaks, corresponding to 5α-cholestane (peak 1) and ergosterol (peak 3), were identified in Ca. albicans (Fig. 2B), whereas only one GC-MS peak, corresponding to the internal control 5α-cholestane, was detected in Py. insidiosum (data not shown). Even when the amount of lyophilized cells from Py. insidiosum was increased 7-fold (1,400 mg), only the 5α-cholestane peak was observed (Fig. 2C).
FIG 2.
Total ion chromatogram of the sterol extracts generated by GC-MS analysis: (A) Standard mixture of sterol end products, (B) sterol extract from the Ca. albicans strain ATCC 90028 (200 mg lyophilized cells), and (C) sterol extract from Py. insidiosum strain Pi-S (1,400 mg lyophilized cells). Peaks 1 to 5 indicate 5α-cholestane (internal control), cholesterol, ergosterol, stigmasterol, and fucosterol, respectively.
Identification of enzymes involved in sterol biosynthesis.
The full-length deduced protein sequences of 22 enzymes involved in the mevalonate pathway (HMG1, HMG2, ERG8, ERG10, ERG12, ERG13, ERG19, and IDI1) and the sterol biosynthetic pathway (ERG1 to -7, ERG9, ERG11, ERG20, and ERG24 to -27) of the model organism Saccharomyces cerevisiae (Fig. 3; Tables 1 to 3), were subject to BLAST search against the translated genomes or transcriptomes of 17 oomycetes (including Py. insidiosum), three algae, two fungi, and one diatom (Table 4). Both fungi (Ca. albicans and Aspergillus fumigatus), two algae (Aurantiochytrium limacinum and Ectocarpus siliculosus), and only one oomycete (Saprolegnia parasitica) harbored the complete set of 8 enzymes in the mevalonate pathway (Table 2). Only the fungi possessed the complete set of 14 enzymes in the sterol biosynthetic pathway (Table 3). ERG5, ERG10, ERG11, and ERG26 homologs were identified in all species included in this study. ERG2 homologs were absent in all nonfungal microorganisms.
FIG 3.
Schematic diagram showing enzymes and substrates in the mevalonate (upper part) and sterol biosynthesis (lower part) pathways. ERG1 and ERG11 are the drug targets of terbinafine and itraconazole, respectively.
TABLE 1.
The best BLAST hits and accession numbers of orthologous enzymes in the mevalonate and sterol biosynthetic pathways presented in the Py. insidiosum genomea
| Protein | Enzymeb | S. cerevisiae enzyme accession no. |
Py. insidiosum genome |
||
|---|---|---|---|---|---|
| Protein ID | Accession no. | E value | |||
| ERG1 | Squalene epoxidase | NP_011691 | |||
| ERG2 | C-8 sterol isomerase | NP_013929 | |||
| ERG3 | C-5 sterol desaturase | NP_013157 | PINS02380006C | GAQ12368.1 | 3.0E−63 |
| ERG4 | C-24(28) sterol reductase | NP_011503 | |||
| ERG5 | C-22 sterol desaturase | NP_013728 | PINS02520001C | GAQ12369.1 | 2.0E−17 |
| ERG6 | Sterol 24-C-methyltransferase | NP_013706 | |||
| ERG7 | Lanosterol synthase | NP_011939 | |||
| ERG8 | Phosphomevalonate kinase | NP_013947 | PINS00060035C | GAQ12361.1 | 6.0E−54 |
| ERG9 | Squalene synthase | NP_012060 | |||
| ERG10 | Acetyl-CoA acyltransferase | NP_015297 | PINS00050004C | GAQ12360.1 | 4.0E−133 |
| ERG11 | Lanosterol 14-α-demethylase | NP_011871 | PINS02520001C | GAQ12369.1 | 2.0E−11 |
| ERG12 | Mevalonate kinase | NP_013935 | |||
| ERG13 | HMG-CoA synthase | NP_013580 | PINS00120058C | GAQ12362.1 | 2.0E−120 |
| ERG19 | Diphosphomevalonate decarboxylase | NP_014441 | PINS01150035C | GAQ12364.1 | 2.0E−92 |
| ERG20 | Farnesyl diphosphate synthase | NP_012368 | PINS01980016C | GAQ12366.1 | 6.0E−102 |
| ERG24 | Delta14-sterol reductase | NP_014119 | PINS01230016C | GAQ12365.1 | 2.0E−38 |
| ERG25 | Methylsterol monooxygenase | NP_011574 | |||
| ERG26 | Sterol-4-α-carboxylate 3-dehydrogenase | NP_011514 | PINS00200056A | GAQ12363.1 | 3.0E−16 |
| ERG27 | 3-Keto-steroid reductase | NP_013201 | |||
| HMG1 | HMG-CoA reductase 1 | NP_013636 | |||
| HMG2 | HMG-CoA reductase 2 | NP_013555 | |||
| IDI1 | Isopentenyl-diphosphate delta-isomerase | NP_015208 | PINS02140012C | GAQ12367.1 | 1.0E−49 |
The best BLAST hits were defined as having a cutoff E value of less than −6. Shown are accession numbers of orthologous enzymes in the mevalonate and sterol biosynthetic pathways presented in the genome of Py. insidiosum strain Pi-S, isolated from a human patient with vascular pythiosis (19). The query sequences were derived from Saccharomyces cerevisiae.
CoA, coenzyme A; HMG, hydroxymethylglutaryl.
TABLE 3.
The best BLAST hits of orthologous enzymes in the sterol biosynthetic pathway presented in the genomes of 17 oomycetes, 3 algae, 2 fungi, and a diatoma
| Organism | Group | Subgroup | E value for enzyme in the sterol biosynthetic pathway |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERG20 | ERG9 | ERG1 | ERG7 | ERG11 | ERG24 | ERG25 | ERG26 | ERG27 | ERG6 | ERG2 | ERG3 | ERG5 | ERG4 | |||
| Ca. albicans | Fungi | Ascomycota | 5.0E−155 | 1.0E−138 | 4.0E−143 | 0.0E+00 | 0.0E+00 | 3.0E−78 | 2.0E−118 | 1.0E−139 | 4.0E−111 | 8.0E−144 | 5.0E−66 | 6.0E−118 | 0.0E+00 | 7.0E−166 |
| As. fumigatus | Fungi | Ascomycota | 2.0E−118 | 8.0E−107 | 2.0E−89 | 0.0E+00 | 5.0E−147 | 2.0E−146 | 2.0E−100 | 2.0E−56 | 5.0E−36 | 1.0E−116 | 8.0E−52 | 3.0E−91 | 2.0E−171 | 2.0E−145 |
| Au. limacinum | Algae | Labyrinthulida | 3.0E−99 | 5.0E−65 | 4.0E−138 | 1.0E−73 | 4.0E−103 | 4.0E−17 | 9.0E−34 | 1.0E−08 | 5.0E−68 | 1.0E−62 | 6.0E−33 | 4.0E−53 | ||
| Au. anophagefferens | Algae | Pelagophyceae | 6.0E−94 | 1.0E−70 | 1.0E−129 | 6.0E−58 | 1.0E−73 | 1.0E−21 | 3.0E−25 | 4.0E−83 | 4.0E−10 | 1.0E−10 | 3.0E−102 | |||
| Ec. siliculosus | Algae | PX clade | 1.0E−118 | 8.0E−91 | 4.0E−65 | 4.0E−79 | 2.0E−66 | 2.0E−100 | 5.0E−09 | 1.0E−37 | 1.0E−99 | 1.0E−16 | 2.0E−17 | 3.0E−55 | ||
| Ph. tricornutum | Diatom | Bacillariophyta | 7.0E−115 | 1.0E−70 | 5.0E−126 | 3.0E−69 | 1.0E−100 | 6.0E−10 | 1.0E−33 | 2.0E−81 | 1.0E−19 | 1.0E−26 | 1.0E−52 | |||
| Ap. euteicheb | Oomycete | Saprolegniales | 3.0E−94 | 2.0E−69 | 4.0E−56 | 4.0E−66 | 8.0E−68 | 2.0E−73 | 1.0E−61 | 2.0E−25 | 2.0E−78 | 6.0E−16 | 1.0E−38 | |||
| Sa. parasitica | Oomycete | Saprolegniales | 9.0E−118 | 3.0E−79 | 4.0E−65 | 2.0E−158 | 1.0E−80 | 2.0E−84 | 2.0E−62 | 2.0E−50 | 5.0E−84 | 1.0E−62 | 1.0E−17 | 3.0E−50 | ||
| Py. insidiosumc | Oomycete | Pythiales | 6.0E−102 | 2.0E−11 | 2.0E−38 | 3.0E−16 | 3.0E−63 | 2.0E−17 | ||||||||
| Py. ultimum | Oomycete | Pythiales | 1.0E−119 | 6.0E−12 | 3.0E−36 | 8.0E−08 | 6.0E−18 | 2.0E−10 | 3.0E−79 | 3.0E−14 | 6.0E−26 | |||||
| Py. arrhenomanes | Oomycete | Pythiales | 6.0E−105 | 3.0E−14 | 1.0E−32 | 2.0E−19 | 7.0E−10 | 3.0E−17 | 2.0E−30 | |||||||
| Py. irregulare | Oomycete | Pythiales | 4.0E−123 | 4.0E−15 | 4.0E−40 | 1.0E−08 | 1.0E−14 | 9.0E−12 | 4.0E−81 | 3.0E−16 | 1.0E−29 | |||||
| Py. iwayamai | Oomycete | Pythiales | 2.0E−120 | 6.0E−17 | 4.0E−31 | 4.0E−07 | 5.0E−14 | 3.0E−10 | 1.0E−71 | 1.0E−15 | 4.0E−22 | |||||
| Py. vexans | Oomycete | Pythiales | 5.0E−95 | 1.0E−11 | 2.0E−36 | 4.0E−14 | 5.0E−12 | 1.0E−78 | 1.0E−14 | 1.0E−26 | ||||||
| Py. aphanidermatum | Oomycete | Pythiales | 6.0E−15 | 5.0E−35 | 6.0E−11 | 2.0E−19 | 6.0E−14 | 2.0E−81 | 9.0E−16 | 4.0E−31 | ||||||
| Ph. sojae | Oomycete | Peronosporales | 1.0E−118 | 3.0E−13 | 4.0E−38 | 4.0E−07 | 4.0E−12 | 9.0E−12 | 2.0E−82 | 2.0E−16 | 6.0E−29 | |||||
| Ph. ramorum | Oomycete | Peronosporales | 4.0E−119 | 2.0E−14 | 6.0E−38 | 3.0E−13 | 2.0E−11 | 7.0E−79 | 2.0E−16 | 5.0E−26 | ||||||
| Ph. parasitica | Oomycete | Peronosporales | 1.0E−115 | 4.0E−16 | 1.0E−39 | 2.0E−07 | 3.0E−14 | 4.0E−14 | 4.0E−79 | 3.0E−16 | 2.0E−26 | |||||
| Ph. capsici | Oomycete | Peronosporales | 1.0E−117 | 7.0E−12 | 8.0E−38 | 2.0E−08 | 9.0E−16 | 4.0E−14 | 8.0E−79 | 7.0E−14 | 1.0E−27 | |||||
| Ph. cinnamomi | Oomycete | Peronosporales | 4.0E−117 | 5.0E−16 | 4.0E−37 | 8.0E−07 | 7.0E−13 | 6.0E−10 | 3.0E−80 | 5.0E−18 | 2.0E−27 | |||||
| Ph. infestans | Oomycete | Peronosporales | 1.0E−115 | 2.0E−13 | 4.0E−39 | 9.0E−08 | 1.0E−14 | 2.0E−13 | 1.0E−74 | 3.0E−14 | 7.0E−28 | |||||
| Hy. arabidopsis | Oomycete | Peronosporales | 3.0E−105 | 2.0E−14 | 1.0E−12 | 4.0E−11 | 5.0E−14 | |||||||||
| Al. laibachii | Oomycete | Albuginales | 2.0E−107 | 2.0E−10 | 6.0E−16 | 4.0E−08 | 2.0E−11 | |||||||||
The best BLAST hits of orthologous enzymes in the sterol biosynthetic pathway are defined as having a cutoff E value of less than −6. The query sequences were derived from Saccharomyces cerevisiae.
Only transcriptome data were available and used for analysis.
Py. insidiosum (strain Pi-S) was isolated from a human patient with vascular pythiosis (19).
TABLE 4.
Results from genome and transcriptome databases for 17 oomycetes, 3 algae, 2 fungi, and a diatom used for the sterol biosynthetic enzyme search
| Organism | Group | Genome size (Mb) | Accession no. or source |
|---|---|---|---|
| Pythium insidiosuma | Oomycetes | 53.24 | BBXB01000001 to BBXB01001192 |
| Pythium ultimum | Oomycetes | 42.8 | http://pythium.plantbiology.msu.edu/ |
| Pythium aphanidermatum | Oomycetes | 35.88 | http://pythium.plantbiology.msu.edu/ |
| Pythium arrhenomanes | Oomycetes | 44.67 | http://pythium.plantbiology.msu.edu/ |
| Pythium irregulare | Oomycetes | 42.97 | http://pythium.plantbiology.msu.edu/ |
| Pythium iwayamai | Oomycetes | 43.2 | http://pythium.plantbiology.msu.edu/ |
| Pythium vexans | Oomycetes | 33.84 | http://pythium.plantbiology.msu.edu/ |
| Phytophthora sojae | Oomycetes | 82.6 | http://www.broadinstitute.org/ |
| Phytophthora ramorum | Oomycetes | 66.65 | http://www.broadinstitute.org/ |
| Phytophthora parasitica | Oomycetes | 82.39 | http://www.broadinstitute.org/ |
| Phytophthora capsici | Oomycetes | 56.04 | http://genome.jgi.doe.gov/ |
| Phytophthora cinnamomi | Oomycetes | 77.97 | http://genome.jgi.doe.gov/ |
| Phytophthora infestans | Oomycetes | 228.54 | http://www.broadinstitute.org/ |
| Hyaloperonospora arabidopsis | Oomycetes | 73.89 | http://protists.ensembl.org/ |
| Albugo laibachii | Oomycetes | 32.76 | http://protists.ensembl.org/ |
| Aphanomyces euteichesb | Oomycetes | http://www.polebio.lrsv.ups-tlse.fr/aphano/ | |
| Saprolegnia parasitica | Oomycetes | 53.13 | http://www.broadinstitute.org/ |
| Phaeodactylum tricornutum | Diatoms | 25.05 | http://genome.jgi.doe.gov/ |
| Aureococcus anophagefferens | Brown tide algae | 56.66 | http://genome.jgi.doe.gov/ |
| Ectocarpus siliculosus | Brown algae | 391.72 | http://bioinformatics.psb.ugent.be/genomes/ |
| Aurantiochytrium limacinum | Microalgae | 60.93 | http://genome.jgi.doe.gov/ |
| Candida albicans | Fungi | 27.56 | http://www.candidagenome.org/ |
| Aspergillus fumigatus | Fungi | 29.39 | http://www.aspgd.org/ |
Py. insidiosum (strain Pi-S) was isolated from a human patient with vascular pythiosis (19).
Only transcriptome data were available and used for analysis.
TABLE 2.
The best BLAST hits of orthologous enzymes in the mevalonate pathway presented in the genomes of 17 oomycetes, 3 algae, 2 fungi, and a diatoma
| Organism | Group | Subgroup | E value for enzyme in the mevalonate pathway |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ERG10 | ERG13 | HMG1 | HMG2 | ERG12 | ERG8 | ERG19 | IDI1 | |||
| Ca. albicans | Fungi | Ascomycota | 2.0E−154 | 0.0E+00 | 0.0E+00 | 0.0E+00 | 8.0E−88 | 6.0E−81 | 6.0E−135 | 1.0E−85 |
| As. fumigatus | Fungi | Ascomycota | 1.0E−137 | 1.0E−164 | 0.0E+00 | 0.0E+00 | 8.0E−68 | 3.0E−53 | 1.0E−115 | 9.0E−73 |
| Ec. siliculosus | Algae | PX clade | 3.0E−136 | 2.0E−125 | 9.0E−98 | 4.0E−101 | 4.0E−45 | 4.0E−45 | 1.0E−100 | 6.0E−52 |
| Au. limacinum | Algae | Labyrinthulida | 2.0E−118 | 8.0E−119 | 3.0E−139 | 1.0E−136 | 2.0E−15 | 3.0E−36 | 2.0E−116 | 3.0E−62 |
| Au. anophagefferens | Algae | Pelagophyceae | 3.0E−114 | 3.0E−109 | 6.0E−09 | 5.0E−41 | 3.0E−53 | |||
| Ph. tricornutum | Diatoms | Bacillariophyta | 3.0E−136 | 6.0E−105 | 5.0E−131 | 2.0E−127 | 8.0E−44 | 7.0E−57 | ||
| Ap. euteicheb | Oomycetes | Saprolegniales | 1.0E−101 | 1.0E−117 | 1.0E−101 | 9.0E−30 | ||||
| Sa. parasitica | Oomycetes | Saprolegniales | 7.0E−137 | 2.0E−119 | 1.0E−134 | 2.0E−133 | 4.0E−34 | 1.0E−45 | 2.0E−95 | 2.0E−59 |
| Py. insidiosumc | Oomycetes | Pythiales | 4.0E−133 | 2.0E−120 | 6.0E−54 | 2.0E−92 | 1.0E−49 | |||
| Py. ultimum | Oomycetes | Pythiales | 2.0E−133 | 2.0E−40 | 4.0E−60 | 4.0E−99 | 3.0E−55 | |||
| Py. arrhenomanes | Oomycetes | Pythiales | 1.0E−133 | 3.0E−118 | 2.0E−35 | 6.0E−07 | 2.0E−106 | 2.0E−52 | ||
| Py. irregulare | Oomycetes | Pythiales | 1.0E−126 | 6.0E−120 | 4.0E−34 | 7.0E−48 | 7.0E−93 | 1.0E−54 | ||
| Py. iwayamai | Oomycete | Pythiales | 1.0E−120 | 3.0E−75 | 7.0E−39 | 3.0E−57 | 6.0E−26 | 4.0E−55 | ||
| Py. vexans | Oomycetes | Pythiales | 4.0E−85 | 3.0E−121 | 2.0E−43 | 2.0E−36 | 2.0E−77 | 1.0E−52 | ||
| Py. aphanidermatum | Oomycetes | Pythiales | 5.0E−133 | 8.0E−121 | 6.0E−42 | 8.0E−14 | 6.0E−89 | 1.0E−51 | ||
| Ph. sojae | Oomycetes | Peronosporales | 9.0E−128 | 1.0E−119 | 7.0E−40 | 7.0E−103 | 1.0E−50 | |||
| Ph. ramorum | Oomycetes | Peronosporales | 1.0E−128 | 9.0E−120 | 2.0E−40 | 7.0E−37 | 6.0E−104 | 5.0E−49 | ||
| Ph. parasitica | Oomycetes | Peronosporales | 5.0E−135 | 1.0E−120 | 5.0E−41 | 2.0E−56 | 6.0E−34 | 1.0E−56 | ||
| Ph. capsici | Oomycetes | Peronosporales | 8.0E−132 | 1.0E−123 | 1.0E−43 | 4.0E−39 | 7.0E−105 | 2.0E−57 | ||
| Ph. cinnamomi | Oomycetes | Peronosporales | 9.0E−129 | 5.0E−117 | 1.0E−38 | 9.0E−38 | 1.0E−100 | 3.0E−56 | ||
| Ph. infestans | Oomycetes | Peronosporales | 4.0E−127 | 3.0E−117 | 9.0E−42 | 3.0E−36 | 1.0E−103 | 2.0E−53 | ||
| Hy. arabidopsis | Oomycetes | Peronosporales | 2.0E−127 | 3.0E−110 | 1.0E−37 | 1.0E−104 | 8.0E−54 | |||
| Al. laibachii | Oomycetes | Albuginales | 1.0E−124 | 6.0E−119 | 6.0E−17 | 4.0E−51 | 2.0E−92 | 3.0E−50 | ||
The best BLAST hits of orthologous enzymes in the mevalonate pathway are defined as having a cutoff E value of less than −6. The query sequences were derived from Saccharomyces cerevisiae.
Only transcriptome data were available and used for analysis.
Py. insidiosum (strain Pi-S) was isolated from a human patient with vascular pythiosis (19).
Regarding the oomycetes, two enzymes in the mevalonate pathway (HMG1 and HMG2) and three enzymes in the sterol biosynthetic pathway (ERG1, ERG7, and ERG9) were absent in all organisms of the non-Saprolegniales subgroups (i.e., Pythiales, Peronosporales, and Albuginales) but present in all organisms of the Saprolegniales subgroup (Tables 2 and 3). Py. insidiosum contained 5 out of 8 enzymes in the mevalonate pathway, including ERG8, ERG10, ERG13, ERG19, and IDI1 (Table 2). In addition, Py. insidiosum harbored genes for only 6 out of 14 sterol biosynthetic enzymes: ERG3, ERG5, ERG11, ERG20, ERG24, and ERG26 (Table 3).
Phylogenetic analysis of ERG11.
ERG11-encoding sequences of all microorganisms included in this study were retrieved for phylogenetic analysis (Table 3). The resulting tree showed two major phylogenetic groups (Fig. 4): group 1 included 3 fungi (S. cerevisiae, Ca. albicans, and Aspergillus fumigatus), 3 algae (Au. limacinum, Aureococcus anophagefferens, and Ec. siliculosus), 2 oomycetes (Aphanomyces euteiches, and Sa. parasitica), and one diatom (Phaeodactylum tricornutum), and group 2 contained the 15 remaining oomycetes (7 Pythium species [Py. insidiosum, Py. ultimum, Py. aphanidermatum, Py. arrhenomanes, Py. irregulare, Py. iwayamai, and Py. vexans], 6 Phytophthora species [Ph. capsici, Ph. infestans, Ph. sojae, Ph. ramorum, Ph. parasitica, and Ph. cinnamomi], Hyaloperonospora arabidopsis, and Albugo laibachii).
FIG 4.
Phylogenetic analysis of ERG11 orthologs of 17 oomycetes, 3 fungi, 3 algae, and a diatom. A phylogenetic tree was reconstructed based on the neighbor-joining algorithm (see Materials and Methods). The branch support values of at least 70% are shown at corresponding nodes. *, oomycete; #, fungus.
DISCUSSION
Inconsistent effects of antifungal drugs in the treatment of pythiosis patients have been reported (2, 12, 15). We have sought to determine whether 30 isolates of Py. insidiosum are susceptible to the antifungal agents and whether the organism harbors the drug target enzymes of the sterol biosynthetic pathway. There is no standardized antibiotic susceptibility test for Py. insidiosum. In our preliminary work to develop such a test, we considered using zoospores or hyphal suspensions as inocula (29, 30). However, in our hands, generation of zoospores from Thai strains is difficult and labor-intensive and did not provide enough zoospores for the susceptibility testing. Likewise, hyphal suspensions, prepared as described by other investigators (15, 30, 31) were not reproducibly uniform in our hands (data not shown). As an alternative, we have used the method of Brown and coworkers (32), starting with agar plugs of active hyphal growth and measuring radial growth of the hyphae. They demonstrated that the radial growth assay was a simple and reproducible method for susceptibility testing of Py. insidiosum and argued that the submerged-growth nature of Py. insidiosum in agar is an advantage in that the organism is kept in direct contact with the antifungal drug. We have found that this assay is simple, reliable, and reproducible. To control for differences in the growth of the different Py. insidiosum isolates, we calculated the percentage of radial growth of each isolate as the growth with exposure to the antifungal drug divided by growth of the same isolate with no drug × 100.
Although RPMI is a commonly used medium for the in vitro susceptibility testing of Py. insidiosum (30), we have chosen to use Sabouraud dextrose (SD) agar for these assays. Since a particular medium could interfere with antifungal drug activity, this may be a concern. However, Stopiglia et al. compared the performances of RPMI and SD agar for in vitro susceptibility testing of a pathogenic fungus against various antifungal drugs (33). They reported no significant differences between the MICs obtained in the two media for many antifungal drugs tested (including itraconazole and terbinafine), with one exception—i.e., amphotericin B. Thus, in the present study, we used SD agar (which is lower cost and easier to prepare) as the medium for in vitro susceptibility testing of Py. insidiosum against itraconazole and terbinafine.
As shown here, two key antifungal drugs (terbinafine and itraconazole) had different impacts on growth of Py. insidiosum. At relatively low concentrations (0.5 to 2.0 mg/liter), both drugs paradoxically slightly enhanced the growth in the majority of Py. insidiosum isolates (Fig. 1). At higher concentrations (≥8 mg/liter), both drugs inhibited the pathogen in a dose-dependent manner (Fig. 1). Terbinafine was more inhibitory than itraconazole against the organism, which is consistent with the published results of Argenta et al. (34) and Fonseca et al. (30). Both of these groups performed comprehensive in vitro susceptibility testing of terbinafine and itraconazole against 30 and 22 Brazilian isolates of Py. insidiosum, respectively. To completely inhibit the growth of 50% of the isolates tested required at least 16 mg/liter of itraconazole and 4 mg/liter of terbinafine in the studies by Argenta et al. (34) and at least 64 mg/liter of itraconazole and 8 mg/liter of terbinafine in those by Fonseca et al. (30), while our group found that more than 128 mg/liter of both drugs was required. The different inhibitory effects of the two drugs against Py. insidiosum may be due to the differences in the strains of Py. insidiosum tested (i.e., Thai versus Brazilian strains) or the differences in the susceptibility methods used. (There is no standardized susceptibility assay available for the organism.)
In the present study, complete growth inhibition of some Py. insidiosum isolates was observed only with terbinafine at extremely high concentrations (64 to 128 mg/liter [Fig. 1A]). Although Py. insidiosum can be inhibited using the high drug concentrations, such concentrations are difficult to achieve in vivo. Kovarik and coworkers studied the pharmacokinetics and tissue distribution of terbinafine and showed that the peak plasma concentration of the drug is ∼1.7 mg/liter, while the peak tissue concentrations in sebum, hair, nail, and skin are ∼20, ∼10, ∼1, and ∼1 mg/liter, respectively (35). These plasma and tissue concentrations (especially for the skin, which is a target site of Py. insidiosum infection) are lower than the minimal drug concentration required to inhibit the organism in our study. Our results explain why Py. insidiosum clinically resists this antifungal drug as observed in most pythiosis patients (2).
In fungi, there are 14 enzymes in the sterol biosynthetic pathway (Fig. 3). The mechanisms of action of terbinafine and itraconazole rely on interfering with the synthesis of sterol at the ERG1 step (converting squalene to squalene epoxide) and the ERG11 step (catalyzing lanosterol to dimethylcholestatrienol), respectively. Based on the genome search, Py. insidiosum contained only six enzymes (ERG3, ERG5, ERG11, ERG20, ERG24, and ERG26) (Table 3) in the sterol biosynthetic pathway. The presence of the drug target ERG11, with an E value of −11, may explain the limited inhibitory effect of itraconazole against Py. insidiosum (Fig. 1B). However, the lack of the drug target ERG1 in the Py. insidiosum genome contradicts the fact that the organism was notably sensitive to terbinafine. There might be another target protein responsible for terbinafine-mediated growth inhibition in Py. insidiosum. This possibility is especially likely in light of the high in vitro concentrations needed to see growth inhibition by terbinafine.
We used GC-MS to detect the sterol end products identified in various organisms—i.e., cholesterol (animals), stigmasterol (plants), ergosterol (fungi), and fucosterol (algae and oomycetes) (13)—to find evidence to support the presence of the sterol biosynthetic pathway in Py. insidiosum. We were able to detect ergosterol in the fungus Ca. albicans. However, no sterol end products were detected in Py. insidiosum, even when the starting amount of sterol extract was 7-fold increased versus that used for Ca. albicans (Fig. 2). Taken together, the bioinformatic and GC-MS findings present a coherent picture that Py. insidiosum lacks a complete set of sterol biosynthetic enzymes, which leads to its failure to produce the sterol end products.
The mevalonate pathway generates dimethylallyldiphosphate, which is a precursor of the sterol biosynthetic pathway. Based on the oomycete genome searches for the homologous enzymes present in the mevalonate and sterol biosynthetic pathways (Fig. 3), we found that the oomycetes in the Saprolegniales subgroup (i.e., Ap. euteiches and Sa. parasitica) possess a nearly complete set of the enzymes in such pathways, whereas the oomycetes in the subgroups Pythiales, Peronosporales, and Albuginales (i.e., Pythium species, Phytophthora species, Hyaloperonospora species, and Albugo species) contain a restricted number of such enzymes (Tables 2 and 3). This finding is consistent with the findings that many oomycetes (including Py. insidiosum) of the non-Saprolegniales subgroups are sterol auxotrophs (11, 13), while the oomycetes of the subgroup Saprolegniales can synthesize their own sterols (i.e., cholesterol and fucosterol), as clearly shown for Ap. euteiches (12). The ability to synthesize sterols in the oomycetes of the Saprolegniales subgroup and the fungi could indicate their derivation from an ancient common ancestor. However, even if so, during the course of evolution, some critical enzymes involved in sterol synthesis (i.e., HMG1, HMG2, ERG1, ERG7, and ERG9) have been lost in the oomycetes of the non-Saprolegniales subgroups. As a compensatory mechanism, the non-Saprolegniales oomycetes increased their capacity to acquire exogenous sterol by greatly expanding the number of genes encoding sterol-binding and -uptake proteins—the elicitin gene family (36–38). In contrast, this elicitin gene expansion is absent in the Saprolegniales oomycetes (36, 38).
In the sterol biosynthetic pathway, ERG1 and ERG11 are the target enzymes of terbinafine and itraconazole, respectively. The enzyme-encoding sequence of ERG11, but not ERG1, was identified in the genomes of all organisms included in this study (Table 3). The ERG11 sequences were then used for investigation of the phylogenetic relationship among oomycetes, fungi, algae, and a diatom. The resulting ERG11-based tree located most of the oomycetes in group 2 and the remaining oomycetes (Ap. euteiches and Sa. parasitica) and all other organisms (fungi, algae, and diatom) in group 1 (Fig. 4). Based on the BLAST search using the query sequence from S. cerevisiae, the E values among the sterol-producing Saprolegniales oomycetes (E value range, −80 to −68) were much lower than that of the sterol auxotrophic, non-Saprolegniales oomycetes (E value range, −17 to −10) (Table 3). This finding suggests that the rate of gene modification (i.e., mutation, insertion, or deletion) of ERG11 since divergence of the oomycetes from the common ancestor with the fungi (represented by S. cerevisiae) was higher for the non-Saprolegniales oomycetes than that for the Saprolegniales oomycetes. This relationship is also found for other enzymes, i.e., ERG6, ERG24, ERG25, and ERG26 (Table 3), and it suggests that there was less selective pressure on the non-Saprolegniales oomycetes to maintain the sterol biosynthetic pathway.
In conclusion, the in vitro susceptibility testing demonstrated that the Py. insidiosum isolates were sensitive to the antifungal agents, but only at concentrations that are difficult or impossible to achieve in vivo. These results explain the clinical resistance of the drugs in the treatment of most pythiosis patients. The genome analysis of Py. insidiosum demonstrated that the organism contains an incomplete set of sterol biosynthetic enzymes. GC-MS analysis did not detect any sterol end products in Py. insidiosum. Clinical resistance of antifungal drugs in Py. insidiosum results from modifications or losses of some of the genes encoding the drug target sterol biosynthetic enzymes.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the Committee on Human Rights Related to Research Involving Human Subjects, at the Faculty of Medicine, Ramathibodi Hospital, Mahidol University (approval no. MURA2011/412/S3). Informed consent was not obtained from patients (from whom microorganisms were obtained) because the data were analyzed anonymously, and the institutional ethics committee waived the need for written informed consent from the participants.
Microorganisms and growth conditions.
Thirty isolates of Py. insidiosum from patients with vascular pythiosis (n = 10), ocular pythiosis (n = 7), cutaneous pythiosis (n = 3), and undefined forms of pythiosis (n = 10) were recruited in this study. The identity of each Py. insidiosum isolate was confirmed by zoospore induction (39, 40) and ribosomal DNA (rDNA) sequence homology analysis (41). All organisms were maintained on SD agar (pH 7.2) at room temperature.
Ten small pieces of SD agar containing a 7-day-old actively growing colony of Py. insidiosum (strain Pi-S) were transferred into 100 ml of SD broth (pH 7.2) and incubated, with shaking (150 rpm), at 37°C for 10 days. The organism was killed with 0.02% (wt/vol) thimerosal (Sigma). The hyphal mat was collected by filtration of the cultured broth through a 0.22-μm-pore-size membrane (Durapore; Merck Millipore). Candida albicans strain ATCC 90028 was cultured in yeast extract-peptone-dextrose medium, with shaking (250 rpm), at 30°C for 3 days. The yeast cells were collected by centrifugation at 6,000 × g for 10 min. Harvested organisms were lyophilized using a Genevac EZ-2 Elite machine and stored at −30°C until used for sterol extraction.
In vitro antifungal susceptibility assay.
The radial growth of Py. insidiosum was determined using the methods described by Brown et al. and Krajaejun et al. (32, 42), with some modifications. Briefly, Py. insidiosum was subcultured on SD agar at 37°C for 5 days. A 5-mm-diameter SD agar plug with actively growing mycelium was cut from the edge of a young Py. insidiosum colony and placed on SD agar (pH 7.2) containing various concentrations (128, 64, 32, 8, 2, 0.5, and 0 [no-drug control] mg/liter) of terbinafine (purity, 100% [Novartis]) or itraconazole (purity, 100% [Janssen Pharmaceutical]). The agar plate was incubated at 37°C for 2 days. Two colony diameters of each Py. insidiosum isolate were measured at day 2. All isolates were tested in duplicate. The average colony diameter was subtracted by 5 mm (the diameter in millimeters of an agar plug) and then divided by 2 to obtain a mean radial growth. The percentage of radial growth of each isolate, after exposure to the antifungal drug, was calculated based on the growth of the same isolate with no drug (control). The percentages of radial growths of Py. insidiosum exposed to different drug concentrations were statistically compared using the GraphPad Prism program version 5.00 (GraphPad Software, USA) and one-way analysis of variance (ANOVA) with Dunnett's posttest.
Sterol extraction.
Sterol extraction was performed using the methods of Axelsson et al. (43) and Madoui et al. (12), with some modifications. Briefly, the lyophilized samples (∼200 mg of either Ca. albicans yeasts or Py. insidiosum hyphae), 10 μg of 5α-cholestane (Sigma [used as an internal control]), and 3 ml of 1% (wt/vol) KOH in methanol were mixed and incubated at 80°C for 90 min. After the reaction mixture was cooled to room temperature, 1 ml of water and 2 ml of hexane were sequentially added to the mixture and incubated on a rocker (BioSan MR-1) overnight at room temperature. The organic phase (upper layer), containing sterols, was transferred to a new glass tube and evaporated using nitrogen gas. The residual material was derivatized at 60°C for 30 min using 20 μl of pyridine (Sigma) and 50 μl of N, O-bis (trimethylsilyl) trifluoroacetamide (Thermo Scientific). The derivatized material was dissolved in 500 μl toluene and subjected to gas chromatography-mass spectrometry (GC-MS).
Four commercially available sterols (including cholesterol [Tokyo Chemical Industry], ergosterol [Acros Organics], stigmasterol [Tokyo Chemical Industry], and fucosterol [Sigma]) were mixed and served as the standard for GC-MS analysis. After addition of 5α-cholestane, the sterol standard mixture was derivatized (as described above) and dissolved in 500 μl toluene.
GC-MS.
The sterol extracts and the sterol standard mixture were analyzed using GC-MS (model 7890A-5975C [Agilent Technologies]; capillary column [model 19091S-433HP-5MS] with 5% phenyl methyl siloxane [size, 30 m by 250 μm by 0.25 μm]). The electro-ionization mass spectrometry was set to the scan mode with an m/z range of 50 to 500. The selected-ion monitoring (SIM) mode was used to analyze trace amounts of sterol compounds: 5α-cholestane masses (m/z of 217, 357, and 372), cholesterol masses (m/z of 129, 329, 368, and 458), ergosterol masses (m/z of 131, 337, 363, and 468), stigmasterol masses (m/z of 83, 255, 394, and 484), and fucosterol masses (m/z of 257, 296, 386, and 484). Temperatures were set as follows: injector, 260°C; oven, 3°C/min increments from 230°C to 239°C, 1°C/min increments to 280°C, and then 280°C for 2 min; MS source, 230°C; and MS quadrupole, 150°C.
Sequence homology analysis.
Genomes or transcriptomes of 17 oomycetes (14, 17, 19, 22–28, 44, 45), two fungi (Candida albicans strain SC5314 and Aspergillus fumigatus strain Af293) (46, 47), and one each of diatom (48), microalga (45), brown tide alga (49), and brown alga (50) were retrieved from public databases (Table 4). The BLAST 2.2.28+ program (http://www.ncbi.nlm.nih.gov/) was locally installed for genome search analysis. The sequences of 22 enzymes (including ERG1 to -13, ERG19, ERG20, ERG24 to -27, HMG1, HMG2, and IDI1) involved in endogenous sterol production of the Saccharomyces cerevisiae strain S288c (51) were obtained from the NCBI database (Fig. 3; Tables 1 to 3) and used to BLAST search against all genomes or transcriptomes included in this study. The cutoff E value was 10−6.
Phylogenetic analysis.
Phylogenetic analysis of the ERG11 homologs, coding for the lanosterol 14-α demethylase (which is the drug target of itraconazole) from all microorganisms, was performed online at www.phylogeny.fr (Table 3) (52). Briefly, the sequences were aligned using the MUSCLE program (53). The phylogenetic relationship was analyzed by neighbor joining with 1,000 bootstraps (54) and the Jones-Taylor-Thornton matrix substitution model (55). A phylogenetic tree was generated by TreeDyn (56).
Sequence accession numbers.
All sequences of orthologous enzymes in the mevalonate and sterol biosynthetic pathways presented in the genome of Py. insidiosum (strain Pi-S) have been submitted to the DNA Data Bank of Japan database, under the accession numbers indicated in Table 1.
ACKNOWLEDGMENTS
We thank Boonmee Sathapatayavongs, Sasisopin Kiertiburanakul, Anuparp Kanchanatame, Angkana Chaiprasert, Somboon Srimuang, Nongnuch Vanittanakom, and Piriyaporn Chongtrakool for suggestions and material support. We also thank Thomas D. Sullivan for review of the manuscript.
This work is supported by the Thailand Research Fund (Theerapong Krajaejun; grant no. BRG5980009 and BRG5680011), the Research Fund from Mahidol University (Theerapong Krajaejun; grant no. E01/2558), and the Royal Golden Jubilee Ph.D. Scholarship Program (Tassanee Lerksuthirat; grant no. PHD/0092/2553). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The authors declare they have no conflicts of interest.
REFERENCES
- 1.Mendoza L, Ajello L, McGinnis MR. 1996. Infection caused by the oomycetous pathogen Pythium insidiosum. J Mycol Med 6:151–164. [Google Scholar]
- 2.Krajaejun T, Sathapatayavongs B, Pracharktam R, Nitiyanant P, Leelachaikul P, Wanachiwanawin W, Chaiprasert A, Assanasen P, Saipetch M, Mootsikapun P, Chetchotisakd P, Lekhakula A, Mitarnun W, Kalnauwakul S, Supparatpinyo K, Chaiwarith R, Chiewchanvit S, Tananuvat N, Srisiri S, Suankratay C, Kulwichit W, Wongsaisuwan M, Somkaew S. 2006. Clinical and epidemiological analyses of human pythiosis in Thailand. Clin Infect Dis 43:569–576. doi: 10.1086/506353. [DOI] [PubMed] [Google Scholar]
- 3.Gaastra W, Lipman LJ, De Cock AW, Exel TK, Pegge RB, Scheurwater J, Vilela R, Mendoza L. 2010. Pythium insidiosum: an overview. Vet Microbiol 146:1–16. doi: 10.1016/j.vetmic.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Thianprasit M, Chaiprasert A, Imwidthaya P. 1996. Human pythiosis. Curr Top Med Mycol 7:43–54. [PubMed] [Google Scholar]
- 5.Kwon-Chung KJ. 1994. Phylogenetic spectrum of fungi that are pathogenic to humans. Clin Infect Dis 19(Suppl):S1–S7. doi: 10.1093/clinids/19.Supplement_1.S1. [DOI] [PubMed] [Google Scholar]
- 6.Kamoun S. 2003. Molecular genetics of pathogenic oomycetes. Eukaryot Cell 2:191–199. doi: 10.1128/EC.2.2.191-199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendoza L, Hernandez F, Ajello L. 1993. Life cycle of the human and animal oomycete pathogen Pythium insidiosum. J Clin Microbiol 31:2967–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendoza L, Newton JC. 2005. Immunology and immunotherapy of the infections caused by Pythium insidiosum. Med Mycol 43:477–486. doi: 10.1080/13693780500279882. [DOI] [PubMed] [Google Scholar]
- 9.dos Santos CEP, Ubiali DG, Pescador CA, Zanette RA, Santurio JM, Marques LC. 2014. Epidemiological survey of equine pythiosis in the Brazilian Pantanal and nearby areas: results of 76 cases. J Equine Vet Sci 34:270–274. doi: 10.1016/j.jevs.2013.06.003. [DOI] [Google Scholar]
- 10.Sathapatayavongs B, Leelachaikul P, Prachaktam R, Atichartakarn V, Sriphojanart S, Trairatvorakul P, Jirasiritham S, Nontasut S, Eurvilaichit C, Flegel T. 1989. Human pythiosis associated with thalassemia hemoglobinopathy syndrome. J Infect Dis 159:274–280. doi: 10.1093/infdis/159.2.274. [DOI] [PubMed] [Google Scholar]
- 11.Marshall JA, Dennis AL, Kumazawa T, Haynes AM, Nes WD. 2001. Soybean sterol composition and utilization by Phytophthora sojae. Phytochemistry 58:423–428. doi: 10.1016/S0031-9422(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 12.Madoui M-A, Bertrand-Michel J, Gaulin E, Dumas B. 2009. Sterol metabolism in the oomycete Aphanomyces euteiches, a legume root pathogen. New Phytol 183:291–300. doi: 10.1111/j.1469-8137.2009.02895.x. [DOI] [PubMed] [Google Scholar]
- 13.Gaulin E, Bottin A, Dumas B. 2010. Sterol biosynthesis in oomycete pathogens. Plant Signal Behav 5:258–260. doi: 10.4161/psb.5.3.10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CM, Dorrance AE, Dou D, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee M-K, McDonald WH, Medina M, Meijer HJ, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JK, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BW, Terry A, Torto-Alalibo TA, Win J, Xu Z, Zhang H, Grigoriev IV, Rokhsar DS, Boore JL. 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 15.Shenep JL, English BK, Kaufman L, Pearson TA, Thompson JW, Kaufman RA, Frisch G, Rinaldi MG. 1998. Successful medical therapy for deeply invasive facial infection due to Pythium insidiosum in a child. Clin Infect Dis 27:1388–1393. doi: 10.1086/515042. [DOI] [PubMed] [Google Scholar]
- 16.Madoui MA, Gaulin E, Mathé C, San Clemente H, Couloux A, Wincker P, Dumas B. 2007. AphanoDB: a genomic resource for Aphanomyces pathogens. BMC Genomics 8:471. doi: 10.1186/1471-2164-8-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaulin E, Madoui M-A, Bottin A, Jacquet C, Mathé C, Couloux A, Wincker P, Dumas B. 2008. Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS One 3:e1723. doi: 10.1371/journal.pone.0001723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krajaejun T, Lerksuthirat T, Garg G, Lowhnoo T, Yingyong W, Khositnithikul R, Tangphatsornruang S, Suriyaphol P, Ranganathan S, Sullivan TD. 2014. Transcriptome analysis reveals pathogenicity and evolutionary history of the pathogenic oomycete Pythium insidiosum. Fungal Biol 118:640–653. doi: 10.1016/j.funbio.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Rujirawat T, Patumcharoenpol P, Lohnoo T, Yingyong W, Lerksuthirat T, Tangphatsornruang S, Suriyaphol P, Grenville-Briggs LJ, Garg G, Kittichotirat W, Krajaejun T. 2015. Draft genome sequence of the pathogenic oomycete Pythium insidiosum strain Pi-S, isolated from a patient with pythiosis. Genome Announc 3:e00574-15. doi: 10.1128/genomeA.00574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krajaejun T, Khositnithikul R, Lerksuthirat T, Lowhnoo T, Rujirawat T, Petchthong T, Yingyong W, Suriyaphol P, Smittipat N, Juthayothin T, Phuntumart V, Sullivan TD. 2011. Expressed sequence tags reveal genetic diversity and putative virulence factors of the pathogenic oomycete Pythium insidiosum. Fungal Biol 115:683–696. doi: 10.1016/j.funbio.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Ascunce MS, Huguet-Tapia JC, Braun EL, Ortiz-Urquiza A, Keyhani NO, Goss EM. 2016. Whole genome sequence of the emerging oomycete pathogen Pythium insidiosum strain CDC-B5653 isolated from an infected human in the USA. Genomics Data 7:60–61. doi: 10.1016/j.gdata.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, Zerillo MM, Beakes GW, Boore JL, Busam D, Dumas B, Ferriera S, Fuerstenberg SI, Gachon CM, Gaulin E, Govers F, Grenville-Briggs L, Horner N, Hostetler J, Jiang RH, Johnson J, Krajaejun T, Lin H, Meijer HJ, Moore B, Morris P, Phuntmart V, Puiu D, Shetty J, Stajich JE, Tripathy S, Wawra S, van West P, Whitty BR, Coutinho PM, Henrissat B, Martin F, Thomas PD, Tyler BM, De Vries RP, Kamoun S, Yandell M, Tisserat N, Buell CR. 2010. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol 11:R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adhikari BN, Hamilton JP, Zerillo MM, Tisserat N, Lévesque CA, Buell CR. 2013. Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes. PLoS One 8:e75072. doi: 10.1371/journal.pone.0075072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamour KH, Mudge J, Gobena D, Hurtado-Gonzales OP, Schmutz J, Kuo A, Miller NA, Rice BJ, Raffaele S, Cano LM, Bharti AK, Donahoo RS, Finley S, Huitema E, Hulvey J, Platt D, Salamov A, Savidor A, Sharma R, Stam R, Storey D, Thines M, Win J, Haas BJ, Dinwiddie DL, Jenkins J, Knight JR, Affourtit JP, Han CS, Chertkov O, Lindquist EA, Detter C, Grigoriev IV, Kamoun S, Kingsmore SF. 2012. Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol Plant Microbe Interact 25:1350–1360. doi: 10.1094/MPMI-02-12-0028-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AM, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JI, Bulone V, Cai G, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, Fischbach MA, Fugelstad J, Gilroy EM, Gnerre S, Green PJ, Grenville-Briggs LJ, Griffith J, Grünwald NJ, Horn K, Horner NR, Hu C-H, Huitema E, Jeong D-H, Jones AM, Jones JD, Jones RW, Karlsson EK, Kunjeti SG, Lamour K, Liu Z, Ma L, Maclean D, Chibucos MC, McDonald H, McWalters J, Meijer HJ, Morgan W, Morris PF, Munro CA, O'Neill K, Ospina-Giraldo M, Pinzón A, Pritchard L, Ramsahoye B, Ren Q, Restrepo S, Roy S, Sadanandom A, Savidor A, Schornack S, Schwartz DC, Schumann UD, Schwessinger B, Seyer L, Sharpe T, Silvar C, Song J, Studholme DJ, Sykes S, Thines M, van de Vondervoort PJ, Phuntumart V, Wawra S, Weide R, Win J, Young C, Zhou S, Fry W, Meyers BC, van West P, Ristaino J, Govers F, Birch PR, Whisson SC, Judelson HS, Nusbaum C. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 26.Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, Thines M, Ah-Fong A, Anderson R, Badejoko W, Bittner-Eddy P, Boore JL, Chibucos MC, Coates M, Dehal P, Delehaunty K, Dong S, Downton P, Dumas B, Fabro G, Fronick C, Fuerstenberg SI, Fulton L, Gaulin E, Govers F, Hughes L, Humphray S, Jiang RH, Judelson H, Kamoun S, Kyung K, Meijer H, Minx P, Morris P, Nelson J, Phuntumart V, Qutob D, Rehmany A, Rougon-Cardoso A, Ryden P, Torto-Alalibo T, Studholme D, Wang Y, Win J, Wood J, Clifton SW, Rogers J, Van den Ackerveken G, Jones JD, McDowell JM, Beynon J, Tyler BM. 2010. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Links MG, Holub E, Jiang RH, Sharpe AG, Hegedus D, Beynon E, Sillito D, Clarke WE, Uzuhashi S, Borhan MH. 2011. De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics 12:503. doi: 10.1186/1471-2164-12-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang RH, de Bruijn I, Haas BJ, Belmonte R, Löbach L, Christie J, van den Ackerveken G, Bottin A, Bulone V, Díaz-Moreno SM, Dumas B, Fan L, Gaulin E, Govers F, Grenville-Briggs LJ, Horner NR, Levin JZ, Mammella M, Meijer HJ, Morris P, Nusbaum C, Oome S, Phillips AJ, van Rooyen D, Rzeszutek E, Saraiva M, Secombes CJ, Seidl MF, Snel B, Stassen JH, Sykes S, Tripathy S, van den Berg H, Vega-Arreguin JC, Wawra S, Young SK, Zeng Q, Dieguez-Uribeondo J, Russ C, Tyler BM, van West P. 2013. Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLoS Genet 9:e1003272. doi: 10.1371/journal.pgen.1003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loreto ES, Tondolo JS, Pilotto MB, Alves SH, Santurio JM. 2014. New insights into the in vitro susceptibility of Pythium insidiosum. Antimicrob Agents Chemother 58:7534–7537. doi: 10.1128/AAC.02680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fonseca AO, Pereira DI, Maia Filho FS, Osório LG, Maroneze BP, Valente JS, Pötter L, Meireles MC. 2014. In vitro susceptibility of zoospores and hyphae of Pythium insidiosum to antifungals. J Antimicrob Chemother 69:1564–1567. doi: 10.1093/jac/dku021. [DOI] [PubMed] [Google Scholar]
- 31.Sekhon AS, Padhye AA, Garg AK. 1992. In vitro sensitivity of Penicillium marneffei and Pythium insidiosum to various antifungal agents. Eur J Epidemiol 8:427–432. doi: 10.1007/BF00158578. [DOI] [PubMed] [Google Scholar]
- 32.Brown TA, Grooters AM, Hosgood GL. 2008. In vitro susceptibility of Pythium insidiosum and a Lagenidium sp to itraconazole, posaconazole, voriconazole, terbinafine, caspofungin, and mefenoxam. Am J Vet Res 69:1463–1468. doi: 10.2460/ajvr.69.11.1463. [DOI] [PubMed] [Google Scholar]
- 33.Stopiglia CD, Marchese DP, Heidrich D, Sorrentino JM, Vieira FJ, Scroferneker ML. 2012. Comparison between two culture media for in vitro evaluation of antifungal susceptibility of the Sporothrix schenckii complex. An Bras Dermatol 87:561–565. doi: 10.1590/S0365-05962012000400007. [DOI] [PubMed] [Google Scholar]
- 34.Argenta JS, Santurio JM, Alves SH, Pereira DI, Cavalheiro AS, Spanamberg A, Ferreiro L. 2008. In vitro activities of voriconazole, itraconazole, and terbinafine alone or in combination against Pythium insidiosum isolates from Brazil. Antimicrob Agents Chemother 52:767–769. doi: 10.1128/AAC.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovarik JM, Mueller EA, Zehender H, Denouel J, Caplain H, Millerioux L. 1995. Multiple-dose pharmacokinetics and distribution in tissue of terbinafine and metabolites. Antimicrob Agents Chemother 39:2738–2741. doi: 10.1128/AAC.39.12.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lerksuthirat T, Lohnoo T, Inkomlue R, Rujirawat T, Yingyong W, Khositnithikul R, Phaonakrop N, Roytrakul S, Sullivan TD, Krajaejun T. 2015. The elicitin-like glycoprotein, ELI025, is secreted by the pathogenic oomycete Pythium insidiosum and evades host antibody responses. PLoS One 10:e0118547. doi: 10.1371/journal.pone.0118547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panabières F, Ponchet M, Allasia V, Cardin L, Ricci P. 1997. Characterization of border species among Pythiaceae: several Pythium isolates produce elicitins, typical proteins from Phytophthora spp. Mycol Res 101:1459–1468. doi: 10.1017/S0953756297004413. [DOI] [Google Scholar]
- 38.Jiang RH, Tyler BM, Whisson SC, Hardham AR, Govers F. 2006. Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol Biol Evol 23:338–351. [DOI] [PubMed] [Google Scholar]
- 39.Chaiprasert A, Samerpitak K, Wanachiwanawin W, Thasnakorn P. 1990. Induction of zoospore formation in Thai isolates of Pythium insidiosum. Mycoses 33:317–323. [DOI] [PubMed] [Google Scholar]
- 40.Mendoza L, Prendas J. 1988. A method to obtain rapid zoosporogenesis of Pythium insidiosum. Mycopathologia 104:59–62. doi: 10.1007/BF00437925. [DOI] [PubMed] [Google Scholar]
- 41.Badenoch PR, Coster DJ, Wetherall BL, Brettig HT, Rozenbilds MA, Drenth A, Wagels G. 2001. Pythium insidiosum keratitis confirmed by DNA sequence analysis. Br J Ophthalmol 85:502–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krajaejun T, Chongtrakool P, Angkananukul K, Brandhorst TT. 2010. Effect of temperature on growth of the pathogenic oomycete Pythium insidiosum. Southeast Asian J Trop Med Public Health 41:1462–1466. [PubMed] [Google Scholar]
- 43.Axelsson BO, Saraf A, Larsson L. 1995. Determination of ergosterol in organic dust by gas chromatography-mass spectrometry. J Chromatogr B Biomed Appl 666:77–84. doi: 10.1016/0378-4347(94)00553-H. [DOI] [PubMed] [Google Scholar]
- 44.Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub E, Studholme DJ, Maclean D, Jones JD. 2011. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol 9:e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, Otillar R, Poliakov A, Ratnere I, Riley R, Smirnova T, Rokhsar D, Dubchak I. 2012. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40:D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, Orvis J, Shah P, Skrzypek MS, Binkley G, Miyasato SR, Wortman JR, Sherlock G. 2010. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res 38:D420–D427. doi: 10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G. 2012. The Candida Genome Database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res 40:D667–D674. doi: 10.1093/nar/gkr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret J-P, Chiovitti A, Choi CJ, Coesel S, Martino AD, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, Roche JL, Lindquist E, Lommer M, Martin-Jézéquel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot Le Secq M-P, Napoli C, Obornik M, Parker MS, Petit J-L, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van de Peer Y, Grigoriev IV. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 49.Gobler CJ, Berry DL, Dyhrman ST, Wilhelm SW, Salamov A, Lobanov AV, Zhang Y, Collier JL, Wurch LL, Kustka AB, Dill BD, Shah M, VerBerkmoes NC, Kuo A, Terry A, Pangilinan J, Lindquist EA, Lucas S, Paulsen IT, Hattenrath-Lehmann TK, Talmage SC, Walker EA, Koch F, Burson AM, Marcoval MA, Tang Y-Z, Lecleir GR, Coyne KJ, Berg GM, Bertrand EM, Saito MA, Gladyshev VN, Grigoriev IV. 2011. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci U S A 108:4352–4357. doi: 10.1073/pnas.1016106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury J-M, Badger JH, Beszteri B, Billiau K, Bonnet E, Bothwell JH, Bowler C, Boyen C, Brownlee C, Carrano CJ, Charrier B, Cho GY, Coelho SM, Collén J, Corre E, Da Silva C, Delage L, Delaroque N, Dittami SM, Doulbeau S, Elias M, Farnham G, Gachon CM, Gschloessl B, Heesch S, Jabbari K, Jubin C, Kawai H, Kimura K, Kloareg B, Küpper FC, Lang D, Le Bail A, Leblanc C, Lerouge P, Lohr M, Lopez PJ, Martens C, Maumus F, Michel G, Miranda-Saavedra D, Morales J, Moreau H, Motomura T, Nagasato C, Napoli CA, Nelson DR, Nyvall-Collén P, Peters AF, Pommier C, Potin P, Poulain J, Quesneville H, Read B, Rensing SA, Ritter A, Rousvoal S, Samanta M, Samson G, Schroeder DC, Ségurens B, Strittmatter M, Tonon T, Tregear JW, Valentin K, von Dassow P, Yamagishi T, Van de Peer Y, Wincker P. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 51.Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry JM, Saccharomyces Genome Database Project. 2006. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23:857–865. doi: 10.1002/yea.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 55.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 56.Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]