ABSTRACT
Extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae strains are increasing in prevalence worldwide. Carbapenem antibiotics are used as a first line of therapy against ESBL-producing Enterobacteriaceae. We examined a cohort of critical care patients for gastrointestinal colonization with carbapenem-resistant ESBL-producing strains (CR-ESBL strains). We cultured samples from this cohort of patients for ESBL-producing Klebsiella spp. and Escherichia coli and then tested the first isolate from each patient for susceptibility to imipenem, doripenem, meropenem, and ertapenem. Multilocus sequence typing was performed on isolates that produced an ESBL and that were carbapenem resistant. Among all patients admitted to an intensive care unit (ICU), 4% were positive for an ESBL-producing isolate and 0.64% were positive for a CR-ESBL strain on surveillance culture. Among the first ESBL-producing E. coli and Klebsiella isolates from the patients' surveillance cultures, 11.2% were carbapenem resistant. Sequence type 14 (ST14), ST15, ST42, and ST258 were the dominant sequence types detected in this cohort of patients, with ST15 and ST258 steadily increasing in prevalence from 2006 to 2009. Patients colonized by a CR-ESBL strain were significantly more likely to receive antipseudomonal and anti-methicillin-resistant Staphylococcus aureus (anti-MRSA) therapy prior to ICU admission than patients colonized by carbapenem-susceptible ESBL-producing strains. They were also significantly more likely to have received a cephalosporin or a carbapenem antibiotic than patients colonized by carbapenem-susceptible ESBL-producing strains. In conclusion, in a cohort of patients residing in intensive care units within the United States, we found that 10% of the isolates were resistant to at least one carbapenem antibiotic. The continued emergence of carbapenem-resistant ESBL-producing strains is of significant concern, as infections due to these organisms are notoriously difficult to treat.
KEYWORDS: ESBL, MLST, carbapenem resistant
INTRODUCTION
Antibiotic-resistant Enterobacteriaceae infections are associated with poor clinical outcomes, including increased rates of morbidity, mortality and increased health care costs (1–3). These antibiotic-resistant bacteria, including carbapenem-resistant Enterobacteriaceae (CRE) and extended-spectrum β-lactamase (ESBL)-producing bacteria, are increasing in prevalence worldwide, despite infection control efforts and antibiotic stewardship efforts (4, 5).
Carbapenem antibiotics with activity against multiple Gram-positive and Gram-negative bacterial pathogens, including members of the Enterobacteriaceae, are used as a first line of therapy against ESBL-producing Enterobacteriaceae and are often used as empirical therapy for the treatment of serious infections or as definitive therapy for antimicrobial-resistant or complex infections. As one of the few options available for the treatment of multiresistant infections, such as those caused by ESBL-producing organisms, it is imperative that we understand the emergence of and risk factors for carbapenem resistance among ESBL-producing organisms.
Gastrointestinal colonization by multidrug-resistant Gram-negative bacteria has been shown to precede clinical infection (6–8), and carbapenems are often used to treat infections cause by ESBL-producing Enterobacteriaceae strains. Furthermore, patients gastrointestinally colonized with unidentified carbapenem-resistant ESBL-producing organisms (CR-ESBL organisms) may serve as an unrecognized reservoir and facilitate the persistent transmission of these organisms to other hospitalized patients. Thus, we examined a cohort of critical care patients for gastrointestinal colonization with CR-ESBL strains.
RESULTS
Patient characteristics.
During the study period there were 15,753 adult admissions to the medical intensive care unit (MICU) and the surgical intensive care unit (SICU). A total of 420 admissions (2.7%) were excluded because the patient had a surveillance culture positive for an ESBL-producing strain on a prior admission during the study period. A total of 1,372 admissions (8.7%) were excluded because a surveillance culture of a perianal swab specimen from the patient was not performed on admission.
The final sample size was 13,961 admissions. Admission and patient characteristics are displayed in Table 1. The mean age was 55.42 years (standard deviation [SD], 15.8 years), and 54.8% of the patients were male. The median length of stay in the intensive care unit (ICU) was 2.9 days (interquartile range [IQR], 1 to 6 days), 19.5% of patients died in the hospital during their admission, and 13.0% of patients died in an ICU. The most prevalent diagnoses or comorbid illnesses were chronic pulmonary disease (20.9%), diabetes (17.7%), and cancer (19.0%).
TABLE 1.
Characteristics of adult patients admitted to medical and surgical ICUsa
| Patient characteristic | Values |
|---|---|
| Mean (SD) age (yr) | 55.42 (15.8) |
| No. (%) of patients of male sex | 7,651 (54.8) |
| No. (%) of patients of white race | 7,387 (52.9) |
| No. (%) of patients who died in hospital | 2,717 (19.5) |
| No. (%) of patients who died in ICU | 1,816 (13.0) |
| No. (%) of patients with: | |
| Surveillance culture positive for ESBL-producing isolate on ICU admission | 557 (4.0) |
| Surveillance culture positive for ESBL-producing isolate >48 h after ICU admission | 247 (1.8) |
| Clinical culture positive for E. coli or Klebsiella spp. at least 48 h after date of transfer into ICU | 77 (0.55) |
| Clinical culture positive for any E. coli or Klebsiella spp. during admission | 67 (0.27) |
| Median (IQR) ICU length of stay (days) | 2.9 (1–6) |
| Mean (SD) Charlson comorbidity index | 2.6 (2.5) |
| Mean (SD) chronic disease score | 7.5 (4.1) |
| No. (%) of patients with the following comorbid illness: | |
| HIV/AIDS | 600 (4.30) |
| Cancer | 2,653 (19.0) |
| Cerebrovascular disease | 1,919 (13.7) |
| Chronic pulmonary disease | 2,929 (20.9) |
| Heart failure | 2,000 (14.3) |
| Dementia | 31 (0.2) |
| Diabetes | 2,467 (17.7) |
| Liver disease (severe) | 942 (6.7) |
| Myocardial infarction | 1,065 (7.6) |
| Peripheral vascular | 941 (6.7) |
| Renal disease | 1,638 (11.7) |
Data are for 13,961 patients.
Four percent (557/13,961) of admissions had a surveillance culture result indicating positivity for an ESBL-producing strain upon ICU admission; 1.8% (247/13,961) of all admissions had a negative culture result on admission but a subsequent surveillance culture result indicating positivity for an ESBL-producing strain result during their ICU stay, indicating the possible acquisition of the ESBL-producing strain during their ICU stay. One percent (144/13,961) of the patients in the cohort had a positive clinical culture result for Escherichia coli or Klebsiella spp. at any point during their admission; 77 (53.5%) of these patients were positive 48 h or more after admission to an ICU. Thirty percent (43/144) of the patients with a positive clinical culture result had a prior surveillance culture result positive for an E. coli or Klebsiella species producing an ESBL.
The prevalence of colonization with a CR-ESBL strain was 0.64% (90/13,961) among all ICU admissions, and among the first ESBL-producing E. coli, Klebsiella pneumoniae, and Klebsiella oxytoca isolates from the patients' surveillance cultures, the prevalence of carbapenem resistance was 11.2% (90/804). The differences between patients colonized with a CR-ESBL strain and patients colonized with a carbapenem-susceptible ESBL-producing strain are displayed in Table 2. Patients colonized with a CR-ESBL strain had a significantly longer median length of stay in an ICU. There were no other significant differences in demographic characteristics, clinical characteristics, or comorbid illnesses between patients colonized with CR-ESBL strains and patients colonized with carbapenem-susceptible ESBL-producing strains. There were, however, several significant differences between these two groups with respect to their antibiotic exposures before their ICU admission. Patients colonized by CR-ESBL strains were significantly more likely to receive any antimicrobial therapy and, specifically, were more likely to receive antipseudomonal, anti-methicillin-resistant Staphylococcus aureus (anti-MRSA), or vancomycin therapy prior to ICU admission than patients colonized by carbapenem-susceptible ESBL-producing strains. They were also significantly more likely to have received a cephalosporin or a carbapenem antibiotic than patients colonized by carbapenem-susceptible ESBL-producing strains.
TABLE 2.
Characteristics of patients positive for CR-ESBL strains compared to those of patients positive for carbapenem-susceptible ESBL-producing isolatesa
| Patient characteristic | Values for patients infected with: |
P valueb | |
|---|---|---|---|
| CR-ESBL strain (n = 90) | Carbapenem-susceptible ESBL-producing strain (n = 714) | ||
| Mean (SD) age (yr) | 56.9 (14.8) | 57.1 (16.3) | 0.89 |
| No. (%) of patients of male sex | 54 (60) | 395 (55.3) | 0.40 |
| No. (%) of patients of white race | 43 (48) | 338 (47.3) | 0.90 |
| Median (IQR) ICU length of stay (days) | 8.9 (17.0) | 5.4 (9.3) | 0.002c |
| Median (IQR) Charlson comorbidity index | 2.0 (2.0) | 2.0 (2.0) | 0.61c |
| No. (%) of patients receiving the following antibiotic before ICU stay: | 56 (62) | 350 (49) | 0.02 |
| Antipseudomonal | 44 (49) | 274 (38) | 0.05 |
| Anti-anerobic organism agent | 41 (46) | 267 (37) | 0.13 |
| Beta-lactam | 44 (49) | 294 (41) | 0.16 |
| Anti-MRSA agent | 39 (43) | 196 (28) | 0.002 |
| Vancomycin | 33 (37) | 173 (24) | 0.01 |
| Antifungals | 11 (12) | 51 (7.1) | 0.09 |
| Imipenem | 11 (12) | 43 (6.0) | 0.03 |
| Carbapenem | 16 (18) | 48 (6.7) | <0.001 |
| Any cephalosporin | 23 (26) | 117 (16) | 0.03 |
| 1st generation | 3 (3.3) | 48 (6.7) | 0.22 |
| 2nd generation | 5 (5.6) | 9 (1.3) | 0.01d |
| 3rd generation | 6 (6.7) | 30 (4.2) | 0.28d |
| Fluoroquinolones | 5 (5.6) | 68 (9.5) | 0.22 |
| Piperacillin-tazobactam | 26 (29) | 188 (26) | 0.60 |
| No. (%) of patients with the following comorbid illness: | |||
| HIV/AIDS | 4 (4.4) | 33 (4.6) | >0.99d |
| Cancer | 10 (11) | 85 (12) | 0.83 |
| Cerebrovascular disease | 7 (7.8) | 54 (7.6) | 0.94 |
| Chronic pulmonary disease | 14 (16) | 150 (21) | 0.23 |
| Heart failure | 19 (21) | 134 (19) | 0.59 |
| Dementia | 1 (1.1) | 2 (0.3) | 0.30d |
| Diabetes | 12 (13) | 136 (19) | 0.19 |
| Liver disease (severe) | 8 (8.9) | 45 (6.3) | 0.35 |
| Myocardial infarction | 7 (7.8) | 39 (5.5) | 0.37 |
| Peripheral vascular disease | 5 (5.6) | 30 (4.2) | 0.58d |
| Renal disease | 21 (23) | 145 (20) | 0.50 |
| Mean (SD) chronic disease score | 8.4 (4.0) | 8.1 (4.2) | 0.56 |
| No. (%) of patient deaths during admission | 24 (27) | 184 (26) | 0.85 |
Data are for 804 patients.
P values were from a chi-square test for categorical variables or Student's t test for continuous variables, except where noted.
Wilcoxon rank sum test.
Fisher's exact test.
Microbiological characteristics.
The prevalence of ESBL-producing E. coli and Klebsiella species in our cohort of ICU patients was 5.8% (804/13,961). From these 804 patients, we detected 891 isolates; 39% (345/891) were E. coli isolates and 61% (546/891) were Klebsiella spp. Data by year are shown in Table 3, which shows a large increase in the prevalence of ESBL-producing strains from 2005 to 2006. Of note, 57 patients (7.1%) were colonized with both an ESBL-producing E. coli strain and an ESBL-producing Klebsiella sp.
TABLE 3.
Number of ESBL-producing E. coli and Klebsiella species isolates from 2001 to 2009
| Organism | No. of isolates |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2001a | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009a | Total | |
| E. coli | 6 | 25 | 40 | 14 | 23 | 89 | 62 | 58 | 28 | 345 |
| Klebsiella spp. | 9 | 27 | 13 | 12 | 35 | 144 | 120 | 135 | 51 | 546 |
| Total | 15 | 52 | 53 | 26 | 58 | 233 | 182 | 193 | 79 | 891 |
Data are from 6 months.
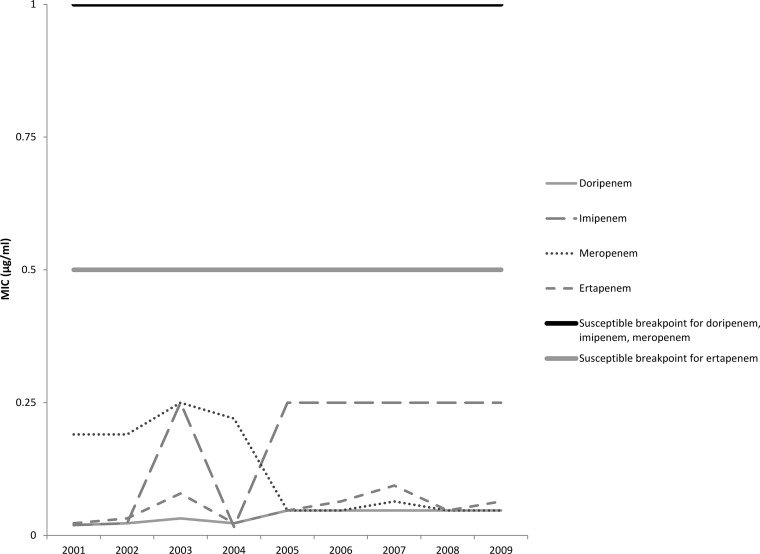
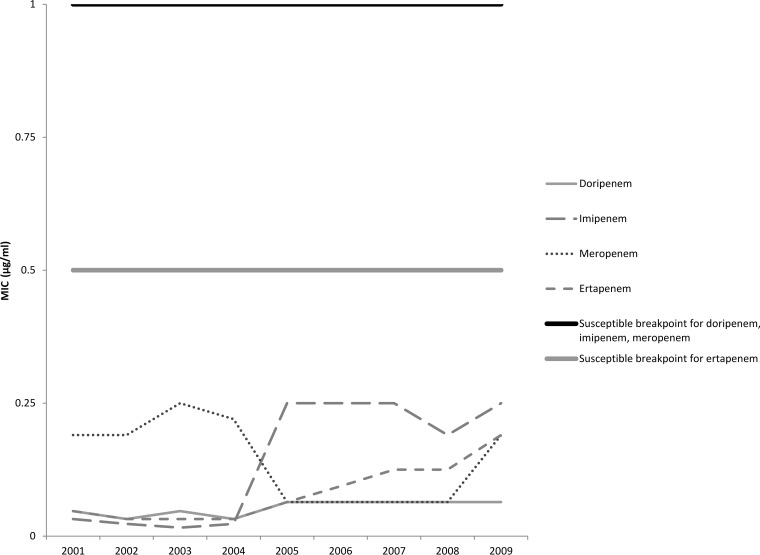
The median MICs for each carbapenem antibiotic by year are shown in Fig. 1 and 2. For both E. coli and Klebsiella spp., the median MICs for all antibiotics except meropenem increased in 2005, which was about the same time that an increase in the prevalence of colonization with ESBL-producing strains was seen. For meropenem, a decrease in the MICs for both E. coli and Klebsiella spp. was noted beginning in 2005; however, the MICs for both groups of isolates began increasing in 2007 and by 2009 the MICs were similar to data reported for 2001 to 2004.
FIG 1.
Median carbapenem MIC per year for ESBL-producing Escherichia coli isolates (n = 345).
FIG 2.
Median carbapenem MIC per year for ESBL-producing Klebsiella spp. (n = 546).
Of the 804 patients, 90 patients (11.2%) were colonized by 91 isolates determined to be CR-ESBL strains, 22 (24%) were colonized by E. coli strains, and 69 (76%) were colonized by Klebsiella spp. One E. coli isolate and eight K. pneumoniae isolates were resistant to all four carbapenem antibiotics (Table 4), with more than half of these isolates being collected in 2009. In addition, 26 K. pneumoniae isolates and 7 E. coli isolates had intermediate resistance to ertapenem but were susceptible to the other three carbapenems. Of the 91 CR-ESBL isolates, 46 (50.5%) were positive for blaKPC (17 were E. coli, 28 were K. pneumoniae, and 1 was K. oxytoca).
TABLE 4.
Characteristics of CR-ESBL strains resistant to all four carbapenems
| Organism | MIC (μg/ml) |
KPC productiona | Yr | MLST ST | |||
|---|---|---|---|---|---|---|---|
| Imipenem | Meropenem | Doripenem | Ertapenem | ||||
| E. coli | >32 | 6 | 12 | 32 | ND | 2009 | ST131 |
| K. pneumoniae | >32 | 8 | 12 | 16 | KPC | 2002 | ST11 |
| K. pneumoniae | 8 | 24 | 12 | >32 | KPC | 2005 | ST228 |
| K. pneumoniae | >32 | 4 | 4 | 12 | KPC | 2008 | ST111 |
| K. pneumoniae | 8 | 4 | 8 | 12 | KPC | 2009 | ST258 |
| K. pneumoniae | 8 | 8 | 6 | 8 | KPC | 2009 | ST258 |
| K. pneumoniae | 12 | >32 | 12 | >32 | KPC | 2009 | ST258 |
| K. pneumoniae | >32 | >32 | >32 | >32 | KPC | 2009 | ST258 |
| K. pneumoniae | >32 | 4 | 8 | 32 | ND | 2009 | ST15 |
KPC, Klebsiella pneumoniae carbapenemase; ND, not detected.
Multilocus sequence typing (MLST) results are displayed for CR-ESBL K. pneumoniae and E. coli isolates by year in Tables 5 and 6, respectively. For K. pneumoniae isolates, sequence type 14 (ST14), ST15, ST42, and ST258 were seen to be the dominant sequence types (STs). The data showed that the ST258 strain was first detected in 2006 and that its incidence increased steadily each year until 2009, when 12 resistant ST258 isolates were detected in a 6-month period. Similar results were observed with ST15. Currently, there is not a standard MLST method for K. oxytoca; therefore, pulsed-field gel electrophoresis (PFGE) was performed. In contrast to the findings for K. pneumoniae, the PFGE results suggest that unrelated CR-ESBL K. oxytoca strains were isolated in 2008. Among the E. coli isolates, ST131, ST167, and ST393 were the dominant sequence types. Unlike the ST258 isolates of K. pneumoniae, the data do not suggest a dominant clone of E. coli that established itself from year to year.
TABLE 5.
Distribution of MLST STs among Klebsiella pneumoniae isolates intermediate or resistant to carbapenems from 2001 to 2009
| ST | No. of isolates |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2001a | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009a | Total | |
| ST11 | 1 | 1 | 2 | |||||||
| ST14 | 4 | 1 | 1 | 6 | ||||||
| ST15 | 1 | 3 | 1 | 3 | 8 | |||||
| ST37 | 1 | 1 | ||||||||
| ST39 | 1 | 1 | 1 | 3 | ||||||
| ST42 | 1 | 6 | 7 | |||||||
| ST111 | 1 | 2 | 3 | |||||||
| ST228 | 1 | 1 | ||||||||
| ST258 | 2 | 5 | 6 | 12 | 25 | |||||
| ST290 | 1 | 1 | ||||||||
| ST327 | 1 | 1 | ||||||||
| ST412 | 1 | 1 | ||||||||
| ST483 | 1 | 1 | ||||||||
| ST567 | 1 | 1 | ||||||||
| ST834 | 1 | 1 | ||||||||
| ST1083 | 1 | 1 | ||||||||
| ST1084 | 1 | 1 | ||||||||
| ST1085 | 1 | 1 | ||||||||
| ST1086 | 1 | 1 | ||||||||
| Undetermined | 1 | 2 | 3 | |||||||
Data are from 6 months.
TABLE 6.
Distribution of MLST STs among E. coli isolates intermediate or resistant to carbapenems from 2001 to 2009
| ST | No. of isolates |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2001a | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009a | Total | |
| ST131 | 1 | 1 | 4 | 2 | 1 | 9 | ||||
| ST167 | 3 | 2 | 1 | 6 | ||||||
| ST354 | 1 | 1 | ||||||||
| ST393 | 2 | 1 | 2 | |||||||
| ST405 | 1 | 1 | ||||||||
| ST410 | 1 | 1 | ||||||||
| ST1114 | 1 | 1 | ||||||||
| Undetermined | 1 | 1 | ||||||||
Data are from 6 months.
DISCUSSION
In the large cohort of ESBL-producing E. coli and Klebsiella spp. isolated from surveillance cultures of perianal swab specimens from critical care patients from 2001 to 2009 evaluated in the present study, we observed that approximately 10% of isolates were resistant to at least one carbapenem antibiotic. The median MICs of all carbapenems except meropenem appeared to increase among Klebsiella spp. and E. coli during the study period, correlating with the increase in the incidence of CR-ESBL strains. Data on health care-associated clinical Klebsiella and E. coli infections reported to the CDC in 2009 and 2010 showed the rates of carbapenem resistance to be 12.8% and 1.9%, respectively (9). These numbers are slightly higher for Klebsiella spp. and lower for E. coli than the numbers that we observed in our study; however, we studied only isolates known to produce ESBLs.
ESBL-producing organisms are prevalent worldwide, and their incidence appears to be increasing (4, 5). Infections due to these organisms, which are associated with poor outcomes, pose a significant challenge, in that multidrug resistance leads to limitations in the number of effective antibiotics that are available. Carbapenem antibiotics are considered the drug of choice for treating infections caused by ESBL-producing organisms. Carbapenem resistance among ESBL-producing strains further compounds the challenge of effective treatment, often leaving clinicians to decide between agents with reduced efficacy and agents with increased toxicity, such as polymyxins or tigecycline. The relatively high rates of carbapenem-resistant ESBL-producing organisms seen in this study are thus of great concern, and research needs to be done in the future to identify strategies aimed at preventing their further emergence. CR-ESBL strains should be considered along with other multidrug-resistant organisms, including extremely drug-resistant (XDR) Acinetobacter baumannii and (XDR) Pseudomonas aeruginosa strains.
Given the high acuity of illness in this population, many patients received antibiotic therapy prior to ICU admission, and several of these antibiotic exposures were associated with carbapenem resistance. Patients colonized by CR-ESBL strains were significantly more likely to receive antipseudomonal and anti-methicillin-resistant Staphylococcus aureus (anti-MRSA) therapy prior to ICU admission than patients colonized by carbapenem-susceptible ESBL-producing strains. They were also significantly more likely to have received a cephalosporin or carbapenem antibiotic than patients colonized by carbapenem-susceptible ESBL-producing strains. Molecular data for the CR-ESBL strains suggest no sporadic strains for E. coli, but among the K. pneumoniae isolates, numerous identical strains were detected in a single year or, once they were detected, became endemic within the ICUs. As such, molecular data suggest that CR-ESBL E. coli and K. oxytoca strains are more likely to be due to antibiotic selective pressure and CR-ESBL K. pneumoniae strains are more likely to be due to patient-to-patient transmission.
This study had limitations. First, isolates were from one tertiary care hospital, so the generalizability to other hospitals might be limited. Specifically, the resistance patterns detected in the hospital that we studied may not be comparable to those detected in other hospitals in other regions of the United States. We also included only carbapenem-resistant isolates that were ESBL positive. The prevalence of carbapenem-resistant isolates and the risk factors for patients who acquire carbapenem resistance could have been different if we had studied carbapenem resistance in all Enterobacteriaceae and not just those that were ESBL positive. The use of ceftazidime in the screening agar may have caused the CTX-M family of β-lactamases to have been missed. However, in previous studies that used this screening agar, we were able to detect bacteria harboring blaCTX-M (10). Lastly, even though we identified several antimicrobial exposures to be risk factors for carbapenem resistance, our dichotomous classification of antimicrobial exposures may have underestimated the magnitude of these and other antimicrobial associations (11).
In conclusion, this is the first study, to our knowledge, to examine the rate of colonization with carbapenem-resistant ESBL-producing E. coli and K. pneumoniae isolates in a cohort of patients residing in intensive care units within the United States, and we found that 10% of isolates were resistant to at least one carbapenem antibiotic. The continued emergence of carbapenem-resistant ESBL strains is of significant concern, as infections due to these organisms are notoriously difficult to treat.
MATERIALS AND METHODS
Study population.
This study utilized surveillance cultures of perianal swab specimens collected from a cohort of adult (age, ≥18 years) patients admitted to the medical and surgical intensive care units of the University of Maryland Medical Center between 1 September 2001 and 30 June 2009. During the study period, the MICU was a 10-bed unit with private rooms that became a 29-bed unit with private rooms in April 2006, and the SICU was a 19-bed unit with private rooms. Prior to study commencement, the University of Maryland, Baltimore, Institutional Review Board approved this study.
Perianal swab specimens for culture were obtained from the patients at ICU admission (within 48 h of admission), weekly, and at the time of ICU discharge according to University of Maryland Medical Center Department of Infection Prevention surveillance practices. Samples for culture were obtained using Staplex II swabs (Staplex, Etobiocoke, Ontario, Canada) and frozen in tryptic soy broth with 15% glycerol using validated methods (12). The rate of compliance with the collection of perianal swab specimens for culture was approximately 90%. For patients who had multiple admissions, each admission could be entered into the cohort until the patient had a culture result indicating positivity for an ESBL-producing organism, after which data on subsequent admissions and associated patient data were excluded from the analysis. Patients were excluded from the study if a specimen for culture was not collected within the first 48 h of admission or if only one swab specimen was collected during their ICU stay.
The characteristics of the eligible patients were obtained using a relational database that contains patients' administrative, pharmaceutical, and microbiological data. These data were validated in previous studies and have positive and negative predictive values in excess of 99% compared to those from paper medical records (13–17).
Bacterial identification.
Perianal swab specimens were plated on MacConkey agar with 1 μg/μl ceftazidime; lactose-fermenting organisms were identified to be Escherichia coli and Klebsiella spp. using standard protocols. Isolates that harbored ESBLs were detected by the use of the ESBL confirmation test according to Clinical and Laboratory Standards Institute (CLSI) guidelines (18) described in a previous study (10). Carbapenem MICs were measured using an Etest (bioMérieux, Durham, NC) for imipenem, doripenem, meropenem, and ertapenem for the first Klebsiella oxytoca, K. pneumoniae, and E. coli isolate from each patient that was confirmed to produce an ESBL. Susceptibility testing was performed following CLSI guidelines (18). Isolates with intermediate resistance or resistance to any of the four carbapenem antibiotics to which the susceptibilities of the isolates were tested were defined to be carbapenem resistant.
Molecular typing.
Multilocus sequence typing was performed on any ESBL-producing E. coli and Klebsiella pneumoniae isolates that were nonsusceptible to a carbapenem. MLST was performed according to previously published protocols (19, 20). The nucleotide sequences of both strands were determined using published primers and compared to existing sequences in the MLST databases (http://mlst.ucc.ie/mlst; http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html) for assignment of allelic numbers. The isolates were assigned a sequence type (ST) number according to their allelic profiles.
Pulsed-field gel electrophoresis (PFGE) was performed on the K. oxytoca isolates following a previously published protocol (21). Interpretation of the results was performed using BioNumerics software (Applied Math, Sint-Martens-Latem, Belgium). A dendrogram comparing the isolates was constructed using the Dice coefficient and unweighted pair group method with arithmetic mean (UPGMA) with a position tolerance of 1. Isolates with >80% similarity were considered similar according to the criteria of Tenover et al. (22).
Molecular detection of KPC.
ESBL-producing E. coli and Klebsiella isolates that were resistant to carbapenems (CR-ESBL isolates) were further tested for the detection of the Klebsiella pneumoniae carbapenemase (KPC) gene following a previous published protocol (23).
Statistical analysis.
Only the first E. coli, K. pneumoniae, or K. oxytoca isolate from each patient that produced an ESBL was used in this study. All data were analyzed using SAS statistical software, version 9.2.1 (SAS Institute, Cary, NC). Univariable analysis utilized means and frequencies to assess distributions. Demographic and antibiotic exposures between patients colonized with ESBL-positive isolates with and without carbapenem resistance were compared using appropriate methods for continuous and categorical variables.
ACKNOWLEDGMENTS
This work was funded by an unrestricted research grant from Merck Pharmaceuticals. This research was also supported by National Institutes of Health grants 1K01AI071015 to J.P.F., R01A1060859 and 5K24AI079040 to A.D.H., IK12RR023250 to J.K.J., and IK23AI082450-03 to K.A.T.
REFERENCES
- 1.Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D, participants of the 3rd World Healthcare-Associated Infections Forum. 2012. Ready for a world without antibiotics? The Pensieres Antibiotic Resistance Call to Action. Antimicrob Resist Infect Control 1:11. doi: 10.1186/2047-2994-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosgrove SE. 2006. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis 42(Suppl 2):S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 3.Slama TG. 2008. Gram-negative antibiotic resistance: there is a price to pay. Crit Care 12(Suppl 4):S4. doi: 10.1186/cc6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbonne A, Arnaud I, Maugat S, Marty N, Dumartin C, Bertrand X, Bajolet O, Savey A, Fosse T, Eveillard M, Senechal H, Coignard B, Astagneau P, Jarlier V, MDRB Surveillance National Steering Group (BMR-Raisin). 2013. National multidrug-resistant bacteria (MDRB) surveillance in France through the RAISIN network: a 9 year experience. J Antimicrob Chemother 68:954–959. doi: 10.1093/jac/dks464. [DOI] [PubMed] [Google Scholar]
- 5.Tacconelli E, Cataldo MA, Dancer SJ, De Angelis G, Falcone M, Frank U, Kahlmeter G, Pan A, Petrosillo N, Rodriguez-Bano J, Singh N, Venditti M, Yokoe DS, Cookson B, European Society of Clinical Microbiology. 2014. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin Microbiol Infect 20(Suppl 1):S1–S55. doi: 10.1111/1469-0691.12427. [DOI] [PubMed] [Google Scholar]
- 6.Donskey CJ. 2006. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis 43(Suppl 2):S62–S69. doi: 10.1086/504481. [DOI] [PubMed] [Google Scholar]
- 7.Hess A, Kleinberg M, Sorkin J, Netzer G, Johnson J, Shardell M, Thom K, Harris A, Roghmann M. 2014. Prior colonization is associated with increased risk of antibiotic-resistant Gram-negative bacteremia in cancer patients. Diagn Microbiol Infect Dis 79:73–76. doi: 10.1016/j.diagmicrobio.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wingard JR, Dick J, Charache P, Saral R. 1986. Antibiotic-resistant bacteria in surveillance stool cultures of patients with prolonged neutropenia. Antimicrob Agents Chemother 30:435–439. doi: 10.1128/AAC.30.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sievert D, Ricks P, Edwards J, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 10.Ajao A, Johnson JK, Harris A, Zhan M, McGregor J, Thom K, Furuno J. 2013. Risk of acquiring extended-spectrum β-lactamase-producing Klebsiella species and Escherichia coli from prior room occupants in the intensive care unit. Infect Control Hosp Epidemiol 34:453–458. doi: 10.1086/670216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyle EP, Gasink LB, Linkin DR, Bilker WB, Lautenbach E. 2007. Use of different thresholds of prior antimicrobial use in defining exposure: impact on the association between antimicrobial use and antimicrobial resistance. J Infect 55:414–418. doi: 10.1016/j.jinf.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Green HP, Johnson JA, Furuno JP, Strauss SM, Perencevich EN, Lautenbach E, Lee D, Harris AD. 2007. Impact of freezing on the future utility of archived surveillance culture specimens. Infect Control Hosp Epidemiol 28:886–888. doi: 10.1086/518843. [DOI] [PubMed] [Google Scholar]
- 13.McGregor JC, Kim PW, Perencevich EN, Bradham DD, Furuno JP, Kaye KS, Fink JC, Langenberg P, Roghmann MC, Harris AD. 2005. Utility of the chronic disease score and Charlson comorbidity index as comorbidity measures for use in epidemiologic studies of antibiotic-resistant organisms. Am J Epidemiol 161:483–493. doi: 10.1093/aje/kwi068. [DOI] [PubMed] [Google Scholar]
- 14.McGregor JC, Perencevich EN, Furuno JP, Langenberg P, Flannery K, Zhu J, Fink JC, Bradham DD, Harris AD. 2006. Comorbidity risk-adjustment measures were developed and validated for studies of antibiotic-resistant infections. J Clin Epidemiol 59:1266–1273. doi: 10.1016/j.jclinepi.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 15.McGregor JC, Rich SE, Harris AD, Perencevich EN, Osih R, Lodise TP Jr, Miller RR, Furuno JP. 2007. A systematic review of the methods used to assess the association between appropriate antibiotic therapy and mortality in bacteremic patients. Clin Infect Dis 45:329–337. doi: 10.1086/519283. [DOI] [PubMed] [Google Scholar]
- 16.Osih RB, McGregor JC, Rich SE, Moore AC, Furuno JP, Perencevich EN, Harris AD. 2007. Impact of empiric antibiotic therapy on outcomes in patients with Pseudomonas aeruginosa bacteremia. Antimicrob Agents Chemother 51:839–844. doi: 10.1128/AAC.00901-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris AD, Johnson JK, Thom KA, Morgan DJ, McGregor JC, Ajao AO, Moore AC, Comer AC, Furuno JP. 2011. Risk factors for development of intestinal colonization with imipenem-resistant Pseudomonas aeruginosa in the intensive care unit setting. Infect Control Hosp Epidemiol 32:719–722. doi: 10.1086/660763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial disk susceptibility testing; twenty-second informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris AD, Perencevich EN, Johnson JK, Paterson DL, Morris JG, Strauss SM, Johnson JA. 2007. Patient-to-patient transmission is important in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae acquisition. Clin Infect Dis 45:1347–1350. doi: 10.1086/522657. [DOI] [PubMed] [Google Scholar]
- 22.Tenover FC, Arbeit RD, Goering RV. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol 18:426–439. [DOI] [PubMed] [Google Scholar]
- 23.Rasheed JK, Biddle JW, Anderson KF, Washer L, Chenoweth C, Perrin J, Newton DW, Patel JB. 2008. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J Clin Microbiol 46:2066–2069. doi: 10.1128/JCM.02038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]