ABSTRACT
We studied in parallel the population structure of 90 carbapenemase-producing and 88 carbapenemase-susceptible Klebsiella pneumoniae isolates collected in 20 Spanish hospitals, in the context of the EuSCAPE project. Fourteen and 50 multilocus sequence types (MLSTs) were detected among the carbapenemase-producing and carbapenem-susceptible isolates, respectively. ST11 and ST15 clones were more frequent in the carbapenemase-producing group than in the carbapenemase-susceptible group (P < 0.0001). Among the members of the carbapenem-suceptible group, the cefotaxime-resistant population showed population parameters that differed between the populations of the wild-type strains and the carbapenemase producers.
KEYWORDS: carbapenem resistance, population structure, MLST, carbapenemases, Klebsiella pneumoniae
TEXT
Carbapenemase-producing Enterobacteriaceae (CPE) have emerged in recent years as being among of the most important threats to public health. CPE have been detected in almost all European countries, although with a highly variable geographical distribution (1). Some high-risk clones of Klebsiella pneumoniae, mainly KPC-producing sequence type 258 (ST258) but also OXA-48-producing ST395 or ST15, have been implicated in the worldwide spread of carbapenemases (2–4).
Previous studies have shown that the population of Escherichia coli strains resistant to amoxicillin-clavulanic acid is less diverse than the susceptible population (5). Little is known about the population structure of carbapenemase-producing K. pneumoniae in comparison with the carbapenem-susceptible population. The aim of this study was to test the hypothesis that the carbapenemase-producing K. pneumoniae strains isolated in Spanish hospitals constitute a distinct and more homogeneous population than the carbapenemase-susceptible isolates.
The European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) is an initiative funded by the European Centre for Disease Prevention and Control (ECDC) and coordinated by the Department of Medical Microbiology of the University Medical Center Groningen (Netherlands) (6). EuSCAPE carried out a structured survey between November 2013 and April 2014 involving national networks of representative laboratories from 35 European countries (7). Each participant laboratory collected up to 10 carbapenem-nonsusceptible clinical isolates of K. pneumoniae or E. coli and 10 susceptible same-species comparator isolates. All clinical specimens were accepted, except for stool and surveillance screening samples; isolates of colonizers obtained from clinical specimens were also included (7). In Spain, 20 hospital laboratories from 16 provinces distributed across the country were enrolled according to EuSCAPE criteria. A total of 116 carbapenem-nonsusceptible K. pneumoniae isolates were collected, 102 (87.9%) of which produced carbapenemases (81 produced OXA-48, 12 VIM, and 9 KPC). A total of 14 carbapenem-nonsusceptible E. coli strains were also detected, 4 (28.5%) of which produced OXA-48 carbapenemases (7).
Our main purpose was to study two groups of representative isolates that were as comparable as possible according to the EUSCAPE project guidelines. Accordingly, a subset of 90 nonduplicated isolates of carbapenemase-producing K. pneumoniae strains was analyzed and compared in detail with 88 carbapenem-susceptible isolates; all of them were selected according to the following criteria: (i) clinical impact (isolates producing infections were prioritized) and (ii) geographic representation (isolates from all 20 participating hospitals were included). An isolate was considered carbapenem susceptible and carbapenemase negative when it was susceptible to all carbapenems according to EUCAST guidelines (8) and when both the Hodge modified test (meropenem disk with 600 μg cloxacillin) and the Carba NP test were negative (9). Carbapenem-susceptible and carbapenemase-producing isolates were matched by hospital and temporal origin. The presence of genes encoding carbapenemases or extended-spectrum β-lactamase (ESBLs) was determined using PCR and DNA sequencing assays (10).
All isolates were subjected to multilocus sequence type (MLST) analysis according to the Institut Pasteur scheme (http://bigsdb.web.pasteur.fr/klebsiella/) (11) A simple-diversity index (SDI) (12) was calculated. The phylogenetic relationships among the different sequence types (STs) were established according to the eBURST program (version 3) (http://eburst.mlst.net).
The statistical analysis was performed using GraphPad Prism software (version 3.02) (GraphPad Software, Inc., San Diego, CA, USA). Population differences were assessed using Fisher's exact test.
Comparisons of clinical and epidemiological data from carbapenemase-producing and carbapenem-susceptible isolates are detailed in Table 1.
TABLE 1.
Comparisons of clinical and epidemiological data of Klebsiella pneumoniae isolates from carbapenemase-producing cases and carbapenem-susceptible controlsa
| Parameter | No. of cases (n = 90) | No. of controls (n = 88) | OR | 95% CI | P |
|---|---|---|---|---|---|
| Age (yrs) | |||||
| <18 | 0 | 3 | 0.32 | 0.03–3.12 | 0.30 |
| >65 | 65 | 44 | 2.6 | 1.49–4.85 | 0.002 |
| Sex | |||||
| Female | 40 | 45 | 0.76 | 0.42–1.38 | 0.37 |
| Clinical relevance | |||||
| Infected | 84 | 86 | 0.32 | 0.64–1.66 | 0.16 |
| Hospital location | |||||
| ICU | 27 | 11 | 3 | 1.38–6.52 | 0.004 |
| Outpatient/emergency department | 22 | 34 | 0.51 | 0.27–0.98 | 0.04 |
| Previous hospital admission in the last 6 mo | 56 | 33 | 2.7 | 1.50–5.03 | 0.001 |
| Sample source | |||||
| Urine | 42 | 47 | 0.76 | 0.42–1.38 | 0.37 |
| Blood | 16 | 22 | 0.65 | 0.31–1.34 | 0.24 |
Boldface indicates statistically significant differences. OR, odds ratio; 95% CI, 95% confidence interval; ICU, intensive care unit.
Of the 90 carbapenemase-producing K. pneumoniae isolates, 70 (77.8%) produced OXA-48, 12 (13.3%) VIM, and 8 (8.9%) KPC. Eighty-four (93.3%) produced clinical infections (mainly urinary tract infections [46.7%], wound infections [20.2%], and bacteremia [17.8%]), and 6 were colonizers. Among the 90 isolates, 14 MLSTs were detected (SDI = 15.6) (Table 2). The mean number of isolates per ST was 6.4 (range, 1 to 31); the most prevalent STs were ST15 (34.4%), ST11 (26.7%), ST405 (13.3%), ST147 (10%), and ST258 (3.3%) (Table 2). The three STs with more than 10 isolates each came from eight hospitals located in six (ST15) and five (ST11 and ST405) provinces.
TABLE 2.
Distribution of different population markers indicating genetic variation between carbapenemase-producing and carbapenem-susceptible Klebsiella pneumoniae isolatesa
| Population marker | Value(s) |
||||||
|---|---|---|---|---|---|---|---|
| Phenotype |
P value |
||||||
| CBP-R | CBP-Sb |
CBP-R vs CBP-S | CBP-R vs WT | CBP-R vs CTX-R/CBP-S | |||
| Total | WT | CTX-R | |||||
| No. of isolates | 90 | 88 | 66 | 22 | |||
| No. of STs | 14 | 50 | 44 | 10 | |||
| Mean no. of isolates per ST (range) | 6.4 (1–31) | 1.8 (1–6) | 1.5 (1–5) | 2.2 (1–5) | |||
| No. (%) of single isolates per ST | 7 (7.8) | 36 (40.9) | 33 (50) | 5 (22.7) | <0.0001 | <0.0001 | 0.13 |
| SDI | 15.6 | 56.8 | 66.7 | 45.5 | |||
| No. (%) of ST15 isolates | 31 (34.4) | 6 (6.8) | 1 (1.5) | 5 (22.7) | <0.0001 | <0.0001 | 0.22 |
| No. (%) of ST11 isolates | 24 (26.7) | 5 (5.7) | 3 (4.5) | 2 (9.1) | 0.0002 | 0.0001 | 0.18 |
| No. (%) of ST405 isolates | 12 (13.3) | 5 (5.7) | 2 (3) | 3 (13.6) | 0.12 | 0.01 | 0.75 |
| No. (%) of ST35 isolates | 0 | 5 (5.7) | 5 (7.6) | 0 | 0.03 | 0.01 | |
| No. (%) of ST147 isolates | 9 (10) | 2 (2.3) | 2 (3) | 0 | 0.06 | 0.13 | 0.20 |
| No. (%) of ST307 isolates | 0 | 5 (5.7) | 1 (1.5) | 4 (18.2) | 0.06 | 0.41 | 0.002 |
CBP-S, carbapenem susceptible isolates; WT, wild-type fully susceptible isolates; CTX-R, cefotaxime-resistant ESBL-producing isolates; CBP-R, carbapenem-resistant carbapenemase-producing isolates; SDI, single-diversity index [(number of STs/total number of isolates) × 100].
CBP-S include WT and CTX-R. Boldface indicates statistically significant differences.
Among the 88 carbapenem-susceptible isolates, 50 STs were detected (SDI = 56.8%) (Table 2). The mean number of isolates determined by STs was 1.8 (range, 1 to 6); the most prevalent STs were ST15 (6.8%), ST11 (5.7%), ST35 (5.7%), ST307 (5.7%), and ST405 (5.7%). eBURST analysis provided a representation of the different population structures of carbapenemase-producing and carbapenem-susceptible K. pneumoniae isolates (Fig. 1).
FIG 1.
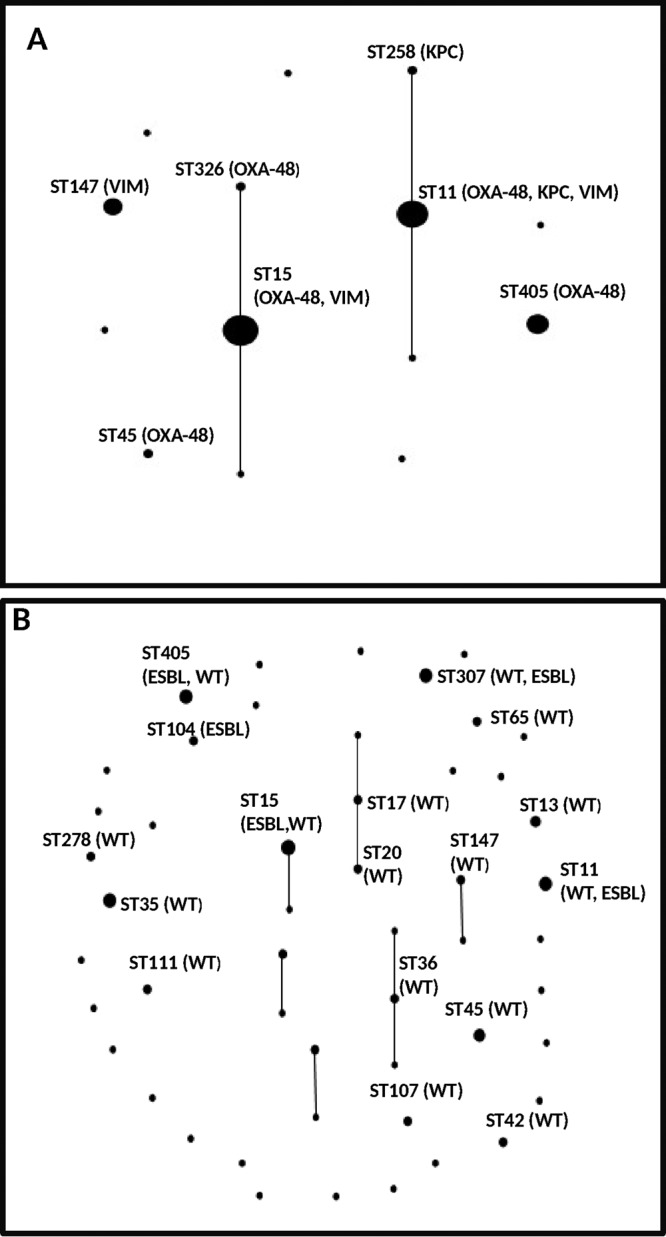
Population snapshot of Klebsiella pneumoniae sequence types (STs) of carbapenemase-producing (n = 90) (A) and carbapenem-susceptible (n = 88) (B) strains. The most frequent STs found in this study are emphasized. In panel A, carbapenemase types are depicted in the most relevant STs. In panel B, ESBL production is depicted in the most relevant STs; also, STs comprising isolates of the ESBL-producing and wild-type (WT) groups of isolates are emphasized.
Only seven STs (ST11, ST15, ST16, ST45, ST104, ST147, and ST405) were identified in both groups, but their prevalences strongly differed between the groups, as they comprised 80 isolates in the carbapenemase-producing group (88.9%) and 25 isolates in the carbapenem-susceptible group (28.4%) (P < 0.0001). The ST11 and ST15 clones were much more predominant among the carbapenemase-producing isolates (55/90, 61.1%) than among the members of the carbapenemase-susceptible group (11/88, 12.5%) (P < 0.0001).
An additional finding of interest was that the 88 carbapenem-susceptible isolates in fact consisted of two subpopulations: a wild-type population fully susceptible to all tested β-lactam antibiotics except ampicillin and ticarcillin (n = 66, 75%) and a cefotaxime-resistant population also producing ESBLs (n = 22, 25%): 14 producing CTX-M-15, 4 SHV-2a, 2 SHV-12, 1 SHV-36, and 1 CTX-M-1. The most prevalent STs among the ESBL-producing isolates were ST15 (3 producing SHV-2a, 1 SHV-12, 1 CTX-M-15), ST405 (3 producing CTX-M-15, 1 SHV-12), ST307 (4 producing CTX-M-15), and ST11 (1 isolate each producing CTX-M-15, CTX-M-1, and SHV-2a). These two subpopulations also showed other important differences (Table 2); for instance, in the fully susceptible subpopulation, the SDI and the prevalence of ST15 were 69.7 and 1.5%, respectively, while the corresponding values were 50 and 22.7%, respectively, in the cefotaxime-resistant subpopulation. Among ESBL-producing K. pneumoniae isolates, the prevalence of successful clones such as ST15 and ST11 has been well documented (13–15).
Our data suggest that carbapenemase-producing K. pneumoniae isolates have a population structure that is different from and less diverse than the population structure of carbapenem-susceptible K. pneumoniae isolates; the latter group in fact consisted of a mixed population of isolates in which ESBL production was relatively frequent (25%). The population structure of carbapenem-susceptible/ESBL-positive isolates showed population parameters that differed between populations of wild-type and carbapenemase producers (Table 2); in fact, some significant differences that obtained between wild-type isolates and carbapenemase-producing isolates were not observed between carbapenem-susceptible/ESBL-positive isolates and carbapenemase producers (Table 2). However, these data should be considered with caution due to the low number of carbapenem-susceptible/ESBL-positive isolates included in the analysis.
The concept of the impact of the consumption of antibiotics on the selection of successful clones is supported by results of a recent study suggesting that different clones of E. coli differ in their responses to antibiotics despite their comparable drug MICs (16). Our results suggest that carbapenem-resistant K. pneumoniae strains have undergone a population shift leading to a marked loss of genetic diversity as a consequence of strong selection pressure; production of ESBLs may be the first step in this process. Therefore, the ecologic and clinical impact of the consumption of antibiotics in the population structure of such pathogen bacteria as K. pneumoniae is of concern beyond the obvious selection of resistant isolates.
ACKNOWLEDGMENTS
We thank Isabel Pérez-Grajera for technical support.
This study was part of the European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) project coordinated by the University Medical Center Groningen and funded by ECDC through a specific framework contract (ECDC/2012/055) following an open call for tender (OJ/25/04/2012-PROC/2012/036). This work was also supported by a grant from the Instituto de Salud Carlos III (grant number MPY 1135/16) and by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III, cofinanced by the European Development Regional Fund “A way to achieve Europe” ERDF, and the Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015).
Members of EuSCAPE-Spain Network are as follows: Teresa Alarcón (Hospital Universitario de La Princesa); Isabel Barba (Hospital General Universitario de Ciudad Real); Emilia Cercenado (Hospital General Universitario Gregorio Marañón); M. Antonia de Miguel (Hospital Universitario de Canarias); Dolores de Miguel (Hospital de Cabueñes); Dionisia Fontanals (Corporació Sanitària Parc Taulí); Ana M. Fleites (Hospital Central de Asturias); Isabel Sánchez-Romero (Hospital Universitario Puerta de Hierro-Majadahonda); Sonia Solís (Hospital Universitario de Guadalajara); Adelina Gimeno (Hospital General Universitario de Alicante); Juan Manuel Hernández (Hospital Regional Universitario Carlos Haya); Jesús Martínez-López (Complejo Hospitalario de Pontevedra); Ferran Navarro (Hospital de la Santa Creu i Sant Pau); Juan Ignacio Alós (Hospital Universitario de Getafe); Marta Arias (Hospital Universitario Rio Hortega); Genoveva Yagüe (Hospital Virgen de la Arrixaca); Caridad Sainz de Baranda (Complejo Hospitalario de Albacete); Luz Albina Cordón (Hospital Txagorritxu); José Leiva (Clínica Universitaria de Navarra); and Mar Olga Pérez-Moreno (Hospital de Tortosa Verge de la Cinta).
REFERENCES
- 1.Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL; European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) working group. 2015. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries. Euro Surveill 20:pii=30062. doi: 10.2807/1560-7917.ES.2015.20.45.30062. [DOI] [PubMed] [Google Scholar]
- 2.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. 2016. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potron A, Kalpoe J, Poirel L, Nordmann P. 2011. European dissemination of a single OXA-48-producing Klebsiella pneumoniae clone. Clin Microbiol Infect 17:E24–E26. doi: 10.1111/j.1469-0691.2011.03669.x. [DOI] [PubMed] [Google Scholar]
- 4.López-Cerero L, Almirante B. 2014. Epidemiology of infections caused by carbapenemase-producing Enterobacteriaceae: reservoirs and transmission mechanisms. Enferm Infecc Microbiol Clin 32(Suppl 4):10–16. doi: 10.1016/S0213-005X(14)70169-7. [DOI] [PubMed] [Google Scholar]
- 5.Oteo J, González-López JJ, Ortega A, Quintero-Zárate JN, Bou G, Cercenado E, Conejo MC, Martínez-Martínez L, Navarro F, Oliver A, Bartolomé RM, Campos J, Spanish Network for Research in Infectious Diseases (REIPI). 2014. Inhibitor-resistant TEM- and OXA-1-producing Escherichia coli isolates resistant to amoxicillin-clavulanate are more clonal and possess lower virulence gene content than susceptible clinical isolates. Antimicrob Agents Chemother 58:3874–8381. doi: 10.1128/AAC.02738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glasner C, Albiger B, Buist G, Tambić Andrasević A, Canton R, Carmeli Y, Friedrich AW, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Nordmann P, Poirel L, Rossolini GM, Seifert H, Vatopoulos A, Walsh T, Woodford N, Donker T, Monnet DL, Grundmann H; European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. 2013. Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Euro Surveill 18:pii=20525. doi: 10.2807/1560-7917.ES2013.18.28.20525. [DOI] [PubMed] [Google Scholar]
- 7.Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasević AT, Cantón R, Carmeli Y, Friedrich AW, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Nordmann P, Poirel L, Rossolini GM, Seifert H, Vatopoulos A, Walsh T, Woodford N, Monnet DL; European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. 17 November 2016. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis pii: S1473-3099(16)30257-2. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 8.EUCAST Subcommittee for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. 2013. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. Version 1.0, December 2013 http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf Accessed September 2016.
- 9.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortega A, Sáez D, Bautista V, Fernández-Romero S, Lara N, Aracil B, Pérez-Vázquez M, Campos J, Oteo J; Spanish Collaborating Group for the Antibiotic Resistance Surveillance Programme. 2016. Carbapenemase-producing Escherichia coli is becoming more prevalent in Spain mainly because of the polyclonal dissemination of OXA-48. J Antimicrob Chemother 71:2131–2138. doi: 10.1093/jac/dkw148. [DOI] [PubMed] [Google Scholar]
- 11.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gastmeier P, Schwab F, Bärwolff S, Rüden H, Grundmann H. 2006. Correlation between the genetic diversity of nosocomial pathogens and their survival time in intensive care units. J Hosp Infect 62:181–186. doi: 10.1016/j.jhin.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Zhou K, Lokate M, Deurenberg RH, Tepper M, Arends JP, Raangs EG, Lo-Ten-Foe J, Grundmann H, Rossen JW, Friedrich AW. 2016. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum β-lactamase producing ST15 Klebsiella pneumoniae. Sci Rep 6:20840. doi: 10.1038/srep20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coque TM, Baquero F, Canton R. 2008. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill 13:pii=19044 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19044. [PubMed] [Google Scholar]
- 15.Oteo J, Cuevas O, López-Rodríguez I, Banderas-Florido A, Vindel A, Pérez-Vázquez M, Bautista V, Arroyo M, García-Caballero J, Marín-Casanova P, González-Sanz R, Fuentes-Gómez V, Oña-Compán S, García-Cobos S, Campos J. 2009. Emergence of CTX-M-15-producing Klebsiella pneumoniae of multilocus sequence types 1, 11, 14, 17, 20, 35 and 36 as pathogens and colonizers in newborns and adults. J Antimicrob Chemother 64:524–528. doi: 10.1093/jac/dkp211. [DOI] [PubMed] [Google Scholar]
- 16.Stewart B, Rozen DE. 2012. Genetic variation for antibiotic persistence in Escherichia coli. Evolution 66:933–939. doi: 10.1111/j.1558-5646.2011.01467.x. [DOI] [PubMed] [Google Scholar]


