Abstract
A 17-year-old female with B-cell precursor acute lymphoblastic leukemia (BCP-ALL) with persistent minimal residual disease (MRD) with standard chemotherapy was found to have a BCR-ABL1-like gene expression pattern. Genome sequencing revealed a JAK2 mutation not previously described in BCP-ALL and a potential therapeutic target. Due to concern for an on-therapy relapse, the JAK2 inhibitor ruxolitinib was incorporated into a modified chemotherapy backbone to achieve complete remission prior to stem cell transplant. Treatment was well tolerated and she had undetectable MRD prior to a matched allogeneic stem cell transplant and remained in remission at day +100.
Keywords: Targeted therapy, personalized medicine, variant of unknown significance, tyrosine kinase inhibitor, TKI
Introduction
Despite advances in the treatment of B-cell precursor acute lymphoblastic leukemia (BCP-ALL), children and young adults who present with clinical and molecular features of high-risk disease remain challenging to cure. Since first described by the Dutch (DCOG) and German (COALL) pediatric cooperative groups in parallel with the St. Jude group, approximately 15 to 20% of patients with BCP-ALL have been found to have gene expression profiles similar to those observed with the BCR-ABL1 fusion, but lack the sentinel t(9;22)(q34;q11.2).1,2 Termed “BCR-ABL1-like”, affected patients present with NCI high-risk features, demonstrate persistent post-induction minimal residual disease (MRD), and harbor a range of genetic alterations that activate kinase signaling.3,4 In case reports where BCR-ABL1-like lesions have been identified early in treatment, imatinib, a tyrosine kinase inhibitor (TKI) targeted against ABL1, ABL2, PDGFRB and other lesions, has been successfully used to treat refractory disease.5 Because BCR-ABL1-like BCP-ALL is more prevalent in adolescents and young adults, this patient population has been identified to potentially benefit greatly from targeted therapies.6,7 In the current treatment era, there are few guidelines regarding the management of BCR-ABL1-like leukemias, including which methods are recommended to identify, characterize and validate novel lesions, when to assess response, or how to integrate TKIs safely into combination therapy. We present a case in which we identified and validated a novel JAK2 activating mutation in a patient with BCR-ABL1-like BCP-ALL and, following the integration of ruxolitinib into a Children’s Oncology Group (COG)-modified BFM regimen, successfully reduced MRD to allow stem cell transplantation in first remission.
Results
Materials, Methods and Case presentation
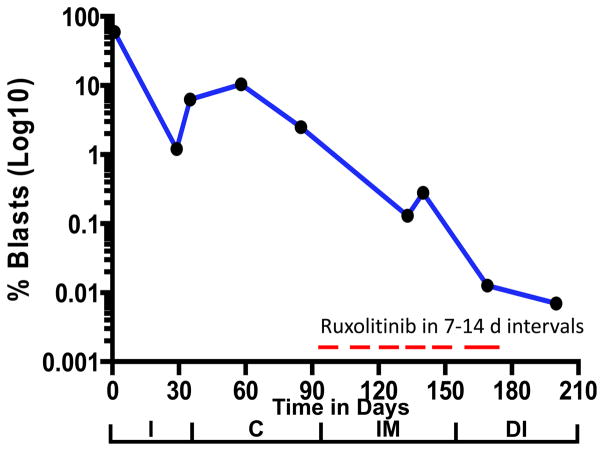
A 17 year-old Native American G1P1 female presented to medical attention at two days post-partum with fever, severe anemia, and post-partum hemorrhage. Initial CBC showed a WBC of 14,300 cells/μL, hemoglobin of 5.9 gm/dL, platelet count of 53,000 cells/μL, and a differential with 68% peripheral blasts. Flow cytometric analyses showed a B lymphoid blast population expressing CD19, CD45 (dim), CD10, TdT, CD79a, CD22, CD34, CD33 (dim subset) and HLA-DR with a ploidy index of 1.0, consistent with diploid BCP-ALL. Bone marrow biopsy showed 95% cellularity with 83% lymphoblasts. FISH was negative for BCR-ABL1 fusion, KMT2A (MLL) rearrangements, and ETV6/RUNX1 fusion. Karyotype showed a stemline and two sidelines with multiple abnormalities: 46, XX, t(7;22)(q32;q11.2), add(9)(p13)[10]/ 48,idem,i(7)(q10),+14, +22[2]/49,idem,+6,+10,+22[1]/ 46,XX[9]. Initial lumbar puncture showed CNS3 disease. In accordance with the Declaration of Helsinki, the patient and her legal guardian provided written informed assent/consent to participate in Whole Genome Sequencing Of Human Cancer Tissues (NCT02105545), and underwent induction therapy in accordance to COG AALL1131 (NCT01406756). CNS remission was achieved the first week of induction. Morphologic remission was achieved by Day 29. End-induction MRD of 1.2% increased to 10.4% by Day 29 of consolidation phase with standard therapy (Fig 1).
Figure 1.
Blast percentage performed by flow cytometry from diagnosis through the end of chemotherapy prior to stem cell transplant. I: Induction, C: Consolidation, IM: Interim Maintenance, DI: Delayed Intensification. MRD timepoints: Induction day 29 (1%), Consolidation day 8 (6.1%), Consolidation day 29 (10.4%), Consolidation day 56 (2.5%), Interim maintenance day 49 (0.13%), Interim maintenance day 56 (0.28%), Delayed intensification day 29 (0.012%), Delayed intensification day 56 (<0.01%). Induction and consolidation were according to COG AALL1131. Interim Maintenance: VCR 1.5 mg/m2 d1,15,29,43; HDMTX 5 g/m2 d1,15,29,43; IT MTX d1,29; Ruxolitinib 40 mg/m2/BID d19–28,33–42,47–56. Delayed Intensification: VCR 1.5 mg/m2 d1,8,15,43,50; CPM 1000 mg/m2 d29; PEG 2500 IU/m2 d4,43; ARAC 75 mg/m2 d29–32, 36–39; DEX 5 mg/m2 d1–7,15–21; DOX 25 mg/m2 d1,8,15; IT MTX d1,29,36; Ruxolitinib 40 mg/m2/BID d8–14,22–28,44–57. Vincristine (VCR) daunorubicin (DAUN), pegylated asparaginase (PEG), prednisone (PRED), intrathecal cytarabine (IT ARAC), intrathecal methotrexate (IT MTX), cyclophosphamide (CPM), cytarabine (ARAC), mercaptopurine (6MP), high dose methotrexate (HDMTX), dexamethasone (DEX), and doxorubicin (DOX).
Leukemia DNA sequencing methods
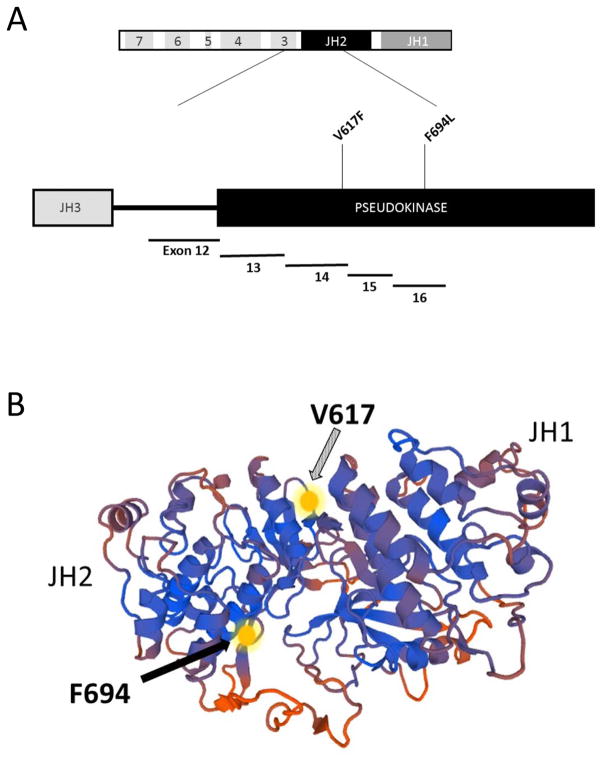
Because persistent MRD beyond Day 29 of induction therapy is highly associated with relapse, we evaluated whether our patient had a BCR-ABL1-like targetable lesion through an iterative cascade of molecular testing.8 As part of NCT02105545, low density array (LDA) screening, deep gene and RNA sequencing was done to further evaluate for a BCR-ABL1-like lesion.4 Using the gene expression assay via LDA card, we found a gene signature consistent with BCR-ABL1-like BCP-ALL with a high expression of CRLF2.3 We next undertook deep sequencing of over more than 400 cancer relevant genes (Ion Ampliseq Comprehensive Cancer Panel, Thermo Fisher) and identified a novel F694L mutation in JAK2, which specifically involved a T to C transition in exon 16 at base-pair locus 5078393, and a cluster of mutations (p.Cys119_Ile121delinsTrpGlyLeu) in IKZF1 in the diagnostic sample (Fig 2). These genomic variants were nearly undetectable in the Day 29 post-induction sample, showing that they were restricted to the lymphoblast population affected by chemotherapy. Unbiased RNA-sequencing confirmed that the JAK2 mutation was present in expressed mRNA. Our findings were validated by CLIA-approved Foundation One Heme (Cambridge, MA), confirming the presence of the molecular variants, in addition to an IGH-CRLF2 translocation (Fig 2). Using CLIA-approved sample acquisition, PCR and Sanger Sequencing techniques, TriCore Reference Laboratories validated the F694L JAK2 mutation in the presentation specimen (Fig 2). At the recommendation of our multidisciplinary molecular tumor board, the patient was removed from protocol therapy and the JAK2 inhibitor ruxolitinib was integrated into conventional cytotoxic therapy, with close monitoring for adverse events.
Figure 2.
Mutation sites within the JAK2 gene, and impact on targeted therapies. Panel A: schematic showing the JH2 pseudokinase domain, including the V617F mutation site in exon 14, and the F694L site in exon 16. The V617F mutation site is commonly identified in patients with myelodysplastic disorders, while the F694L site has not been previously described to occur in ALL. Panel B: the F694L mutation is predicted to interfere with signal transduction in the location shown by the solid arrow.18
Because there is no precedent for the incorporation of ruxolitinib into dose-intensified conventional therapy, we chose to use 40 mg/m2/day BID for two weeks on, two weeks off, based on the safety phase findings of COG ADVL1011.6 It was delayed until interim maintenance as the JAK2 mutation was confirmed in two CLIA labs and authorization was obtained from insurance. Our patient had expected and manageable toxicities related to the consolidation, interim maintenance and delayed intensification phases of COG AALL1131. She also experienced delays in treatment related to persistent thrombocytopenia and neutropenia, which were attributable to the inclusion of ruxolitinib to her treatment schedule. Ruxolitinib was held when the ANC <750 or Platelets <75,000. Our patient’s MRD continued to decline during these phases of therapy (Fig 1). There was a small transient rise in MRD level during interim maintenance which was felt to be due to several held doses due to admission for febrile neutropenia. When her MRD had diminished to <0.01%, she was taken to an allogeneic transplant with a fully matched sibling. Molecular remission was unable to be ascertained due to the undetectable level of blasts. She was found to be 100% engrafted with donor cells at day 100 post-transplant.
Discussion
Among the many prognostic factors that have been developed to predict relapse, the persistence of minimal residual disease (MRD) has emerged as one of the strongest and most reliable.8–10 The international BFM and GIMEMA cooperative groups reported that children and adults with Day 29 MRD levels of greater than 1% were at a very high risk of relapse. In the International BFM study, patients who had MRD levels of 10−2 or greater had five to ten-fold higher relapse rates at 3 years (39% – 86%).9 In the GIMEMA study, the probability of relapse at 2 years for MRD-positive patients at preconsolidation was 81.5% vs. 38.9% for MRD-negative patients.11 In the last COG high-risk ALL study, the presence of MRD at the end of consolidation had a DFS of only 39% versus 79% in patients without MRD. 8 Because our patient had post-induction MRD that was rising, we were compelled to augment her care with a therapeutic approach that might prevent relapse.
The utilization of targeted therapies provides a novel opportunity to prevent relapsed disease, but targets must be identified and implemented early in the treatment course to be of benefit. Using an iterative approach to our patient’s molecular diagnostics, we identified and characterized a novel JAK2 lesion in a highly conserved region of the enzyme which provided an opportunity to select a TKI for incorporation into our patient’s treatment regimen. 12
Targeted therapies as single agents against kinome deregulated leukemias are alone insufficient for cure in the relapsed setting, calling for a rapid and accurate identification of lesions.13,14 In the setting of BCR-ABL positive leukemias, the introduction of integrated TKI therapies, starting during induction, has achieved remarkable treatment success.15,16 Preclinical data have demonstrated that ruxolitinib is effective in BCR-ABL1-like ALL. 17 Pending COG study AALL1521 will investigate whether adding a JAK2 inhibitor to chemotherapy for patients with CRLF2-rearranged or JAK-mutated ALL will improve outcome. Our findings demonstrate the promise of genome sequencing in acute lymphoblastic leukemia in a patient who demonstrated persistent high MRD without targeted therapy. With the inclusion of a lesion-specific TKI into a dose-intensified, COG-modified BFM regimen, she achieved an MRD negative remission, and remains in CR1 more than 100 days post transplant.
Acknowledgments
Financial Support
S.A. Ness: NIH grants 5R01CA170250, 5R01DE023222, 5P30CA118100
The authors acknowledge the outstanding technical support from Jennifer Woods, Maggie Cyphery, Jamie Padilla and Jason Byars. We also thank Drs. I-Ming Chen, Richard Harvey and Cheryl Willman for their assistance and guidance. Some experiments used the facilities or services provided by the Analytical and Translational Genomics Shared Resource, which is supported by the State of New Mexico and the UNM Comprehensive Cancer Center P30CA1181
List of Abbreviations
- ALL
Acute lymphoblastic leukemia
- ANC
Absolute neutrophil count
- BCP-ALL
B-cell precursor acute lymphoblastic leukemia
- BFM
Berlin-Frankfurt-Münster
- BID
“bis in die” / twice a day
- CBC
Complete blood count
- CLIA
Clinical Laboratory Improvement Amendments
- COG
Children’s Oncology Group
- CR1
First complete response
- DNA
Deoxyribonucleic acid
- FISH
Fluorescence in situ hybridization
- GIMEMA
Gruppo Italiano Malattie e Matologiche dell’Adulto (Italian Group for Adult Hematologic Diseases)
- M2
Meter-squared
- MG
Milligram
- MRD
Minimal residual disease
- mRNA
Messenger ribonucleic acid
- NCI
National Cancer Institute
- PCR
Polymerase chain reaction
- RNA
Ribonucleic acid
- WBC
White blood count
- TKI
Tyrosine kinase inhibitor
Footnotes
Conflicts of Interest
None.
References
- 1.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. The lancet oncology. 2009;10(2):125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115(26):5312–5321. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey RC, Kang H, Roberts KG, et al. Development and Validation Of a Highly Sensitive and Specific Gene Expression Classifier To Prospectively Screen and Identify B-Precursor Acute Lymphoblastic Leukemia (ALL) Patients With a Philadelphia Chromosome-Like (“Ph-like” or “BCR-ABL1-Like”) Signature For Therapeutic Targeting and Clinical Intervention. Blood. 2013;122:826. [Google Scholar]
- 5.Weston BW, Hayden MA, Roberts KG, et al. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(25):e413–416. doi: 10.1200/JCO.2012.47.6770. [DOI] [PubMed] [Google Scholar]
- 6.Loh ML, Tasian SK, Rabin KR, et al. A phase 1 dosing study of ruxolitinib in children with relapsed or refractory solid tumors, leukemias, or myeloproliferative neoplasms: A Children’s Oncology Group phase 1 consortium study (ADVL1011) Pediatr Blood Cancer. 2015;62(10):1717–1724. doi: 10.1002/pbc.25575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts KG, Li Y, Payne-Turner D, et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N Engl J Med. 2014;371(11):1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borowitz MJ, Wood BL, Devidas M, et al. Prognostic significance of minimal residual disease in high risk B-ALL: a report from Children’s Oncology Group study AALL0232. Blood. 2015;126(8):964–971. doi: 10.1182/blood-2015-03-633685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Dongen JJ, Seriu T, Panzer-Grumayer ER, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet. 1998;352(9142):1731–1738. doi: 10.1016/S0140-6736(98)04058-6. [DOI] [PubMed] [Google Scholar]
- 10.Borowitz MJ, Devidas M, Hunger SP, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children’s Oncology Group study. Blood. 2008;111(12):5477–5485. doi: 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krampera M, Vitale A, Vincenzi C, et al. Outcome prediction by immunophenotypic minimal residual disease detection in adult T-cell acute lymphoblastic leukaemia. Br J Haematol. 2003;120(1):74–79. doi: 10.1046/j.1365-2141.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhao L, Dong H, Zhang CC, et al. A JAK2 interdomain linker relays Epo receptor engagement signals to kinase activation. J Biol Chem. 2009;284(39):26988–26998. doi: 10.1074/jbc.M109.011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344(14):1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 14.Piccaluga PP, Paolini S, Martinelli G. Tyrosine kinase inhibitors for the treatment of Philadelphia chromosome-positive adult acute lymphoblastic leukemia. Cancer. 2007;110(6):1178–1186. doi: 10.1002/cncr.22881. [DOI] [PubMed] [Google Scholar]
- 15.Schultz KR, Bowman WP, Aledo A, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children’s oncology group study. J Clin Oncol. 2009;27(31):5175–5181. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz KR, Carroll A, Heerema NA, et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia. 2014;28(7):1467–1471. doi: 10.1038/leu.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maude SL, Tasian SK, Vincent T, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120(17):3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]