Abstract
Humans adapt their behavior to their external environment in a process often facilitated by learning. Efforts to describe learning empirically can be complemented by quantitative theories that map changes in neurophysiology to changes in behavior. In this review we highlight recent advances in network science that offer a sets of tools and a general perspective that may be particularly useful in understanding types of learning that are supported by distributed neural circuits. We describe recent applications of these tools to neuroimaging data that provide unique insights into adaptive neural processes, the attainment of knowledge, and the acquisition of new skills, forming a network neuroscience of human learning. While promising, the tools have yet to be linked to the well-formulated models of behavior that are commonly utilized in cognitive psychology. We argue that continued progress will require the explicit marriage of network approaches to neuroimaging data and quantitative models of behavior.
Learning as a Network Phenomenon
Human learning is a complex phenomenon requiring the acquisition of knowledge and the flexibility to adapt existing neural processes to drive new patterns of desired behavior [1]. Learning can occur during explicit instruction – such as in classroom environments – or can occur implicitly by experience as we perceive statistical regularities in our external world [2]. In cognitive psychology, we often separately study explicit versus implicit learning [3], and we also often separate learning that occurs with or without feedback on the correctness or effectiveness of one’s actions, which we refer to as reinforcement [4]. Learning can be accompanied by a change in neural processes [5], an alteration in neuronal synapses [6], a signal of meaningful reward [7], and a reframing of one’s expectations [8], to name a few.
At a fundamental level, however, learning is about forming, removing, or altering associations. These associations can be of many types, such as between a word and a list, a particular face and its name, a specific food and reward, an image and a motor response, and even between one muscle and another as required for any type of complex motor sequence. Obviously, images, words, rewards, and motor responses all have underlying neural representations, and it is therefore no surprise that learning can also be cast purely in terms of associations in the brain. Nonetheless, while the majority of studies to date have focused on associations at the microscale, such as the formation of new synapses or the strengthening and weakening of existing ones, a new literature is emerging that focuses on the effects of learning at a coarser level – between entire brain regions. The neuroscience of learning, therefore, could benefit from theories and methods that can describe and probe these associations dynamically as learning unfolds in time.
Empirically, studies do indeed suggest that some types of learning (Box 1) can be characterized by relatively local changes in underlying neuroanatomy, such as visual perceptual learning supported by orientation tuning in V1 [9] or one-shot episodic memory formation supported by synaptic plasticity in hippocampus [10]. Even so, other types of learning – such as the acquisition of new visuomotor skills – are accompanied by broad-scale changes in neurophysiological dynamics across distributed neural circuits or networks subserving executive function, visual processing, and motor response [1]. For example, while motor cortex, visual cortex, basal ganglia, precuneus, dorsolateral prefrontal cortex, and cerebellum all show changes in their activity levels during motor skill learning [5], the changes in one area are not always independent of the changes in another area. Instead, computations that occur in one brain region impact on the computations in other brain regions, and the collective pattern of interactions, communication, or information transmission forms a circuit (or network) that directly enables a behavioral output [11,12]. When learning produces such distributed network changes – reflected either in anatomy or function – it is useful to consider quantitative methods that can not only describe that network architecture but also predict its dynamics.
Box 1. Commonly Studied Categories of Learning.
In response to external stimuli, the central nervous system selects motor commands that optimize the organism’s chances for survival [143]. Nevertheless, owing to the complexity of the stimuli, the nervous system may not utilize a simple rule-book, but instead implements a host of adaptive processes to modulate behavior [144]. Such adaptation, in addition to knowledge acquisition, facilitates learning.
When learning is influenced by an external driver that provides information about the course of knowledge or skill acquisition, the process is referred to as instructed learning [145]. When that influence includes information about whether the learner is using the correct map from the stimulus to the behavioral response, the learning is said to be supervised [145,146]. If instead the influence contains only information about whether the response was correct (a binary signal) or good (a continuous signal), the process is referred to as reinforcement learning [147]. Note that in some – but not all – cases, the instruction signal is offered in the form of language [148]. If no instructor is present, the learning process is often slower, and is referred to as unsupervised (other common terms for this type of learning include non-instructed or discovery learning) [149]. Both instructors and learners can be humans or animals (in the most natural use of the terms), or they can be brain regions or neural signals.
Learning can be a conscious process (explicit learning) or an unconscious process (implicit learning), driven by the constant stream of complex stimuli from the environment [150]. When that learning builds on observed statistical relationships between stimuli, the process is referred to as statistical learning, and offers rules for predicting broader structures or features of the environment [2,151]. While inference is a useful tool in learning, so too is chunking, or the ability to combine fine-scale solutions into bigger sets of solutions. For example, one can combine a series of motor responses into a single response [140,152], thereby reducing the complexity of the required input–output mapping.
In this review we describe recent advances in the field of network science that offer a unique approach to describing neural systems in terms of associations in the brain [13 – 15], and the reconfigurations underlying its adaptive processes [16]. We focus on processes at the macro-spatial scale, highlighting the application of these tools to region- and voxel-level data from non-invasive neuroimaging of adult humans. We place particular emphasis on functional magnetic resonance imaging (fMRI), a technique able to capture changes in region-to-region interactions in the brain. Although the framework we describe is fairly general, we illustrate its utility in the context of a specific example in which individuals learn a new visuomotor skill. Although not the focus of the review, we also mention a few important considerations in expanding the tools for applications in other human cohorts, across other learning tasks, or in data acquired from non-human primates and other animals. Finally, we discuss how network neuroscience could provide a quantitative framework that complements existing models of learning by cohesively accounting for network structure in neurophysiological and behavioral data [17].
Network Neuroscience
Network science is a subfield of complex systems science [18] which mathematically codifies systems whose function can be parsimoniously described by the patterns of interactions between components [19,20] or by the dynamics of such patterns. This field draws on tools from graph theory in mathematics, algorithm development in computer science, statistical mechanics in physics, and systems engineering. Traditionally, the field has been developed to understand social systems [21,22]. Nevertheless, the tools are simply foundational from a mathematical perspective, and can be flexibly shifted to other fields to understand complex functions of biological [23-26], technological [27,28], and physical [29-31] systems.
The use of network science tools to address neuroscientific hypotheses has now come to be called ’network neuroscience’ [32-34]. This burgeoning field encompasses the use of network science in understanding connectivity patterns in genes, neurons, organisms, and social groups in both human and animals, and moreover to understand the impact of these connectivity patterns on animal behavior [35] (Figure 1A,B). The potential utility of network neuroscience in informing a quantitative theory of human learning (particularly if combined with explicit models of behavior) lies in its mathematical principles [36]. These principles can be used to parsimoniously describe the architecture of relational data to be learned [37], as well as the patterns of interactions between neural components that enable behavioral adaptation [1].
Figure 1.
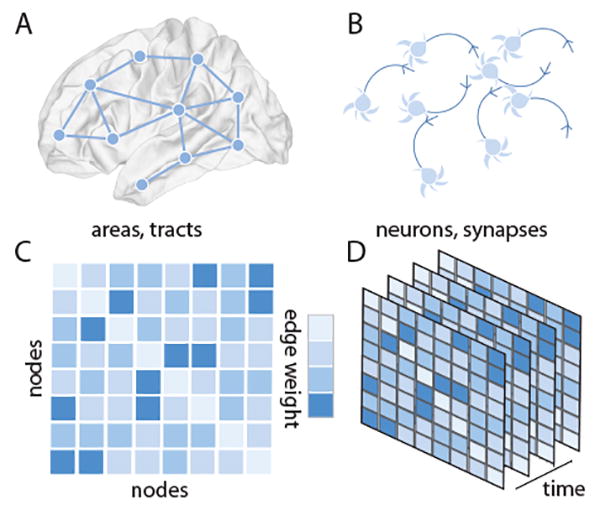
Network Models of Neural Systems. Network models represent complex systems by their fundamental units or elements, and by the pattern of connections, interactions, or relationships between those elements. In neural systems, networks can be defined across many spatial scales. (A) At the large scale, brain areas can be represented as network nodes, and white matter tracts or functional interactions between areas can be represented as network edges. (B) At the small scale, neurons, glia, or other cells can be represented as network nodes, and gap junctions or chemical synapses can be represented as network edges. (C) We represent the network as a graph, and encode the connectivity information in an adjacency matrix that stores the weight of each edge connecting each pair of nodes. The representation here is a weighted graph representation where the notion of neighbors becomes graded: the nearest neighbor of a node is the node to which it most strongly connects. Importantly, the order of nodes presented in a visualization is independent of the topology of the network. (D) To study learning, we string together connectivity patterns (or adjacency matrices) as they evolve through time.
Mathematically, a network is a graph G composed of V nodes and E edges [38,39]; each edge connects two nodes forming a dyad (Figure 1C), and these dyads can change in their strength over time (Figure 1D). In the simplest form of a network, these edges are unweighted (taking on either values of 1 to indicate existence or 0 to indicate nonexistence), and undirected (if the edge from node i to node j exists, so does the edge from node j to node i).
The architecture of the graph G can be probed and characterized quantitatively using a battery of summary statistics ranging from measures of local structure to measures of mesoscale and global structure [19]. A quintessential example of a local measure is the clustering coefficient of a graph, which measures the fraction of the neighbors of a node that are connected with one another [40]. Similarly, a common measure of mesoscale structure is the modularity of a graph, which measures the presence and strength of local clusters of densely interconnected nodes [41]. Commonly used measures for characterizing network structure are summarized in Box 2.
Box 2. Commonly Used Measures for Characterizing Network Structure.
As the field of network science has grown, so too has the number of measures that can be calculated to characterize the structure of a network. While a comprehensive account of all these measures is beyond the scope of this piece, we briefly describe here a few commonly used measures which can be categorized according to the topological scale to which they are most sensitive [153].
A commonly applied measure of local network structure is the clustering coefficient, which can be defined in a binary graph as the number of connected triangles divided by the number of connected triples [40]. By contrast, a commonly applied measure of global network structure is the average shortest path length, defined as the shortest path between any pair of nodes, averaged over all node pairs. The inverse of the harmonic mean of the shortest path is also referred to as the network efficiency, another common measure of global network structure [154]. This notion of network efficiency can also be calculated on a subgraph of connections surrounding a single node, thereby providing a measure of local efficiency [155]. Two common notions of mesoscale structure are community structure [82,81] and core–periphery structure [156,157]. Briefly, community structure refers to the presence of groups of nodes, where nodes within a group are more densely interconnected to other nodes in their group than to nodes in other groups. Core–periphery structure refers to the presence of a densely connected core and a sparsely connected periphery.
For more information, one can turn to several recent textbooks. For example, Networks: An Introduction by Mark Newman (MIT Press, 2010) [19] is an excellent introduction to the field of network science, Networks of the Brain by Olaf Sporns (MIT Press, 2011) [44] is a thoughtful introduction to the utility of these ideas for addressing questions in neuroscience, and Fundamentals of Brain Network Analysis (Academic Press, 2015) [158] from Fornito, Zalesky, and Bullmore is a useful textbook describing the application of network science tools to neuroimaging data. For particularly useful software, we refer the reader to the MATLAB-based, open-source Brain Connectivity Toolbox [159].
Graphs can display a range of different structures, and each of these structures can have very distinct implications for how the system functions. For example, a random graph, in which each node has an equal probability of connecting to any other node in the network, may transmit information quickly but may not be capable of performing any local information integration (Figure 2A). A regular graph, in which each node connects to an equal number of neighbors, does allow local information integration but is not optimized for global transmission of information across the network (Figure 2B). A modular graph is optimized for evolvability, and contains groups of nodes that can change or adapt their function without perturbing other groups (Figure 2C). Finally, a hierarchical modular network (similar to that observed in the human brain [42]) facilitates specialization of nested functions as well as their adaptability of time (Figure 2D).
Figure 2.
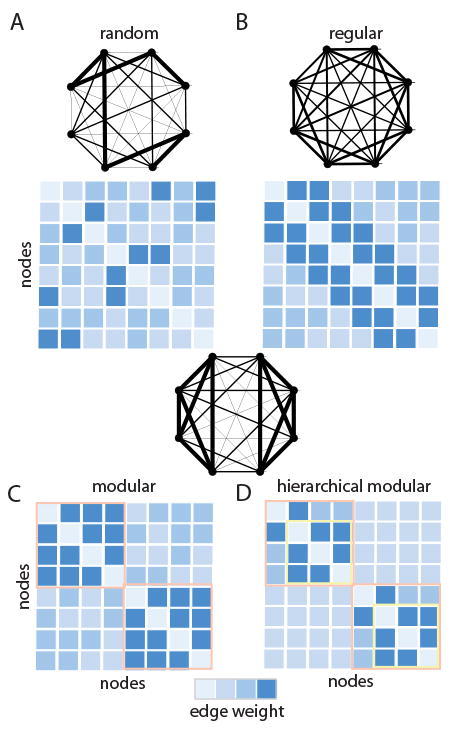
Neural Systems Can Display Different Connectivity Patterns. (A) In a random graph, each node has an equal probability of connecting to any other node. (B) In a regular graph, every node has an equal degree, k; in a regular ring lattice, every node has the same number of edges, and those edges link each node to its k nearest neighbors on the ring. (C) In a modular graph, groups of nodes are densely interconnected in clusters. (D) In a hierarchically modular graph, large modules are composed of smaller modules.
While the purview of network neuroscience is broad – ranging from genes to social groups [26,35], it is useful to briefly illustrate the use of this framework in the context of the human neuroimaging techniques traditionally used to probe facets of human learning [13-15,43–47]. For example, if one considers fMRI, magnetoencephalography (MEG), or electroencephalography (EEG) data collected as a subject is learning a new skill, then the network nodes can represent brain regions or voxels, and the network edges can represent functional connectivity [48] between nodes. This so-called ’functional brain network’ then represents a pattern of putative interactions between brain regions. Such patterns of functional connectivity can be used to infer the presence of distributed circuits supporting both intrinsic and task-evoked neurophysiological processes.
In contrast to functional neuroimaging, diffusion-weighted imaging data can also be represented fruitfully as a network [49,50]. In this case, the network nodes still represent brain regions or voxels, but the network edges now represent the estimated strength of white matter tracts linking pairs of nodes [51,52]. These so-called ’structural brain networks’ then represent the pattern of white matter microstructure supporting both baseline function [51,53,54] and its adaptability in response to task demands [55].
Dynamic Networks During Learning
The utility of network neuroscience in understanding human learning has recently become particularly clear with the development of an underlying mathematics for temporal or dynamic networks [56]. A temporal network is an ensemble of graphs in which each graph represents an interaction pattern in a single time-window, and therefore the ensemble of graphs represents the evolution of interaction patterns over time. Temporal graphs are therefore an ordered set of graphs, sometimes represented as a multilayer network [57], where a node (brain region) in one time-window is connected to itself in neighboring time-windows using an interlayer link. While traditional graph statistics are constructed to address adjacency matrices, new statistics are needed to address these adjacency tensors. Recent efforts from the applied mathematics community have developed a battery of dynamic extensions of traditional graph measures that can be used to evaluate and describe the architecture of temporal graphs, and the types of reconfigurations that they display [57].
In the case of brain networks, temporal graphs can be extracted from small time-windows during task performance [58] or over longer timescales using longitudinal imaging [1]. Examination of within-scan changes is only relevant for functional neuroimaging, while examination of across-scan changes is relevant for both functional and structural imaging. Indeed, structural changes have been observed in non-invasive imaging of white matter microstructure in humans following the learning of content as varied as a second language [59] or a new motor skill [60], suggesting that the edges within structural brain networks are malleable. It is intuitively plausible that these structural changes that occur both at the macro scale, and presumably also at the microscale, impact on the possible dynamic repertoire of the brain [61]. Consistent with this intuition, studies have shown that baseline neurophysiological dynamics as measured by resting-state blood oxygen-level dependent (BOLD) signal acquired with fMRI displays nontrivial changes following learning [62]. For example, local efficiency, a measure strongly correlated with local clustering, shows an increase in frontal cortex following motor learning [63], and an increase in superior temporal cortex following spoken language learning [64]. Importantly, these changes differ between individuals, and their magnitude is correlated with individual differences in learning success [64].
Based on the principles of Hebbian learning, these changes in both structural connectivity and baseline functional network architecture are presumably – at least in part – driven by patterns of activity and connectivity elicited by task performance. Indeed, perhaps the largest body of literature describing changes in network architecture related to learning has focused on the patterns of functional connectivity that are evident while a human engages in the task. For example, evidence suggests that network architecture can be used to (i) detect differential effects of memory rehabilitation training interventions following stroke [65], and to (ii) predict the future ability of an individual to learn a new motor skill [1] or the words of an artificial spoken language [66].
To make these advances more concrete, we focus on motor skill learning – that is addressed by the majority of literature examining dynamic functional networks as learning takes place. A particularly early study observed that functional connectivity of the sensorimotor network EEG could be modulated by focusing attention on the movements involved in ambulation [67]. Using functional near-infrared spectroscopy (fNIRS), a separate group demonstrated that functional network organization tracked cognitive burden, rising strongly in the middle phase of learning [68]. Indeed, even imagining movements – a common mechanism to drive brain computer interfaces for motor rehabilitation – produces significant changes in the functional connectivity of the default mode network [69] (a constellation of areas known to be active while a subject rests [70]). Importantly, some – but not all – of these changes in functional connectivity persist for some period of time after training [62,71]. Indeed, network reconfiguration accompanying learning can refer to structural network changes that may last over timescales of months or longer, or to functional network changes that may only characterize patterns of activity that are elicited during task performance.
Reconfiguration of Network Modules During Learning
A particularly crucial question to ask concerns whether learning is accompanied by some common property of plasticity that is observable with statistics that quantify network reconfiguration. In the context of motor skill learning, many network statistics have been observed to change: increased clustering coefficients, higher number of network connections, increased connection strength, shorter communication distances, and changes in network centrality [72,73]. To distill this apparent complexity into an intuition that could frame a quantitative theory, it has been posited that learning requires a change in the modular organization of brain network architecture [1] (Figure 3). This proposition builds on the now well-validated observation that the pattern of connections observed in structural brain networks, as well as in functional brain networks constructed from both resting and task-based data, displays modular organization [74]. A modular network is one that contains local clusters of densely interconnected nodes [41]. In empirical data, these clusters show beautiful overlap with known cognitive systems [75-79], including motor, visual, auditory, default mode, salience, attention, and executive systems, as well as with the group of subcortical areas. In the context of learning, evidence suggests that both the recruitment of and integration between these systems (or network modules) is altered as humans adapt their behavior [80].
Figure 3.
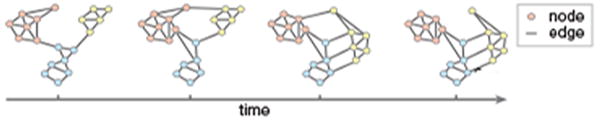
Reconfiguration of Network Modules. A schematic illustrating how patterns of connectivity can change over time as someone learns. For example, network modules can separate (as the pink and yellow modules do) or coalesce (as the blue and yellow modules do). Reconfiguration can also occur at the level of single nodes, which might initially be part of one module, and then change to be part of another module (as does the node at the top of this temporal graph, which starts by being affiliated with the pink module and ends being affiliated with the yellow module).
To mathematically codify how network modules may explicitly support human learning, several studies have capitalized on a recent extension of community detection techniques [81,82] to uncover modules in time-evolving networks [83]. While a methodologically nontrivial endeavor [84], the application of these tools to neuroimaging data has demonstrated that motor-learning, in particular, is accompanied by a consistent recruitment and growing autonomy of motor and visual modules, as well as by a disengagement of cognitive control regions [80]. These reconfigurations are more broadly characteristic of a flexible brain network organization [85,86], and individual differences in this flexibility of modular architecture predicts future learning rate [1]. Interestingly, this flexibility is produced by sets of edges that change in strength with one another instead of independently of one another [87], collectively linking a relatively stable core of regions that are thought to be necessary for task performance and a relatively flexible periphery of regions thought to be only supportive of task performance [88].
This growing literature supports the notion that brain network modularity plays a fundamental role in behavioral adaptability. Indeed, the general conceptual link between modularity and adaptability is not a stranger to scientific inquiry [89]; instead, it has been suggested as a nearly fundamental law of evolution and development [90,91]. The inherent compartmentalization of modular systems confers both robustness and the ability to evolve to external demands by changing modules relatively independently from one another [92,93]. The fundamental link between modularity and adaptivity offers explanations for beak development in Darwin’s finches [94] and the heterochrony of mammalian skull bones [95].
In light of this broader context, one might naturally ask whether modularity (and flexible modular rearrangement) is a marker of high cognitive function more generally [96], and is perhaps not solely related to learning. Initial evidence suggests that that is indeed the case: functional brain network modularity correlates with individual differences in working memory [97], and flexible rearrangement of modules correlates with individual differences in memory accuracy [98] and cognitive flexibility [98]. Moreover, the modules that are most flexible differ according to task demands [99], suggesting the potential to map the dynamics of modular organization in a task-dependent manner [100]. Moreover, modularity is thought to support the development of new skills without the forgetting of old ones [101], and has been shown to be positively correlated with improvement in attention and executive function after cognitive training [102].
Challenges and Opportunities
Although these initial studies offer a foundation on which to build a deeper understanding of human learning, many crucial challenges and promising opportunities remain. Perhaps the simplest set of questions surrounds the set of neurophysiological processes or environmental cues that may modulate brain network reconfiguration. Is brain network reconfiguration a state or a trait [103]? Do patterns of reconfiguration depend on the type of learning that participants engage in? Can one predict flexible network reconfiguration observed during task execution from baseline resting state measurements [103]? – or from underlying white matter microstructure [104]? Are brains that have more or less structural network control more or less flexible [105,106]? Can one modulate brain network flexibility with mood induction [85] or pharmacological intervention [107]? Is flexible network reconfiguration modulated by NMDA [107], norepinephrine [108], or other neurotransmitters?
Any such links could help to inform the development of generative network models of dynamic reconfiguration. A generative model is one that posits a set of parameterized wiring rules that can produce network architectures consistent with the observed data [109,110]. In recent applications to brain networks, these models have been exercised in the context of static network representations in both health [111] and disease [112], and in both humans and non-human animals [113]. Extending these tools into the temporal domain is a particularly exciting prospect which could offer fundamental insights into the mechanisms of network reconfiguration, and alterations in those mechanisms that may accompany normative neurodevelopment [114], healthy aging [115], or aberrant dynamics in neurological disease [116-118] or psychiatric disorders [107,119,120] that impact on learning. Classical network models are summarized in Box 3.
Box 3. Classical Network Models.
Building network models is an important component of general mathematical inquiry as well as of the application of network science tools to real-world systems. Particularly in the context of the latter, network models can form important benchmarks against which to compare the real networks, and similarities or differences between models and real data can offer insights into potential organizational principles in the specific application domain. We briefly describe here a few common network models as well as some of their interesting properties.
One of the common instantiations of the Erdős–Rényi random graph model is the G(n,p) model where each edge exists with a probability p that is independent from every other edge [38,39]. This graph has a binomial degree distribution P(k) where ki is the number of nodes connected with node i. By contrast, the degree distribution of a regular graph is a Dirac delta function [19]: in a regular graph, each vertex has the same degree k. The concepts of regular and random graphs can be related to one another through the Watts–Strogatz small-world graph model [40]. In this model, one begins with a regular ring lattice, where each node is connected to its k nearest neighbors on the ring. Edges are then randomly rewired with probability p. For intermediate values of p, the graph has a relatively high clustering coefficient (similar to that of a ring lattice) and a relatively short path length (similar to that of an Erdős–Rényi random graph). Importantly, the Erdős–Rényi, regular lattice, and Watts–Strogatz graph models all display narrower degree distributions than those often observed in real-world networks. A model that displays a broader degree distribution (indeed, a power-law degree distribution) is the Barabási–Albert model [160], which is built on the historical notion of preferential attachment [161-163]. In this model, one starts with a network of m0 connected nodes, and then adds new nodes one at a time by connecting a new node to m ≤ m0 existing nodes with a probability p that is proportional to the degree of the existing nodes.
A generative model that takes into account community (or modular) structure is the stochastic block model. In some ways, this model can be thought of as an extension of the Erdős–Rényi random graph model, G(n,p), where instead of p being a constant throughout the entire network, p now takes on different values within each community, and between each pair of communities. Extensions of these and similar notions to temporal graphs are a growing area of research [139,164].
While generative models may be a key first step towards a mechanistic theory of network reconfiguration accompanying learning, they will still fall short (in their current formulation) of addressing the perhaps more pervasive question – of how network organization relates to other neurophysiological measurements frequently studied in the literature. For example, learning can be studied by examining patterns of regional activation in MRI or patterns of signal power in EEG/MEG [5], both representing node-level properties. Even so, graphs in their simplest form contain nodes but do not allow properties to be assigned to nodes, other than the property of having an edge. This simplification limits the capabilities of the graph approach to integrate with alternative models of learning that include predictions about the anatomical locations of activation and the magnitude of that activation.
To overcome this challenge, one particularly promising approach is to utilize so-called annotated graphs (also known as tagged graphs) [121], which are extensions of network frameworks that could be used to incorporate any additional variables of interest associated with brain regions. These graphs allow one to assign properties to a node, and, in the context of dynamic networks during learning, one could imagine assigning properties to brain regions including activity magnitudes [122], oscillatory information [123], grey matter volumes [124], and dopamine levels [12] that together would offer a more comprehensive picture of the neurophysiological processes accompanying learning. Tools are currently being developed to extend the network metrics described earlier for use in annotated graphs. For example, methods now exist to quantify the degree to which nodes with similar annotations are located in the same or different network modules [121]. In the context of learning, such tools are being used to better understand the relationship between regional activation (e.g., decreases in BOLD magnitude) and dynamic patterns of connectivity (e.g., increasingly autonomous modules) that accompany behavioral change [125].
Towards a Network-Based Theory of Learning
Building on our growing ability to describe network-level changes that can accompany learning, we are faced with the question of whether we can extend these descriptions to predictions, thereby providing a richer quantitative theory of learning [126]. We may wish for such a theory to be biophysical in the sense that it goes beyond descriptions of BOLD magnitudes, explaining also neurophysiological data [127], and to be mechanistic in the sense that we understand how a change in network-level neurophysiological processes will lead to a change in behavior [128,129]. We suggest that a network-based theory of human learning will require models that explicitly bridge network models of brain function with mathematical models of human behavior (Figure 4). These would constitute formal mathematical models that can be fit to the data, and from which model parameters can be estimated [130]. Moreover, the models would be predictive in the sense that one has the potential to carefully perturb, manipulate, and indeed control the system with an explicit knowledge of the likely outcomes [35]. Common types of brain networks are summarized in Box 4.
Figure 4.
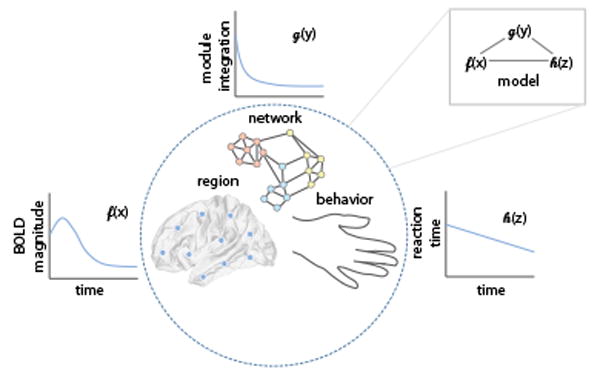
A Network-Based Theory of Learning. We suggest that a network-based theory of human learning will require models that explicitly bridge network models of brain function with mathematical models of human behavior. In this conceptual schematic, we illustrate the idea that changes in regional activation, network architecture, and behavioral measures can be linked mathematically in a formal modeling framework. We are careful to note that, although we suggest example variables of interest [blood oxygen level-dependent (BOLD) signal magnitude, module integration, and reaction time] and example temporal trends of these variables with learning, the particular functions may depend explicitly on the neuroscientific question of interest.
Box 4. Common Types of Brain Networks.
In applying concepts and tools from network science to neuroimaging data, the investigator is faced with the fact that network representations can be useful to describe several different types of inter-area or inter-neuron relationships. For example, the most commonly studied types of brain networks are referred to as structural networks and functional networks [15,44]. A structural network is one in which neural units (cells, columns, areas) are connected to one another by anatomical links (synapses, axonal projections, white matter tracts). By contrast, a functional network is one in which the same neural units are connected to one another with estimates of statistical similarities in their activity. Common measures of functional connectivity include a Pearson correlation coefficient, coherence, and synchronization. Although somewhat less common, brain morphometry can also be studied using morphometric networks in which the connectivity between regions is given by the correlation in a morphometric variable (grey matter density, cortical thickness, surface area, curvature) over subjects [165].
When one thinks of quantitative theories of learning, one probably turns to elegant mathematical models of human behavior. The drift-diffusion model is an example of a useful mathematical model of a behavior (decision making) that is important for many types of learning, particularly the illustrative type – motor skill learning – discussed at some length here. In this model, evidence is integrated until a decision threshold is reached, a process that has proven to be a good fit to both accuracy and reaction times in human data for two-alternative forced choice tasks [131], as well as to neural data [132,133]. In the specific context of learning itself, one might think of the Rescorla–Wagner model of reinforcement learning in which the strength of prediction of an unconditioned stimulus can be represented as the summed associative strengths of all conditioned stimuli [134]. While both models have been extended in various ways, and indeed the development of such models is an ongoing area of research [135,136], the beauty of these types of models is that they truly are quantitative theories in that a simple mathematical equation can be used to explain and predict human behavior. However, these theories tend to be limited to a single spatial scale of description, and it is often difficult to comprehend how theories at one scale relate to theories at another.
What would a region-scale theory look like that bridged network models of brain function with mathematical models of human behavior? One natural place to start would be to use probabilistic temporal network models such as stochastic block models [137,138]. Hypothetically speaking, these approaches would allow one to write down a model that could be used to predict how edges in a network change as a function of time and as a function of the modular architecture of the network [139]. Next, one would wish to expand the model to include annotations on individual network nodes, perhaps representing the level of BOLD activation [121]. Together, the annotated, dynamic stochastic block model would provide a prediction of regional levels of activity and inter-regional strengths of connectivity at future time-points based on an initial state. The model parameters could be fit to acquired data for validation (for the spatial scale, cohort, and type of learning under study), and one could test whether parameter values would predict individual differences in behavior.
Moving beyond correlations with behavior, one would turn to incorporating mathematical models of behavior into the annotated network models of neurophysiology. For example, in the context of a reinforcement learning paradigm, one could do this explicitly by using the learning rate α, which measures the extent to which the value of a choice is updated by feedback from a single trial. Higher (lower) α indicates more rapid (or slower) updating based on few (or more) trials. One might hypothesize that this α value estimated from behavior could be used to parameterize the stochastic block model, with higher α being associated with a swifter change in modular architecture, and lower α being associated with slower change in modular architecture. It would also be of interest to move beyond single-variable summaries of behavior by, for example, considering higher-order dependencies between actions [140], which can also be represented as networks. Indeed, if both brain and behavior were represented by network models, one could imagine linking the two in multilayer network representations [57]. Important open questions include which behavioral variables would be considered essential [141], and could we incorporate an ensemble of behavioral variables to better build and train models that could predict the behavioral responses of an individual to training? Answering these questions will serve to expand our understanding of the complexity of human learning.
Concluding Remarks and Future Directions
While we have focused this review on the application of network neuroscience tools to non-invasive neuroimaging data from adult humans, particularly collected while individuals are learning a new visuomotor skill, it will also be of interest in future to extend these ideas to non-adult cohorts where patterns of network reconfiguration may display distinct trajectories [114]. In addition, it will be important to understand the degree to which the reconfiguration of modules is an important biomarker of other types of learning, and whether these same biomarkers are characteristic of neural networks constructed at much finer spatial scales [142], such as where neurons are treated as network nodes, and synapses or similarities in spiking or calcium transients are treated as network edges. More generally, the concepts that we have discussed throughout this paper motivate and encourage future efforts that explicitly marry network approaches to neuroimaging data and quantitative theories of behavior. We anticipate that these efforts will also be informed by the complexity of the content to be learned [17], with particular regimes of model dynamics being predictive of the learning of relational data characterized by specific graph architectures.
Outstanding Questions.
What would a quantitative theory of learning built on network neuroscience have to offer?
Is brain network reconfiguration a state or a trait?
Do patterns of reconfiguration depend on the type of learning that participants engage in?
Can one predict flexible network reconfiguration observed during task execution from baseline resting state measurements? Or from underlying white matter microstructure?
Are brains that have more or less structural network control more or less flexible?
Can one modulate brain network flexibility with mood induction or pharmacological intervention?
Is flexible network reconfiguration modulated by glutamate, dopamine, norepinephrine, or other neurotransmitters?
Would a more extensive and comprehensive network neuroscience of human learning constitute a quantitative theory?
Which behavioral variables should be incorporated into network neuroscience models? and can we incorporate an ensemble of behavioral variables that together offer predictive power?
Trends Box.
Recent advances in network science offer a sets of tools and a general perspective that may be particularly useful in understanding types of learning that are supported by distributed neural circuits.
Recent applications of these tools to neuroimaging data provide unique insights into adaptive neural processes, the attainment of knowledge, and the acquisition of new skills, forming a network neuroscience of human learning.
Continued progress will require an explicit marriage between network approaches to neuroimaging data and well-formulated quantitative models of behavior commonly utilized in cognitive psychology.
Acknowledgments
We thank Elisabeth Karuza, Andrew Murphy, Ari Kahn, and Jonathan Soffer for helpful comments on earlier version of this manuscript. D.S.B. acknowledges support from the John D. and Catherine T. MacArthur Foundation, the Alfred P. Sloan Foundation, the Army Research Laboratory and the Army Research Office through contract numbers W911NF-10-2-0022 and W911NF-14-1-0679, the National Institute of Mental Health (2-R01-DC-009209-11), the National Institute of Child Health and Human Development (1R01HD086888-01), the Office of Naval Research, and the National Science Foundation (BCS-1441502, BCS-1430087, NSF PHY-1554488, and BCS-1631550). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies. The piece was inspired by discussions that occurred at a 2014 National Science Foundation workshop on Quantitative Theories of Learning, Memory, and Prediction (funded jointly by Betty Tuller in Behavioral and Cognitive Sciences, and Krastan Blagoev in Physics of Living Systems) that was attended by Robert Ajemian (MIT), Danielle Bassett (University of Pennsylvania), Matthew Botvinick (Princeton), Steve Bressler (Florida Atlantic University), Uri Eden (Boston University), Ila Fiete (University of Texas at Austin), Randy Gallistel (Rutgers University), Surya Ganguli (Stanford University), Kalanit Grill-Spector (Stanford University), Stephanie Jones (Brown University), Shella Keilholz (Georgia Tech), Nancy Kopell (Boston University), Randy McIntosh (Baycrest Health Sciences), Leslie Osborne (University of Chicago), Petra Ritter (Bernstein Center for Computational Neuroscience, Berlin), Nicole Rust (University of Pennsylvania), Daphna Shohamy (Columbia University), Roger Traub (IBM), Van Wedeen (Harvard/MGH), Fred Wolf (Max Planck, Goettingen), and Vijay Balasubramanian (University of Pennsylvania). The content is solely the responsibility of the authors and does not necessarily represent the views of any of these individuals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bassett DS, et al. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci U S A. 2011;108:7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslin RN, Newport EL. Statistical learning: from acquiring specific items to forming general rules. Curr Dir Psychol Sci. 2012;21:170–176. doi: 10.1177/0963721412436806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chrysikou EG, et al. A matched filter hypothesis for cognitive control. Neuropsychologia. 2014;62:341–355. doi: 10.1016/j.neuropsychologia.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shteingart H, Loewenstein Y. Reinforcement learning and human behavior. Curr Opin Neurobiol. 2014;25:93–98. doi: 10.1016/j.conb.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Dayan E, Cohen LG. Neuroplasticity subserving motor skill learning. Neuron. 2011;72:443–454. doi: 10.1016/j.neuron.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smolen P, et al. The right time to learn: mechanisms and optimization of spaced learning. Nat Rev Neurosci. 2016;17:77–88. doi: 10.1038/nrn.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keiflin R, Janak PH. Dopamine prediction errors in reward learning and addiction: from theory to neural circuitry. Neuron. 2015;88:247–263. doi: 10.1016/j.neuron.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholl J, et al. The good, the bad, and the irrelevant: neural mechanisms of learning real and hypothetical rewards and effort. J Neurosci. 2015;35:11233–11251. doi: 10.1523/JNEUROSCI.0396-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogels R. Mechanisms of visual perceptual learning in macaque visual cortex. Top Cogn Sci. 2010;2:239–250. doi: 10.1111/j.1756-8765.2009.01051.x. [DOI] [PubMed] [Google Scholar]
- 10.Nakazawa K, et al. Hippocampal CA3 NMDA receptors are crucial for memory acquisition of one-time experience. Neuron. 2003;38:305–315. doi: 10.1016/s0896-6273(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 11.Friston K. Beyond phrenology: what can neuroimaging tell us about distributed circuitry? Annu Rev Neurosci. 2002;25:221–250. doi: 10.1146/annurev.neuro.25.112701.142846. [DOI] [PubMed] [Google Scholar]
- 12.Happel MF. Dopaminergic impact on local and global cortical circuit processing during learning. Behav Brain Res. 2016;299:32–41. doi: 10.1016/j.bbr.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006;12:512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- 14.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 15.Bullmore ET, Bassett DS. Brain graphs: graphical models of the human brain connectome. Annu Rev Clin Psychol. 2011;7:113–140. doi: 10.1146/annurev-clinpsy-040510-143934. [DOI] [PubMed] [Google Scholar]
- 16.Bassett DS, et al. Adaptive reconfiguration of fractal small-world human brain functional networks. Proc Natl Acad Sci U S A. 2006;103:19518–19523. doi: 10.1073/pnas.0606005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karuza EA, et al. Local patterns to global architectures: influences of network topology on human learning. Trends Cogn Sci. 2016;20:629–640. doi: 10.1016/j.tics.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassett DS, Gazzaniga MS. Understanding complexity in the human brain. Trends Cogn Sci. 2011;15:200–209. doi: 10.1016/j.tics.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman MEJ. Networks: An Introduction. MIT Press; 2010. [Google Scholar]
- 20.Newman MEJ. Complex systems: a survey. Am J Phys. 2011;79:800–810. [Google Scholar]
- [21].Granovetter MS. The strength of weak ties. Am J Sociol. 1973;78:1360–1380. [Google Scholar]
- [22].Travers J, Milgram S. An experimental study of the small world problem. Sociometry. 1969;32:425–443. [Google Scholar]
- [23].Steinway SN, et al. Inference of network dynamics and metabolic interactions in the gut microbiome. PLoS Comput Biol. 2015;11:e1004338. doi: 10.1371/journal.pcbi.1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Baldassano SN, Bassett DS. Topological distortion and reorganized modular structure of gut microbial co-occurrence networks in inflammatory bowel disease. Sci Rep. 2016;6:26087. doi: 10.1038/srep26087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zanudo JG, Albert R. Cell fate reprogramming by control of intracellular network dynamics. PLoS Comput Biol. 2015;11:e1004193. doi: 10.1371/journal.pcbi.1004193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Conaco C, et al. Functionalization of a protosynaptic gene expression network. Proc Natl Acad Sci U S A. 2012;109:10612–10618. doi: 10.1073/pnas.1201890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Onnela JP, Rauch SL. Harnessing smartphone-based digital phenotyping to enhance behavioral and mental health. Neuropsychopharmacology. 2016;41:1691–1696. doi: 10.1038/npp.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Torous J, et al. New tools for new research in psychiatry: a scalable and customizable platform to empower data driven smartphone research. JMIR Ment Health. 2016;3:e16. doi: 10.2196/mental.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bassett DS, et al. Extraction of force-chain network architecture in granular materials using community detection. Soft Matter. 2015;11:2731–244. doi: 10.1039/c4sm01821d. [DOI] [PubMed] [Google Scholar]
- [30].Bassett DS, et al. Influence of network topology on sound propagation in granular materials. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;86:041306. doi: 10.1103/PhysRevE.86.041306. [DOI] [PubMed] [Google Scholar]
- [31].Lopez JH, et al. Jamming graphs: a local approach to global mechanical rigidity. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;88:062130. doi: 10.1103/PhysRevE.88.062130. [DOI] [PubMed] [Google Scholar]
- [32].Muldoon SF, Bassett DS. Why network neuroscience? Compelling evidence and current frontiers. Comment on ‘Understanding brain networks and brain organization’ by Luiz Pessoa. Phys Life Rev. 2014;11:455–457. doi: 10.1016/j.plrev.2014.06.006. [DOI] [PubMed] [Google Scholar]
- [33].Medaglia JD, et al. Cognitive network neuroscience. J Cogn Neurosci. 2015;27:1471–1491. doi: 10.1162/jocn_a_00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Misic B, Sporns O. From regions to connections and networks: new bridges between brain and behavior. Curr Opin Neurobiol. 2016;40:1–7. doi: 10.1016/j.conb.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassett DS, Sporns O. Network neuroscience. Nature Neuroscience. 2016 doi: 10.1038/nn.4502. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hermundstad AM, et al. Learning, memory, and the role of neural network architecture. PLoS Comput Biol. 2011;7:e1002063. doi: 10.1371/journal.pcbi.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schapiro A, et al. Neural representations of events arise from temporal community structure. Nat Neurosci. 2013;16:486–492. doi: 10.1038/nn.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bollobás B. Random Graphs. Academic Press; 1985. [Google Scholar]
- [39].Bollobás B. Graph Theory: An Introductory Course. Springer-Verlag; 1979. [Google Scholar]
- [40].Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- [41].Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bassett DS, et al. Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Comput Biol. 2010;6:e1000748. doi: 10.1371/journal.pcbi.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bassett DS, Bullmore ET. Human brain networks in health and disease. Curr Opin Neurol. 2009;22:340–347. doi: 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sporns O. Networks of the Brain. MIT Press; 2011. [Google Scholar]
- [45].Reijneveld JC, et al. The application of graph theoretical analysis to complex networks in the brain. Clin Neurophysiol. 2007;118:2317–2331. doi: 10.1016/j.clinph.2007.08.010. [DOI] [PubMed] [Google Scholar]
- [46].Fornito A, et al. Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage. 2013;80:426–444. doi: 10.1016/j.neuroimage.2013.04.087. [DOI] [PubMed] [Google Scholar]
- [47].Kaiser M. A tutorial in connectome analysis: topological and spatial features of brain networks. Neuroimage. 2011;57:892–907. doi: 10.1016/j.neuroimage.2011.05.025. [DOI] [PubMed] [Google Scholar]
- [48].Friston KJ. Functional and effective connectivity: a review. Brain Connect. 2011;1:13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- [49].Hagmann P, et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bassett DS, et al. Conserved and variable architecture of human white matter connectivity. Neuroimage. 2011;54:1262–1279. doi: 10.1016/j.neuroimage.2010.09.006. [DOI] [PubMed] [Google Scholar]
- [51].Hermundstad AM, et al. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc Natl Acad Sci U S A. 2013;110:6169–6174. doi: 10.1073/pnas.1219562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Behrens TE, Sporns O. Human connectomics. Curr Opin Neurobiol. 2012;22:144–153. doi: 10.1016/j.conb.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Honey C, et al. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Goñi J, et al. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc Natl Acad Sci U S A. 2014;111:833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hermundstad AM, et al. Structurally-constrained relationships between cognitive states in the human brain. PLoS Comput Biol. 2014;10:e1003591. doi: 10.1371/journal.pcbi.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Holme P, Saramaki J. Temporal networks. Phys Rep. 2012;519:97–125. [Google Scholar]
- [57].Kivel M, et al. Multilayer networks. J Complex Netw. 2014;2:203–271. [Google Scholar]
- 58.Telesford QK, et al. Detection of functional brain network reconfiguration during task-driven cognitive states. Neuroimage. 2016;142:198–210. doi: 10.1016/j.neuroimage.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Schlegel AA, et al. White matter structure changes as adults learn a second language. J Cogn Neurosci. 2012;24:1664–1670. doi: 10.1162/jocn_a_00240. [DOI] [PubMed] [Google Scholar]
- [60].Scholz J, et al. Training induces changes in white-matter architecture. Nat Neurosci. 2009;12:1370–1371. doi: 10.1038/nn.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Senden M, et al. Rich club organization supports a diverse set of functional network configurations. Neuroimage. 2014;96:174–182. doi: 10.1016/j.neuroimage.2014.03.066. [DOI] [PubMed] [Google Scholar]
- [62].Sami S, Miall RC. Graph network analysis of immediate motor-learning induced changes in resting state BOLD. Front Hum Neurosci. 2013;7:166. doi: 10.3389/fnhum.2013.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ma L, et al. Changes occur in resting state network of motor system during 4 weeks of motor skill learning. Neuroimage. 2011;58:226–233. doi: 10.1016/j.neuroimage.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Deng Z, et al. Resting-state low-frequency fluctuations reflect individual differences in spoken language learning. Cortex. 2016;76:63–78. doi: 10.1016/j.cortex.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Toppi J, et al. Time varying effective connectivity for describing brain network changes induced by a memory rehabilitation treatment. Conf Proc IEEE Eng Med Biol Soc. 2014;2014:6786–6789. doi: 10.1109/EMBC.2014.6945186. [DOI] [PubMed] [Google Scholar]
- [66].Sheppard JP, et al. Large-scale cortical network properties predict future sound-to-word learning success. J Cogn Neurosci. 2012;24:1087–1103. doi: 10.1162/jocn_a_00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sacco K, et al. Reorganization and enhanced functional connectivity of motor areas in repetitive ankle movements after training in locomotor attention. Brain Res. 2009;1297:124–134. doi: 10.1016/j.brainres.2009.08.049. [DOI] [PubMed] [Google Scholar]
- [68].James DR, et al. Cognitive burden estimation for visuomotor learning with fNIRS. Med Image Comput Comput Assist Interv. 2010;13:319–326. doi: 10.1007/978-3-642-15711-0_40. [DOI] [PubMed] [Google Scholar]
- [69].Ge R, et al. Motor imagery learning induced changes in functional connectivity of the default mode network. IEEE Trans Neural Syst Rehabil Eng. 2015;23:138–148. doi: 10.1109/TNSRE.2014.2332353. [DOI] [PubMed] [Google Scholar]
- [70].Raichle ME. The brain’s default mode network. Annu Rev Neurosci. 2015;38:433–447. doi: 10.1146/annurev-neuro-071013-014030. [DOI] [PubMed] [Google Scholar]
- [71].Zhang H, et al. Parallel alterations of functional connectivity during execution and imagination after motor imagery learning. PLoS One. 2012;7:5. doi: 10.1371/journal.pone.0036052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Heitger MH, et al. Motor learning-induced changes in functional brain connectivity as revealed by means of graph-theoretical network analysis. Neuroimage. 2012;61:633–650. doi: 10.1016/j.neuroimage.2012.03.067. [DOI] [PubMed] [Google Scholar]
- [73].Mantzaris AV, et al. Dynamic network centrality summarizes learning in the human brain. J Complex Networks. 2013;1:83–92. [Google Scholar]
- [74].Sporns O, Betzel RF. Modular brain networks. Annu Rev Psychol. 2016;67:613–640. doi: 10.1146/annurev-psych-122414-033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Chen ZJ, et al. Revealing modular architecture of human brain structural networks by using cortical thickness from MRI. Cereb Cortex. 2008;18:2374–2381. doi: 10.1093/cercor/bhn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Meunier D, et al. Age-related changes in modular organization of human brain functional networks. Neuroimage. 2009;44:715–723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- [77].Power JD, et al. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yeo BT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gordon EM, et al. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb Cortex. 2016;26:288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bassett DS, et al. Learning-induced autonomy of sensorimotor systems. Nat Neurosci. 2015;18:744–751. doi: 10.1038/nn.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Porter MA, et al. Communities in networks. Notices AMS. 2009;56:1082–1097. [Google Scholar]
- [82].Fortunato S. Community detection in graphs. Phys Rep. 2010;486:75–174. [Google Scholar]
- [83].Mucha PJ, et al. Community structure in time-dependent, multiscale, and multiplex networks. Science. 2010;328:876–878. doi: 10.1126/science.1184819. [DOI] [PubMed] [Google Scholar]
- [84].Bassett DS, et al. Robust detection of dynamic community structure in networks. Chaos. 2013;23:013142. doi: 10.1063/1.4790830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Betzel RF, et al. A positive mood, a flexible brain. arXiv. 2016;1601:07881. [Google Scholar]
- 86.Mattar M, et al. The flexible brain. Brain. 2016;139:2110–2112. doi: 10.1093/brain/aww151. [DOI] [PubMed] [Google Scholar]
- [87].Bassett DS, et al. Cross-linked structure of network evolution. Chaos. 2014;24:013112. doi: 10.1063/1.4858457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bassett DS, et al. Task-based core-periphery organization of human brain dynamics. PLoS Comput Biol. 2013;9:e1003171. doi: 10.1371/journal.pcbi.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Simon H. The architecture of complexity. Am Philos Soc. 1962;106:467–482. [Google Scholar]
- [90].Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci U S A. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc Natl Acad Sci U S A. 2005;102:13773–13778. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Wagner GP, Altenberg L. Complex adaptations and the evolution of evolvability. Evolution. 1996;50:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- [93].Schlosser G, Wagner GP. Modularity in Development and Evolution. University of Chicago Press; 2004. [Google Scholar]
- [94].Mallarino R, et al. Two developmental modules establish 3D beak-shape variation in Darwin’s finches. Proc Natl Acad Sci U S A. 2011;108:4057–4062. doi: 10.1073/pnas.1011480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Koyabu D, et al. Mammalian skull heterochrony reveals modular evolution and a link between cranial development and brain size. Nat Commun. 2014;5:3625. doi: 10.1038/ncomms4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kitzbichler MG, et al. Cognitive effort drives workspace configuration of human brain functional networks. J Neurosci. 2011;31:8259–8570. doi: 10.1523/JNEUROSCI.0440-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Stevens AA, et al. Functional brain network modularity captures inter- and intra-individual variation in working memory capacity. PLoS One. 2012;7:e30468. doi: 10.1371/journal.pone.0030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Braun U, et al. Dynamic reconfiguration of frontal brain networks during executive cognition in humans. Proc Natl Acad Sci U S A. 2015;112:11678–11683. doi: 10.1073/pnas.1422487112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Mattar MG, et al. A functional cartography of cognitive systems. PLoS Comput Biol. 2015;11:e1004533. doi: 10.1371/journal.pcbi.1004533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chai L, et al. Functional network dynamics of the language system. Cerebral Cortex. 2016 doi: 10.1093/cercor/bhw238. Published online October 17, 2016 http://dx.doi.org/10.1093/cercor/bhw238. [DOI] [PMC free article] [PubMed]
- [101].Ellefsen KO, et al. Neural modularity helps organisms evolve to learn new skills without forgetting old skills. PLoS Comput Biol. 2015;11:e1004128. doi: 10.1371/journal.pcbi.1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Arnemann KL, et al. Functional brain network modularity predicts response to cognitive training after brain injury. Neurology. 2015;84:1568–1574. doi: 10.1212/WNL.0000000000001476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mattar M, et al. Predicting future learning from baseline network architecture. bioRXiv. 2016 doi: 10.1016/j.neuroimage.2018.01.037. Published online June 3, 2016 http://doi.org/10.1101/056861. [DOI] [PMC free article] [PubMed]
- [104].Kahn AE, et al. Structural pathways supporting swift acquisition of new visuo-motor skills. arXiv. 2016;1605:04033. doi: 10.1093/cercor/bhw335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Gu S, et al. Controllability of structural brain networks. Nat Commun. 2015;6:8414. doi: 10.1038/ncomms9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Betzel RF, et al. Optimally controlling the human connectome: the role of network topology. Sci Rep. 2016;6:30770. doi: 10.1038/srep30770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Braun U, et al. Dynamic reconfiguration of brain networks: a potential schizophrenia genetic risk mechanism modulated by NMDA receptor function. Proc Natl Acad Sci USA. 2016;113:12568–12573. doi: 10.1073/pnas.1608819113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Nassar MR, et al. Rational regulation of learning dynamics by pupil-linked arousal systems. Nat Neurosci. 2012;15:1040–1046. doi: 10.1038/nn.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zanin M, et al. From phenotype to genotype in complex brain networks. Sci Rep. 2016;6:19790. doi: 10.1038/srep19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Simpson SL, et al. Exponential random graph modeling for complex brain networks. PLoS One. 2011;6:e20039. doi: 10.1371/journal.pone.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Betzel RF, et al. Generative models of the human connectome. Neuroimage. 2016;124:1054–1064. doi: 10.1016/j.neuroimage.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Vertes PE, et al. Simple models of human brain functional networks. Proc Natl Acad Sci U S A. 2012;109:5868–5873. doi: 10.1073/pnas.1111738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Vertes PE, et al. Generative models of rich clubs in Hebbian neuronal networks and large-scale human brain networks. Philos Trans R Soc Lond B Biol Sci. 2014;369:1653. doi: 10.1098/rstb.2013.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chai L, et al. Evolution of brain network dynamics in neurodevelopment. Network Neuroscience. 2017 doi: 10.1162/NETN_a_00001. Published online January 6, 2017 http://dx.doi.org/10.1162/NETN_a_00001. [DOI] [PMC free article] [PubMed]
- [115].Betzel RF, et al. Functional modules reconfigure at multiple scales across the human lifespan. arXiv. 2016;1510:08045. [Google Scholar]
- [116].Burns SP, et al. Network dynamics of the brain and influence of the epileptic seizure onset zone. Proc Natl Acad Sci U S A. 2014;111:E5321–E5330. doi: 10.1073/pnas.1401752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Khambhati AN, et al. Dynamic network drivers of seizure generation, propagation and termination in human neocortical epilepsy. PLoS Comput Biol. 2015;11:e1004608. doi: 10.1371/journal.pcbi.1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Khambhati A, et al. Virtual cortical resection reveals push-pull network control preceding seizure evolution. Neuron. 2016;91:1170–1182. doi: 10.1016/j.neuron.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Rashid B, et al. Classification of schizophrenia and bipolar patients using static and dynamic resting-state fMRI brain connectivity. Neuroimage. 2016;134:645–657. doi: 10.1016/j.neuroimage.2016.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Siebenhuhner F, et al. Intra- and inter-frequency brain network structure in health and schizophrenia. PLoS One. 2013;8:e72351. doi: 10.1371/journal.pone.0072351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Newman MEJ, Clauset A. Structure and inference in annotated networks. Nat Commun. 2016;7:11863. doi: 10.1038/ncomms11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Garvert MM, et al. Learning-induced plasticity in medial prefrontal cortex predicts preference malleability. Neuron. 2015;85:418–428. doi: 10.1016/j.neuron.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Buzsáki G, Draguhn A. Neuronal oscillations in cortical networks. Science. 2004;304:1926–1929. doi: 10.1126/science.1099745. [DOI] [PubMed] [Google Scholar]
- 124.Wenger E, et al. Repeated structural imaging reveals nonlinear progression of experience-dependent volume changes in human motor cortex. Cereb Corte. 2016 doi: 10.1093/cercor/bhw141. Published online May 25, 2016 http://dx.doi.org/10.1093/cercor/bhw141. [DOI] [PubMed]
- [125].Murphy AC, et al. Explicitly linking regional activation and function connectivity: community structure of weighted networks with continuous annotation. arXiv. 2016;1611:07962. [Google Scholar]
- [126].Valiant LG. What must a global theory of cortex explain? Curr Opin Neurobiol. 2014;25:15–19. doi: 10.1016/j.conb.2013.10.006. [DOI] [PubMed] [Google Scholar]
- [127].Jones SR. When brain rhythms aren’t ‘rhythmic’: implication for their mechanisms and meaning. Curr Opin Neurobiol. 2016;40:72–80. doi: 10.1016/j.conb.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Malanowski S, Craver CF. The spine problem: finding a function for dendritic spines. Front Neuroanat. 2014;8:95. doi: 10.3389/fnana.2014.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Craver CF. Beyond reduction: mechanisms, multifield integration and the unity of neuroscience. Stud Hist Philos Biol Biomed Sci. 2005;36:373–395. doi: 10.1016/j.shpsc.2005.03.008. [DOI] [PubMed] [Google Scholar]
- [130].Fitch WT. Toward a computational framework for cognitive biology: unifying approaches from cognitive neuroscience and comparative cognition. Phys Life Rev. 2014;11:329–364. doi: 10.1016/j.plrev.2014.04.005. [DOI] [PubMed] [Google Scholar]
- [131].Bogacz R, et al. The physics of optimal decision making: a formal analysis of models of performance in two-alternative forced-choice tasks. Psychol Rev. 2006;113:700–765. doi: 10.1037/0033-295X.113.4.700. [DOI] [PubMed] [Google Scholar]
- [132].Shadlen MN, Newsome WT. Motion perception: seeing and deciding. Proc Natl Acad Sci U S A. 1996;93:628–633. doi: 10.1073/pnas.93.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Platt ML, Glimcher PW. Neural correlates of decision variables in parietal cortex. Nature. 1999;400:233–238. doi: 10.1038/22268. [DOI] [PubMed] [Google Scholar]
- 134.Rescorla RA, Wagner AR. A theory of Pavlovian conditioning: variations in the effectiveness of reinforcement and nonreinforcement. In: Black AH, Prokasy WF, editors. Classical Conditioning II: Current Research and Theory. Appleton-Century-Crofts; 1972. pp. 64–99. [Google Scholar]
- [135].Dayan P. How to set the switches on this thing. Curr Opin Neurobiol. 2012;22:1068–1074. doi: 10.1016/j.conb.2012.05.011. [DOI] [PubMed] [Google Scholar]
- [136].Maia TV, Frank MJ. From reinforcement learning models to psychiatric and neurological disorders. Nat Neurosci. 2011;14:154–162. doi: 10.1038/nn.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Aicher C, et al. Learning latent block structure in weighted networks. J Complex Networks. 2014;3:221–248. [Google Scholar]
- [138].Abbe E, et al. Exact recovery in the stochastic block model. IEEE Trans Info Theory. 2015;62:471–487. [Google Scholar]
- [139].Zhang X, et al. Random graph models for dynamic networks. arXiv. 2016;1607:07570. [Google Scholar]
- [140].Wymbs NF, et al. Differential recruitment of the sensorimotor putamen and frontoparietal cortex during motor chunking in humans. Neuron. 2012;74:936–946. doi: 10.1016/j.neuron.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Acuna DE, et al. Multifaceted aspects of chunking enable robust algorithms. J Neurophysiol. 2014;112:1849–1856. doi: 10.1152/jn.00028.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Bettencourt LM, et al. Functional structure of cortical neuronal networks grown in vitro. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;75:021915. doi: 10.1103/PhysRevE.75.021915. [DOI] [PubMed] [Google Scholar]
- 143.Sejnowski TJ. The Computational Brain. MIT press; 1994. [Google Scholar]
- 144.Schunk DH. Learning Theories, An Educational Perspective. 2. Merrill; 1996. [Google Scholar]
- [145].Knudsen E. Supervised learning in the brain. J Neurosci. 1994;14:3985–3997. doi: 10.1523/JNEUROSCI.14-07-03985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ayodele TO. Types of machine learning algorithms. In: Zhang Y, editor. New Advances in Machine Learning. InTech; 2010. pp. 19–48. [Google Scholar]
- [147].Niv Y. Reinforcement learning in the brain. J Math Psychol. 2009;53:139–154. [Google Scholar]
- [148].Cole MW, et al. Rapid instructed task learning: a new window into the human brain’s unique capacity for flexible cognitive control. Cogn Affect Behav Neurosci. 2013;13:1–22. doi: 10.3758/s13415-012-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Barlow HB. Unsupervised learning. Neural Comput. 1989;1:295–311. [Google Scholar]
- [150].Seger CA. Implicit learning. Psychol Bull. 1994;115:163. doi: 10.1037/0033-2909.115.2.163. [DOI] [PubMed] [Google Scholar]
- [151].Turk-Browne NB, et al. The automaticity of visual statistical learning. J Exp Psychol Gen. 2005;134:552. doi: 10.1037/0096-3445.134.4.552. [DOI] [PubMed] [Google Scholar]
- [152].Ramkumar P, et al. Chunking as the result of an efficiency computation trade-off. Nat Commun. 2016;7:12176. doi: 10.1038/ncomms12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Betzel RF, Bassett DS. Multi-scale brain networks. Neuroimage. 2016 doi: 10.1016/j.neuroimage.2016.11.006. Published online November 11, 2016. http://dx.doi.org/10.1016/j.neuroimage.2016.11.006. [DOI] [PMC free article] [PubMed]
- [154].Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001;87:198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- [155].Achard S, Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Comput Biol. 2007;3:e17. doi: 10.1371/journal.pcbi.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Karrer B, Newman ME. Stochastic blockmodels and community structure in networks. Phys Rev E. 2011;83:016107. doi: 10.1103/PhysRevE.83.016107. [DOI] [PubMed] [Google Scholar]
- [157].Borgatti SP, Everett MG. Models of core/periphery structures. Soc Networks. 1999;21:375–395. [Google Scholar]
- 158.Fornito A, et al. Fundamentals of Brain Network Analysis. Academic Press; 2015. [Google Scholar]
- [159].Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- [160].Albert R, Barabási A-L. Statistical mechanics of complex networks. Rev Mod Phys. 2010;74:47–97x. [Google Scholar]
- [161].Yule GU. A mathematical theory of evolution based on the conclusions of Dr. J. C. Willis, F.R.S. J R Stat Soc. 1925;88:433–436. [Google Scholar]
- [162].Simon HA. On a class of skew distribution functions. Biometrika. 1955;42:425–440. [Google Scholar]
- 163.Price DJDS. A general theory of bibliometric and other cumulative advantage processes. J Am Soc Inf Sci. 1976;27:292–306. [Google Scholar]
- [164].Bazzi M, et al. Generative benchmark models for mesoscale structures in multilayer networks. arXiv. 2016;1608:06196. [Google Scholar]
- [165].Alexander-Bloch A, et al. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 2013;14:322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]


