Abstract
All happy families are alike; each unhappy family is unhappy in its own way.
--Leo Tolstoy, Anna Karenina
Keywords: Telomeres, Recombination, ALT, survivors, fungi, Ustilago maydis, C-circles
This review provides a summary of and comparisons between several mechanistically distinct telomere recombinational pathways that have been described in human cancer cells and fungal mutants. The maintenance of an adequate amount of telomere repeats at chromosome ends is essential for genome stability and cell proliferation. While the majority of proliferating cells use telomerase, a reverse transcriptase, to compensate for telomere loss, some telomerase-negative cells (including a subset of cancer cells) employ a recombination-based mechanism for telomere addition. Understanding the mechanisms of telomere recombination may therefore lead to targeted, telomere-based therapies for a subset of cancers. In contrast to telomerases, whose essential core components are conserved in diverse organisms, the telomere recombination pathways are characterized by distinct mechanisms, genetic requirements, and reaction products. The engagement of telomeres with recombination factors is often associated with defects in telomere structural integrity as well as gene expression profiles indicative of cellular stress. In this brief survey, we summarize the major telomere recombination pathways that have been described in cancer cells and various fungi. We also describe a newly characterized pathway in the basidiomycete Ustilago maydis, which in comparison to other fungal models, exhibits greater similarities to the pathway in cancer cells with respect to its genetic requirements and telomere structures. In view of the distinguishing features of the telomere and recombination machinery in the various systems, it is tempting to speculate that differences in the make-up of these machinery account for the mechanistic distinctions between the different telomere recombination pathways.
Eukaryotic chromosome termini, or telomeres, typically consist of a repetitive DNA sequence covered by a complex array of proteins (de Lange 2009). The maintenance of an adequate amount of telomeric DNAs at chromosome ends is required for genome stability and for sustaining the “replicative immortality” of a cell population. Owing to the “end-replication” problem, telomeric DNAs suffer progressive loss with each cell division (Olovnikov 1973). Once the lengths of telomeres fall below a threshold, the shortened chromosomes become recognizable by components of the DNA damage response machinery, which prevents the cells from progressing through the cell cycle and recruits recombination and repair proteins to act on the perceived damage (d'Adda di Fagagna, et al. 2003, Enomoto, et al. 2002).
Proliferating cells need a compensatory mechanism to replenish telomeres and overcome the end replication problem. In particular, a hallmark of cancer cells is replicative immortality, which is sustained by active telomere maintenance mechanisms (Hanahan and Weinberg 2011). The most prevalent telomere maintenance mechanism is mediated by telomerase, a special reverse transcriptase dedicated to the synthesis of telomere repeat units (Autexier and Lue 2006, Blackburn and Collins 2011). Telomerase is conserved in the vast majority of eukaryotes and consists of two core components (TERT and TER), each essential for catalytic activity. In contrast to normal somatic cells, which possess little or no telomerase activity, ~ 85% of cancer cells exhibit substantial telomerase activity due to transcriptional up-regulation of the normally repressed TERT gene (Heidenreich, et al. 2014, Kim, et al. 1994). Indeed, promoter mutations that enhance TERT transcription represent one of the most frequent mutations found in all cancers (Heidenreich, et al. 2014). However, telomerase is not the only telomere maintenance mechanism in cancer cells; ~10–15% of cancer cells possess no detectable telomerase activity and utilize an aberrant recombination pathway named ALT to achieve replicative immortality (Cesare and Reddel 2010, Shay, et al. 2012). ALT is a complex pathway; telomere elongation in ALT cells has been proposed to involve numerous factors. Interestingly, studies of budding and fission yeast have also uncovered mutants that utilize recombination in lieu of telomerase to maintain telomeres (Lundblad 2002, McEachern and Haber 2006). While these fungal mutants do not recapitulate all the telomere phenotypes of ALT cells, they have provided general insights on the variety of recombination factors and pathways that can be triggered at telomeres. More recently, a fungal model that exhibits greater similarity to ALT than previous models was reported in Ustilago maydis (de Sena-Tomas, et al. 2015, Yu, et al. 2015). U. maydis is a basidiomycete that is distantly related to budding and fission yeasts, and features of the U. maydis ALT model provide insights on the reasons that may underlie mechanistic distinctions and resemblances between different telomere recombination pathways in different organisms.
In this brief survey, we discuss the current understanding of the ALT pathway, and compare features of ALT cells to models of telomere recombination in various fungal mutants. These comparisons spotlight the existence of mechanistically distinct recombination pathways in different organisms, as illustrated by their distinctive genetic requirements and telomere structural aberrations. The potential reasons for the mechanistic diversity are then discussed. Some aspects of these topics have been covered in more detail in recent reviews of telomere recombination mechanisms (Dehe and Cooper 2010, Dilley and Greenberg 2015, Eckert-Boulet and Lisby 2010, Pickett and Reddel 2015).
ALT cancer cells
ALT was initially described in immortalized human cell lines that lack telomerase activity (for reviews, see (Pickett and Reddel 2015, Shay, et al. 2012)). Consistent with active recombination at chromosome ends, an ectopically introduced marker was observed to spread from one telomere to others (Dunham, et al. 2000). The telomeric DNAs in ALT cells are quite abnormal, and are characterized by extreme length heterogeneity, accumulation of unpaired telomere C-strands, and a substantial increase in extra-chromosomal telomere repeats (ECTRs), which exist in both linear and circular forms. Circular ECTRs have attracted considerable attention because of their plausible role in promoting telomere extension by serving as substrates for rolling-circle replication ((Nosek, et al. 2005, Tomaska, et al. 2004, Tomaska, et al. 2009)) (Fig. 1). Depending on the detection methods and structures, these circular telomeric DNAs have been named t-circles, G-circles, and C-circles. T-circles, or telomere circles, were initially identified in 2-D gels and electron microscopy (Cesare and Griffith 2004, Cesare, et al. 2008, Groff-Vindman, et al. 2005, Wang, et al. 2004). Subsequently, an assay based on phi29-mediated amplification of circular DNA was developed. Subsets of circular telomeric DNAs are further defined as G- or C-circles based on the continuity of particular strands. G-circles consist of a continuous G-strand and nicks or gaps in the complementary C-strand, whereas C-circles have the opposite characteristics. The free 3’-OH groups at the nicks or gaps can serve as primers for rolling circle amplification of the G- and C-circles by Phi29 polymerase, enabling highly sensitive detection of these structures (Henson, et al. 2009). Among the ECTRs, C-circles (telomere circles that consist of a continuous C-strand and discontinuous G-strands) are considered to be a specific marker for ALT (Henson, et al. 2009). How C-circles are generated is not well understood, but it is tempting to speculate that this structure may allow telomeres in ALT cells to be elongated through a rolling-circle mechanism. Besides abnormal telomere DNA structures, ALT is associated with genome rearrangements, marked micronucleation, defects in the G2/M checkpoint, and altered double-strand break (DSB) repair (Lovejoy, et al. 2012). Despite extensive efforts, including large-scale hunts for driver mutations in ALT cancer cell lines, the mechanisms that underlie ALT induction and maintenance remain incompletely understood (Lovejoy, et al. 2012). Factors implicated in repressing or promoting the ALT pathway include TERRA (telomere repeat-containing RNA), regulators of chromatin structure (e.g., ATRX, DAXX, and ASF1), nuclear receptors, and repair/recombination factors (Dilley and Greenberg 2015, Pickett and Reddel 2015). Among the factors that repress ALT, regulators of chromatin structure have received growing interests. In particular, mutations in the chromatin remodeling protein ATRX are frequently found in ALT cancers, and forced expression of ATRX can partially suppress ALT activity (Clynes, et al. 2015). In addition, knockdown of the histone chaperone ASF1a and ASF1b is evidently sufficient to trigger the ALT pathway (O'Sullivan, et al. 2014). Among the factors implicated in promoting ALT, most are known to participate in some aspects of DNA damage response and repair. However, quite a few such studies relied on co-localization of particular factors with telomeres and did not present strong functional evidence. DNA processing factors that have been functionally implicated in telomere metabolism in ALT cells include the MRE11-RAD50-NBS1 complex and the BLM helicase (Henson, et al. 2009, O'Sullivan, et al. 2014, Stavropoulos, et al. 2002, Zhong, et al. 2007). Curiously, several well-characterized homologous recombination factors such as RAD51 and RAD52 do not appear to be essential (Henson, et al. 2009, Oganesian and Karlseder 2011, Potts and Yu 2007).
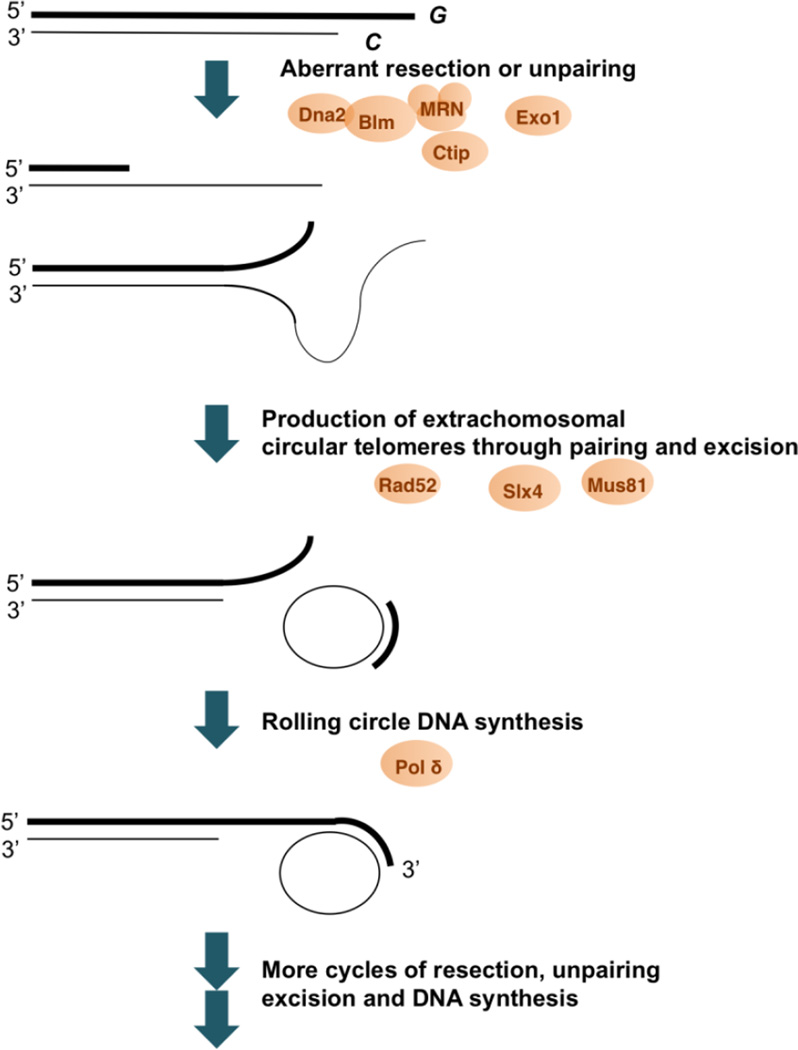
Figure 1. Potential recombination steps in the S. cerevisiae type II survivors, S. pombe linear survivors, U. maydis ku mutants and ALT cancer cells.
When telomeres are uncapped due to excessive shortening or defects in the chromatin structure, they become substrates for helicases and/or nucleases (e.g., MRN, Ctip1, Blm, Dna2, Exo1, etc.). The actions of these enzymes will lead to high levels of single stranded DNAs, which can then undergo annealing or strand invasion to generate recombination intermediates. A likely form of intermediates is circular extrachromosomal telomere DNA, which may be generated through the actions of structure-specific nucleases such as Mus81 or Slx4. Interestingly, different forms of circular DNA (i.e., t-circles, G-circles and C-circles) accumulate in different pathways; this could be due to distinctive interactions between the telomere nucleoprotein complex and recombination factors in different organisms. Notably, both t- and C-circles are capable of serving as template for rolling circle DNA synthesis of G-strand tails (e.g., by DNA polymerase δ).
Budding Yeast Survivors
In S. cerevisiae, the loss of telomerase activity results in progressive telomere loss, leading eventually to replicative senescence (Lundblad and Szostak 1989). However, about 0.01 percent of the cells in the population become survivors, which utilize recombinational telomere elongation to extend their telomere repeat tracts. Two types of survivors, distinguished from each other by their telomere structures and genetic requirements, have been described (Eckert-Boulet and Lisby 2010, Lundblad 2002, McEachern and Haber 2006) (Table 1). Type I survivors are characterized by multiple copies of the subtelomeric Y’ elements at chromosome ends (Lundblad and Blackburn 1993), and are dependent on Rad51, Rad52, Rad54, Rad55 and Rad57 (Chen, et al. 2001). The telomere addition mechanism in these survivors thus requires both the central yeast recombination protein Rad52 (critical for all recombination events in budding yeast) and the canonical strand exchange protein Rad51. Notably, the telomeres of type I survivors are capped by relatively short TG1–3 repeat tracts (the budding yeast terminal repeats at chromosome ends), which may account for their slower growth. In contrast, type II survivors possess long and heterogeneous TG1–3 tracts at their chromosome ends and exhibit faster growth rate than type I survivors (Teng, et al. 2000, Teng and Zakian 1999). Moreover, the propagation of type II survivors requires a distinct set of factors that includes Rad52, the MRX (Mre11-Rad50-Xrs2) complex, Sgs1 (BLM ortholog), and Rad59, but not Rad51. Since Rad51-mediated strand exchange is not essential for type II recombination, the current model for this pathway invokes the production of single stranded DNA (ssDNA) (produced by e.g., Sgs1 and MRX) and subsequent annealing between complementary sequences (mediated by e.g., Rad52 and Rad59) to generate recombination intermediates, which can then be further processed by polymerases and other enzymes to effect telomere elongation (Fig. 1). An important substrate in this pathway is thought to be extra-chromosomal telomeric circles, which are elevated in type II survivors, and may anneal to chromosome ends to enable elongation of telomeric repeat tracts through rolling circle DNA synthesis (Larrivee and Wellinger 2006, Lin, et al. 2005) (Fig. 1). Notably, the telomere recombination mechanism in type II survivors is believed to mimic that in ALT cells better than type I survivors. However, some discrepancies are clear. For example, ALT does not seem to require RAD52 whereas the type II pathway does. In addition, the most reliable marker of ALT activity, namely C-circles, has never been reported in type II survivor. Furthermore, the heterogeneous telomeres of type II survivors are relatively stable and undergo progressive shortening during passage, just like pre-senescent telomerase null cells (Teng, et al. 2000). In other words, the telomere elongation events in the survivors are quite infrequent. In contrast, ALT-based telomere maintenance appears to be constitutively active in cancer cells—i.e., the long telomeres in ALT cells do not appear to undergo progressive shortening during passage.
Table 1.
Comparisons of telomere recombination pathways
| Pathway | Activating Factors |
Repressing Factors |
Telomere structures |
|---|---|---|---|
| ALT | BLM MRE11 NBS1 RAD50 |
ATRX DAXX1 ASF1 |
heterogeneous terminal tract lengths unpaired G and C-strand at chromosome ends ECTRs C-circles, G-circles and t-circles |
|
S. cerevisiae type I survivor |
Rad51 Rad52 Rad54 Rad55 Rad57 |
increased Y’ elements at chromosome ends short terminal TG1–3 |
|
|
S. cerevisiae type II survivor |
Mre11 Rad50 Rad52 Rad59 Xrs2 Sgs1 Tel1 Mec1 |
heterogeneous terminal tract lengths t-circles |
|
|
S. pombe linear survivor |
Rad22 Rad32 Rad50 Nbs1 Rap1 |
Taz1 Trt1 Est1 Ku70 |
heterogeneous terminal tract lengths |
| U. maydis ALT-like | Blm Mre11 |
Ku70 Ku80 |
heterogeneous terminal tract lengths unpaired C-strand at chromosome ends linear and circular ECTRs t-circles, C-circles, and G-circles |
While the survivors are capable of indefinite proliferation in the absence of telomerase, they are likely to be experiencing stress. Transcriptional profiling of telomerase-negative mutants during passage showed, not surprisingly, that a large collection of stress-related genes are activated or repressed during senescence (Nautiyal, et al. 2002). Importantly, a significant subset of these genes does not return to normal expression levels even after the emergence of survivors and the restoration of growth rate. Thus, the long and heterogeneous telomeres of type II survivors (which were the survivors investigated in the gene expression study) can still be sensed by the cells as aberrant and induce a stress response. In addition, both type I and type II survivors exhibit accelerated replicative aging (i.e., giving rise to fewer daughter cells before permanent mitotic arrest), a feature that could be related to persistent cellular stress (Peng, et al. 2015).
Fission yeast survivors
The fission yeast telomerase-negative mutant has a broader repertoire of survival strategies (for a review see (Dehe and Cooper 2010)), only one of which bears some resemblance to ALT cells. The ALT-like survivors in S. pombe, named the linear survivors, have genetic requirements and telomere structures that are similar to the type II survivors in budding yeast (Subramanian, et al. 2008). That is, the linear survivors are dependent on Rad22 (the fission yeast ortholog of budding yeast Rad52) and the MRN (Rad32–Rad50-Nbs1 in the S. pombe gene nomenclature) complex, and harbor heterogeneously sized telomeres that consist of terminal repeats (i.e., the sequence G2–8-TTAC(A) rather than subtelomeric DNAs) (Subramanian, et al. 2008). Interestingly, the S. pombe linear survivor pathway is both positively and negatively regulated by components of the shelterin complex, the multi-protein assembly that caps chromosome ends. Specifically, the duplex telomere-binding protein Taz1 strongly inhibits the formation of linear survivors in telomerase mutants, suggesting that it can reduce the accessibility of telomeres to recombination proteins. In contrast, another telomeric protein Rap1 has a positive regulatory function for this pathway (Subramanian, et al. 2008). The resemblance of S. pombe linear survivors to the S. cerevisiae type II survivor suggests that similar recombination factors and mechanisms are responsible. However, the genetic requirements for the linear survivors have not been as thoroughly characterized. It is not known, for example, whether the S. pombe Rqh1 (the ortholog of BLM and Sgs1) is essential. Nor is it known whether t-circles are elevated in the linear survivors.
The other two groups of S. pombe survivors have no budding yeast equivalents. In contrast to budding yeast survivors, which invariably maintain some telomere repeats at chromosome ends, these two groups of S. pombe survivors can proliferate in the complete absence of telomere sequence by (i) circularizing all three chromosomes in the cell, or (ii) retaining heterochromatic DNA elements (e.g., subtelomeric DNA elements (STE) or rDNA repeats) at chromosome ends. The circularization pathway does not depend on classic NHEJ factors such as pKu70/80 and Lig4, and has been postulated to be mediated through single-strand annealing of a conserved, ~270 bp sequence near chromosome ends (Wang and Baumann 2008). Notably, once circularized, the chromosomes of these survivors will cease to engage in telomere-related recombination. In contrast, cells utilizing the heterochromatic DNA-based survivor pathway (named HAATI) harbor chromosomes that terminate in STEs or rDNAs, and these heterochromatic DNA elements likely undergo frequent recombination with one another (Jain, et al. 2010). However, the precise recombination mechanisms, as well as the factors required to sustaining the growth of HAATI survivors, have not been characterized in detail. The broad repertoire of survival strategies in S. pombe highlights the diverse means that the cell can use to solve the end replication and protection problem.
Fungal telomere recombination in the presence of active telomerase
While telomere recombination in fungi was first discovered in telomerase-negative cells, subsequent studies revealed activation of recombination in the absence of telomere attrition and senescence. Telomere recombination pathways in these mutants are triggered instead by mutations in telomere proteins. Examples have been described in S. cerevisiae, K. lactis and C. albicans, and several such mutants harbor mutations in the Cdc13-Stn1-Ten1 complex, a complex that binds to telomere G-strand and mediates diverse functions in telomere protection and maintenance (Giraud-Panis, et al. 2010, Price, et al. 2010). For instance, the S. cerevisiae cdc13-1 mutant, which is temperature sensitive, can “survive” when combined with a checkpoint mutation, and these survivors exhibits telomere phenotypes of type II survivors (Grandin, et al. 2001). Notably, these mutants possess normal telomerase genes and did not experience telomere loss prior to the activation of recombination, indicating that telomere loss is not an absolute pre-requisite for inducing telomere recombination. In K. lactis and C. albicans (both budding yeast that are evolutionarily close to S. cerevisiae), a point mutation or a complete knockout of the Stn1 component of the CST complex in the presence of active telomerase is sufficient to cause heterogeneous telomeres and high levels of telomere circles, which are characteristic of type II survivors (Iyer, et al. 2005, Sun, et al. 2009). The CST complex therefore appears to play a major role in suppressing the type II recombination pathway. Interestingly, deletion of the C. albicans RAP1 (the major duplex telomere binding protein) gene also triggers the formation of type II survivor-like telomeres (Yu, et al. 2010). This is reminiscent of the role of S. pombe Taz1 in suppressing linear survivor formation. Thus multiple components of the telomere nucleoprotein complex appear to play a role in suppressing telomere recombination. However, an interesting shared function for the genes referenced in this section is that they have all been reported to contribute to telomere replication. Thus, an alternative and more parsimonious hypothesis is that defects in telomere replication may promote recombination at telomeres.
A Ustilago maydis model of the ALT pathway
In both the S. cerevisiae type II survivor and the S. pombe linear survivors, long and heterogeneous telomere repeat tracts and extra-chromosomal telomere DNAs reminiscent of ALT cells were detected, suggesting similar underlying mechanisms of recombination (Fig. 1). However, other findings point to significant differences. For example, Rad52/rad22+ is essential for survivor formation in both fungi, but is evidently dispensable for ALT (O'Sullivan, et al. 2014, Oganesian and Karlseder 2011). In addition, neither fungal survivor exhibits a high level of C-circles, which is pathognomonic of ALT (Table 1). What could account for these differences? One reason may be the distinct recombination machinery in these organisms. Unlike mammalian cells, in which RAD52 plays a non-essential role in resistance to genotoxic stress and in recombinational repair (because of functional redundancy with BRCA2) (Feng, et al. 2011, Yamaguchi-Iwai, et al. 1998), budding and fission yeasts are severely deficient in all types of recombination when RAD52/rad22+ is deleted (Paques and Haber 1999). Also in yeast, the critical mammalian recombination repair protein BRCA2 is conspicuously absent, its function in promoting RAD51 filament formation having been assumed by Rad52 (Holloman 2011). Another source of difference is likely to be the distinct telomere nucleoprotein structures in different systems (Bianchi and Shore 2008, Lue 2010). The budding yeast telomere complex, in particular, contains duplex and G-strand bindings proteins (i.e., Rap1 and Cdc13) that are quite distinct from those in mammalian cells (i.e., TRFs and Pot1) (Lue 2010). In addition, the interactions between recombination factors and telomere components may also have diverged, leading to differences in how de-protected telomere DNAs are processed. Finally, it is worth noting that neither the budding nor the fission yeast possesses the same telomere repeat sequence as that in mammals; unlike the regular 6-bp telomere repeat unit in mammals (TTAGGG), both the budding yeast repeat (TG1–3) and the fission yeast repeat (G2–8-TTAC(A)) are irregular. The possibility that the different telomere DNAs can form distinct structures that interact differently with recombination factors should not be discounted.
Recently, a fungal telomere recombination pathway that more closely mimic the ALT pathway was discovered in U. maydis. As discussed below, the greater accuracy of the Ustilago model for ALT may be due to the stronger resemblance between the telomere and recombination machinery in Ustilago and humans. Ustilago maydis belongs to the phylum Basidiomycota, which branched off early from both Saccharomyces and Schizosaccharomyces in fungal phylogeny. U. maydis was developed decades ago by Robin Holliday as an experimental system to study DNA repair (Holloman, et al. 2008). Subsequent investigations revealed greater commonality between the recombination systems of this fungus and mammals, as evidenced by the shared presence of BRCA2 homologs. Also, unlike budding yeast but similar to fission yeast, U. maydis has a mammal-like telomere nucleoprotein complex (Yu, et al. 2013). Notably, initial analysis of the U. maydis telomerase-null mutant trt1Δ did not reveal an ALT-like pathway, but rather survivors that resemble the S. cerevisiae type I survivors ((Bautista-Espana, et al. 2014) and E.Y.Y., unpublished data). Instead, the Ustilago ALT-like pathway was discovered in the course of investigating Ku function. The Ku70/80 complex is a widely conserved heterodimeric protein complex that plays critical roles in both non-homologous end joining (NHEJ) and telomere protection (Downs and Jackson 2004). It had been shown earlier that human KU86 (the human Ku80 gene) is essential and that KU86-null cancer cells rapidly lose viability due to massive loss of telomere repeats from chromosome ends (Wang, et al. 2009). Remarkably, these observations were recently shown to hold true for the U. maydis ku70 and ku80 genes (even though other vertebrate and mammalian KU mutants are known to be viable) (de Sena-Tomas, et al. 2015, Yu, et al. 2015). The non-viability of the Ustilago Ku70- and Ku80-deficient cells can be suppressed by atr1 or chk1 deletion, and these ku/checkpoint double mutants exhibit all the hallmark telomere aberrations of ALT cancer cells, including telomere length heterogeneity and high levels of ECTRs. Most importantly, both the ku and ku/checkpoint mutants possess high levels of C-circles that are diagnostic of ALT activation. Thus the U. maydis mutants appear to be engaged in ALT-like recombination reactions, which involve both the telomere repeats at chromosome ends and those in extra-chromosomal forms.
The similarities between ALT cancer cells and the U. maydis model extend beyond the aberrant structures to the recombination factors involved in these pathways. In the U. maydis model, the most critical factors that have been identified thus far are Mre11 and Blm, both of which have been strongly implicated in ALT (O'Sullivan, et al. 2014, Stavropoulos, et al. 2002, Zhong, et al. 2007). Also resembling ALT cancer cells, the Rad51 recombinase is clearly dispensable in the U. maydis model. Notably, many factors previously suggested to play a role in ALT (e.g., the 9-1-1 complex and MUS81 (Nabetani, et al. 2004, Zeng, et al. 2009)) are not essential in the U. maydis model. Because these factors were implicated in ALT based on co-localization or partial effects of knockdown, it remains unclear whether they are indispensable. An apparent discrepancy between ALT and the Ustilago model is in regard to the role of Top3: knockdown of human TOP3α inhibited proliferation of and reduced telomere stability in ALT cells (Temime-Smaali, et al. 2008, Tsai, et al. 2006), but deletion of top3 enhanced telomere length heterogeneity and C-circle formation in U. maydis (Yu, et al. 2015). While this discrepancy may point to significant mechanistic differences between the model and ALT cancer cells, an alternative explanation should be considered. Specifically, knockdown of TOP3α was shown to drastically reduce the BLM protein level (Temime-Smaali, et al. 2008), and may thus have abolished the pathway through an indirect effect. Clearly, more studies are required to assess the similarities and differences between the model and ALT caner cells. In particular, the use of gene-editing technologies to completely eliminate specific gene functions in ALT cells will simplify the interpretation of results and provide a better frame of reference for the Ustilago findings.
Why should the Ustilago model show greater resemblance to the ALT pathway than the previously characterized S. cerevisiae type II survivors and S. pombe linear survivors? As noted before, in regard to the telomere sequence, the telomere nucleoprotein complex, and the recombination repair machinery, U. maydis exhibit either comparable or greater similarity to the human system than budding or fission yeast. The distinct roles of RAD52 in recombination in different organisms could very well explain their participation or lack of participation in the different recombination pathways. Current evidence suggest that yeast Rad52 mediates multiple steps of the recombination process (e.g., Rad51 filament assembly and the annealing of DNA strands during second end capture) (Paques and Haber 1999), whereas human RAD52 is more limited in its function (Feng, et al. 2011, Liu and Heyer 2011). The lack of RAD52 requirement for ALT could therefore reflect its more limited role in recombination in human cell. Alternatively, the aforementioned functional redundancy between RAD52 and BRCA2 may render each protein dispensable for ALT recombination. As for the distinct telomere structures in the survivors relative to ALT cells and the Ustilago model, the interplay between telomere proteins and recombination proteins that give rise to these structures are probably responsible. For example, mammalian telomere proteins (including the duplex binding proteins TRF1 and TRF2 as well as the G-strand binding protein POT1) have been reported to physically interact with BLM and regulate its activity (Lillard-Wetherell, et al. 2004, Opresko, et al. 2005, Stavropoulos, et al. 2002), but comparable interactions and regulation have not been reported for the yeast telomere proteins and either the S. cerevisiae Sgs1 or S. pombe Rqh1. Along these lines, it will be interesting to determine if this type of interactions and regulation exists in U. maydis.
Conclusion
Of the two broad categories of telomere maintenance mechanisms, telomerase-mediated extension entails the activity of a universally conserved ribonucleoprotein reverse transcriptase, whereas recombination-based extension is mediated by mechanistically distinct factors and pathways. Since recombination is generally repressed by the normal nucleoprotein structure at telomeres, the triggering of a recombination pathway requires some degree of uncapping due either to short telomeres or protein alterations. The nature of the uncapping is likely to determine the access and activity of recombination proteins, whose distinct make-up and activities in different organisms in turn dictate the reactions and the resulting telomere structures. Hence, a detailed understanding of ALT and other telomere recombination pathways will require understanding of the telomere machinery, the recombination machinery, and the interplay between the two groups of factors.
REFERENCES
- Autexier C, Lue NF. The Structure And Function of Telomerase Reverse Transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Bautista-Espana D, Anastacio-Marcelino E, Horta-Valerdi G, Celestino-Montes A, Kojic M, Negrete-Abascal E, Reyes-Cervantes H, Vazquez-Cruz C, Guzman P, Sanchez-Alonso P. The telomerase reverse transcriptase subunit from the dimorphic fungus Ustilago maydis. PloS one. 2014;9:e109981. doi: 10.1371/journal.pone.0109981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. doi: S1097-2765(08)00431-0 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a003558. 3cshperspect.a003558 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol Cell Biol. 2008;28:20–29. doi: 10.1128/MCB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. doi: nrg2763 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes D, Jelinska C, Xella B, Ayyub H, Scott C, Mitson M, Taylor S, Higgs DR, Gibbons RJ. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nature communications. 2015;6:7538. doi: 10.1038/ncomms8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. nature02118. [DOI] [PubMed] [Google Scholar]
- de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. doi: 326/5955/948 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sena-Tomas C, Yu EY, Calzada A, Holloman WK, Lue NF, Perez-Martin J. Fungal Ku prevents permanent cell cycle arrest by suppressing DNA damage signaling at telomeres. Nucleic Acids Res. 2015;43:2138–2151. doi: 10.1093/nar/gkv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehe PM, Cooper JP. Fission yeast telomeres forecast the end of the crisis. FEBS Lett. 2010;584:3725–3733. doi: 10.1016/j.febslet.2010.07.045. [DOI] [PubMed] [Google Scholar]
- Dilley RL, Greenberg RA. ALTernative Telomere Maintenance and Cancer. Trends Cancer. 2015;1:145–156. doi: 10.1016/j.trecan.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. nrm1367. [DOI] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Eckert-Boulet N, Lisby M. Regulation of homologous recombination at telomeres in budding yeast. FEBS Lett. 2010;584:3696–3702. doi: 10.1016/j.febslet.2010.05.037. [DOI] [PubMed] [Google Scholar]
- Enomoto S, Glowczewski L, Berman J. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2626–2638. doi: 10.1091/mbc.02-02-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci U S A. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E. CST meets shelterin to keep telomeres in check. Mol Cell. 2010;39:665–676. doi: 10.1016/j.molcel.2010.08.024. doi: S1097-2765(10)00634-9 10.1016/j.molcel.2010.08.024. [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 2001;20:6127–6139. doi: 10.1093/emboj/20.21.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groff-Vindman C, Cesare AJ, Natarajan S, Griffith JD, McEachern MJ. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol Cell Biol. 2005;25:4406–4412. doi: 10.1128/MCB.25.11.4406-4412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. TERT promoter mutations in cancer development. Curr Opin Genet Dev. 2014;24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. doi: nbt.1587 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol. 2011;18:748–754. doi: 10.1038/nsmb.2096. doi: nsmb.2096 10.1038/nsmb.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman WK, Schirawski J, Holliday R. The homologous recombination system of Ustilago maydis. Fungal Genet Biol. 2008;45(Suppl 1):S31–S39. doi: 10.1016/j.fgb.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Chadha A, McEachern M. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol Cell Biol. 2005;25:8064–8073. doi: 10.1128/MCB.25.18.8064-8073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature. 2010;467:223–227. doi: 10.1038/nature09374. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Larrivee M, Wellinger RJ. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat Cell Biol. 2006;8:741–747. doi: 10.1038/ncb1429. [DOI] [PubMed] [Google Scholar]
- Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- Lin C, Chang H, Wu K, Tseng S, Lin C, Lin C, Teng S. Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:327–336. doi: 10.1128/EC.4.2.327-336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Heyer WD. Who's who in human recombination: BRCA2 and RAD52. Proc Natl Acad Sci U S A. 2011;108:441–442. doi: 10.1073/pnas.1016614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. PGENETICS-D-12-00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem Sci. 2010;35:8–17. doi: 10.1016/j.tibs.2009.08.006. doi: S0968-0004(09)00176-5 10.1016/j.tibs.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. Telomere maintenance without telomerase. Oncogene. 2002;21:522–531. doi: 10.1038/sj.onc.1205079. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Yokoyama O, Ishikawa F. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J Biol Chem. 2004;279:25849–25857. doi: 10.1074/jbc.M312652200. [DOI] [PubMed] [Google Scholar]
- Nautiyal S, DeRisi JL, Blackburn EH. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:9316–9321. doi: 10.1073/pnas.142162499. 142162499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosek J, Rycovska A, Makhov AM, Griffith JD, Tomaska L. Amplification of telomeric arrays via rolling-circle mechanism. J Biol Chem. 2005;280:10840–10845. doi: 10.1074/jbc.M409295200. [DOI] [PubMed] [Google Scholar]
- O'Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol. 2014;21:167–174. doi: 10.1038/nsmb.2754. doi: nsmb.2754 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L, Karlseder J. Mammalian 5' C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell. 2011;42:224–236. doi: 10.1016/j.molcel.2011.03.015. doi: S1097-2765(11)00215-2 10.1016/j.molcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov A. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, He MH, Duan YM, Liu YT, Zhou JQ. Inhibition of telomere recombination by inactivation of KEOPS subunit Cgi121 promotes cell longevity. PLoS Genet. 2015;11:e1005071. doi: 10.1371/journal.pgen.1005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett HA, Reddel RR. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat Struct Mol Biol. 2015;22:875–880. doi: 10.1038/nsmb.3106. [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–3165. doi: 10.4161/cc.9.16.12547. doi: 12547 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Reddel RR, Wright WE. Cancer. Cancer and telomeres--an ALTernative to telomerase. Science. 2012;336:1388–1390. doi: 10.1126/science.1222394. doi: 336/6087/1388 10.1126/science.1222394. [DOI] [PubMed] [Google Scholar]
- Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- Subramanian L, Moser BA, Nakamura TM. Recombination-based telomere maintenance is dependent on Tel1-MRN and Rap1 and inhibited by telomerase, Taz1, and Ku in fission yeast. Mol Cell Biol. 2008;28:1443–1455. doi: 10.1128/MCB.01614-07. doi: MCB.01614-07 10.1128/MCB.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2–Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. doi: 23/24/2900 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temime-Smaali N, Guittat L, Wenner T, Bayart E, Douarre C, Gomez D, Giraud-Panis MJ, Londono-Vallejo A, Gilson E, Amor-Gueret M, Riou JF. Topoisomerase IIIalpha is required for normal proliferation and telomere stability in alternative lengthening of telomeres. EMBO J. 2008;27:1513–1524. doi: 10.1038/emboj.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Chang J, McCowan B, Zakian V. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- Teng S, Zakian V. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaska L, McEachern MJ, Nosek J. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 2004;567:142–146. doi: 10.1016/j.febslet.2004.04.058. [DOI] [PubMed] [Google Scholar]
- Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HJ, Huang WH, Li TK, Tsai YL, Wu KJ, Tseng SF, Teng SC. Involvement of topoisomerase III in telomere-telomere recombination. J Biol Chem. 2006;281:13717–13723. doi: 10.1074/jbc.M600649200. [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Wang X, Baumann P. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol Cell. 2008;31:463–473. doi: 10.1016/j.molcel.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. doi: 0903362106 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Buerstedde JM, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Kojic M, Holloman WK, Lue NF. Brh2 and Rad51 promote telomere maintenance in Ustilago maydis, a new model system of DNA repair proteins at telomeres. DNA Repair (Amst) 2013;12:472–479. doi: 10.1016/j.dnarep.2013.04.027. doi: S1568-7864(13)00121-3 10.1016/j.dnarep.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Perez-Martin J, Holloman WK, Lue NF. Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient Ustilago maydis. PLoS Genet. 2015;11:e1005570. doi: 10.1371/journal.pgen.1005570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Yen WF, Steinberg-Neifach O, Lue NF. Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol Cell Biol. 2010;30:1254–1268. doi: 10.1128/MCB.00986-09. doi: MCB.00986-09 10.1128/MCB.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, Yang Q. Telomere recombination requires the MUS81 endonuclease. Nat Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong ZH, Jiang WQ, Cesare AJ, Neumann AA, Wadhwa R, Reddel RR. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. doi: M701413200 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]