Abstract
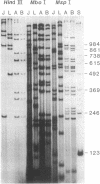
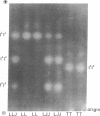
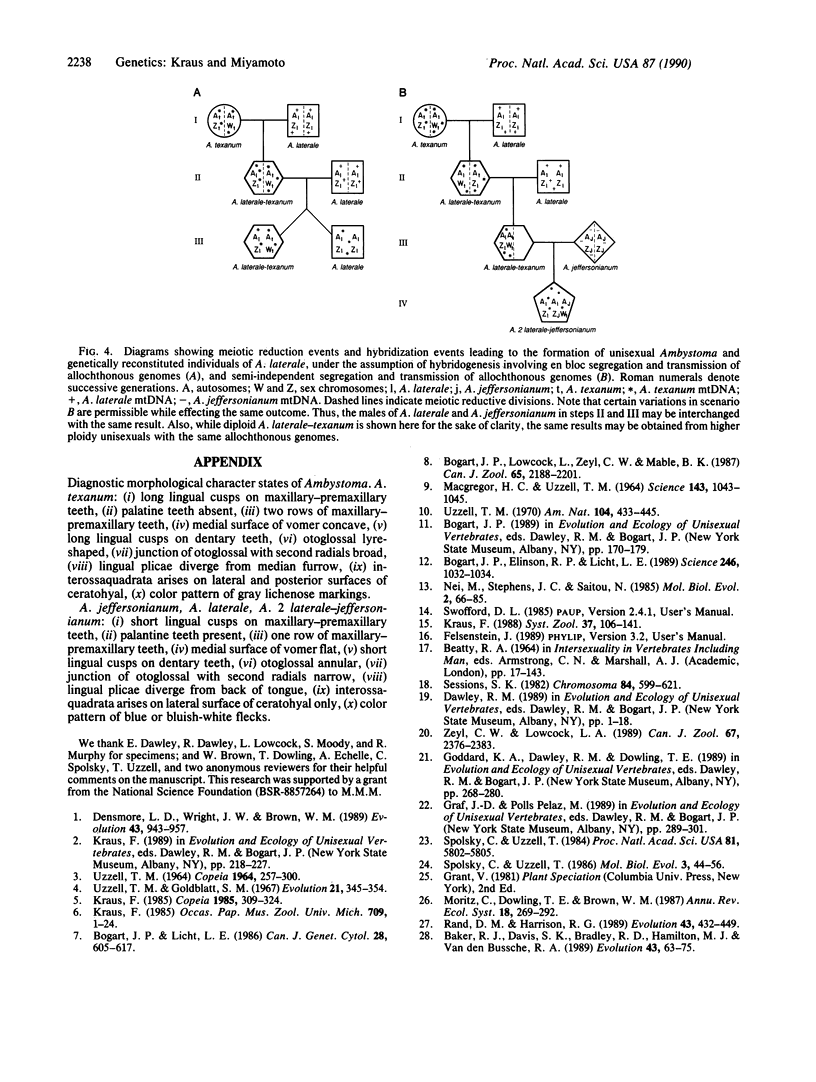
We examined mitochondrial DNA (mtDNA), enzyme, and morphological variation among 17 unisexual Ambystoma of hybrid origin. Electrophoretic comparison of diagnostic enzymes indicates that these unisexuals are triploid with two nuclear genomes from the bisexual species Ambystoma laterale and one from Ambystoma jeffersonianum; however, according to restriction analysis, the mtDNAs of these specimens derive from a third species, Ambystoma texanum. This unusual situation is apparently due to a partially independent segregation event in an ancestor of these unisexuals. This situation highlights the potential importance of molecules with different inheritance patterns in elucidating complex cases of reticulate evolution.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bogart J. P., Elinson R. P., Licht L. E. Temperature and sperm incorporation in polyploid salamanders. Science. 1989 Nov 24;246(4933):1032–1034. doi: 10.1126/science.2587986. [DOI] [PubMed] [Google Scholar]
- Bogart J. P., Licht L. E. Reproduction and the origin of polyploids in hybrid salamanders of the genus Ambystoma. Can J Genet Cytol. 1986 Aug;28(4):605–617. doi: 10.1139/g86-089. [DOI] [PubMed] [Google Scholar]
- MACGREGOR H. C., UZZELL T. M., Jr GYNOGENESIS IN SALAMANDERS RELATED TO AMBYSTOMA JEFFERSONIANUM. Science. 1964 Mar 6;143(3610):1043–1045. doi: 10.1126/science.143.3610.1043. [DOI] [PubMed] [Google Scholar]
- Nei M., Stephens J. C., Saitou N. Methods for computing the standard errors of branching points in an evolutionary tree and their application to molecular data from humans and apes. Mol Biol Evol. 1985 Jan;2(1):66–85. doi: 10.1093/oxfordjournals.molbev.a040333. [DOI] [PubMed] [Google Scholar]
- Spolsky C., Uzzell T. Evolutionary history of the hybridogenetic hybrid frog Rana esculenta as deduced from mtDNA analyses. Mol Biol Evol. 1986 Jan;3(1):44–56. doi: 10.1093/oxfordjournals.molbev.a040376. [DOI] [PubMed] [Google Scholar]
- Spolsky C., Uzzell T. Natural interspecies transfer of mitochondrial DNA in amphibians. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5802–5805. doi: 10.1073/pnas.81.18.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]