Abstract
Stressor exposure significantly affects the colonic mucosa-associated microbiota, and exacerbates Citrobacter rodentium-induced inflammation, effects that can be attenuated with probiotic Lactobacillus reuteri. This study assessed the structure of the colonic mucosa-associated microbiota in mice exposed to a social stressor (called social disruption), as well as non-stressed control mice, during challenge with the colonic pathogen C. rodentium. Mice were exposed to the social stressor or home cage control conditions for six consecutive days and all mice were challenged with C. rodentium immediately following the first exposure to the stressor. In addition, mice received probiotic L. reuteri, or vehicle as a control, via oral gavage following each stressor exposure. The stressor-exposed mice had significant differences in microbial community composition compared to non-stressed control mice. This difference was first evident following the six-cycle exposure to the stressor, on Day 6 post-C. rodentium challenge, and persisted for up to 19 days after stressor termination. Mice exposed to the stressor had different microbial community composition regardless of whether they were treated with L. reuteri or treated with vehicle as a control. These data indicate that stressor exposure affects the colonic microbiota during challenge with C. rodentium, and that these effects are long-lasting and not attenuated by probiotic L. reuteri.
The human gastrointestinal (GI) tract is the site of many chronic inflammatory illnesses including the inflammatory bowel diseases (IBD), i.e., ulcerative colitis and Crohn’s disease1. The exact origins of these illnesses have not been fully explicated. The GI tract has a unique micro-environment that consists of monitoring immune and epithelial cells in close proximity to a constant source of external stimuli and luminal antigen, which can stem in part from the expansive intestinal microbiota that co-exists adjacently2. There is normal bidirectional communication between host immune cells sampling the periphery and the microbiota, and disruptions in the microbiota have been associated with negative health outcomes3,4,5. As such, the conditions that skew the composition of luminal antigen or the activity and response of resident host GI cells could be factors that associate with IBD. Psychological stress is one such factor.
Psychological stressor exposure affects GI functioning and symptoms in both healthy and diseased individuals. For example, psychological stress is associated with elevated inflammation, bleeding, and pain in both IBD and enteric infections6,7,8. Although the mechanisms by which psychological stressor exposure leads to heightened inflammatory responses are unknown, previous studies have shown that stressor exposure can affect the GI microbiota in a number of different mammalian hosts, including humans, non-human primates, and rodents9,10,11. Affected bacterial groups included lactic acid bacteria and other health-promoting groups, which were reduced after exposure to stress11,12. Recently, we have shown that mice exposed to social disruption (SDR), a social stressor that involves aggressive interactions between mice, have significant changes to the mucosa-associated colonic microbiota community structure10. The stressor also reduces the absolute abundance of beneficial commensal groups like Lactobacillus and Parabacteroides. These previous observations were made in healthy, uninfected mice even though studies indicate that stressor-induced changes in the microbiota impact the colonic inflammatory response to C. rodentium6,13,14. For example, exposure to stress prior to oral challenge with C. rodentium changed gut microbiota composition and increased subsequent colonic inflammatory responses to C. rodentium6. Transplanting the microbiota from stressor-exposed mice to germfree mice prior to challenge with C. rodentium resulted in an exaggerated colonic inflammatory response compared to germfree mice that received microbiota from non-stressed donors15, demonstrating the impact that the effects of stress on the microbiota can have on the susceptibility to and severity of C. rodentium infection. However, because inflammation in the intestines can significantly change microbial community composition16, it is not immediately clear whether stressor-induced changes in gut microbiota composition are still evident in mice with C. rodentium-induced colonic inflammation, and whether the effects of the stressor are evident in probiotic-treated animals.
Probiotic bacteria, as defined by the World Health Organization, are living microbes that can confer a health benefit upon a host when given in adequate numbers. Lactobacillus reuteri is an immunomodulatory probiotic that can ameliorate the severity of colonic infection17 and can down-regulate CCL2, TNF-α, and iNOS mRNA levels in SDR-exposed C. rodentium-infected mice, as well as abrogate the heightened colonic pathology in stressor-exposed mice18. Probiotic microbes like L. reuteri can act directly upon host immunity, such as by modulating phagocytosis and cytokine release by macrophages and monocytes or intestinal epithelial cells13,18, but they can also affect overall microbiome diversity, which is associated with host health14,19,20,21. Thus, it is possible that L. reuteri prevents the exacerbating effects of stressor exposure on C. rodentium-induced intestinal inflammation by preventing stressor-induced dysbiosis. The purpose of this study was to determine whether the effects of stressor exposure on microbial community composition were evident throughout the course of C. rodentium infection and extend beyond termination of the stressor. A secondary objective was to determine whether the effects of the stressor on microbial community composition were evident in probiotic-treated animals.
Materials and Methods
Mice
Male C57Bl/6 mice (age 6–8 weeks) were obtained from Charles River (Raleigh, NC), housed three to a cage, and allowed to habituate in an approved Ohio State University vivarium for one week upon arrival. Mice were given food and water ad libitum and kept on a 12:12 hour light:dark cycle, with lights on from 0600 to 1800 hr. All procedures were carried out in accordance with guidelines by Office of Laboratory Animal Welfare at the National Institutes of Health and were approved by the Animal Care and Use Committee at the Ohio State University.
Bacteria
Citrobacter rodentium, DBS120, was grown for 18 hr at 37 °C in lysogeny broth. Prior to infection, C. rodentium was diluted to a final stock concentration of 3–5 × 107 CFU/mL in PBS. To measure C. rodentium in shed stool pellets, stool was homogenized in a slurry in PBS, then plated in serial dilutions in MacConkey Agar with 40 μg/mL of kanamycin added. Lactobacillus reuteri, ATCC 23272, was grown for 18 hr at 37 °C at 5% CO2 in MRS broth. Lactobacillus reuteri was prepared to a stock concentration of 1 × 109 CFU/mL. Each mouse received a total inoculum of 1 × 108 CFU of L. reuteri.
Stress and Infection Study
Test mice were exposed to social disruption stress (SDR), wherein an aggressive CD-1 retired breeder male mouse is placed in a cage with the smaller and younger test mice. The aggressive intruder attacks and defeats the test mice over the course of two hours as previously described15,22,23,24. This process is repeated for a total of six evenings, from 1700 to 1900 hr, the beginning of the mouse active cycle. A group termed home cage control (HCC) mice were left undisturbed for the duration of the stressor. The SDR and HCC mice were infected with C. rodentium (Cr) immediately following the first cycle of SDR. Each mouse received 100 μl of the C. rodentium stock for a total of 3–5 × 106 colony-forming-units (CFU)/mouse. All infected mice had food and water removed for two hours post infection. In addition, following each of the six cycles of SDR, half of the SDR and HCC mice received 1 × 108 CFU of L. reuteri (Lr), while the other half of the SDR and HCC mice received PBS vehicle (Veh). In sum, there were four experimental groups: HCC-Cr-Veh, HCC-Cr-Lr, SDR-Cr-Veh, and SDR-Cr-Lr.
Sacrifice
Mice from the four experimental groups (HCC-Cr-Veh, HCC-Cr-Lr, SDR-Cr-Veh, and SDR-Cr-Lr) were sacrificed at 1, 6, 12, and 24 days post infection (DPI). Colons were collected for Illumina sequencing analysis, while stool was collected for the purpose of C. rodentium quantification. Colons were snap frozen in liquid nitrogen and stored at −80 °C until DNA was isolated for sequencing. An initial experiment was performed, as well as three experimental repeats, for four total experimental runs. The experimental design is shown in Fig. 1A. Total sample sizes at the four time points (1, 6, 12, 24 DPI) varied from 9 to 12 for each experimental group at each time point after combining the four experimental runs. There were a total of 5 uninfected mice, split over two cages, for descriptive comparisons.
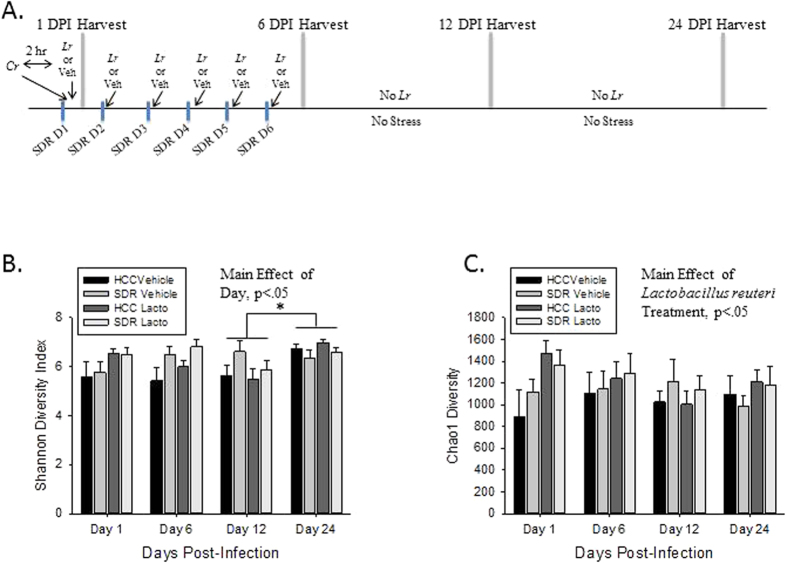
Figure 1. Alpha diversity is affected by probiotic L. reuteri treatment and progression of C. rodentium infection.
(A) A timeline of the experimental design. Mice were exposed to SDR from 0 DPI to 5 DPI. All mice were infected w/C. rodentium immediately following the first cycle of SDR (0 DPI), and half of the mice were gavaged w/L. reuteri following all six cycles of SDR. The other half received a PBS vehicle gavage as control. Mice were harvested at 1, 6, 12, and 24 DPI. (B) There is an effect of DPI upon the Shannon Diversity Index. Post-hoc testing indicated that the alpha diversity of mice at 24 DPI were significantly increased over those at 12 DPI. (C) There is also an effect of L. reuteri treatment upon the Chao1 Richness Index over all DPI.
Semi-Quantitative Real-Time PCR
Total RNA was isolated from the distal portion of the colon using Trizol reagent as per manufacturer’s instructions (Invitrogen, Carlsbad, CA), and RNA was reverse transcribed to make complementary DNA using a commercially available kit (Promega, Madison, WI). Real-time PCR primers and probes were synthesized by Applied Biosystems with the sequences as previously reported8. Real-time PCR reactions were performed as previously reported6. The change in fluorescence was measured using an Applied Biosystems 7000 Sequence Detector and analyzed using Sequence Detector version 1.0 software. In all cases, 18S was used as a housekeeping gene, and the relative amount of transcript was determined using the comparative cycle threshold (Ct) method as described by the manufacturer.
DNA Extraction and Library Preparation
DNA was extracted from the proximal section of the colon (~10 mg tissue) using a QIAgen DNA Mini Kit, following manufacturer’s instructions with slight modifications. In summary, colon contents were removed via direct excision, and colon tissues were briefly washed in a PBS bath, so as not to disturb the mucosal layer. Tissues were incubated for 45 mins at 37 °C in lysozyme buffer (20 mg/mL lysozyme, 20 mM TrisHCl, 2 mM EDTA, 1.2% Triton-X, pH 8.0), then bead-beat for 150 sec with 0.7 mm zirconia beads. Samples were incubated at 56 °C for 2 hr with Buffer ATL and Proteinase K, then incubated at 56 °C for 30 mins and 95 °C for 10 mins upon addition of Buffer AL. From this point, the Qiagen DNA Mini Kit isolation protocol was followed from the ethanol step forward. Samples were quantified with the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) using the dsDNA Broad Range Assay Kit. Samples were standardized to at least 5 ng/μl before being sent to the Molecular and Cellular Imaging Center (MCIC) in Wooster, OH for library preparation. The V1–V3 hypervariable region of the 16S rRNA gene was targeted in this study. To amplify and sequence the V1–V3 hypervariable region of the 16S rRNA gene, we used primers that contain a heterogeneity spacer in line with the targeted sequence. Four sets of spacers of different lengths were used to compensate for the low nucleotide diversity of the amplicons; since accurate base-calling on Illumina platforms and generation of high-quality data requires sequence diversity at each nucleotide position before the clustering occurs. For the targeted region, we used well-known universal primers that were modified to include degenerate bases for maximal inclusiveness25.
Libraries were prepared in two rounds of PCR amplification. The first round amplified the locus of interest and added a portion of the Illumina adapter sequence; and the second round completed the Illumina adapter sequence which contained a unique dual combination of the Nextera indices for individual tagging of each sample. Twenty nanograms of each genomic DNA was used as input for the first PCR reaction and 3 μl of the clean PCR 1 product was used as input for PCR 2 reaction. PCR amplifications were carried out as follows: initial denaturation at 96 °C for 3 min, followed by 25 (PCR 1) or 8 (PCR 2) cycles each of 96 °C for 30 sec, 55 °C for 30 sec and 72 °C for 30 sec, and a final extension at 72 °C for 5 min. The PCR products were purified after each PCR amplification using the Agencourt AMPure XP beads (Beckman Coulter Life Sciences, Indianapolis, IN, USA). All the steps for library preparation and cleaning were carried out on the epMotion5075 automated liquid handler (Eppendorf, Hamburg, Germany). The purified amplicon libraries were quantified and pooled at equimolar ratios before sequencing. The final pool was also purified using the Pippin Prep size selection system (Sage Science, Beverly, MA, USA) to discard the presence of any primer dimers.
Sequencing
The amplicon libraries were sequenced at the Molecular and Cellular Imaging Center (MCIC) in Wooster, OH using the MiSeq sequencing platform (Illumina) at a final concentration of 15.4 pM. A genomic library of well-known diversity previously sequenced in the lab was combined with the pool of amplicon libraries for the sequencing run (expected at 20%). The run was clustered to a density of 1131 k/mm2 and the libraries were sequenced using a 300PE MiSeq sequencing kit with the standard Illumina sequencing primers. Image analysis, base calling and data quality assessment were performed on the MiSeq instrument.
Data Analysis
Forward and reverse ends were demultiplexed using Sabre (website: http://github.com/najoshi/sabre), with 1 allowed barcode mismatch. Barcodes were removed and sequences were trimmed for equal lengths using FastX Trimmer (website: http://hannonlab.cshl.edu/fastx_toolkit). Sequences were joined with Fastq-Join, with 10% allowed differences within the overlap region. Quality filtering was performed with the following parameters: quality score of 20, 0 allowed N characters, 1.5 allowed barcode errors, 3 consecutive low quality bases allowed. qiime_tools (website: http://github.com/smdabdoub/phylotoast) was used for closed reference OTU picking against the 13_8 GreenGenes database26. Briefly, the complete dataset was split into smaller.fasta files, and OTUs were picked in parallel on the Ohio Supercomputer using parallel BLAST OTU picking27.
Statistical Analysis
Alpha diversity was measured using the Shannon Diversity index metric, and Chao1 methods. Beta diversity was measured with the unweighted UniFrac distance metric28. Alpha and beta diversity were analyzed using Quantitative Insights Into Microbial Ecology (QIIME)29. Differences in alpha diversity were calculated with 3 factor ANOVA with DPI, stress group, and probiotic treatment as the between subjects factors, while beta diversity shifts were calculated with adonis, which permutationally analyzes variance in distance matrices30. Taxonomic abundances at the phyla and genera levels were normalized by finding the arcsin of the square root of the proportion for each taxonomic classification. The relative abundances were compared using three factor ANOVA with DPI, stress group, and probiotic treatment as the between subjects variables using SPSS v. 21 (IBM, Chicago, IL). Post-hoc LSD tests were used when appropriate. The Benjamini-Hochberg method31 was used to correct p values for multiple-tests.
Results
Stressor exposure and Infection were both associated with significant alterations to the microbiota
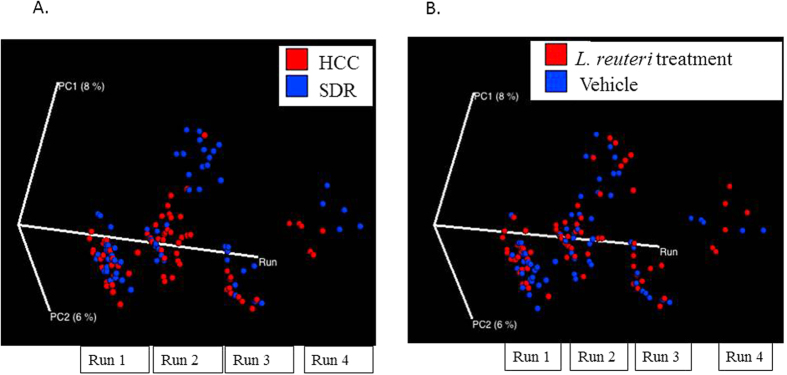
Alpha diversity was estimated using the Shannon Diversity Index and Chao1. There was a significant effect of DPI on Shannon Diversity Index (p < 0.05), with post-hoc testing indicating that 24 DPI was higher than 12 DPI (p < 0.05; Fig. 1B). No other DPI were significantly different. When Chao1 was assessed, there was a significant main effect of L. reuteri treatment in the 3 factor ANOVA (p < 0.05; Fig. 1C), indicating that L. reuteri in general increased Chao1 alpha diversity across the different DPI. Changes in beta diversity of the microbiota community were assessed using the adonis test statistic and the unweighted UniFrac distance metric. When the data were collapsed across all of the experimental runs, the adonis statistic indicated that exposure to the stressor (p = 0.001, R2 = 0.01705) and C. rodentium infection (p = 0.001, R2 = 0.02176) were associated with significant differences in microbial community composition, whereas probiotic gavage with L. reuteri did not affect the microbiota structure in the overall community (p = 0.258, R2 = 0.00779). However, the adonis statistic also indicated that microbial community composition was significantly different across each of the experimental runs, regardless of treatment condition (p = 0.001, R2 = 0.02517). Therefore, in order to visualize the effects of experimental treatments on community composition using a PCoA plot, a custom axis for experimental run was added at the expense of the third PCoA axis. This strategy indicated that exposure to the stressor impacted community composition based on sample clustering in experimental runs 2, 3, and 4 (Fig. 2A), whereas clustering was not present in any of the four experimental runs as a function of probiotic treatment (Fig. 2B).
Figure 2. Stressor exposure significantly disturbs microbiota community structure in the overall sample, while probiotic treatment has no effect.
(A) Mice exposed to SDR had significantly altered microbial profiles, as indicated on a principal coordinate analysis that used unweighted UniFrac distances. This clustering was found to be significant using the adonis statistic. Because the repeated experiments had a significant effect on community structure, the PCoA shows the effect of SDR in each of four repeats of the study on the third axis of the PCoA. (B) Probiotic L. reuteri treatment was not associated with unique clustering of microbial communities.
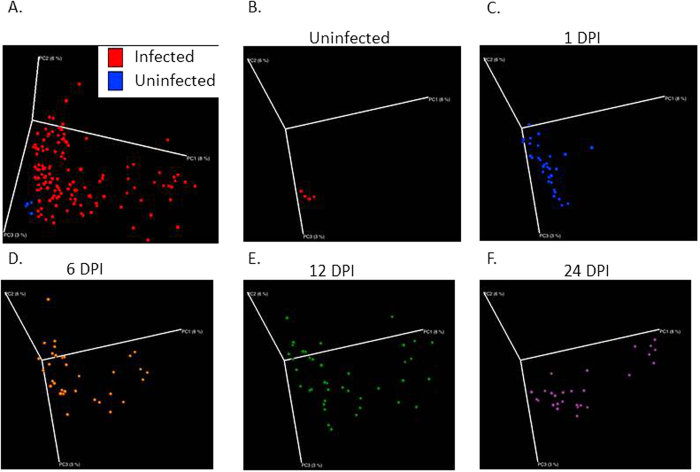
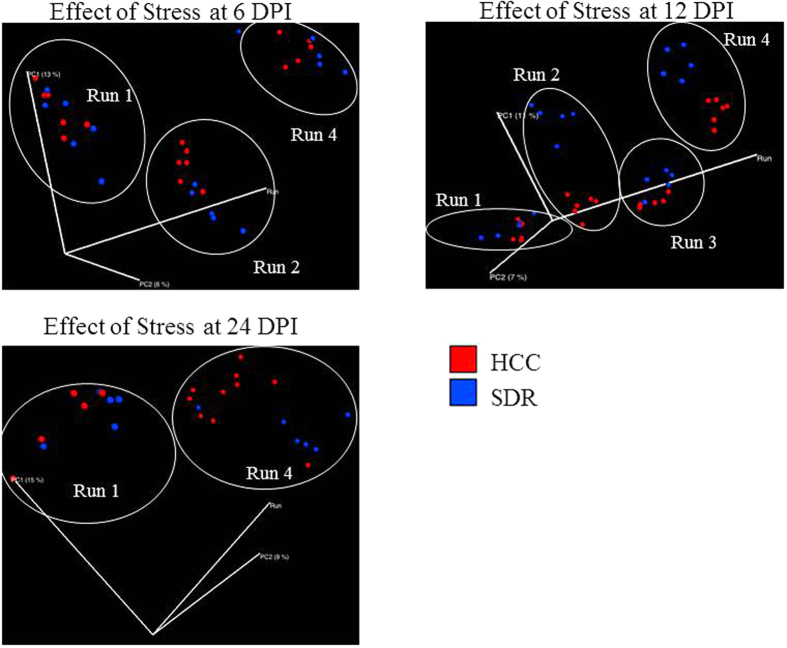
A PCoA produced with QIIME indicated that throughout the time course, the dispersion of the samples increased as a function of DPI (p = 0.001, R2 = 0.02405) (Fig. 3). In order to examine how stress and probiotic use affected the microbiota as the infection progressed, samples were filtered based on DPI. At 1 DPI, the L. reuteri probiotic had a significant effect upon the structure of the mucosal-associated microbiota (p = 0.005, R2 = 0.05243) (Fig. 4), while stressor exposure did not. As the infection proceeded to 6 DPI, the SDR stressor began to impact microbial community composition, as by this point, the mice had undergone six consecutive cycles of the stressor (p = 0.013, R2 = 0.0478). Probiotic gavage no longer significantly associated with changes in the microbiota. The effect of the SDR stressor was observed at 12 DPI (p = 0.001, R2 = 0.05282) and 24 DPI (p = 0.029, R2 = 0.05725) (Fig. 5), despite the cessation of exposure to the stressor. L. reuteri treatment did not have an effect at either of these two time points.
Figure 3. As infection progresses, microbial profiles become increasingly spread along the 3D PCoA space.
(A) Uninfected mice cluster separately from all infected mice on a principal coordinate analysis (PCoA) based upon unweighted UniFrac distances, which was confirmed with the adonis statistic. (B–F) Separating each timepoint indicates dispersion of microbial communities along the first PCoA as the infection continues to 24 DPI.
Figure 4. Probiotic treatment significantly shifts the colonic mucosal microbiota at 1 DPI.
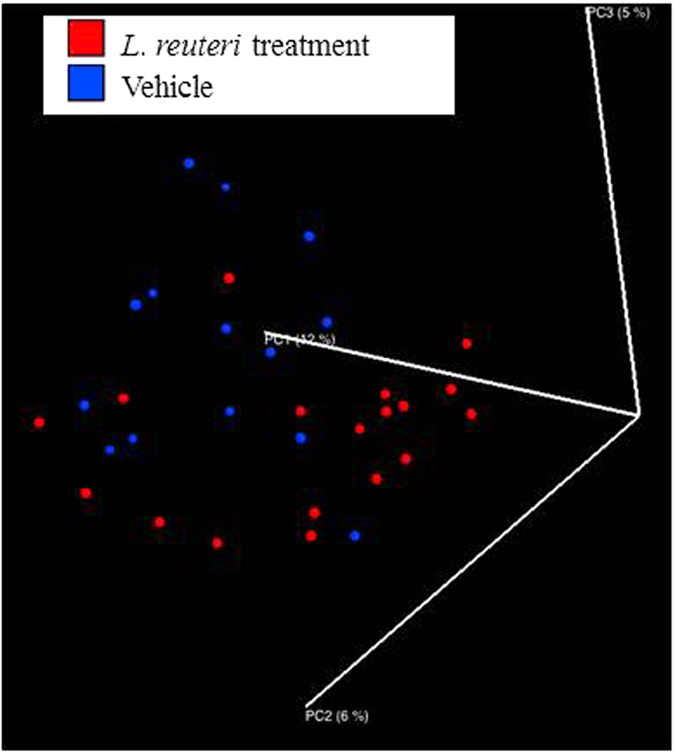
Unweighted UniFrac distances indicate significant clustering of colons treated with probiotic L. reuteri. No later timepoints exhibited clustering based upon probiotic treatment.
Figure 5. Exposure to SDR affects colonic mucosal microbiota structure regardless of DPI up to 19 days after cessation of exposure.
(A–C) PCoAs based on unweighted UniFrac were filtered based upon experimental repeat to illustrate the continued effect of stress upon the microbiota at 6, 12, and 24 DPI.
Major taxonomic changes were associated with stress and infection
Firmicutes were reduced across the different DPI, as indicated by a significant main effect in the ANOVA (p < 0.001). This main effect was due to significant reductions on 6, 12, and 24 DPI compared with 1 DPI (p < 0.01). In contrast, Bacteroidetes were increased across the different DPI (p < 0.001) with significant increases observed on 6, 12, and 24 DPI compared with 1 DPI (p < 0.01). Interestingly, the relative abundances of these phyla were also impacted by stressor exposure. There was a main effect of stress in the ANOVA (p < 0.01) indicating that stressor-exposure significantly reduced the relative abundance of Firmicutes. Similarly, stressor exposure significantly affected Bacteroidetes (main effect of stress, p < 0.001; Table 1). Stressor exposure did not affect the relative abundance of any other phyla. Moreover, treatment with L. reuteri did not significantly affect the relative abundance of any bacterial phyla (Table 1).
Table 1. Phyla Relative Abundances.
|
SDR |
HCC |
SDR |
HCC |
|||||
|---|---|---|---|---|---|---|---|---|
| L.r. | Veh | L.r. | Veh | L.r. | Veh | L.r. | Veh | |
| Day 1 | Day 6 | |||||||
| Firmicutes*,† | 85.70 ± 2.86 | 88.10 ± 2.12 | 89.96 ± 1.34 | 78.18 ± 6.27 | 73.53 ± 6.00 | 72.21± | 80.52 ± 2.46 | 75.00 ± 5.87 |
| Bacteroidetes*,† | 9.66 ± 2.02 | 6.66 ± 1.40 | 6.56 ± 1.18 | 8.24 ± 1.98 | 20.85 ± 6.70 | 14.14 ± 2.99 | 10.88 ± 2.25 | 13.18 ± 3.83 |
| Proteobacteria | 3.78 ± 0.96 | 4.62 ± 1.44 | 2.51 ± 0.54 | 11.50 ± 4.62 | 4.75 ± 1.18 | 10.81 ± 5.77 | 7.18 ± 2.84 | 9.98 ± 5.43 |
| Deferribacteres | 0.56 ± 0.10 | 0.48 ± 0.22 | 0.48 ± 0.19 | 1.79 ± 1.23 | 0.36 ± 0.10 | 2.54 ± 1.42 | 0.96 ± 0.76 | 1.46 ± 1.18 |
| Actinobacteria | 0.15 ± 0.04 | 0.08 ± 0.02 | 0.30 ± 0.21 | 0.15 ± 0.09 | 0.30 ± 0.09 | 0.20 ± 0.07 | 0.15 ± 0.03 | 0.13 ± 0.03 |
| Verrucomicrobia | 0.06 ± 0.02 | 0.03 ± 0.02 | 0.08 ± 0.03 | 0.03 ± 0.02 | 0.02 ± 0.02 | 0.01 ± 0.01 | 0.21 ± 0.09 | 0.16 ± 0.12 |
| Day 12 | Day 24 | |||||||
| Firmicutes | 66.73 ± 6.74 | 64.54 ± 7.21 | 76.49 ± 5.24 | 82.57 ± 2.46 | 62.27 ± 5.60 | 72.60 ± 5.99 | 74.62 ± 6.58 | 77.37 ± 3.31 |
| Bacteroidetes | 19.94 ± 3.90 | 15.94 ± 3.22 | 8.62 ± 2.65 | 11.36 ± 2.06 | 31.20 ± 5.00 | 18.62 ± 4.91 | 16.13 ± 6.09 | 13.22 ± 2.67 |
| Proteobacteria | 8.04 ± 3.02 | 17.34 ± 5.38 | 13.54 ± 4.71 | 4.43 ± 0.55 | 4.34 ± 1.12 | 7.83 ± 2.22 | 7.17 ± 2.73 | 7.07 ± 1.45 |
| Deferribacteres | 0.27 ± 0.10 | 0.35 ± 0.21 | 0.85 ± 0.41 | 0.36 ± 0.09 | 0.11 ± 0.04 | 0.17 ± 0.09 | 0.95 ± 0.45 | 1.14 ± 0.65 |
| Actinobacteria | 4.25 ± 2.65 | 1.41 ± 1.07 | 0.18 ± 0.07 | 1.00 ± 0.49 | 1.28 ± 0.59 | 0.27 ± 0.14 | 0.72 ± 0.58 | 0.70 ± 0.36 |
| Verrucomicrobia | 0.27 ± 0.11 | 0.08 ± 0.06 | 0.25 ± 0.09 | 0.19 ± 0.10 | 0.36 ± 0.19 | 0.02 ± 0.01 | 0.11 ± 0.05 | 0.16 ± 0.06 |
*Indicates a main effect of DPI in the 3 factor ANOVA.
†Indicates a main effect of stress in the 3 factor ANOVA.
L. reuteri treatment did not affect relative abundances.
At lower taxonomic levels, S24-7 (p < 0.001) and Enterobacteriaceae (p < 0.05) were significantly increased in infected mice, whereas Prevotella (p < 0.05) and Parabacteroides (p < 0.005) were reduced in those mice. Post-hoc testing indicated that S24-7 was increased on 6, 12, and 24 DPI compared with 1 DPI (p < 0.001), whereas the Enterobacteriaceae were increased on 6 and 12 DPI compared to 1 DPI. Parabacteroides levels were reduced on 6 compared to 1 DPI and reduced on 12 DPI compared to 1 and 24 DPI (p < 0.001) (Table 2). Prevotella levels were significantly reduced at 12 DPI (p < 0.001) (Table 2).
Table 2. Top 15 Most Abundant Genera.
| SDR |
HCC |
SDR |
HCC |
|||||
|---|---|---|---|---|---|---|---|---|
| L.r. | Veh | L.r. | Veh | L.r. | Veh | L.r. | Veh | |
| Day 1 | Day 6 | |||||||
| Unclassified Clostridiales | 46.86 ± 4.10 | 37.30 ± 5.36 | 44.61 ± 4.36 | 29.00 ± 5.68 | 42.63 ± 5.10 | 40.49 ± 5.99 | 39.00 ± 4.28 | 29.10 ± 4.21 |
| Lactobacillus | 18.54 ± 5.35 | 32.69 ± 8.67 | 24.12 ± 4.74 | 33.04 ± 9.82 | 12.67 ± 3.38 | 14.23 ± 3.66 | 24.97 ± 5.66 | 32.59 ± 7.98 |
| Lachnospiraceae | 7.96 ± 1.04 | 7.07 ± 0.80 | 7.03 ± 0.58 | 5.50 ± 1.20 | 7.32 ± 1.20 | 7.26 ± 1.11 | 6.61 ± 1.18 | 4.80 ± 0.72 |
| S24-7 | 0.01 ± 0.01 | 0.33 ± 0.30 | 0.05 ± 0.02 | 2.15 ± 1.45 | 16.77 ± 6.17 | 9.58 ± 2.14 | 2.70 ± 1.50 | 5.19 ± 2.77 |
| Bacteroides | 6.01 ± 1.31 | 3.85 ± 0.67 | 3.94 ± 0.61 | 3.42 ± 0.70 | 1.62 ± 0.32 | 2.07 ± 0.57 | 5.49 ± 1.56 | 3.46 ± 0.96 |
| Oscillospira | 3.05 ± 0.26 | 3.26 ± 0.73 | 3.48 ± 0.34 | 2.17 ± 0.36 | 2.87 ± 0.86 | 3.21 ± 0.61 | 2.56 ± 0.29 | 2.47 ± 0.40 |
| Shewanella | 1.40 ± 0.38 | 1.76 ± 0.59 | 0.97 ± 0.20 | 5.40 ± 2.32 | 1.35 ± 0.27 | 4.00 ± 2.42 | 1.93 ± 0.81 | 1.69 ± 0.39 |
| UnclassifiedRuminococcaceae | 2.27 ± 0.25 | 2.40 ± 0.54 | 3.36 ± 1.13 | 1.55 ± 0.26 | 2.38 ± 0.60 | 2.10 ± 0.20 | 1.97 ± 0.32 | 1.65 ± 0.22 |
| Lachnospiraceae; Ruminococcus | 2.66 ± 0.50 | 1.37 ± 0.18 | 2.63 ± 0.58 | 1.32 ± 0.33 | 2.25 ± 0.38 | 2.13 ± 0.48 | 1.94 ± 0.90 | 1.94 ± 0.45 |
| Unclassified Halomonadaceae | 1.20 ± 0.29 | 1.56 ± 0.48 | 0.97 ± 0.18 | 4.29 ± 1.78 | 1.17 ± 0.19 | 2.60 ± 1.36 | 1.45 ± 0.46 | 1.28 ± 0.23 |
| Halomonas | 1.10 ± 0.27 | 1.40 ± 0.39 | 0.94 ± 0.17 | 4.04 ± 1.76 | 1.09 ± 0.16 | 3.27 ± 2.07 | 1.39 ± 0.43 | 1.32 ± 0.33 |
| Parabacteroides | 1.68 ± 0.44 | 1.21 ± 0.33 | 1.51 ± 0.25 | 1.04 ± 0.20 | 0.46 ± 0.14 | 0.33 ± 0.10 | 1.11 ± 0.17 | 1.67 ± 0.87 |
| Prevotella | 1.55 ± 0.43 | 1.49 ± 0.28 | 0.93 ± 0.20 | 1.01 ± 0.25 | 1.22 ± 0.28 | 1.05 ± 0.18 | 1.14 ± 0.27 | 2.25 ± 0.96 |
| Enterobacteriaceae | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.60 ± 0.57 | 0.36 ± 0.24 | 1.29 ± 1.22 | 3.01 ± 2.99 |
| Ruminococcus | 1.13 ± 0.15 | 1.35 ± 0.39 | 1.07 ± 0.17 | 1.02 ± 0.19 | 0.99 ± 0.25 | 0.96 ± 0.14 | 0.92 ± 0.13 | 0.70 ± 0.11 |
| Day 12 | Day 24 | |||||||
| Unclassified Clostridiales | 28.46 ± 6.06 | 32.56 ± 5.06 | 34.90 ± 5.11 | 36.20 ± 2.33 | 27.67 ± 3.54 | 34.29 ± 8.09 | 46.57 ± 6.93 | 44.66 ± 3.19 |
| Lactobacillus | 20.07 ± 6.75 | 15.31 ± 5.75 | 25.51 ± 6.49 | 25.61 ± 5.70 | 18.03 ± 2.76 | 21.98 ± 8.77 | 10.26 ± 3.58 | 15.37 ± 2.05 |
| Lachnospiraceae | 5.22 ± 0.87 | 7.03 ± 1.14 | 6.81 ± 0.92 | 8.13 ± 1.25 | 4.30 ± 0.60 | 6.39 ± 0.66 | 7.08 ± 1.08 | 6.24 ± 0.54 |
| S24-7 | 12.05 ± 2.60 | 8.37 ± 2.80 | 2.31 ± 1.55 | 4.99 ± 1.92 | 18.76 ± 4.68 | 11.44 ± 3.93 | 8.24 ± 5.62 | 0.98 ± 0.44 |
| Bacteroides | 1.59 ± 0.28 | 3.49 ± 1.62 | 3.82 ± 0.98 | 3.78 ± 1.29 | 6.23 ± 2.07 | 2.81 ± 1.34 | 3.62 ± 1.44 | 6.47 ± 1.46 |
| Oscillospira | 2.47 ± 0.56 | 2.67 ± 0.48 | 2.59 ± 0.36 | 2.56 ± 0.33 | 2.23 ± 0.22 | 2.02 ± 0.48 | 2.68 ± 0.36 | 2.76 ± 0.34 |
| Shewanella | 1.75 ± 0.50 | 4.36 ± 1.90 | 2.09 ± 0.46 | 1.30 ± 0.15 | 1.44 ± 0.43 | 2.82 ± 0.82 | 2.70 ± 1.17 | 2.64 ± 0.52 |
| UnclassifiedRuminococcaceae | 2.22 ± 0.51 | 2.23 ± 0.26 | 1.78 ± 0.25 | 1.90 ± 0.28 | 1.45 ± 0.23 | 2.35 ± 0.74 | 2.53 ± 0.47 | 2.97 ± 0.55 |
| Lachnospiraceae; Ruminococcus | 2.23 ± 0.71 | 1.72 ± 0.39 | 1.44 ± 0.21 | 2.07 ± 0.37 | 1.86 ± 0.27 | 1.55 ± 0.14 | 1.74 ± 0.19 | 1.95 ± 0.35 |
| Unclassified Halomonadaceae | 1.34 ± 0.30 | 3.68 ± 1.50 | 1.69 ± 0.32 | 1.12 ± 0.13 | 1.37 ± 0.36 | 2.19 ± 0.57 | 1.83 ± 0.59 | 2.23 ± 0.47 |
| Halomonas | 1.42 ± 0.34 | 3.47 ± 1.50 | 1.54 ± 0.30 | 1.08 ± 0.10 | 1.22 ± 0.32 | 2.45 ± 0.87 | 2.06 ± 0.83 | 2.13 ± 0.46 |
| Parabacteroides | 1.21 ± 0.43 | 0.47 ± 0.09 | 1.43 ± 0.37 | 1.44 ± 0.42 | 0.92 ± 0.16 | 0.99 ± 0.20 | 1.42 ± 0.46 | 3.21 ± 0.77 |
| Prevotella | 0.74 ± 0.28 | 1.06 ± 0.41 | 0.63 ± 0.16 | 0.51 ± 0.13 | 1.48 ± 0.47 | 1.50 ± 0.47 | 0.87 ± 0.23 | 1.85 ± 0.53 |
| Enterobacteriaceae | 1.84 ± 1.74 | 3.01 ± 2.14 | 4.50 ± 2.84 | 0.49 ± 0.20 | 0.03 ± 0.01 | 0.04 ± 0.04 | 0.24 ± 0.20 | 0.01 ± 0.00 |
| Ruminococcus | 0.74 ± 0.15 | 0.81 ± 0.16 | 0.93 ± 0.13 | 0.88 ± 0.11 | 0.68 ± 0.10 | 0.83 ± 0.22 | 1.08 ± 0.19 | 1.12 ± 0.09 |
When the effects of stressor exposure and of L. reuteri treatment on the relative abundance of bacterial taxa were tested, it was evident that stressor exposure, but not L. reuteri treatment impacted the microbiota (Table 2). Mice exposed to the SDR stressor during C. rodentium challenge affected the relative abundance of Clostridales (DPI x group interaction, p < 0.05), but this difference did not remain significant after correction for multiple tests. Stressor exposure had a more predominant effect on S24-7 relative abundance which was significantly increased in SDR-Cr mice across the duration of the experiment regardless of whether they were treated with vehicle or with L. reuteri (main effect of stress, p < 0.001). Parabacteroides and Lactobacillus levels were also significantly reduced by stressor exposure throughout the experiment (main effect of stress, p < 0.001), and there was a tendency for L. reuteri treatment to normalize Parabacteroides (main effect of treatment, p = 0.02 (not significant after correcting for multiple comparisons)) (Table 2).
Redundancy analysis (RDA) using CANOCO further highlighted these results. OTUs with <10 total observations were removed for RDA analysis. DPI (p < 0.01, F = 4.2), Stress (p < 0.01, F = 3.4), and Infection (p < 0.05, F = 1.8) were all significant on the RDA plot. OTUs that accounted for greater than 20% of the variance explained by the two axes were included on the RDA triplot, wherein SDR associated with S24-7 OTUs, and no stress identified with Bacteroides, Parabacteroides, and Adlercreutzia OTUs. Clostridiales had a negative association with the temporal variable of DPI (Fig. 6).
Figure 6. DPI and stressor exposure are associated with specific OTUs on an RDA biplot.
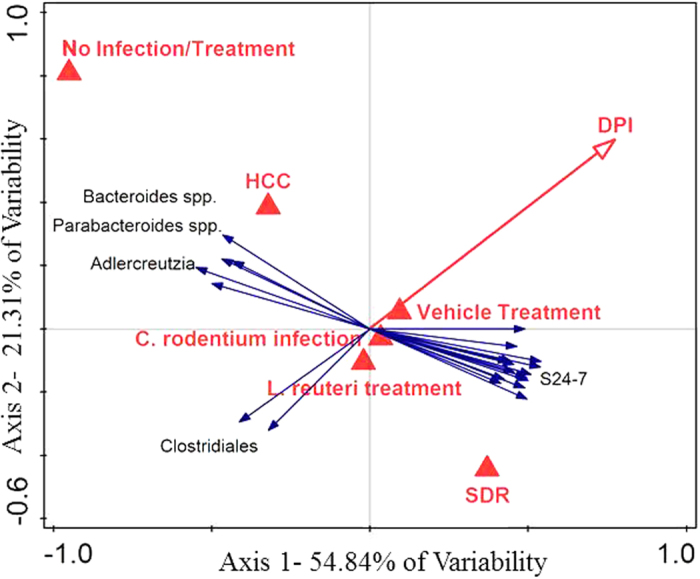
OTUs that were identified in Clostridiales negatively associated with increasing DPI. OTUs in Bacteroides spp., Parabacteroides spp., and Adlercreutzia spp. associated with HCC and OTUs in S24-7 associated with SDR. Canoco 5 software was used to construct an RDA plot. OTUs that contributed to at least 20% of the variance were included on the plot.
Stressor-induced increases in the severity of C. rodentium infection were attenuated by probiotic L. reuteri
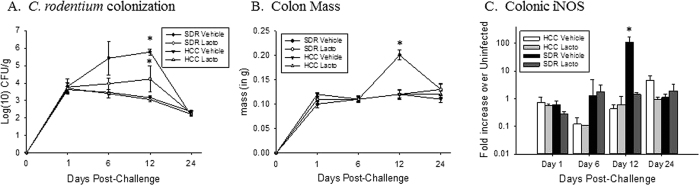
Because L. reuteri had only a modest effect on alpha diversity and failed to significantly impact beta diversity of the gut microbiota of SDR-Cr-Lr, C. rodentium load and markers of colonic inflammation were assessed to verify that exposure to the SDR stressor enhanced the colonic inflammatory response to C. rodentium, and that administration of probiotic L. reuteri attenuated the effects of the stressor on colonic inflammation as we have previously reported. Consistent with our previous studies, exposure to the stressor significantly increased the amount of C. rodentium in the infected mouse stool (Fig. 7A). In the statistical analysis, there was no three-way interaction between Stress, Treatment and DPI, but there was an interaction effect between Stress*DPI (p < 0.05). This indicates that regardless of treatment with probiotic or vehicle, C. rodentium levels were significantly increased at 12 DPI in SDR mice over HCC (p < 0.005) (Fig. 7A). Likewise, an interaction effect existed between Stress*Treatment (p = 0.05) indicating that over the course of infection, SDR-Cr-Veh mice had significantly more C. rodentium than non-stressed HCC-Cr-Veh control mice treated with vehicle (p < 0.005). SDR-Cr-Lr mice had marginally increased pathogenic burden over HCC-Cr-Lr mice that received the probiotic (p = 0.06).
Figure 7. Probiotic Lactobacillus reuteri treatment abrogates stressor-induced increase in C. rodentium-induced colonic inflammation.
(A) Mice that were exposed to SDR had significantly higher shed C. rodentium in the stool at 12 DPI than control mice. Treatment with L. reuteri blocked this increase in C. rodentium abundance in the stool. *p < 0.05 vs. HCC Vehicle on Day 12 post-challenge. All groups n = 3 (B) Mice exposed to SDR had significantly higher colon mass, an indicator of colitic pathology, at 12 DPI. Mice given L. reuteri did not exhibit the same increase in colon mass. *p < 0.05 vs. HCC Vehicle on Day 12 post-challenge. All groups n = 9–12 (C) Stressor exposure significantly increased C. rodentium-induced colonic iNOS mRNA levels. Treatment with L. reuteri attenuated this effect. All groups n = 3. *p < 0.05 vs. all other groups.
We have previously demonstrated that stressor-induced increases in C. rodentium levels are associated with increases in C. rodentium-induced colonic pathology, and that L. reuteri can attenuate the effects of the stressor on colonic pathology18. Thus, markers of colonic inflammation were assessed to verify that the L. reuteri attenuated the effects of the stressor in the current study. We previously found that colon mass strongly correlates with colonic histopathology18. Thus, we assessed colon mass as a surrogate marker of colonic pathology. Colon mass was significantly increased in mice exposed to the SDR stressor as indicated by a significant interaction between Stress and DPI (p < 0.001). Post-hoc tests indicate that mice exposed to SDR had significantly greater colon mass at 12 DPI (p < 0.005), and marginally increased colon mass at 1 DPI (p = 0.069) and 24 DPI (p = 0.064) (Fig. 7B). There was also an interaction effect between Treatment*DPI (p < 0.001), with post-hoc tests indicating that mice given vehicle treatment had significantly higher colon mass over probiotic-treated mice at 12 DPI (p < 0.005). Similar results were evident when cytokine and inflammatory mediator mRNA levels in the colon were assessed. iNOS levels were significantly higher in mice exposed to the SDR stressor, specifically on 12 DPI (p < 0.0001) (Fig. 7C). This effect was only evident in SDR-Cr-Veh mice; stressor-exposed SDR-Cr-Lr mice did not have elevations in iNOS mRNA levels in comparison to non-stressed controls on any day post-challenge. Stressor-exposed mice also had increases in TNF-α gene expression on 12 DPI (p < 0.0001); SDR-Cr-Lr mice had lower levels of TNF-α than did SDR-Cr-Veh mice, but this difference was not statistically significant (data not shown).
Discussion
Exposure to psychological stressors can alter microbial community structures, turning a normally stable microenvironment into a dysbiotic profile of volatility9,10,32. Disruptions in the microbiota can have serious consequences on host physiology and immunity, and those induced by stress may be associated with aggravation of colonic inflammation. For example, C. rodentium-induced colonic inflammation was greater in germfree mice that were colonized with microbiota from donor mice exposed to a prolonged restraint stressor when compared to germfree mice colonized with microbiota from non-stressed control donors15. Thus, it is now evident that stressor exposure changes the composition of the gut microbiota that can then lead to exacerbations of colonic inflammation. However, whether changes in the gut microbiota persist and are observed throughout the duration of a colonic inflammatory challenge is not yet known. Thus, we determined whether microbiota community composition was different in stressor-exposed vs. non-stressed mice during ongoing C. rodentium challenge.
The current results confirm previous studies demonstrating that the SDR stressor can impact microbiota composition during the 6 day period of stressor exposure10,24, and extends these previous studies by demonstrating that the stressor effects are long-lasting. Here, the residual impact of the stressor upon the microbiota is evident, as the effect of SDR upon the microbiota can be observed up to 24 DPI (19 days after cessation of SDR). It is possible that the effects of the stressor upon the microbiota at 12 DPI is associated with the infection, because the inflammatory response to the C. rodentium challenge peaked at 12 DPI. However, the prolonged effects of the stressor on the microbiota on 24 DPI is not likely due to the C. rodentium infection, because C. rodentium levels were below the level of detection on 24 DPI and markers of colonic inflammation were not elevated in stressor-exposed mice on 24 DPI. Thus, the effects of the stressor on microbial community composition extended past effects of the stressor on colonic inflammation. This finding has important implications, because many physiological and behavioral effects of the SDR stressor that have been associated with changes to the microbiota are evident up to 14 days after termination of the stressor. For example, both enhanced immune system activity and anxiety-like behavior are evident up to 14 days after termination of the stressor23,33, and both enhanced immune system activity and anxiety-like behavior have been linked to the microbiota22,34. Thus, it is possible that the protracted effects of the stressor on microbiota composition contribute to the continuation of immune and behavioral changes after termination of stressor exposure. The long-term relationships between stressor exposure, host physiology and commensal microbes warrant further testing.
Measures of alpha diversity were affected over the 24 day C. rodentium challenge (Shannon Diversity) and by L. reuteri treatment (Chao1). Alpha diversity was significantly different between 12 and 24 DPI, which overall is the peak of colonic inflammation (12 DPI) and the return to baseline (24 DPI). The effects of L. reuteri on alpha diversity were transient, and were only evident on 1 DPI. L. reuteri treatment ended on 5 DPI, and by 6 DPI (as well as later time points), Chao1 measures were not different in mice that had been treated with L. reuteri. This study also corroborated an earlier study that showed no change in the alpha diversity of the colonic mucosa-associated microbiota as a function of SDR10. Interestingly, these findings contrast sharply with previously published research that indicates that chronic restraint stress and water avoidance stress shift the alpha diversity of the colonic mucosa and distal ileum respectively35,36. Restraint stress and SDR share downstream effects, including increases in anxiety-like behavior in exposed mice. However, considerable psychological and physiological effects differ between the two stressors. Restraint stress is known to induce depressive-like behavior in mice, a hallmark not observed in SDR-exposed mice37,38. Further, SDR is defined by activation of multiple arms of the immune response, including macrophage oxidative burst and myeloid cell trafficking, while restraint is often associated with immune repression39,40. Thus, changes to the mucosa-associated microbiota alpha diversity as a function of stress is likely tied to host responses to different types of stressors. Further investigation is required to determine how unique stressors may differentially affect the microbiota.
The RDA plot indicated that the HCC group associates with Parabacteroides, Adlercreutzia, and Bacteroides, whereas SDR stressor associates with increased S24-7. In addition, consistent with previous studies10, exposure to the SDR stressor was associated with significant reductions in the relative abundances of Parabacteroides and Lactobacillus. Both genera have anti-inflammatory properties8,19,41, and their reduction by SDR may be associated with stressor-induced increases in colitis during C. rodentium infection. To better understand how SDR-induced disruptions in the microbiota adversely affect host immune responses, the involvement of these microbial groups in immune maintenance and why they are specifically targeted by social stress must be explored in future studies. The importance of Adlercreutzia and Bacteroides for maintaining health are not completely understood, however some species of Bacteroides, namely B. fragilis, are known to promote Foxp3+ T regulatory cells that can limit intestinal inflammation42. Thus, it is possible that higher levels of Bacteroides in non-stressed control mice contribute to increased intestinal homeostasis during pathogen challenge.
Treatment with probiotic L. reuteri had little effect on the composition of the gut microbiota. Administering probiotic L. reuteri only affected microbial populations at 1 DPI, even though L. reuteri administration was repeated through 6 DPI. The repeated L. reuteri administration was not sufficient to significantly impact microbial community structure. This result is surprising, because severe inflammation within the GI microenvironment is known to lead to dysbiosis16,43. This led us to assess whether L. reuteri reduced colonic inflammation in animals exposed to the stressor during C. rodentium challenge. As with our previous study18, probiotic L. reuteri significantly attenuated the exacerbating effects of stressor exposure on markers of C. rodentium-induced colonic inflammation including colon mass (which we have found closely reflects overall colonic histopathology18), and colonic iNOS gene expression. Our finding that L. reuteri reduces colonic inflammation, but not microbial community composition, is intriguing, since it is known that inflammation alone can cause changes in microbial community composition16. Although it is not yet known why changes in the microbiota were not evident in L. reuteri-treated mice, it is possible that stressor exposure leads to changes in microbial composition that are similar to those observed during inflammatory states. This explanation is plausible, since, many stressor-induced changes in microbial community composition, such as changes to Lactobacillus and Parabacteroides, have also been observed during periods of intestinal inflammation44,45. Thus, future studies should assess similarities between stressor-induced and inflammation-induced changes in the composition of the gut microbiota.
The current finding that L. reuteri reduces colonic inflammation, but not microbial community composition, also suggests that probiotic L. reuteri amelioration of pathogen-induced inflammation in stressor exposed animals is not mediated through manipulation of the commensal microbiota. This is an important finding, since several strains of L. reuteri have been shown to reduce colonic inflammation13,18,46,47,48,49, but mechanisms by which this occurs are not completely known. Several strains of L. reuteri have been shown to reduce cytokine and chemokine production by monocytes, as well as epithelial cells13,18, thus it is more likely that probiotic L. reuteri attenuates pathogen-induced colitis through direct effects on mucosal immune responses rather than mediating an effect through the microbiota. It is also possible that microbial community function, as opposed to microbial community structure, that is truly impacted by L. reuteri; others have found that changes in microbial community function can occur with relatively minor changes to microbial community structure50. These hypotheses warrant further investigation in future studies.
There is now substantial evidence that the gut microbiota are linked to gastrointestinal illness, such as IBD and irritable bowel syndrome (IBS), as well as systemic diseases, such as diabetes and obesity. And, there are a growing number of studies indicating that the symptoms of many of these same illnesses can be exacerbated during stressful periods. As an example, low social support, perceived distress, the experience of the disease as traumatic, and low quality of life were predictive of disease exacerbation in IBD patients51. Moreover, recent, major life events are more common in patients with severe IBD vs. those patients that are in remission52. The mechanisms linking stress with exacerbation of these diseases are not completely understood, but the results of this study are consistent with others from our group indicating that stressor-induced changes in the composition of the gut microbiota contribute to dysregulation of mucosal inflammatory responses15. Future studies involving assessment of microbial community functions are needed to better understand how stressor-induced alterations of the gut microbiota contribute to dysregulation of colonic inflammation and whether microbial community functions are involved with the beneficial effects of probiotic L. reuteri in stressor-exposed animals.
Additional Information
How to cite this article: Galley, J. D. et al. Stressor exposure has prolonged effects on colonic microbial community structure in Citrobacter rodentium-challenged mice. Sci. Rep. 7, 45012; doi: 10.1038/srep45012 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
Research reported in this publication was supported by the National Center for Complementary and Integrative Health of the National Institutes of Health under Award Number RO1AT006552 (to Michael T. Bailey). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Jeffrey Galley was supported by T32 training grant DE014320.
Footnotes
The authors declare no competing financial interests.
References
- Zhang Y. Z. & Li Y. Y.. Inflammatory bowel disease: pathogenesis. World journal of gastroenterology 20, 91–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N. & Nunez G.. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology 146, 1477–1488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassis C. M., Theriot C. M. & Young V. B.. Alteration of the Murine Gastrointestinal Microbiota by Tigecycline Leads to Increased Susceptibility to Clostridium difficile Infection. Antimicrobial agents and chemotherapy (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie C. G., Jarchum I., Equinda M., Lipuma L., Gobourne A., Viale A., Ubeda C., Xavier J. & Pamer E. G.. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infection and immunity 80, 62–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. A., Gu W., Lee I. A., Joh E. H. & Kim D. H.. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PloS one 7, e47713 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. T., Dowd S. E., Parry N. M., Galley J. D., Schauer D. B. & Lyte M.. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infection and immunity 78, 1509–1519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein C. N., Singh S., Graff L. A., Walker J. R., Miller N. & Cheang M.. A prospective population-based study of triggers of symptomatic flares in IBD. The American journal of gastroenterology 105, 1994–2002 (2010). [DOI] [PubMed] [Google Scholar]
- Mackos A. R., Eubank T. D., Parry N. M. & Bailey M. T.. Probiotic Lactobacillus reuteri attenuates the stressor-enhanced severity of Citrobacter rodentium infection. Infection and immunity 81, 3253–3263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. T. & Coe C. L.. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Developmental psychobiology 35, 146–155 (1999). [PubMed] [Google Scholar]
- Galley J. D., Nelson M. C., Yu Z., Dowd S. E., Walter J., Kumar P. S., Lyte M. & Bailey M. T.. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC microbiology 14, 189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles S. R., Nelson E. A. & Palombo E. A.. Investigating the role of perceived stress on bacterial flora activity and salivary cortisol secretion: a possible mechanism underlying susceptibility to illness. Biological psychology 77, 132–137 (2008). [DOI] [PubMed] [Google Scholar]
- Bailey M. T., Lubach G. R. & Coe C. L.. Prenatal stress alters bacterial colonization of the gut in infant monkeys. Journal of pediatric gastroenterology and nutrition 38, 414–421 (2004). [DOI] [PubMed] [Google Scholar]
- Lin Y. P., Thibodeaux C. H., Pena J. A., Ferry G. D. & Versalovic J.. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflammatory bowel diseases 14, 1068–1083 (2008). [DOI] [PubMed] [Google Scholar]
- Liu Y., Fatheree N. Y., Mangalat N. & Rhoads J. M.. Lactobacillus reuteri strains reduce incidence and severity of experimental necrotizing enterocolitis via modulation of TLR4 and NF-kappaB signaling in the intestine. American journal of physiology. Gastrointestinal and liver physiology 302, G608–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galley J. D., Parry N. M., Ahmer B. M., Fox J. G. & Bailey M. T.. The commensal microbiota exacerbate infectious colitis in stressor-exposed mice. Brain, behavior, and immunity (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C., Robertson M. L., Wickham M. E., Sekirov I., Champion O. L., Gaynor E. C. & Finlay B. B.. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe 2, 119–129 (2007). [DOI] [PubMed] [Google Scholar]
- Schreiber O., Petersson J., Phillipson M., Perry M., Roos S. & Holm L.. Lactobacillus reuteri prevents colitis by reducing P-selectin-associated leukocyte- and platelet-endothelial cell interactions. American journal of physiology. Gastrointestinal and liver physiology 296, G534–542 (2009). [DOI] [PubMed] [Google Scholar]
- Mackos A. R., Galley J. D., Eubank T. D., Easterling R. S., Parry N. M., Fox J. G., Lyte M. & Bailey M. T.. Social stress-enhanced severity of Citrobacter rodentium-induced colitis is CCL2-dependent and attenuated by probiotic Lactobacillus reuteri. Mucosal immunology 9, 515–526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan R. L., Yeh K. C., Hsieh M. H., Lin Y. L., Kao H. F., Li P. H., Chang Y. S. & Wang J. Y.. Lactobacillus gasseri suppresses Th17 pro-inflammatory response and attenuates allergen-induced airway inflammation in a mouse model of allergic asthma. The British journal of nutrition 108, 130–139 (2012). [DOI] [PubMed] [Google Scholar]
- Preidis G. A., Saulnier D. M., Blutt S. E., Mistretta T. A., Riehle K. P., Major A. M., Venable S. F., Finegold M. J., Petrosino J. F., Conner M. E. & Versalovic J.. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 26, 1960–1969 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K., He F., Kawase M., Miyazawa K. & Hiramatsu M.. Oral administration of Lactobacillus gasseri TMC0356 stimulates peritoneal macrophages and attenuates general symptoms caused by enteropathogenic Escherichia coli infection. Journal of microbiology, immunology, and infection=Wei mian yu gan ran za zhi 47, 81–86 (2014). [DOI] [PubMed] [Google Scholar]
- Allen R. G. et al. The intestinal microbiota are necessary for stressor-induced enhancement of splenic macrophage microbicidal activity. Brain Behav. Immun. 26, 371–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitsur R., Stark J. L., Dhabhar F. S., Padgett D. A. & Sheridan J. F.. 2002. Social disruption-induced glucocorticoid resistance: kinetics and site specificity. J. Neuroimmunol. 124, 54–61 (2002). [DOI] [PubMed] [Google Scholar]
- Bailey M. T., Dowd S. E., Galley J. D., Hufnagle A. R., Allen R. G. & Lyte M.. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain, behavior, and immunity 25, 397–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Chen P. H. & Yu Z.. Spatial and temporal variations of microbial community in a mixed plug-flow loop reactor fed with dairy manure. Microbial biotechnology 7, 332–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J. J., Koren O., Hugenholtz P., DeSantis T. Z., Walters W. A., Caporaso J. G., Angenent L. T., Knight R. & Ley R. E.. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. The ISME journal 6, 94–103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J.. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Lozupone C. & Knight R.. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J. & Knight R.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J. B. F., Kindt R., Legendre R., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H. & Wagner H. Vegan: community ecology package, p. R package version 2, 0–3 (2012). [Google Scholar]
- Benjamini Y. Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 57, 12 (1995). [Google Scholar]
- Hoffmann C., Hill D. A., Minkah N., Kirn T., Troy A., Artis D. & Bushman F.. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infection and immunity 77, 4668–4678 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb E. S., McKim D. B., Shea D. T., Powell N. D., Tarr A. J., Sheridan J. F. & Godbout J. P.. Re-establishment of anxiety in stress-sensitized mice is caused by monocyte trafficking from the spleen to the brain. Biological psychiatry 75, 970–981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumeyrolle-Arias M., Jaglin M., Bruneau A., Vancassel S., Cardona A., Dauge V., Naudon L. & Rabot S.. Absence of the gut microbiota enhances anxiety-like behavior and neuroendocrine response to acute stress in rats. Psychoneuroendocrinology 42, 207–217 (2014). [DOI] [PubMed] [Google Scholar]
- Galley J. D., Yu Z., Kumar P., Dowd S. E., Lyte M. & Bailey M. T.. The structures of the colonic mucosa-associated and luminal microbial communities are distinct and differentially affected by a prolonged murine stressor. Gut microbes 5, 748–760 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Arase S., Nagaoka N., Kawai M. & Matsumoto S.. Chronic Psychological Stress Disrupted the Composition of the Murine Colonic Microbiota and Accelerated a Murine Model of Inflammatory Bowel Disease. PloS one 11, e0150559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim D. B., Patterson J. M., Wohleb E. S., Jarrett B. L., Reader B. F., Godbout J. P. & Sheridan J. F.. Sympathetic Release of Splenic Monocytes Promotes Recurring Anxiety Following Repeated Social Defeat. Biological psychiatry 79, 803–813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M. D., Godbout J. P. & Sheridan J. F.. Repeated Social Defeat, Neuroinflammation, and Behavior: Monocytes Carry the Signal. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett D. A. & Glaser R.. How stress influences the immune response. Trends in immunology 24, 444–448 (2003). [DOI] [PubMed] [Google Scholar]
- Reader B. F., Jarrett B. L., McKim D. B., Wohleb E. S., Godbout J. P. & Sheridan J. F.. Peripheral and central effects of repeated social defeat stress: monocyte trafficking, microglial activation, and anxiety. Neuroscience 289, 429–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kverka M., Zakostelska Z., Klimesova K., Sokol D., Hudcovic T., Hrncir T., Rossmann P., Mrazek J., Kopecny J., Verdu E. F. & Tlaskalova-Hogenova H.. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clinical and experimental immunology 163, 250–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesford K. M., Yan W., Ochoa-Reparaz J., Pant A., Kircher C., Christy M. A., Begum-Haque S., Kasper D. L. & Kasper L. H.. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut microbes 6, 234–242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson A., Tormo-Badia N., Baridi A., Xu J., Molin G., Hagslatt M. L., Karlsson C., Jeppsson B., Cilio C. M. & Ahrne S.. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clinical and experimental medicine (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen K. L., Malfair D., Gray D., Doyle J. S., Jewell L. D. & Fedorak R. N.. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflammatory bowel diseases 5, 262–270 (1999). [DOI] [PubMed] [Google Scholar]
- Tyler A. D., Knox N., Kabakchiev B., Milgrom R., Kirsch R., Cohen Z., McLeod R. S., Guttman D. S., Krause D. O. & Silverberg M. S.. Characterization of the gut-associated microbiome in inflammatory pouch complications following ileal pouch-anal anastomosis. PloS one 8, e66934 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemarajata P., Gao C., Pflughoeft K. J., Thomas C. M., Saulnier D. M., Spinler J. K. & Versalovic J.. Lactobacillus reuteri-specific immunoregulatory gene rsiR modulates histamine production and immunomodulation by Lactobacillus reuteri. Journal of bacteriology 195, 5567–5576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. E. & Versalovic J.. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC microbiology 9, 35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. E., Whitehead K., Saulnier D., Thomas C. M., Versalovic J. & Britton R. A.. Cyclopropane fatty acid synthase mutants of probiotic human-derived Lactobacillus reuteri are defective in TNF inhibition. Gut microbes 2, 69–79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Hong T., van Pijkeren J. P., Hemarajata P., Trinh D. V., Hu W., Britton R. A., Kalkum M. & Versalovic J.. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PloS one 7, e31951 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. D., Compher C., Chen E. Z., Smith S. A., Shah R. D., Bittinger K., Chehoud C., Albenberg L. G., Nessel L., Gilroy E., Star J., Weljie A. M., Flint H. J., Metz D. C., Bennett M. J., Li H., Bushman F. D. & Lewis J. D.. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 65, 63–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler G. & Vavricka S.. Exposome in IBD: recent insights in environmental factors that influence the onset and course of IBD. Inflammatory bowel diseases 21, 400–408 (2015). [DOI] [PubMed] [Google Scholar]
- Tocchi A., Lepre L., Liotta G., Mazzoni G., Costa G., Taborra L. & Miccini M.. Familial and psychological risk factors of ulcerative colitis. Italian journal of gastroenterology and hepatology 29, 395–398 (1997). [PubMed] [Google Scholar]