Abstract
The mevalonate (MEV) cascade is responsible for cholesterol biosynthesis and the formation of the intermediate metabolites geranylgeranylpyrophosphate (GGPP) and farnesylpyrophosphate (FPP) used in the prenylation of proteins. Here we show that the MEV cascade inhibitor simvastatin induced significant cell death in a wide range of human tumor cell lines, including glioblastoma, astrocytoma, neuroblastoma, lung adenocarcinoma, and breast cancer. Simvastatin induced apoptotic cell death via the intrinsic apoptotic pathway. In all cancer cell types tested, simvastatin-induced cell death was not rescued by cholesterol, but was dependent on GGPP- and FPP-depletion. We confirmed that simvastatin caused the translocation of the small Rho GTPases RhoA, Cdc42, and Rac1/2/3 from cell membranes to the cytosol in U251 (glioblastoma), A549 (lung adenocarcinoma) and MDA-MB-231(breast cancer). Simvastatin-induced Rho-GTP loading significantly increased in U251 cells which were reversed with MEV, FPP, GGPP. In contrast, simvastatin did not change Rho-GTP loading in A549 and MDA-MB-231. Inhibition of geranylgeranyltransferase I by GGTi-298, but not farnesyltransferase by FTi-277, induced significant cell death in U251, A549, and MDA-MB-231. These results indicate that MEV cascade inhibition by simvastatin induced the intrinsic apoptosis pathway via inhibition of Rho family prenylation and depletion of GGPP, in a variety of different human cancer cell lines.
Tumor cells undergo significant metabolic reprogramming to serve their increased needs for energy and macromolecules1,2. Cholesterol is essential for eukaryotic cells and critical for the propagation of tumor cells which utilize de novo cholesterol biosynthesis via fatty acid synthesis and the mevalonate (MEV) cascade3,4,5. The MEV pathway also generates non-sterol end products called isoprenoids, including isopentenyl pyrophosphate, farnesyl and geranylgeranyl isoprenoids, dolichol, ubiquinone, and isopentenyl adenine5,6. In eukaryotes, the MEV pathway is the only metabolic pathway capable of generating the isoprenoids FPP and GGPP. In tumor cells, the MEV metabolism is dysregulated7,8 and glucose, glutamine, and acetate serve as substrates to fuel an anabolic MEV pathway5,9,10. 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) is the rate-limiting enzyme in the biosynthesis of MVA and cholesterol. Considered a new member of the family of metabolic oncogenes11, this key enzyme is frequently dysregulated in tumor cells12,13,14,15.
Statin drugs inhibit HMGCR and this can modulate several cellular signaling pathways relevant to tumor formation, including angiogenesis, cellular proliferation, cell cycle regulation, gene expression, metastatic potential, and cell death5,16,17,18. Statins deplete cellular FPP, GGPP, and cholesterol by decreasing MEV levels5,19,20, cause cell cycle arrest by up-regulation of cyclin-dependent kinase inhibitors p2121 and p2722,23, induce apoptosis21,24,25,26 and modulate proteasome activity27,28. While statin-mediated inhibition of small Rho GTPase prenylation (geranylgeranylation and farnesylation) and cholesterol depletion are important contributing factors, the exact mechanisms of statin-induced apoptosis in tumor cells has not yet been fully elucidated.
The increased demand on the MEV pathway and its products during enhanced proliferation sensitizes tumor cells to inhibition of the MEV pathway by statins. Several recent epidemiological investigations confirmed a possible role of statins as anti-cancer agents29,30,31. In recent phase II clinical trials, lovastatin or pravastatin only showed limited inhibition of tumor growth21,32. However, their combination with different chemotherapeutic drugs produced more effective anti-tumor effects in several preclinical models32,33. According to www.clinicaltrials.gov there are approximately 153 clinical trials evaluating the use of statins as adjunctive or co-treatment for various types of cancers. The most common statins used in these studies is simvastatin (53) followed by atorvastatin (37), rosuvastatin (21), pravastatin (18), lovastatin (11), and fluvastatin (3). This adds up to 143 studies, and the remaining 10 studies are looking at “statin use” and are inclusive of all statins.
This raises the important question of which cancers, which statins, and with which chemotherapeutic or radiation combination will we produce the desired result of selective or preferential cancer cell apoptosis while reducing mortality. Therefore, elucidating these mechanisms remains of paramount importance if we hope to develop potentially more specific and effective targeted therapies based on the anti-cancer effects of statins. Our present study provides promising avenues to pursue this line of thinking.
In the present study, we investigated the ability of simvastatin to induce apoptosis in a broad range of human tumor cell lines of different origin. We hypothesized that statin-induced depletion of prenylation intermediates in the mevalonate cascade is responsible for apoptosis in cancer cells. Here we show that simvastatin-induced apoptosis was independent of cholesterol but required the prenylation intermediates GGPP and FPP.
Results
Simvastatin induces concentration- and time-dependent cell death in human brain, lung, and breast cancer cell lines
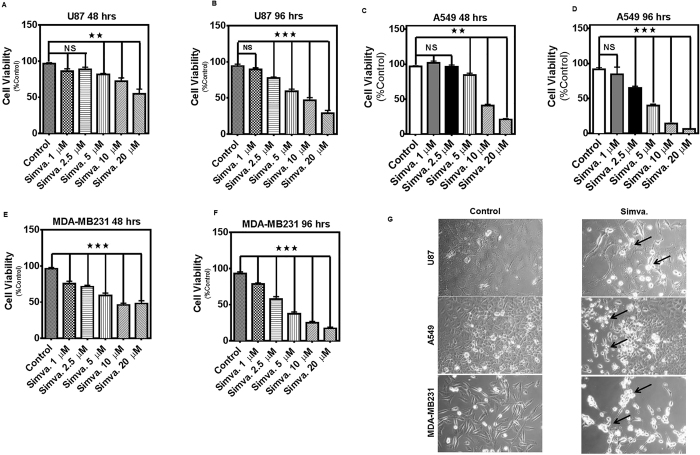
We initially tested the concentration (0–20 μM) and time (0–96 h) effects of simvastatin on viability of glioblastoma (U87 (ATCC- HTB-14™), U251 (ATCC)), neuroblastoma (SH-SY5Y (ATCC-CRL-2266™)), lung adenocarcinoma (A549 (ATCC- CCL-185™), H460 (ATCC- HTB-177™), H1650 (ATCC-CRL-5883™), H1975 (ATCC-CRL-5908™)), breast cancer (MCF-7 (ATCC-HTB-22™), MDA-MB-231 (ATCC-HTB-26™)), human astrocyte (Sciencell-1800), human HBE1(Gift from Dr. Amir Zeki lab, UC Davis), and human MCFD10A (Gift from Dr. Amir Zeki lab, UC Davis) (Fig. 1A–F) (Supplementary Fig. 1A–S). Simvastatin induces significant cell death in U87 (Fig. 1A,B) [(48 h, P < 0.01 for concentrations ≥5 μM), (96 h, P < 0.001 for concentrations ≥2.5 μM)], U251 (Fig. 1C,D) [(48 h, 96 h P < 0.001 for concentrations ≥2.5 μM)], SH-SY5Y (Fig. 1E,F) [(96 h, P < 0.001 for concentrations ≥2.5 μM)], A549 (G, H) [(48 h, P < 0.01 for concentrations ≥5 μM), (96 h, P < 0.001 for concentrations ≥2.5 μM)], H460 (I, J) [(48 h, P < 0.01 for concentrations ≥10 μM), (96 h, P < 0.001 for concentrations ≥2.5 μM)], H1650 (K, L) [(48 h, P < 0.01 for concentrations ≥5 μM), (96 h, P < 0.001 for concentrations ≥1 μM)], H1975 (M, N) [(48 h, P < 0.01 for concentrations ≥5 μM), (96 h, P < 0.001 for concentrations ≥1 μM)], MCF-7 (O, P) [(48 h, P < 0.01 for concentrations ≥2.5 μM), (96 h, P < 0.001 for concentrations ≥2.5 μM)], MDA-MB-231 (Q, R) [(48 h, P < 0.001 for concentrations ≥1 μM), (96 h, P < 0.0001 for concentrations ≥1 μM)]. The morphological changes of U87, A549, and MDA-MB213 cells observed when treated with simvastatin (10 μM, 60 h) are shown in Fig. 1G. Our results also showed that simvastatin induced both time- and concentration-dependent significant cell death (P < 0.0001) in human astrocytes (Supplementary Figure 1E and F), HBE1 (human bronchial epithelial cells) (Supplementary Figure 1M and N), and MCF10A (non-tumorigenic epithelial breast cell line) (Supplementary Figure 1Q and S). In summary, simvastatin induced a concentration- and time-dependent cell death in a broad range of human tumor and non-malignant cell lines.
Figure 1. Simvastatin induces cell death in glioblastoma, non-small lung cancer cell, and breast cancer cell lines.
(A,B) U87 cells were treated with simvastatin (1, 2.5, 5, 10 or 20 μM) and cell viability was assessed 48 and 96 hrs thereafter by MTT assay. Control cells for each time point were treated with the solvent control (DMSO). Results are expressed as percentage of corresponding time point control and represent the means ± SD of 15 replicates in three independent experiments (**P < 0.01; ***P < 0.001). (C,D) A549 cells were treated with simvastatin (1, 2.5, 5, 10 or 20 μM) and cell viability was assessed 48 and 96 hrs thereafter by MTT assay. Control cells for each time point were treated with the solvent control (DMSO). Results are expressed as percentage of corresponding time point control and represent the means ± SD of 15 replicates in 3 independent experiments (**P < 0.01; ***P < 0.001). (E,F) MDA-MB-231 cells were treated with simvastatin (1, 2.5, 5, 10 or 20 μM) and cell viability was assessed 48 and 96 hrs thereafter by MTT assay. Control cells for each time point were treated with the solvent control (DMSO). Results are expressed as percentage of corresponding time point control and represent the means ± SD of 15 replicates in three independent experiments (***P < 0.001). (G) U87, A549, and MDA-MB231 cells treated with 10 μM simvastatin for 60 hrs were then photographed under phase contrast microscopy settings. Arrows indicate partially detached cells with condensed morphology.
Simvastatin induces the intrinsic apoptotic pathway in human cancer cells
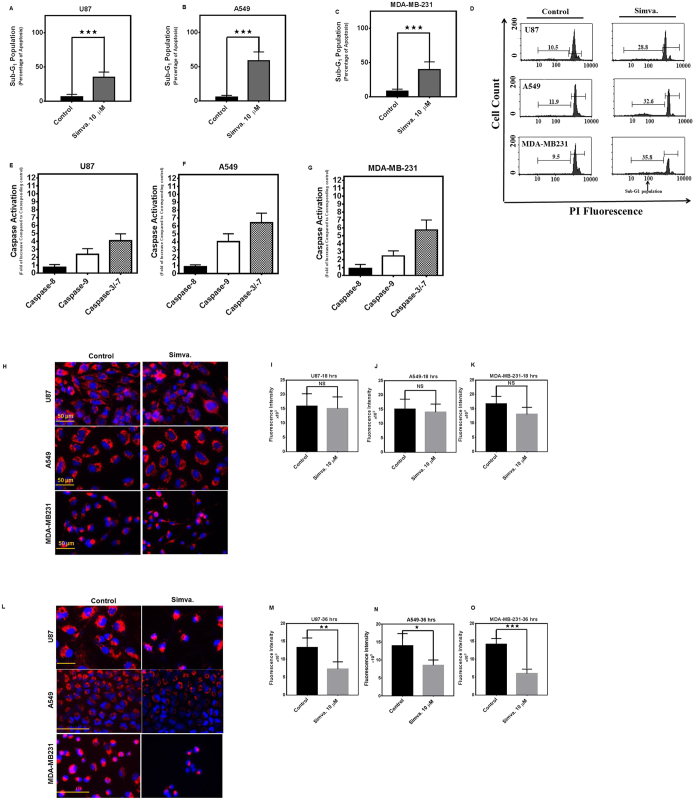
We confirmed that simvastatin (10 μM, 60 h) significantly induced apoptotic cell death in U87, U251, SH-SY5-Y, A549, H460, H1650, H1975, MCF-7, and MDA-MB-231 cell lines (Fig. 2A–D) (Supplementary Fig. 2A–F). As shown in Fig. 2D, an apoptotic cell population was identified as sub-G1 population in flow cytometry as had been described by us previously27. Simvastatin (10 μM, 48 h) failed to activate caspase-8 but did activate caspase-9 and caspase-3/-7 in all cell lines tested at 36 h (Fig. 2E–G) (Supplementary Fig. 2G–L). These findings indicated that MEV cascade inhibition induced the intrinsic apoptotic pathways in all tumor cell lines tested. The involvement of the intrinsic apoptosis pathway was further confirmed by measuring mitochondrial membrane potential in simvastatin-treated (10 μM, 36 h) cells (U87, A549, MDA-MB231) (Fig. 2L–O). Simvastatin significantly reduced mitochondrial membrane potential in these tumor cells (U87, P < 0.01, A549, P < 0.05, and MDA-MB231, P < 0.001). We also showed that shorter treatment of simvastatin (10 mΜ, 18 h) did not change mitochondrial membrane potential in these cells (Fig. 2H–K) (P > 0.05).
Figure 2. Simvastatin induces intrinsic apoptosis in glioblastoma, non-small lung cancer cell, and breast cancer cell lines.
Percent sub-G1 (A) U87, (B) A549, (C) MDA-MB-231 abundance induced by simvastatin (10 μM) or DMSO solvent control after 60 hrs. Results represent the means ± SD of 9 replicates in three independent experiments. *P < 0.05; and ***P < 0.001 compared to time-matched control. Representative figures of the flow cytometry histogram for U87, A549 and MDA-MB-231 are shown (D). Effects of simvastatin (10 μM) treatment (36 hrs) on caspase-8, caspase-3/-7, and caspase-9 enzymatic activity, as detected by Caspase-Glo® luminometric assay in U87 (E), A549 (F), MDA-MB-231 (G). Caspase activity normalized to that measured for solvent-only treated cultures is represented on the Y-axis. The data represent mean ± SD of triplicate experiments performed on 3 independent experiments. U87, A549, and MDA-MB231 cells were treated with 10 μM simvastatin for 18 and 36 hrs. Control cells were treated with media and vehicle control (DMSO). Cells were stained with TMRM, Hoechst, and imaged by standard fluorescence techniques. Simvastatin (10 μM, 18 hrs) did not significantly change TMRM fluorescence intensity in U87, A549, and MDA-MB231 cells (H–K),while simvastatin (10 μM, 36 hrs) significantly decreased TMRM florescence intensity in U87 (P < 0.01), A549 (P < 0.05), and MDA-MB231 (P < 0.001) (L–O) which indicates the decrease of mitochondrial membrane potential in simvastatin-treated cells. Data represent the average values from triplicates of three independent experiments.
Simvastatin-induced cell death in human cancer cells is not rescued by cholesterol but is dependent on depletion of FPP and GGPP
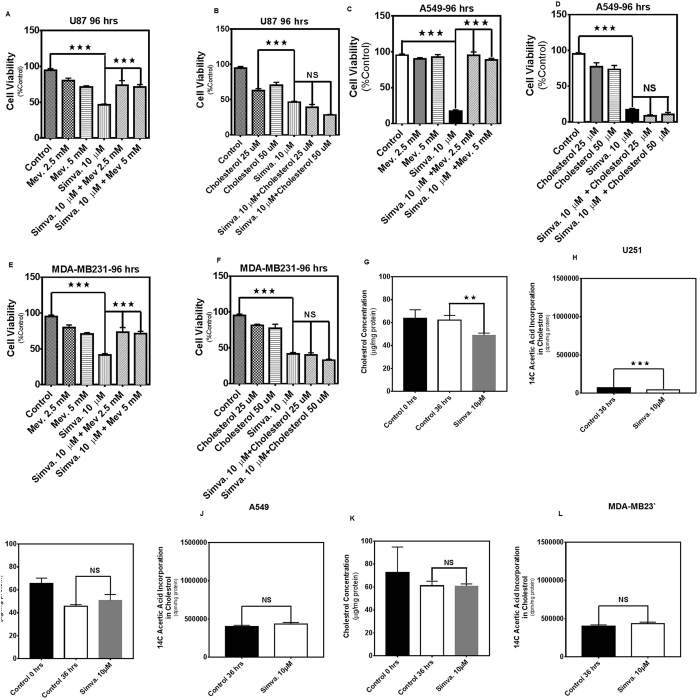
Statin-induced apoptosis is due to a loss of cell membrane cholesterol and/or depletion of the polyisoprene cholesterol precursors, FPP and GGPP, which are essential lipid anchors for active small GTPase proteins in cells5,34,35. We used different approaches to identify the mechanism by which MEV cascade inhibition caused apoptosis. First, we performed treatment of tumor cells (U87, U251, A549, H460, H1650, H1975, MCF-7, MDA-MB-231) with simvastatin (10 μM, 96 h) in the presence of MEV (2.5, 5 mM), cholesterol (25, 50 μM), and next used FPP (7.5, 15 μM), and GGPP (7.5, 15 μM) pre-treatment and co-treatment in the cell models. MEV (Fig. 3A,C,E and Supplementary Fig. 3A,C,E,G,I,K), FPP (Fig. 4A,C,E, and Supplementary Fig. 4A,C,E,G,I,K), and GGPP (Fig. 4B,D,F, and Supplementary Fig. 4B,D,F,H,J,L) were each able to significantly inhibit (P < 0.001) simvastatin-induced cell death in all cell lines tested. However, cholesterol did not have any significant effect on simvastatin-induced cell death (Fig. 3B,D,F, and Supplementary Fig. 3B,D,F,H,J). Our results also showed that simvastatin significantly decreased total cholesterol (P < 0.01) and de novo cholesterol biosynthesis (P < 0.001) in U251 cells (Fig. 3G,H). Simvastatin did not significantly change total cholesterol or de novo cholesterol biosynthesis (P > 0.05) in A549 (Fig. 3I,J) and MDA-MB231 cells (Fig. 3K,L).
Figure 3. Simvastatin-induced cell death in glioblastoma, non-small lung cancer cell, and breast cancer cell lines is independent of cholesterol.
2.5, and 5 mM MEV or, 25, 50 μM cholesterol, were added to the cells 4 hrs prior to treatment with simvastatin (10 μM, 96 hrs). Cell death was measured by MTT assay in U87 (A,B), A549 (C,D), and MDA-MB-231 (E,F). For each experiment control cells were treated with simvastatin solvent (DMSO) alone (control) or with both DMSO and the appropriate solvent (i.e. ethanol for “mevalonate control”). Results are expressed as mean ± SD of 9 replicate in 3 independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001). U87, A549, and MDA-MB231 cells were treated with simvastatin (10 μM) and after 36 hrs total and de novo cholesterol content in cells were measured. Both total and de novo cholesterol were significantly decreased in U251 cells (G,H) after simvastatin treatment. However, there was no significant change in the amount of total and de novo cholesterol for A549 (I,J) and MDA-MB231 cells (K,L). For each experiment control cells were treated with DMSO. Results are expressed as mean ± SD of 3 replicate 3 independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001).
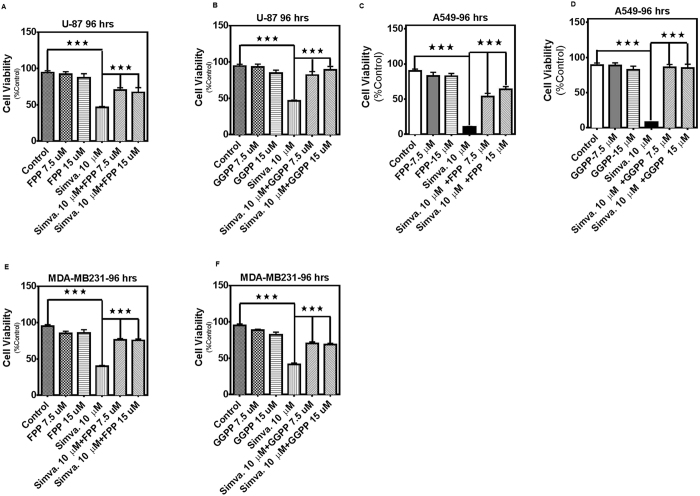
Figure 4. Simvastatin-induced cell death in glioblastoma, non-small lung cancer cell, and breast cancer cell lines dependents on FPP and GGPP.
7.5, 15 μM FPP, or 7.5, 15 μM GGPP were added to the cells 4 hrs prior to treatment with simvastatin (10 μM, 96 hrs) on cell death, measured by MTT assay in U87 (A,B), A549 (C,D), and MDA-MB-231 (E,F). For each experiment control cells were treated with simvastatin solvent (DMSO) alone (control) or with both DMSO and the appropriate solvent for each cholesterol precursor (i.e. DMSO for “FPP control” and “GGPP control”). Results are expressed as mean ± SD of 9 replicate in 3 independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001).
Simvastatin blocks membrane translocation of Rho family small GTPases
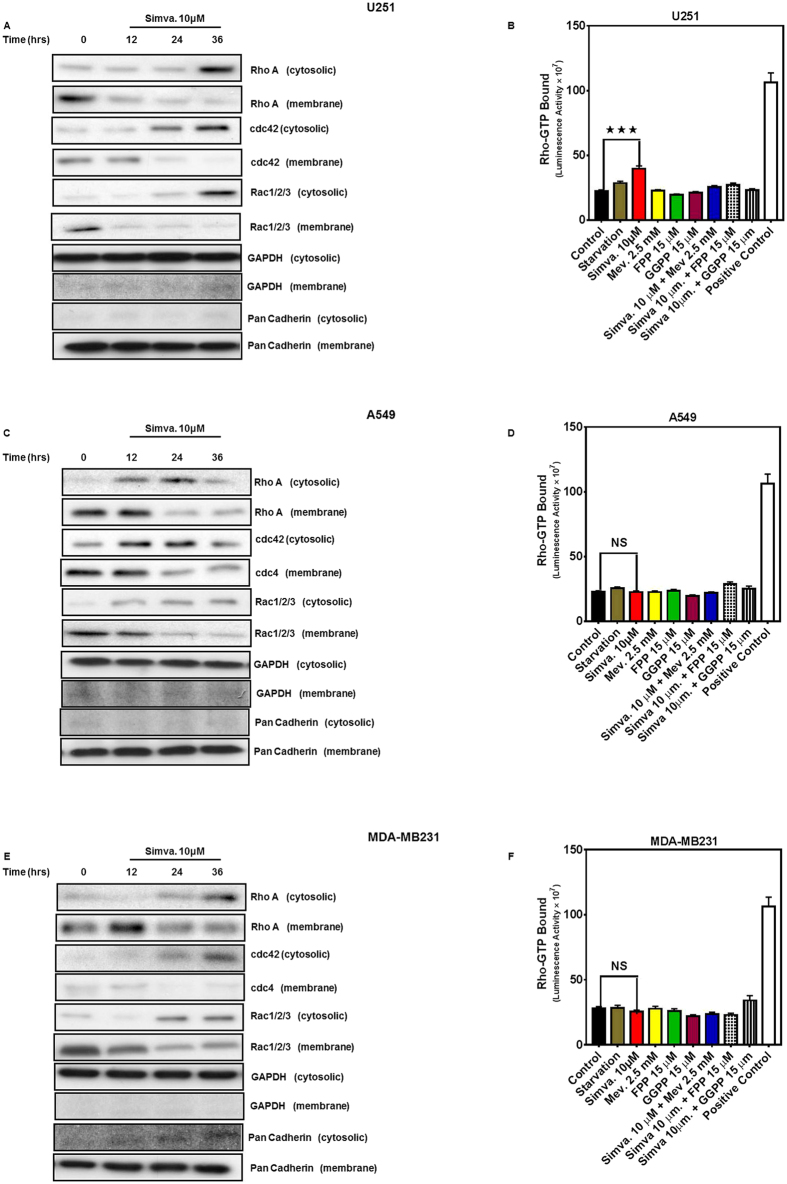
We investigated the ability of simvastatin to affect the translocation of small Rho GTPases from the plasma membrane to the cytosol in U251 (Fig. 5A), A549 (Fig. 5C), and MDA-MB231 (Fig. 5E). Simvastatin decreased membrane localization of RhoA, Cdc42, and Rac/1/2/3 and their cytosolic cellular fraction suggesting simvastatin-mediated block in prenylation of small GTPases and poor membrane translocation. We measured GTP-bound Rho protein and showed that simvastatin significantly (P < 0.05) increased GTP-bound Rho in U251 (Fig. 5B), whereas mevalonate, FPP, and GGPP co-treatment decreased GTP-bound Rho to untreated levels. Simvastatin did not significantly change GTP-bound Rho protein in A549 (Fig. 5D) and MDA-MB-231 (Fig. 5F). Thus, simvastatin-induced small GTPase protein prenylation and intracellular localization may vary among different tumor cell lines.
Figure 5. Simvastatin promotes cytosolic localization of RhoA, CDC42, and Rac1/2/3.
U87, A549, and MDA-MB231 were treated with simvastatin (10 μM, 12, 24, 36 hrs) and the abundance of RhoA, cdc42, and Rac1/2/3 in membrane and cytosolic fractions obtained from U251 (A), A549 (C), and MDA-MB-231 (E). GAPDH and Pan-Cadherin abundance was also assessed to control for loading in cytosolic and membrane fractions, and to confirm lack of cytosolic contamination in membrane fractions. Data are typical of 3 independent experiments using different primary cultures. Cropped representative of blots have been showed. G-LISA assay was done to evaluate the GTP-bound Rho protein in U251, A549, and MDA-MB231. Different conditions were tested for 36 hrs including starvation, Simva. (10 μM), Mev (2.5 mM), FPP, and GGPP (15 μM), Simva. + Mev, Simva. + FPP, and Simva. + GGPP. Simva. significantly increased GTP-bound Rho in U251 cells (B) while Mev, FPP, and GGPP co-treatment decreased GTP-bound Rho compared to control. none of the treatments significantly change GTP-bound protein in A549 (D) and MDA-MB231 (F) cells. For each experiment a positive control provided in the kit was used and control cells were treated with the reagent in the kit. Results are expressed as mean ± SD of 2 replicates in an independent experiment (***P < 0.001).
Inhibitors of Geranygeranyltransferase and Farnesyltransferase Induce Differential Cell Death in Cancer Cells
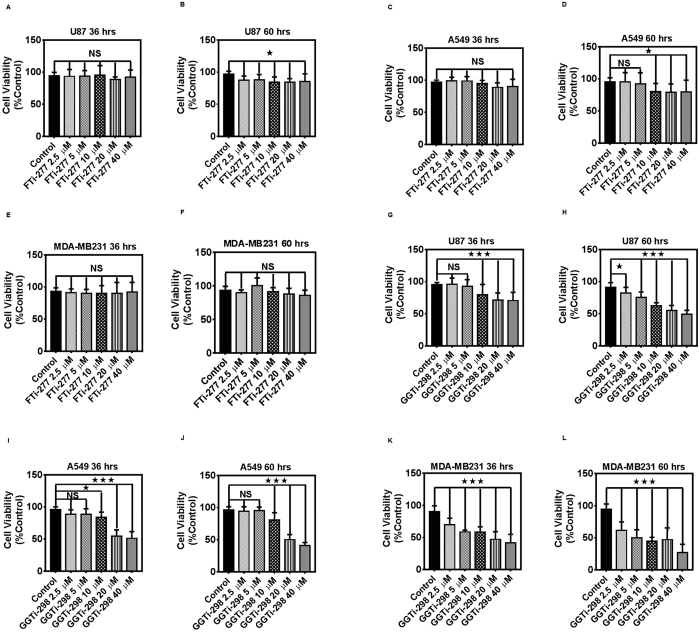
Simvastatin-induced cell death in cancer cells was further investigated using farnesyl transferase inhibitor (FTi-277) and geranylgeranyl transferase I inhibitor (GGTi-298) in U87, A549, and MDA-MB231 cells. FTi-277 (0–40 μM, 36, 60 h) had no significant effect (P > 0.05) on cell death in all three cells in 36 h (Fig. 6A,C,E) and induced less than 15% death in U87 and A549 cells at 60 h (Fig. 6B,D). GGTi-298 (>5 μM) induced significant (P > 0.001) cell death in U87, A549, and MDA-MB231 cells (36 and 60 h, Fig. 6G–L), indicating that gernylgernalytion was the leading mechanism in simvastatin-induced cell death in these cancer cells.
Figure 6. FTi-277 and GGTi-298 induces differential cell death in cancer cells.
U87, A549, and MDA-MB231 cells were treated with FTi-277 (0–40 μM, 36, 60 hrs) (A–F) and GGTi-298 (0–40 μM, 36, 60 hrs) (G–L) and the cytotoxic effects were measured using MTT assay. For each experiment control cells were treated with GGTi solvent (DMSO) and FTi-277 solvent (distilled water) alone (control). Results are expressed as mean ± SD of 15 replicate in 3 independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001).
Discussion
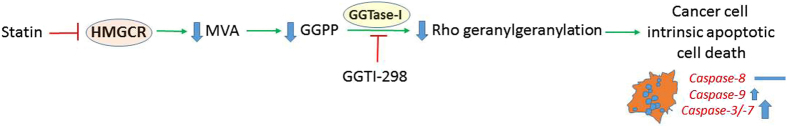
In the present study, we found that MEV cascade inhibition by the HMGCR inhibitor simvastatin induced intrinsic apoptosis cell death and decreased mitochondrial membrane potential in a broad range of human tumor cell lines, including glioma (U87, U251), neuroblastoma (SH-SY5Y), lung (A549, H460, H1650, H1975), and breast cancer cells (MCF7, MDA-MB231). Inhibition of the MEV cascade prevented the membrane translocation of the small Rho GTPases RhoA, Cdc42 and Rac1/2/3 and this simvastatin effect was independent of cellular cholesterol but partially rescued by FPP and GGPP. Inhibition of FTase had no effect on cell death and FPP supplementation caused partial reversal of the simvastatin-mediated effects. FPP can also be converted into GGPP and the addition of FPP can restore farnesylation and geranylgeranylation. We also showed that, at least in U251 brain tumor cell line, the membrane localization of prenylated Rho GTPases was important for GTPase activity. Our findings are summarized in Fig. 7.
Figure 7. Summary of the mechanism involved in statin-induced cell death in cancer cells. MEV cascade inhibitors induce the intrinsic apoptotic pathway which is regulated by gernaylgenralyation of small Rho GTPAse protein.
Simvastatin (10 μM) is known to induce mitochondrial-dependent apoptotic cell death in brain tumor cell lines (U87 and U251) with subsequent activation of caspase-336. Lovastatin (5 μM) can induce GGPP- and FPP-dependent apoptosis in U87 and U251 cells which is regulated by the expression of Bcl2 pro-apoptotic protein (Bim) in the absence of any changes in Blc2 anti-apoptotic protein expression37. Interestingly, lovastatin (10 μM)-induced caspase-dependent apoptosis in SH-SY5Y cells was inhibited using caspase-3 (Z-DEVD-FMK) (50 μM), and caspase-9 (Z-LEHD-FMK) (50 μM) inhibitors37. Thus, statin mediates its cytotoxic effects via caspases of the intrinsic apoptotic pathway.
Simvastatin also induces pro-apoptotic Bcl2 family members Bcl2 and Bax in MCF-7 and MDA-MB231 cells in a time- and dose-dependent manner38,39. These effects were also caspase-dependent since simvastatin increased caspase-3 and -9 activity in MDA-MB231 cells39. Similarly, fluvastatin and atorvastatin, were also reported to induce dose- and time-dependent apoptosis in MCF-7 and MDA-MB-231 breast cancer cell lines40,41. Several mechanisms have been proposed for statin-induced cell death in breast cancer cells including an increase of reactive oxygen species (ROS)42, the downregulation of survivin expression43, increased nitric oxide synthase activity (iNOS or NOS II) via geranylgeranylation44, and G1/S cell cycle arrest due to an increase in p21(Waf1/Cip1)45.
Simvastatin was shown to induce caspase-dependent apoptosis in A549 cells which is regulated by the Bcl2 family proteins and ROS and can be reversed by treatment with N-acetylcysteine46,47. Other investigators have reported that simvastatin can induce apoptotic pathways in A549 and H460 cells by blocking the cell cycle and down-regulating cyclin D1 and cyclin dependent kinases (CDKs) expression48,49. In A549 cells, simvastatin-induced cell death was associated with decreased expression of survivin50, like in breast cancer cells.
Our previous investigations in human airway smooth muscle and fibroblasts and in human atrial fibroblasts showed that simvastatin induces the intrinsic apoptosis pathways in a small GTPase prenylation dependent way. This statin effect was not reversible with cholesterol co-treatment27,28,51. It has been also shown that GGTI-298, but not FTI-277, can mimic the cytotoxic effects of statins, indicating that statins induce cell death by inactivating Rho/Rac GTPase activities52. These results were also confirmed by translocation of RhoA, Rac1/2/3, and Cdc42 to cytosol in simvastatin-treated cells. In eukaryotic cells, prenylation is carried out by three different prenyl transferases: farnesyl transferase (FT), geranylgeranyl transferase I (GGTI) and Rab geranylgeranyl transferase (Rab GGT or GGTII)53. FT is responsible for prenylation of proteins such as Ras and lamins. The GGTI catalyses the geranylgeranylation of proteins in the Rho and Rac family, whereas the Rab GGT is responsible for the geranylgeranylation of the Rab protein family54. Simvastatin increases Rac GTP loading in THP-1 monocyte cells while decreasing prenylation of Rac in the presence of amyloid beta stimulation, and decreasing the induction of inflammation in these cells55. In addition, T-cell function is not affected by Rho GTP loading, whereas geranylgeranylation of these small Rho GTPases is the determining step that affects their function56,57. Our results showed that statin-induced Rho protein GTP loading is dependent to specific cell type and Rho protein localization and geranylgeranylation is the determining step in regulation of their function (Figs 5, 6 and 7).
One of the signaling proteins involved in transmitting extracellular stimuli to intracellular components is the Ras (Rat sarcoma) superfamily of small GTPases5,58. In normal cellular and biological conditions these proteins play essential functions in the regulation of pathways critical to cytoskeletal reorganization, cell survival and proliferation, transformation, and vesicular trafficking59,60. One of the major subgroups in Ras superfamily of small GTPases is Rho (Ras homologous) family GTPases61. The members of small Rho GTPase (Rho, Rac, and Cdc42) are well known for their key functions in regulating actin cytoskeleton controlling actin stress fibers62,63,64. The function of small GTPases is very tightly regulated by different molecular switches, including prenylation and guanosine triphosphate (GTP) binding. When bound to GTP and prenylated, small GTPases not only translocate to the membrane but also undergo a conformational change to engage effectors that promote downstream signaling pathways5,65.
In addition to their role in normal physiological and developmental processes, Rho GTPases can contribute to pathological processes including cancer cell migration, invasion, metastasis, and inflammation66,67. Activating mutations in Ras proteins (such as K-Ras, N-Ras, and H-Ras) are found in 15 to 30% of human tumors68. Several Rho proteins are upregulated in different human tumor types, including RhoA69,70, RhoC71, Rac169,70, Rac272, Rac373, and Cdc4269,70. Rac1 is overexpressed in breast70, lung74, oral squamous cell75, testicular76, and gastric carcinoma77, whereas RhoA is also overexpressed in breast70, colon78, lung64, gastric77, head and neck79, bladder80, and testicular carcinomas76,77,79,80,81. Cdc42 is overexpressed in breast69 and testicular cancers76. Therefore, inhibition of these over-expressed Rho small GTPases might improve current therapeutic approaches in the treatment these cancers.
There is increasing interest in repurposing statins for use in the treatment of human cancers. An epidemiologic study has shown that statin use in patients with cancer was associated with reduced cancer-related mortality82. Our results showed that the HMGCR inhibitor simvastatin induces intrinsic apoptotic cell death in different cancer cell models via a unique small Rho GTPase-dependent pathway which prevents small Rho GTPase prenylation and inhibits subsequent translocation to the membrane, thus, effectively deactivating Rho GTPases. Interestingly, our current investigation suggests that cholesterol depletion is not involved in simvastatin-induced apoptosis in glioblastoma, neuroblastoma, non-small lung cancer cells, and breast cancer cell lines.
In our forthcoming studies, we are studying how the combination of simvastatin with different chemotherapeutic agents may synergize to enhance cancer cell killing. We will explore mechanisms of cell fate that could mediate this beneficial statin effect, thereby supplementing and enhancing the therapeutic effects of cancer medications.
Materials and Methods
Reagents
Cell culture plastic ware, media, penicillin, streptomycin, and fetal bovine serum (FBS) were obtained from VWR (Toronto, ON, Canada). Secondary anti-rabbit and anti-mouse antibodies, propidium iodide (PI), simvastatin, mevalonate (Mev), farnesylpyrophosphate (FPP), geranylgeranylpyrophosphate (GGPP), GGTi-298, FTi-277, cholesterol, and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) (MTT), were obtained from Sigma (Sigma-Aldrich, Oakville, CA). Rabbit anti-human/mouse Rac1/2/3, RhoA, and Cdc42 were purchased from Cell Signaling (Canada). Casapase-Glo®-3/7, Caspase-Glo®-8 and Caspase-Glo®-9 assay were purchased from Promega (Toronto, ON, Canada). Tetramethylrhodamine, Methyl Ester, Perchlorate (TMRM) was purchased from Biotium (Hayward, CA, USA). Thin layer chromatographic plates (silica gel G, 0.25-mm thickness) were from Fisher (Winnipeg, Manitoba, Canada). Ecolite scintillant was from ICN Biochemicals (Montreal, Quebec, Canada). Unlabeled cholesterol standard was from Sigma-Aldrich (Oakville, ON, Canada). [1-14C]Acetate was from American Radiolabeled Chemicals (St. Louis, MO, USA). All other biochemicals were American Chemical Society grade and were obtained from either Sigma-Aldrich or Fisher Scientific (Winnipeg, MB, Canada).
Cell Culture
For all experiments we used the following human cancer cell lines: (U87 (ATCC- HTB-14™), U251 (ATCC)), neuroblastoma (SH-SY5Y (ATCC-CRL-2266™)), lung adenocarcinoma (A549 (ATCC- CCL-185™), H460 (ATCC- HTB-177™), H1650 (ATCC-CRL-5883™), H1975 (ATCC-CRL-5908™)), breast cancer (MCF-7 (ATCC-HTB-22™), MDA-MB-231 (ATCC-HTB-26™)), human astrocyte (Sciencell-1800), human HBE1(Gift from Dr. Amir Zeki lab, UC Davis), and human MCFD10A (Gift from Dr. Amir Zeki lab, UC Davis). Cancer cells were cultured in DMEM (high glucose) supplemented with FBS (10%), penicillin (1%), and streptomycin (1%). Human astrocytes were cultured in DMEM supplemented with FBS (10%), penicillin (1%), and streptomycin (1%). HBE1 cells were grown to 90% confluency on a 100 mm cell culture plate in serum-free medium containing Ham’s F12/DMEM (1:1), 15 mM NaHCO3, 15 mM Hepes (pH 7.4), with these factors: transferrin (5 μg/mL), insulin (5 μg/mL), cholera toxin (10 ng/mL), epidermal growth factor (10 ng/mL), dexamethasone (0.1 μM), bovine hypothalamus extract (15 μg/mL). Cells were then transferred to 6-well plates in submerged media conditions. MCF10A were grown in Lonza CC-3150 MEGM per ATCC’s recommendation without gentamicin/amphotericinplus 100 ng/mL cholera toxin. Cells were then transferred to 6-well plates.
Cell Viability Assay
We measured the viability of different cancer cells under various treatment conditions, as described previously, using MTT assay28,83,84,85. Briefly, U87, U251, SH-SY5Y, A549, H460, H1650, H1975, MCF7, MDA-MB-231n normal astrocytes, HBE1, and MCF10A cells were treated with different concentrations of simvastatin for different time points (0–20 μM, 0–96 h). Relative cell viability (percent of control) was calculated using the equation: (mean OD (570) of treated cells/mean OD (570) of control cells) × 100. For each time point, the treated cells were compared with control cells that had been treated with dimethyl sulfoxide (DMSO) vehicle only. In experiments investigating the role of MEV, GGPP, FPP, and cholesterol in mediating the cytotoxic effects of simvastatin on tumor cells, MEV (2.5 and 5 mM), GGPP (7.5 and 15 μM), FPP (7.5 and 15 μM), and cholesterol (25 and 50 μM) were added to culture media 4 h before subsequent simvastatin co-treatment (10 μM, 96 h). The cytotoxic effect of GGTi-298 and FTi-277 (0–40 μM) in U87, A549, and MDA-MB-231, was evaluated using the same protocol as described for the simvastatin-induced cytotoxic assay.
Analysis of cellular morphology
To assess cell viability based on gross cellular appearance, U87, A549, and MDA-MB-231 cells were grown on 6 well plates, treated with simvastatin (10 μM, 60 h) and assessed by phase contrast microscopy (Zeiss Axioverts 100) using a Olympus DP10 CCD digital camera to capture images27.
Measurement of Apoptosis by Flow Cytometry
The Nicoletti method was used to measure cellular apoptosis86,87. Briefly, cells cultured in 12 well plates were treated with simvastatin (10 μM, 48 h). Cells were detached using EDTA buffer and harvested by centrifugation at 1500 g for 5 min at 4 °C. Cells were washed once in PBS before resuspending in a hypotonic PI lysis buffer (0.1% Triton X-100, 1% sodium citrate, 0.5 mg/ml RNase A, 40 μg/ml propidium iodide). Cell nuclei were incubated at 37 °C for 30 min and analyzed by flow cytometry. Apoptotic nuclei were located on the left side of the G1 peak and contained hypo-diploid DNA.
Luminometric Caspase Assay
For the proteolytic activity of caspase-8 (IETD-ase), -3/-7 (DEVD-ase), -9 (LEHDase), Caspase-Glo®-3/-7, -8, and -9 (Promega) were determined in luminometric assays according to the manufacturer’s instructions and our previous report51,87. Briefly, cells were grown in 96-well plate (15,000 cells/well) and treated with simvastatin (10 μM, 36 h). Fresh caspase reagents were prepared containing z-LETD-Luciferin, z-DEVD-Luciferin or z-LEHD-Luciferin and whole protein cell lysate extract buffer. Cells treated only with medium and reagent blank (negative controls) were included in each experiment. Plates were shaken gently at 300–500 rpm for 30 sec and then incubated for 90 min at room temperature. The solution was transferred to a white-well plate and then the luminescence was measured for each sample and compared to negative control values51,87.
TMRM Staining for Mitochondrial Membrane Potential Measurement
U87, A549, and MDA-MB231 cells were cultured in 6 well plates and were treated with simvastatin (10 μM, 36 h) and then were stained with (Tetramethylrhodamine, Methyl Ester, Perchlorate TMRM (100 nM) and Hoechst (10 μM) nuclear stain at 37 °C for 30 minutes. After washing, the cells they were imaged using florescence microscope and the fluorescence was quantified on ImageJ software in at least 50 individual cells in different views (NIH, Bethesda, MD, USA)88.
Membrane anchoring of Rho GTPases
To determine membrane anchoring of prenylated Rho and Rac GTPases, U251, A549, and MDA-MB-231 were cultured in DMEM with high glucose/10% FBS in the presence or absence of simvastatin (10 μM, 36 h). Upon washing, cells were scraped in ice cold buffer (10 mM Tris–HCl, pH 7.5, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, and protease inhibitor cocktail), sonicated on ice three times for 5 sec and cell homogenate was separated into cytoplasmic and membrane fractions by ultracentrifugation (100,000 × g for 35 min)89. Membrane fractions were solubilized in dissociation buffer (50 mM Tris–HCl, pH 7.5, 0.15 M NaCl, 1 mM dithiothreitol, 1% SDS, 1 mM EDTA, 1 mM EGTA, protease inhibitor cocktail), and subsequently size fractioned by SDS–PAGE (15%) for immunoblot analysis using primary antibodies to Rac1/2/3, Cdc42, and RhoA (all Cell Signaling)27.
Measurement of Rho GTPase activity
Rho-GTP bound was measured in snap-frozen cell lysates harvested from cells cultured in medium without FBS and treated with simvastatin (10 μM), mevalonate (2.5 mM), FPP, and GGPP (15 μM), Simva. + mevalonate, simvastatin + FPP, and simvastatin + GGPP for 36 h. We used a luminometric-based G-LISA Rho –GTP bound assay (Cytoskeleton, Inc, Denver, Colo) for U251, A549, and MDA-MB231 cells Briefly, cell lysates were subjected to Rho binding domain in a Rho-GTP affinity 96-well plate (Cytoskeleton, Inc, Denver, Colo). Rho-GTP was detected with specific primary antibody, followed by horseradish peroxidase-conjugated secondary antibody detection and development with a chemiluminescent reagent. A constitutively active Rho-GTP provided in the kit was used as positive control in all experiments90.
Cholesterol mass measurement
U87, A549, and MDA-MB231 cells were treated with simvastatin (10 μM, 36 h) and subsequently cholesterol content of cells was determined by colorimetric reaction using the Amplex Red Cholesterol assay kit (Invitrogen) as per the manufacturer’s instructions. Lipids were extracted and cholesterol isolated on thin layer chromatography plates prior to analysis as described27,91,92,93. All isolates were measured immediately after drying down under N2.
Cholesterol labeling experiment
Cells were incubated with 0.1 μM [1-14C]acetate (10 μCi/dish) for 24 h. Lipids were extracted and cholesterol isolated on thin layer chromatography plates as above. Spots corresponding to cholesterol standards were removed, and radioactivity incorporated into cholesterol determined by liquid scintillation counting as described92.
Immunoblotting
Western blotting was used to detect Cdc42, Rac1/2/3 and RhoA in membrane and cytosolic fractions as described previously51,94. Pan-cadherin and GAPDH were used to confirm membrane and cytosolic fraction purity, respectively. Briefly, cells protein extracts were prepared in lysis buffer (20 mM Tris-HCl (pH 7.5), 0.5 mM PMSF, 0.5% Nonidet P-40, 100 μM β-glycerol 3-phosphate and 0.5% protease inhibitor cocktail). Supernatant protein content was measured by Lowry protein assay after centrifugation at 13,000 g for 10 min. Proteins were separated by SDS-PAGE and transferred to nylon membranes under reducing conditions. Membranes were blocked with non-fat dried milk and Tween 20 followed by overnight incubation with the primary antibodies at 4 °C, followed by incubation with HRP-conjugated secondary antibody for 1 h at room temperature. Blots were developed by enhanced chemiluminescence (ECL) detection (Amersham-Pharmacia Biotech).
Statistical Analysis
The results were expressed as means ± SEM and statistical differences were evaluated by one-way or two-way ANOVA followed by Tukey’s or Bonferroni’s post hoc testing, using Graph Pad Prism 7.0. A p-value < 0.05 was considered statistically significant. For all experiments data were collected in five replicates and in three separate experiments.
Additional Information
How to cite this article: Alizadeh, J. et al. Mevalonate Cascade Inhibition by Simvastatin Induces the Intrinsic Apoptosis Pathway via Depletion of Isoprenoids in Tumor Cells. Sci. Rep. 7, 44841; doi: 10.1038/srep44841 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Suzanne Miyamoto, PhD, for donating lung cancer cell lines from University of California, Davis. We also thank Mr. Sean Ott (University of California, Davis) and Mrs. Ilnaz Badbezanchi (University of Manitoba) for conducting experiments using lung cancer, breast cancer, and glioblastoma cells line culture. Saeid Ghavami was supported by University of Manitoba start-up grant, URGP, Health Science Foundation Operating grant and Manitoba Medical Service Foundation. Amir A. Zeki was supported by 1K08HL114882-01A1 and Divisional start-up funds. Javad Alizadeh stipend was supported by University of Manitoba start-up grant. Sandipan Tewary’s salary was supported by the MITACS Globalink program in Canada. Adel Rezaei Moghaddam stipend was supported by the Graduate Enhancement of Tri-council Stipends (GETS) program at the University of Manitoba. Joseph W Gordon is supported by NSERC Canada. Sabine Hombach-Klonisch was supported by Research Manitoba, Canadian Research Society (CRS), and a Department of Surgery Research Fund. Thomas Klonisch was supported by NSERC, Canadian Research Society (CRS), and Department of Surgery Research Fund. Eftekhar Eftekharpour was supported by Health Science Foundation Centre (Winnipeg) operating grant. Pandian Nagakanan was supported by a Research Manitoba PhD studentship award. Grant M Hatch is supported by a Heart and Stroke Foundation of Canada Grant and is a Canada Research Chair in Molecular Cardiolipin Metabolism.
Footnotes
The authors declare no competing financial interests.
Author Contributions Javad Alizadeh (J.A.) and Saeid Ghavami (S.G.) wrote the manuscript. J.A. performed experiments and analyzed data. Nima Mirzaei (N.M.), Sandipan Tewary (S.T.), Adel Rezaei Moghaddam (A.R.M.), Alexandra Glogowska (A.G.), Jared Field and Nagakanan Pandian (N.P.) performed experiments and analyzed data. Eftekhar Eftekharpour (E.E.) critically revised manuscript and helped in analyzing data for neuroblastoma cells. Additional experiments of cell viability assays were also conducted in Amir A. Zeki’s (A.A.Z.) lab. A.A.Z., Nicholas J. Kenyon (N.J.K.), and Ken Y. Yoneda (K.Y.Y.) critically revised and edited the manuscript and helped analyze data for HBE1 and non-small lung cancer cells. Mohammad Hashemi (M.H.) critically revised manuscript and helped in analyzing data and did the statistical analysis. Thomas Klonisch (T.K.) and Sabine Hombach-Klonisch (S.H.K.) critically revised manuscript and helped in analyzing data for breast cancer and glioblastoma cell. Emilia Weichec (E.W.) critically revised manuscript and helped in analyzing data for apoptosis. Joseph W. Gordon (J.W.G.) critically revised manuscript and helped in analyzing data mitochondrial membrane potential. Fred Y. Xu performed cholesterol analysis experiments. Grant M. Hatch critically revised manuscript and helped in analyzing data for cholesterol metabolism. A.A.Z. and Saeid Ghavami (S.G.) designed experiments, analyzed data, critically revised manuscript and secured financial support for this work.
References
- Zong W. X., Rabinowitz J. D. & White E. Mitochondria and Cancer. Mol Cell 61, 667–676, doi: 10.1016/j.molcel.2016.02.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost F., Decoux-Poullot A. G., Tanti J. F. & Clavel S. Energy disruptors: rising stars in anticancer therapy? Oncogenesis 5, e188, doi: 10.1038/oncsis.2015.46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima T., Seimiya H. & Tsuruo T. De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. British journal of cancer 100, 1369–1372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N., Swinnen J. V. & Smans K. ATP-citrate lyase: a key player in cancer metabolism. Cancer research 72, 3709–3714 (2012). [DOI] [PubMed] [Google Scholar]
- Yeganeh B. et al. Targeting the mevalonate cascade as a new therapeutic approach in heart disease, cancer and pulmonary disease. Pharmacol Ther 143, 87–110, doi: 10.1016/j.pharmthera.2014.02.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P., Hemmerlin A., Bach T. J. & Chye M. L. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol Adv, doi: 10.1016/j.biotechadv.2016.03.005 (2016). [DOI] [PubMed] [Google Scholar]
- Brown D. N. et al. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci Rep 6, 19435, doi: 10.1038/srep19435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendening J. W. et al. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci USA 107, 15051–15056, doi: 10.1073/pnas.0910258107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T. et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 159, 1603–1614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford S. A. et al. Acetate dependence of tumors. Cell 159, 1591–1602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendening J. W. et al. Dysregulation of the mevalonate pathway promotes transformation. Proceedings of the National Academy of Sciences 107, 15051–15056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W. W.-L. et al. Determinants of sensitivity to lovastatin-induced apoptosis in multiple myeloma. Molecular cancer therapeutics 6, 1886–1897 (2007). [DOI] [PubMed] [Google Scholar]
- Tatidis L., Gruber A. & Vitols S. Decreased feedback regulation of low density lipoprotein receptor activity by sterols in leukemic cells from patients with acute myelogenous leukemia. Journal of lipid research 38, 2436–2445 (1997). [PubMed] [Google Scholar]
- Vitols S., Björkholm M., Gahrton G. & Peterson C. Hypocholesterolaemia in malignancy due to elevated low-density-lipoprotein-receptor activity in tumour cells: evidence from studies in patients with leukaemia. The Lancet 326, 1150–1154 (1985). [DOI] [PubMed] [Google Scholar]
- Chen Y. & Hughes‐Fulford M. Human prostate cancer cells lack feedback regulation of low‐density lipoprotein receptor and its regulator, SREBP2. International journal of cancer 91, 41–45 (2001). [DOI] [PubMed] [Google Scholar]
- Likus W. et al. Could drugs inhibiting the mevalonate pathway also target cancerstem cells? Drug Res. Update 25, 13–25, doi: 10.1016/j.drup.2016.02.001 (2016). [DOI] [PubMed] [Google Scholar]
- Zhao T. T., Trinh D., Addison C. L. & Dimitroulakos J. Lovastatin inhibits VEGFR and AKT activation: synergistic cytotoxicity in combination with VEGFR inhibitors. PLoS One 5, e12563, doi: 10.1371/journal.pone.0012563 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. et al. Simvastatin-induced breast cancer cell death and deactivation of PI3K/Akt and MAPK/ERK signalling are reversed by metabolic products of the mevalonate pathway. Oncotarget 7, 2532–2544, doi: 10.18632/oncotarget.6304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S. & Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. Journal of lipid research 21, 505–517 (1980). [PubMed] [Google Scholar]
- Kita T., Brown M. S. & Goldstein J. L. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in livers of mice treated with mevinolin, a competitive inhibitor of the reductase. Journal of Clinical Investigation 66, 1094 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W., Dimitroulakos J., Minden M. & Penn L. HMG-CoA reductase inhibitors and the malignant cell: the statin family of drugs as triggers of tumor-specific apoptosis. Leukemia 16, 508–519 (2002). [DOI] [PubMed] [Google Scholar]
- Laufs U., Marra D., Node K. & Liao J. K. 3-Hydroxy-3-methylglutaryl-CoA reductase inhibitors attenuate vascular smooth muscle proliferation by preventing rho GTPase-induced down-regulation of p27 Kip1. Journal of Biological Chemistry 274, 21926–21931 (1999). [DOI] [PubMed] [Google Scholar]
- Rao S., Lowe M., Herliczek T. W. & Keyomarsi K. Lovastatin mediated G1 arrest in normal and tumor breast cells is through inhibition of CDK2 activity and redistribution of p21 and p27, independent of p53. Oncogene 17, 2393–2402 (1998). [DOI] [PubMed] [Google Scholar]
- Garwood E. R. et al. Fluvastatin reduces proliferation and increases apoptosis in women with high grade breast cancer. Breast cancer research and treatment 119, 137–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendening J. W. et al. Exploiting the mevalonate pathway to distinguish statin-sensitive multiple myeloma. Blood 115, 4787–4797 (2010). [DOI] [PubMed] [Google Scholar]
- Xia Z. et al. Blocking protein geranylgeranylation is essential for lovastatin-induced apoptosis of human acute myeloid leukemia cells. Leukemia 15, 1398–1407 (2001). [DOI] [PubMed] [Google Scholar]
- Ghavami S. et al. Statin-triggered cell death in primary human lung mesenchymal cells involves p53-PUMA and release of Smac and Omi but not cytochrome c. Biochim Biophys Acta 1803, 452–467, doi: 10.1016/j.bbamcr.2009.12.005 (2010). [DOI] [PubMed] [Google Scholar]
- Ghavami S. et al. Apoptosis, autophagy and ER stress in mevalonate cascade inhibition-induced cell death of human atrial fibroblasts. Cell Death Dis 3, e330, doi: 10.1038/cddis.2012.61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern T. P. et al. Statin prescriptions and breast cancer recurrence risk: a Danish nationwide prospective cohort study. Journal of the National Cancer Institute 103, 1461–1468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana V., Bejjanki H. R., Caldito G. & Owens M. W. Statins reduce the risk of lung cancer in humans: a large case-control study of US veterans. CHEST Journal 131, 1282–1288 (2007). [DOI] [PubMed] [Google Scholar]
- Farwell W. R. et al. The association between statins and cancer incidence in a veterans population. Journal of the National Cancer Institute 100, 134–139 (2008). [DOI] [PubMed] [Google Scholar]
- Chan K. K., Oza A. M. & Siu L. L. The statins as anticancer agents. Clinical cancer research 9, 10–19 (2003). [PubMed] [Google Scholar]
- Gaist D., Hallas J., Friis S., Hansen S. & Sorensen H. T. Statin use and survival following glioblastoma multiforme. Cancer Epidemiol 38, 722–727, doi: 10.1016/j.canep.2014.09.010 (2014). [DOI] [PubMed] [Google Scholar]
- van de Donk N. W., Kamphuis M. M., van Kessel B., Lokhorst H. M. & Bloem A. C. Inhibition of protein geranylgeranylation induces apoptosis in myeloma plasma cells by reducing Mcl-1 protein levels. Blood 102, 3354–3362, doi: 10.1182/blood-2003-03-09702003-03-0970 (2003). [DOI] [PubMed] [Google Scholar]
- Zhuang L., Kim J., Adam R. M., Solomon K. R. & Freeman M. R. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J Clin Invest 115, 959–968, doi: 10.1172/JCI19935 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. et al. Effect of simvastatin on glioma cell proliferation, migration, and apoptosis. Neurosurgery 65, 1087–1096, discussion 1096–1087, doi: 10.1227/01.NEU.0000360130.52812.1D (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Zheng X., Lytle R. A., Higashikubo R. & Rich K. M. Lovastatin-induced up-regulation of the BH3-only protein, Bim, and cell death in glioblastoma cells. J Neurochem 89, 168–178, doi: 10.1111/j.1471-4159.2004.02319.x (2004). [DOI] [PubMed] [Google Scholar]
- Shen Y., Du Y., Zhang Y. & Pan Y. Synergistic effects of combined treatment with simvastatin and exemestane on MCF-7 human breast cancer cells. Mol Med Rep 12, 456–462, doi: 10.3892/mmr.2015.3406 (2015). [DOI] [PubMed] [Google Scholar]
- Shen Y. Y., Yuan Y., Du Y. Y. & Pan Y. Y. Molecular mechanism underlying the anticancer effect of simvastatin on MDA-MB-231 human breast cancer cells. Mol Med Rep 12, 623–630, doi: 10.3892/mmr.2015.3411 (2015). [DOI] [PubMed] [Google Scholar]
- Goard C. A. et al. Identifying molecular features that distinguish fluvastatin-sensitive breast tumor cells. Breast Cancer Res Treat 143, 301–312, doi: 10.1007/s10549-013-2800-y (2014). [DOI] [PubMed] [Google Scholar]
- Kwok S. C., Samuel S. P. & Handal J. Atorvastatin activates heme oxygenase-1 at the stress response elements. J Cell Mol Med 16, 394–400, doi: 10.1111/j.1582-4934.2011.01324.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C. A. et al. Statin-induced inhibition of MCF-7 breast cancer cell proliferation is related to cell cycle arrest and apoptotic and necrotic cell death mediated by an enhanced oxidative stress. Cancer Invest 26, 698–707, doi: 10.1080/07357900701874658 (2008). [DOI] [PubMed] [Google Scholar]
- Moriai R., Tsuji N., Moriai M., Kobayashi D. & Watanabe N. Survivin plays as a resistant factor against tamoxifen-induced apoptosis in human breast cancer cells. Breast Cancer Res Treat 117, 261–271, doi: 10.1007/s10549-008-0164-5 (2009). [DOI] [PubMed] [Google Scholar]
- Kotamraju S., Williams C. L. & Kalyanaraman B. Statin-induced breast cancer cell death: role of inducible nitric oxide and arginase-dependent pathways. Cancer Res 67, 7386–7394, doi: 10.1158/0008-5472.CAN-07-0993 (2007). [DOI] [PubMed] [Google Scholar]
- Denoyelle C. et al. Cerivastatin, an inhibitor of HMG-CoA reductase, inhibits the signaling pathways involved in the invasiveness and metastatic properties of highly invasive breast cancer cell lines: an in vitro study. Carcinogenesis 22, 1139–1148 (2001). [DOI] [PubMed] [Google Scholar]
- Hwang K. E. et al. Enhanced apoptosis by pemetrexed and simvastatin in malignant mesothelioma and lung cancer cells by reactive oxygen species-dependent mitochondrial dysfunction and Bim induction. Int J Oncol 45, 1769–1777, doi: 10.3892/ijo.2014.2584 (2014). [DOI] [PubMed] [Google Scholar]
- Hwang K. E. et al. Synergistic induction of apoptosis by sulindac and simvastatin in A549 human lung cancer cells via reactive oxygen species-dependent mitochondrial dysfunction. Int J. Oncol 43, 262–270, doi: 10.3892/ijo.2013.1933 (2013). [DOI] [PubMed] [Google Scholar]
- Yu X., Pan Y., Ma H. & Li W. Simvastatin inhibits proliferation and induces apoptosis in human lung cancer cells. Oncol Res 20, 351–357, doi: 10.3727/096504013X13657689382897 (2013). [DOI] [PubMed] [Google Scholar]
- Park I. H., Kim J. Y., Choi J. Y. & Han J. Y. Simvastatin enhances irinotecan-induced apoptosis in human non-small cell lung cancer cells by inhibition of proteasome activity. Invest New Drugs 29, 883–890, doi: 10.1007/s10637-010-9439-x (2011). [DOI] [PubMed] [Google Scholar]
- Hwang K. E. et al. Apoptotic induction by simvastatin in human lung cancer A549 cells via Akt signaling dependent down-regulation of survivin. Invest New Drugs 29, 945–952, doi: 10.1007/s10637-010-9450-2 (2011). [DOI] [PubMed] [Google Scholar]
- Ghavami S. et al. Airway mesenchymal cell death by mevalonate cascade inhibition: integration of autophagy, unfolded protein response and apoptosis focusing on Bcl2 family proteins. Biochim Biophys Acta 1843, 1259–1271, doi: 10.1016/j.bbamcr.2014.03.006 (2014). [DOI] [PubMed] [Google Scholar]
- Kanugula A. K. et al. Statin-induced inhibition of breast cancer proliferation and invasion involves attenuation of iron transport: intermediacy of nitric oxide and antioxidant defence mechanisms. FEBS J 281, 3719–3738, doi: 10.1111/febs.12893 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang F. L. & Casey P. J. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 65, 241–269, doi: 10.1146/annurev.bi.65.070196.001325 (1996). [DOI] [PubMed] [Google Scholar]
- Ageberg M. et al. Inhibition of geranylgeranylation mediates sensitivity to CHOP-induced cell death of DLBCL cell lines. Exp Cell Res 317, 1179–1191, doi: 10.1016/j.yexcr.2011.02.006 (2011). [DOI] [PubMed] [Google Scholar]
- Cordle A., Koenigsknecht-Talboo J., Wilkinson B., Limpert A. & Landreth G. Mechanisms of statin-mediated inhibition of small G-protein function. J Biol Chem 280, 34202–34209, doi: 10.1074/jbc.M505268200 (2005). [DOI] [PubMed] [Google Scholar]
- Waiczies S., Bendix I. & Zipp F. Geranylgeranylation but not GTP-loading of Rho GTPases determines T cell function. Sci Signal 1, pt3, doi: 10.1126/stke.112pt3 (2008). [DOI] [PubMed] [Google Scholar]
- Waiczies S. et al. Geranylgeranylation but not GTP loading determines rho migratory function in T cells. J Immunol 179, 6024–6032 (2007). [DOI] [PubMed] [Google Scholar]
- Johnson C. W. & Mattos C. The allosteric switch and conformational states in Ras GTPase affected by small molecules. Enzymes 33, Pt A, 41–67, doi: 10.1016/B978-0-12-416749-0.00003-8 (2013). [DOI] [PubMed] [Google Scholar]
- Wennerberg K., Rossman K. L. & Der C. J. The Ras superfamily at a glance. J Cell Sci 118, 843–846 (2005). [DOI] [PubMed] [Google Scholar]
- Goldfinger L. E. Choose your own path: specificity in Ras GTPase signaling. Mol Biosyst 4, 293–299, doi: 10.1039/b716887j (2008). [DOI] [PubMed] [Google Scholar]
- Timmerman I., Daniel A. E., Kroon J. & van Buul J. D. Leukocytes Crossing the Endothelium: A Matter of Communication. Int Rev Cell Mol Biol 322, 281–329, doi: 10.1016/bs.ircmb.2015.10.005 (2016). [DOI] [PubMed] [Google Scholar]
- Wennerberg K. & Der C. J. Rho-family GTPases: it’s not only Rac and Rho (and I like it). Journal of cell science 117, 1301–1312 (2004). [DOI] [PubMed] [Google Scholar]
- Wherlock M. & Mellor H. The Rho GTPase family: a Racs to Wrchs story. Journal of cell science 115, 239–240 (2002). [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. & Hall A. Rho GTPases in cell biology. Nature 420, 629–635 (2002). [DOI] [PubMed] [Google Scholar]
- Jaffe A. B. & Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 (2005). [DOI] [PubMed] [Google Scholar]
- Aspenström P., Fransson Å. & Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochemical Journal 377, 327–337 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J. Rho proteins and cancer. Breast cancer research and treatment 84, 13–19 (2004). [DOI] [PubMed] [Google Scholar]
- Forbes S. A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Research 39, 945–950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz G., Brachetti C., Bahlmann F., Schmidt M. & Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. British journal of cancer 87, 635–644 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz G., Just I. & Kaina B. Rho GTPases are over‐expressed in human tumors. International journal of cancer 81, 682–687 (1999). [DOI] [PubMed] [Google Scholar]
- Van Golen K. L., Wu Z.-F., Qiao X. T., Bao L. W. & Merajver S. D. RhoC GTPase, a novel transforming oncogene for human mammary epithelial cells that partially recapitulates the inflammatory breast cancer phenotype. Cancer research 60, 5832–5838 (2000). [PubMed] [Google Scholar]
- Hwang S.-L. et al. Rac2 expression and mutation in human brain tumors. Acta neurochirurgica 147, 551–554 (2005). [DOI] [PubMed] [Google Scholar]
- Culig Z. & Bartsch G. Androgen axis in prostate cancer. Journal of cellular biochemistry 99, 373–381 (2006). [DOI] [PubMed] [Google Scholar]
- Shieh D. B. et al. Cell motility as a prognostic factor in stage I nonsmall cell lung carcinoma. Cancer 85, 47–57 (1999). [DOI] [PubMed] [Google Scholar]
- Liu S.-Y., Yen C.-Y., Yang S.-C., Chiang W.-F. & Chang K.-W. Overexpression of Rac-1 small GTPase binding protein in oral squamous cell carcinoma. Journal of oral and maxillofacial surgery 62, 702–707 (2004). [DOI] [PubMed] [Google Scholar]
- Kamai T. et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clinical Cancer Research 10, 4799–4805 (2004). [DOI] [PubMed] [Google Scholar]
- Pan Y. et al. Expression of seven main Rho family members in gastric carcinoma. Biochemical and biophysical research communications 315, 686–691 (2004). [DOI] [PubMed] [Google Scholar]
- DerMardirossian C. & Bokoch G. M. GDIs: central regulatory molecules in Rho GTPase activation. Trends in cell biology 15, 356–363 (2005). [DOI] [PubMed] [Google Scholar]
- Abraham M. T. et al. Motility‐Related Proteins as Markers for Head and Neck Squamous Cell Cancer. The Laryngoscope 111, 1285–1289 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamai T. et al. RhoA is associated with invasion and lymph node metastasis in upper urinary tract cancer. BJU international 91, 234–238 (2003). [DOI] [PubMed] [Google Scholar]
- Kamai T., Arai K., Tsujii T., Honda M. & Yoshida K. Overexpression of RhoA mRNA is associated with advanced stage in testicular germ cell tumour. BJU international 87, 227–231 (2001). [DOI] [PubMed] [Google Scholar]
- Nielsen S. F., Nordestgaard B. G. & Bojesen S. E. Statin use and reduced cancer-related mortality. N Engl J Med 367, 1792–1802, doi: 10.1056/NEJMoa1201735 (2012). [DOI] [PubMed] [Google Scholar]
- Ghavami S. et al. Autophagy regulates trans fatty acid-mediated apoptosis in primary cardiac myofibroblasts. Biochim Biophys Acta 1823, 2274–2286, doi: 10.1016/j.bbamcr.2012.09.008 (2012). [DOI] [PubMed] [Google Scholar]
- Ghavami S. et al. Geranylgeranyl transferase 1 modulates autophagy and apoptosis in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol 302, L420–428, doi: 10.1152/ajplung.00312.2011 (2012). [DOI] [PubMed] [Google Scholar]
- Ghavami S. et al. Role of BNIP3 in TNF-induced cell death–TNF upregulates BNIP3 expression. Biochim Biophys Acta 1793, 546–560, doi: 10.1016/j.bbamcr.2009.01.002 (2009). [DOI] [PubMed] [Google Scholar]
- Hashemi M., Ghavami S., Eshraghi M., Booy E. P. & Los M. Cytotoxic effects of intra and extracellular zinc chelation on human breast cancer cells. Eur J Pharmacol 557, 9–19, doi: 10.1016/j.ejphar.2006.11.010 (2007). [DOI] [PubMed] [Google Scholar]
- Ghavami S. et al. Brevinin-2R(1) semi-selectively kills cancer cells by a distinct mechanism, which involves the lysosomal-mitochondrial death pathway. J Cell Mol Med 12, 1005–1022, doi: 10.1111/j.1582-4934.2008.00129.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughal W. et al. A conserved MADS-box phosphorylation motif regulates differentiation and mitochondrial function in skeletal, cardiac, and smooth muscle cells. Cell Death Dis 6, e1944, doi: 10.1038/cddis.2015.306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D. et al. Simvastatin potentiates tumor necrosis factor alpha-mediated apoptosis of human vascular endothelial cells via the inhibition of the geranylgeranylation of RhoA. Life Sci 79, 1484–1492, doi: S0024-3205(06)00357-210.1016/j.lfs.2006.04.019 (2006). [DOI] [PubMed] [Google Scholar]
- Movassagh H. et al. Neuronal chemorepellent Semaphorin 3E inhibits human airway smooth muscle cell proliferation and migration. J Allergy Clin Immunol 133, 560–567, doi: 10.1016/j.jaci.2013.06.011 (2014). [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M. & Sloane Stanley G. H. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226, 497–509 (1957). [PubMed] [Google Scholar]
- Hauff K. D. & Hatch G. M. Reduction in cholesterol synthesis in response to serum starvation in lymphoblasts of a patient with Barth syndrome. Biochem Cell Biol 88, 595–602, doi: 10.1139/O09-186 (2010). [DOI] [PubMed] [Google Scholar]
- Schaafsma D. et al. The mevalonate cascade as a target to suppress extracellular matrix synthesis by human airway smooth muscle. Am J Respir Cell Mol Biol 44, 394–403, doi: 10.1165/rcmb.2010-0052OC (2011). [DOI] [PubMed] [Google Scholar]
- Cieslar-Pobuda A. et al. Human induced pluripotent stem cell differentiation and direct transdifferentiation into corneal epithelial-like cells. Oncotarget, doi: 10.18632/oncotarget.9791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.